Abstract
Several different mRNAs can be produced from a given pre-mRNA by regulated alternative splicing, or as the result of deregulations that may lead to pathological states. Analysing splicing patterns is therefore of importance to describe and understand developmental programs, cellular responses to internal or external cues, or human diseases. We describe here a method, Pyrosequencing Analysis of Splicing Patterns (PASP), that combines RT–PCR and pyrosequencing of PCR products. We demonstrated that: (i) Ratios of two pure RNAs mixed in various proportions were accurately measured by PASP; (ii) PASP can be adapted to virtually any splicing event, including mutually exclusive exons, complex patterns of exon skipping or inclusion, and alternative 3′ terminal exons; (iii) In extracts from different organs, the proportions of RNA isoforms measured by PASP reflected those measured by other methods. The PASP method is therefore reliable for analysing splicing patterns. All steps are done in 96-wells microplates, without gel electrophoresis, opening the way to high-throughput comparisons of RNA from several sources.
INTRODUCTION
Gene expression in eukaryotes consists of several coupled steps that take place in the nucleus and the cytoplasm. The nucleus is the place where pre-mRNA are transcribed from DNA matrices and matured to mRNA. Maturation consists of 5′ end capping, 3′ end cleavage and polyadenylation and removal of internal introns associated with exons splicing. Furthermore, different mRNAs can be obtained from the same pre-mRNA due to a process referred to as alternative splicing. In extreme cases, such as Drosophila Dscam, several thousands of different mRNAs may be generated from a given pre-mRNA (1). The term alternative splicing encompasses mutually exclusive exons (where splicing leads to the inclusion of either of two exons), exon skipping, intron retention, alternative 5′ or 3′ splice sites (leading to retention of all or only part of an exon or an intron), or alternative initial or terminal exons (2).
Alternative splicing is responsible for a considerable variety of mature, cytoplasmic mRNAs, greatly expanding the proteome. It is generally regulated in a cell or tissue-specific manner, often during developmental programmes. In addition to regulated alternative splicing, defects in constitutive splicing may also generate some diversity in mature mRNAs. These splicing defects may lead to deleterious effects. It is estimated that a large part (up to 50%) of the mutations causing human diseases alter mRNA splicing (3).
These observations highlight the need for methods to analyse mRNA-splicing patterns. A number of methods have been published, and all of them have advantages and limitations. Here, we sought to test if a quantification of alternative mRNA isoforms can be achieved by pyrosequencing. In contrast to dideoxynucleotide-based methods for DNA sequencing, pyrosequencing is a quantitative method, in as much as the heights of the peaks generated from pyrosequencing a mixture of several DNAs are strictly proportional to the molar ratios of these DNAs (4). These properties were used to develop quantitative analyses on DNA, such as characterization of methylation patterns (5,6) and determination of Single Nucleotide Polymorphism (SNP) allele frequencies in a population (7,8). Pyrosequencing also allowed measurements of mRNA ratios after reverse-transcription, for instance to demonstrate allele-specific transcript expression of imprinted genes (9,10) or to compare gene expression levels from different sources following sequence-tagged RT-PCR (11).
Pyrosequencing was used in a limited number of cases to analyse splicing patterns. Exon 3 of human stimulatory G protein Gαs can be spliced or skipped. Furthermore, there exists an alternative 3′ splice site in intron 3, leading to shortening of exon 4 by 3 nt. Consequently, two alternative 5′ ends of exon 4 can be spliced to either exon 2 or exon 3, generating four splice isoforms. Frey et al. quantified these isoforms in human tissues by pyrosequencing (12). Also in several tissues, Szafranski et al. measured the frequencies of alternative 3′ splice sites usage by pyrosequencing (13). However, these two articles did not examine other types of splicing events, like mutually exclusive exons, alternative terminal exons or consecutive alternative exons potentially leading to complex patterns. We develop here a RT–PCR-pyrosequencing based method that we termed Pyrosequencing Analysis of Splicing Patterns (PASP). PASP is applicable to analysing virtually any splicing pattern. Furthermore, the majority of the steps can be automated, allowing several hundreds of splicing events to be analysed within a limited time scale and in cost-effective conditions.
MATERIALS AND METHODS
General biological and biochemical methods
Anatomical pieces from euthanized mice or Xenopus were harvested in liquid nitrogen. Total RNA was extracted using Tri-Reagent (Euromedex). Animals were treated following European rules for animal welfare. Xenopus embryos were obtained by in vitro fertilization following standard procedures.
Pure RNA containing either the 6A or the 6B exon of the Xenopus α-tropomyosin gene were obtained by in vitro transcription (Promega) from the XTMα24 and XTMO54 plasmids (14) and mixed in various proportions. Pure RNA or RNA extracted from animal organs or embryos were reverse-transcribed using random primers (except 3′RACE), 0.5 mM dNTP and Superscript II Reverse Transcriptase (Invitrogen) following standard procedures. For 3′RACE, reverse transcriptions were made with using an anchored oligo(dT) primer (GGCCAGGGTTTTCCCAGTCACGACT16V, V = A, C or G) as described (15).
For radioactive analysis of splicing patterns, the α-tropomyosin forward primer (sequence CAGAGGAACGTGCTGAACTTT) was radiolabelled using T4 polynucleotide kinase (Promega) and γ32P-ATP. cDNAs were amplified using that radiolabelled forward primer and a reverse primer (sequence ATTTGTCCTCCTTCTGGGAGTA) with the following program: 90 s at 94°C, then (1 min at 94°C, 30 s at 58°C, 30 s at 72°C) for 30 cycles. The resulting PCR mixtures were divided into three parts, one of which was directly electrophorized on 12% polyacrylamide gels, and the two other parts were digested with BstNI or AvaII before electrophoresis. Dried gels were submitted to STORM Phosphorimager (Molecular Dynamics) analysis and quantified with the Imagequant software.
Pyrosequencing
PCR and sequencing primers were designed using the PSQ assay design software and purchased from Sigma Proligo or Eurogentec. The PCR conditions were the same as above except for the number of cycles. The sequences of PCR and sequencing primers are given in Table 1. Pyrosequencing was done following the instructions of the manufacturer of the pyrosequencing device (Biotage). Briefly, the resulting amplimeres were bound to agarose–streptavidin beads (GE Healthcare) for 15 min under continuous agitation. Beads were loaded on the Biotage Workstation under vacuum for successive treatments with 70% ethanol, alkaline denaturation buffer and neutralization buffer. Finally, beads were resuspended in the pyrosequencing plate in wells containing 0.4 µM of sequencing primer and heat-denatured (2 min, 85°C). The content of the plate was pyrosequenced on a Biotage PSQ 96 MA device and analysed using the Pyromark software. The nucleotide dispensation order and the formulae used to calculate the percentages of the different isoforms are given in Table 1.
Table 1.
Primer sequences and formulae
| mRNA | PCR primers | Sequencing primer | Sequences of each isoform | Nucleotide dispensation order | Formulae |
|---|---|---|---|---|---|
| α-tropo, 6A/6B | b-fwd CAGAGGAACGTGCTGAACTTT | s TCCTTCTGGGAGTACTT | 6A: CTCTTCTG | ACTCTCGTGC | (6B)%=100 × G7/(G7 + G9) |
| rev ATTTGTCCTCCTTCTGGGAGTA | 6B: CTCTGCC | (6A)%=100 × G9/(G7 + G9) | |||
| α-tropo, | Fwd AGGCTGAAACACGTGCTGAG | s1 GTCCATTGATGATTTAGA | 9A:AGATGA | CAGAGTGA | (O5)%=100 × G5/(G5 + G7) |
| terminal exons | b-rev CGCCAGGGTTTTCCCAGTCACGAC | O5:AGAGAAA | (9A)%=100 × G7/(G5 + G7) | ||
| s2 CTCTCAATGACATGACTTC | 9′ :AATGTAAATTC | GATGATATCG | (9′)%=100 × G4/(G4 + G10) | ||
| 9B:AATATAAAATG | (9B)%=100 × G10/(G4 + G10) | ||||
| Tnnt2 | fwd GGAGAGCCGAGAGCATGT | s1 GGAGTACGAGGAGGAAC | 4 and 9: AGGAAGAAGCTG | GAGAGAGAGCTGAG | (4 + 9)%=100 × G12/(G9 + G12 + G14) |
| b-rev TCAGGACCAACCTCTTCTACG | 5: AGGAAGAGGAAGA | (5)%=100 × G9/(G9 + G12 + G14) | |||
| ref: AGGAAGAGCAAG | (ref)%=100 × G14/(G9 + G12 + G14) | ||||
| s2 AACAGGAAGAAGCTGTG | 4: GAAGAGCAAGA | TGAGAGCAGAC | (4)%=100 × C7/(C7 + C11) | ||
| 9: GAAGAGGAAGAC | (9)%=100 × C11/(C7 + C11) |
b, biotinylated; fwd, forward primer; rev, reverse primer; s, sequencing primer.
RESULTS
Principe of analysis of splicing pattern by pyrosequencing
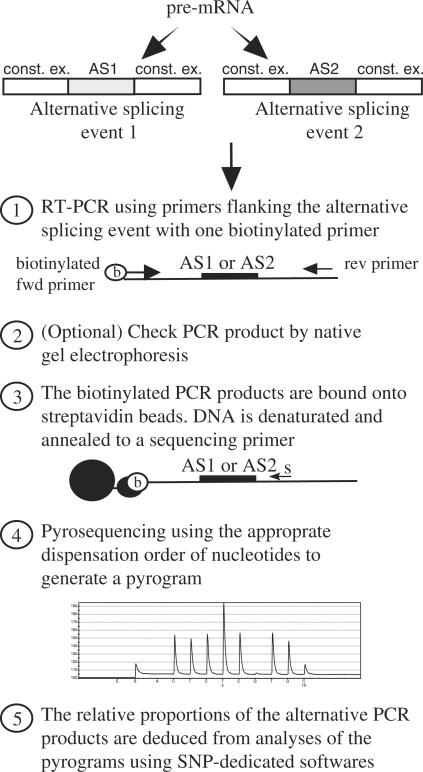
The flow chart of the PASP method is shown in Figure 1. Total RNA is extracted and reverse-transcribed following standard procedures. The cDNAs are PCR-amplified using primers complementary to conserved regions flanking the alternative-splicing event to be analysed (indicated by alternative-splicing event 1 or 2 on Figure 1). This results in a mixture of DNAs whose relative proportions are related to those of the initial mRNA pool. These proportions can be measured by pyrosequencing as would be done in a SNP analysis by determining, for a population of molecules, the relative proportions of each nucleotide at a given genomic position. Briefly, correct amplification can be checked by native electrophoresis (but this is not always necessary, see below). Next, PCR products are bound to streptavidin beads via the biotin moiety that is present at the 5′ end of one of the PCR primers. Bound PCR products are denatured by alkaline treatment. A sequencing primer is then annealed to the resulting single-stranded DNA fragment bound to the beads. This primer must be designed to hybridize to a region that is conserved between all the PCR products, but should be adjacent (1–3 nt) to the alternative region. Finally, pyrosequencing is done using a nucleotide dispensation order permitting calculations of the relative proportions of the alternative PCR products (see examples below), using software dedicated to SNP analyses. Importantly, most of the PASP steps can be automated, greatly increasing throughput.
Figure 1.
Flow-chart of the PASP method. See text for details.
Mutually exclusive exons
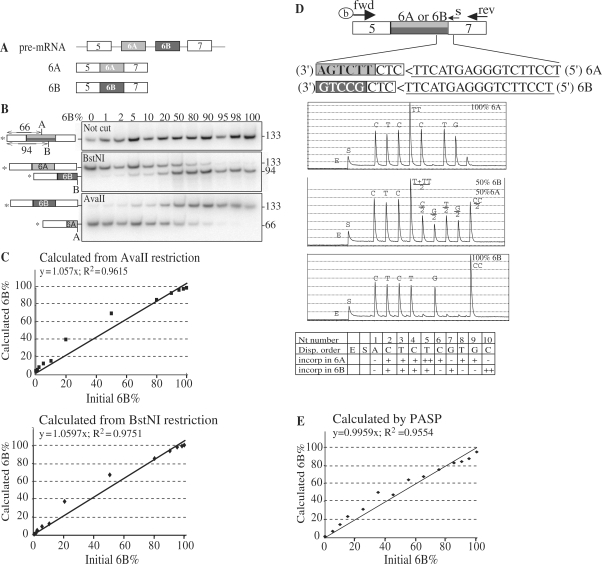
We decided to use the Xenopus α-tropomyosin mRNA to provide a proof-of-principle of the method. Both exons 5 and 7 are constitutive, and two mutually exclusive exons 6 (6A and 6B) contain the same number of nucleotides despite different sequences (Figure 2A) (14). A method to determine the relative proportions of these two exons within a population of mRNA is to run a PCR using primers hybridizing in the exons 5 and 7, one of which is radiolabelled, then to separate the PCR products by electrophoresis and to analyse them by autoradiography. One such analysis is shown in Figure 2A, upper panel. RNAs were obtained by in vitro transcription from α-tropomyosin templates containing either the 6A or the 6B exon. These RNAs were mixed in various proportions. After reverse transcription, they were PCR-amplified using a forward 32P-labelled reverse primer. Due to the fact that exons 6A and 6B have the same size, this yielded a unique, 133-bp long, PCR product, irrespective of the proportions of exons 6A and 6B (Figure 2B, upper panel). To discriminate between the two PCR products, restriction with AvaII or BstNI was used. The BstNI site is only present in the 6B exon whereas the AvaII site is only present in the 6A exon. Hence the proportions of BstNI-restricted PCR product reflect those containing exon 6B (middle panel) and the proportions of AvaII-restricted PCR product reflect those containing exon 6A (lower panel). The ratios of exons 6A to 6B in the populations, calculated from the proportions of the differentially restricted cDNAs, were in close agreement with the expected ratios (Figure 2C). Very similar results were obtained when 10 times or 100 times more RNA were used as RT–PCR matrices (data not shown).
Figure 2.
Comparison of radioactive and PASP methods to analyse splicing patterns of two mutually exclusive exons. (A) Schematic of Xenopus α-tropomyosin pre-mRNA and the two alternative mature isoforms (6A and 6B). Exons are represented by boxes and introns by lines. (B) RNAs containing either the exon 6A or the exon 6B of Xenopus α-tropomyosin were obtained by in vitro transcription and mixed in the indicated proportions (6B%). In all mixes, the total amount of RNA was 1 ng. RNAs were reverse-transcribed using random primers and PCR-amplified using a radiolabelled (as indicated by the star) forward primer complementary to exon 5 and a reverse primer complementary to exon 7. One-third of the PCR products was electrophoresed in a native acrylamide gel (upper panel), and the other two-thirds were digested by either BstNI or AvaII before electrophoresis (middle and lower panels, respectively). On the sides of the gels are indicated the positions of the restriction sites BstNI (B, in exon 6B) and AvaII (A, in exon 6A) as well as the sizes of the radiolabelled restriction fragments. (C) The percentages of exon 6B calculated from the proportion of DNA left intact after AvaII restriction (upper panel) or cut by BstNI (lower panel) were plotted versus the proportion of exon 6B in the initial RNA mixture. (D) Positions of the biotinylated forward (fwd), reverse (rev) and sequencing (s) primers used for PASP of α-tropomyosin. Exons 6A and 6B are in light and dark grey, respectively. Inverse complementary of the sequencing primer is underlined (note that 5′–3′ orientation is from right to left). Boxed CTC is a trinucleotide that is present in both 6A and 6B PCR products and is immediately downstream of the sequencing primer. Three representative pyrograms corresponding to the indicated proportions of 6A and 6B RNA are shown with the nucleotide (Nt) dispensation order. E and S stand for enzyme and substrate, respectively. (E) One nanogram of total RNA containing various proportions of 6A and 6B isoforms was reverse transcribed, PCR amplified for 35 cycles, and submitted to PASP analysis. The proportions of exon 6B calculated from the ratios of peak heights in pyrograms [100 × G7/(G7 + G9)] were plotted versus the proportions of exon 6B in the initial RNA mixture.
For the PASP method, cDNAs were PCR-amplified using a biotinylated forward primer complementary to exon 5 and a reverse primer complementary to exon 7 (Figure 2D). The PCR products were pyrosequenced using an antisense sequencing primer (s) complementary to the 5′ end of exon 7 (just 3′ of the 6A or 6B exon). With this primer, the beginning of the sequence read from a 6A matrix would be CTCTTCTGA, whereas that from a 6B matrix would be CTCTGCCTG. The dispensation order and typical pyrograms, corresponding to different percentages of exon 6B, are shown in Figure 2D. The first dispensed nucleotide (A1) is present in neither of the two sequences, and was used to estimate the background signal. The following three nucleotides (C2, T3 and C4) are present in both isoforms. As expected, they were detected at similar levels irrespective of the percentage of 6B. Nucleotides 5–10 discriminate between the two isoforms (Figure 2D).
Peak heights of the pyrograms, as provided by the Pyromark software, were used to calculate the percentages of 6B mRNA. The yields of the different nucleotides were not exactly the same. For example, in Figure 2D, the peak for T3 is slightly smaller than the peaks for C2 and C4, although these three peaks correspond to constitutive nucleotides and should be similarly incorporated. Since these differences might introduce a bias if different nucleotides were used to calculate isoforms ratios, we used the same nucleotide that is present in both isoforms at different positions for the calculations. G7 and G9 correspond to the 6B and 6A isoforms respectively (Figure 2D), hence the percentage of 6B equals to 100× G7/(G7 + G9). Figure 2E demonstrates a close agreement between the percentage of 6B calculated by this method and the percentage of 6B in the initial RNA mixture. Furthermore, the PASP results from 30 or 35 PCR cycles were indistinguishable, as well as the results obtained from various initial amounts of mixed mRNAs (data not shown), showing that the PASP method is largely insensitive to variations in these reaction conditions. Comparison of Figures 2C and 2E indicates that the radioactive and PASP methods are equally precise in quantifying the proportions of mutually exclusive exons in a population.
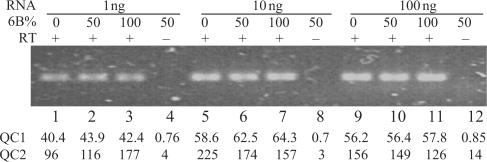
In order to reduce the time of the PASP analysis, we evaluated whether checking the PCR amplification by gel electrophoresis was mandatory. We observed that inefficient PCR amplifications, resulting in too low an amount of product to be sequenced, yielded low-quality pyrograms, and incorrect estimations of the 6B percentage (data not shown). To eliminate the electrophoresis step, we therefore developed quality controls of the pyrograms so as to be able to discard potentially non-reliable results due to bad amplifications. We examined the predictive values for reliable results of two putative quality controls (QC). As a first quality control (QC1), we examined the height of the first constitutive nucleotide (C2), reasoning that a high peak value should reveal a large amount of amplified DNA. Quality control 2 (QC2) was calculated as the ratio between C2 and A1 (respectively, first constitutive nucleotide and nucleotide absent from the two isoforms). This ratio was expected to correspond to a signal/noise ratio; hence it should be as high as possible. To determine the values of QC1 and QC2 relative to the quality of PCR amplification, PCR were performed on mixtures of 6A and 6B RNA either with (Figure 3, lanes 1–3, 5–7 and 9–11) or without (lanes 4, 8 and 12) previous reverse-transcription. As expected, correct PCR products were present only in the presence of reverse-transcriptase (Figure 3). When PCR products were present, the values for QC1 (strength of signal), were above 40, and the values for QC2 (signal/background) were above or close to 100. In contrast, when amplification failed, QC1 values were less than 1, and QC2 values were no more than 14. Several experiments correlated correct amplifications (as tested by gel electrophoresis) with high QC1 and QC2 values (data not shown). We found that values higher than 40 and 80 for QC1 and QC2, respectively, accurately reflected correct amplifications and PASP. Hence, checking PCR by gel electrophoresis can be replaced by adequate QCs of PASP.
Figure 3.
Quality controls of PASP. RNAs containing exons 6A or 6B were mixed in the indicated proportions (6B%) to a total amount of 1, 10 and 100 ng and reverse-transcribed, except for lanes 4, 8 and 12 where the reverse transcriptase was omitted. The resulting cDNAs were PCR-amplified for 25 cycles in a total volume of 50 µl. Five microlitres of the PCR products were electrophorised on a native agarose gel in the presence of Syber-Green and the gel was photographed. The remaining PCR products were submitted to PASP with the same dispensation order as in Figure 2D. QCs 1 and 2 were calculated as described in text and the values are given below each lane.
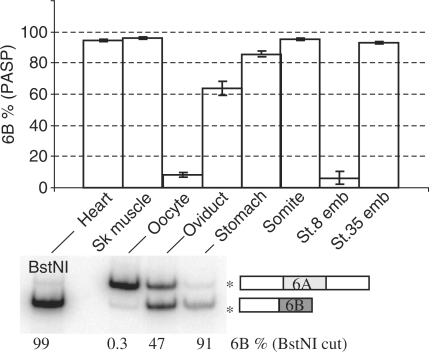
The above results were obtained from mixes of pure RNAs produced by in vitro transcription. It was important therefore to check that the PASP method also applies to complex RNA mixtures extracted from cells or tissues. It was shown previously that exon 6A is essentially retained in mRNA transcribed in oocytes, whereas exon 6B is essentially retained in embryonic somite (presumptive muscles) or adult skeletal and cardiac muscle (14). By PASP, oocytes and early embryos (Stage 8) were found mainly to express a 6A isoform, whereas adult heart, skeletal muscle or stomach, or whole late embryos (Stage 35) and dissected somites were found to express mainly a 6B isoform. An intermediate situation was found for oviducts that contain muscles but are frequently contaminated by oocytes (Figure 4, upper panel). These results were similar to those obtained by radioactive RT–PCR (Figure 4, lower panel) and to previous data (14).
Figure 4.
PASP of α-tropomyosin in Xenopus embryos and organs. RNAs were extracted from oocytes, stage 8 or 35 embryos, dissected embryonic somites or the indicated adult organs, and α-tropomyosin-splicing patterns were analysed by PASP. The percentages of exon 6B (mean ± SD of six independent RNA preparations) are shown (upper panel). Adult heart, oviduct stomach, and oocyte RNA were also analysed by radioactive RT–PCR as in Figure 2B (lower panel). The percentages of 6B exon (restricted by BstNI) are indicated under the gel.
Complex combination of exons skipping and inclusion
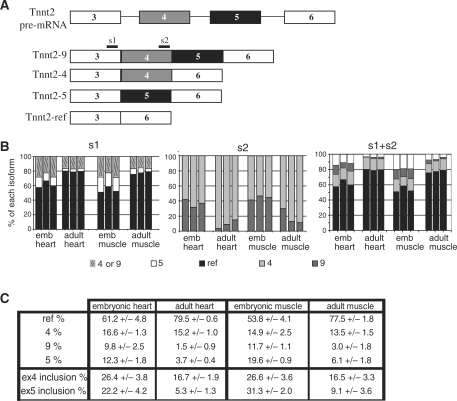
Mutually exclusive exons are only part of the alternative-splicing events that can occur on a pre-mRNA. One characteristic is that they may produce alternatively spliced products that have the same size (if the mutually exclusive exons have the same size, as for exons 6A and 6B of Xenopus α-tropomyosin). In contrast, splicing products of different sizes are obtained from other alternative-splicing events [exon skipping, intron retention, alternative 5′ or 3′ sites (2)]. In addition, more than two isoforms can be produced by alternative splicing. We evaluated therefore whether the PASP method could apply to these complex-splicing events. We choosed murine Tnnt2 (troponin T type 2, also named cTNT for cardiac troponin T) as a model to test this issue. Two constitutive exons (3 and 6) flank two alternative exons (4 and 5). In muscular cells, each alternative exon can be either included or skipped, so that four isoforms of different sizes can be obtained from the same pre-mRNA (Figure 5A) (16).
Figure 5.
PASP analysis of Tnnt2 splicing in mice. (A) Schematic drawing of mouse Tnnt2 pre-mRNA and of the alternative isoforms of Tnnt2 mRNA. The positions of the primers for pyrosequencing are shown. Primer s1 hybridizes in the constitutive exon 3 and sequences either exon 4 (isoforms 4 and 9), exon 5 (isoform 5) or constitutive exon 6 (isoform ref, reference). Primer s2 hybridizes in exon 4 and sequences either exon 5 or exon 6, hence it discriminates between isoforms 4 and 9. (B) PASP of Tnnt2 in the hearts or the skeletal muscles of three different adult or foetal mice using primer s1 or s2 as indicated (two left panels). The right panel (s1 + s2) combines the data shown in the two left panels to calculate the percentages of the four Tnnt2 isoforms. (C) Mean ± SD values were calculated from the above data. Inclusion of exon 4 corresponds to isoforms 4 and 9, and inclusion of exon 5 corresponds to isoforms 5 and 9.
For PASP analysis, total RNA extracted from mouse heart or skeletal muscle was reverse-transcribed, PCR-amplified using a biotinylated reverse primer, and pyrosequenced using two different sequencing primers (Figure 5A). Sequencing primer s1 hybridizes to exon 3. It allows sequencing of the exon spliced 3′ to exon 3 and discriminates between isoforms 4 + 9 (exon 4), 5 (exon 5) and ref (reference, exon 6). It does not discriminate between isoforms 4 and 9, which is achieved by sequencing primer s2. Organs from three different mice (adults or foetuses) were analysed. Results of pyrosequencing using primers s1 and s2 are shown in Figure 5B, left and middle panels, respectively. The percentages of each of the four isoforms were calculated from these series of data (right panel). The results obtained from the three different mice were very similar, and average values were calculated (Figure 5C). These results show that, whereas the splicing patterns in skeletal muscle and heart are similar, the percentages of alternative exons inclusion strongly decreased between embryos and adults. Inclusion of exon 5 decreased more than 3-fold (from 22.2 and 31.3% down to 5.3 and 9.1% in heart and muscle, respectively). A similar decrease in exon 5 inclusion has already been observed (16). Consequently, PASP analysis successfully reproduced results obtained with other methods.
Alternative 3′ terminal exons
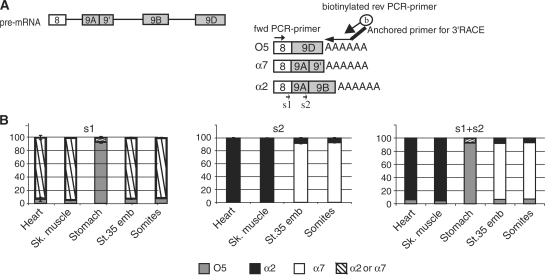
We next used PASP to analyse alternative 3′ terminal exons. In this case, there is no constitutive exon 3′ to the alternative-splicing event that can be targeted by a PCR primer. In 3′RACE, reverse transcription is made with an oligo(dT) primer containing an additional 5′ extension (‘anchor’). Next, a primer corresponding to the anchor is used together with a gene-specific primer for PCR (15). To test 3′RACE-PASP on Xenopus α-tropomyosin mRNA, we made RTs using RNAs from several different Xenopus tissues with an oligo(dT)-anchor, then a PCR with a biotinylated anchor and a forward primer specific for α-tropomyosin exon 8 (Figure 6A). α-Tropomyosin mRNA may have three different terminal exons as depicted in Figure 6A (17). We used two sequencing primers to discriminate between the three different isoforms. Primer s1 hybridizes in exon 8 and discriminates O5 versus α2 and α7, s2 hybridizes in exon 9A and discriminates α2 versus α7.
Figure 6.
Combined PASP and 3′RACE to analyse splicing of alternative 3′ terminal exons. (A) Schematic drawing of the 3′ terminal part of Xenopus α-tropomyosin pre-mRNA. Constitutive exon 8 can be spliced to exon 9D to generate isoform O5. Alternatively, it can be spliced to exon 9A9′. Exon 9A9′ can be used as the terminal exon (yielding isoform α7) or can be spliced to exon 9B via an alternative internal 5′ splice site to create isoform α2. In 3′RACE-PASP, an anchored oligo(dT) primer is used for RT, and a biotinylated reverse oligonucleotide corresponding to the anchor is used for PCR together with a forward primer in constitutive exon 8. Sequencing primer s1 hybridizes within constitutive exon 8, to discriminate between exon 9D, isoform O5, and 9A, isoforms α2 plus α7. Sequencing primer s2 hybridizes within exon 9A, to discriminate between exon 9′, isoform α7, and 9B, isoform α2. (B) 3′RACE-PASP of α-tropomyosin in the indicated tissues. Shown are the results obtained using sequencing primers 1 and 2 (mean ± SD of three different animals), and the results calculated from combining these two sequencings.
Results obtained with sequencing primer s1 are shown in Figure 6B, left panel. Striated muscles, as well as embryos and embryonic somites, mainly spliced exon 9A9′ to exon 8. In contrast, in stomach, isoform O5 was clearly predominant. Results of sequencing with primer s2 (Figure 6B, middle) showed a difference between adult organs that produced mainly α2, and embryonic organs that produced α7. Due to the small amount of exon 9A (binding site for primer s2) in stomach, the sequencing using primer s2 failed and it was not possible to discriminate α2 from α7 isoforms in that organ. Combining the analyses with s1 and s2 (right panel) showed that the main isoforms were O5 in stomach, α2 in adult striated muscles and α7 in embryos, again consistent with previous results (17).
DISCUSSION
Several methods aimed at analysing splicing patterns have been published. Northern blotting may be considered if the isoforms to be discriminate have sufficiently different sizes to be resolved by electrophoresis. An advantage of northern blotting is that the RNA to be analysed are directly detected, avoiding amplification bias that may arise with PCR. However, this approach is virtually no longer used due to a very limited sensitivity (it requires ten to several tens µg of total RNA), and an incapacity to analyse splicing events that lead to isoforms of similar sizes. RNase protection assays (RPA) share with northern blotting the lack of requirement for amplification. The sensitivity of RPA is better than that of northern, although still low (it requires 1–10 µg of total RNA), and RPA is able to analyse more subtle splicing patterns [for instance (18)]. However, the method is time-consuming. Among non-automatable steps, it requires synthesis of labelled RNA probes, hybridization and RNase treatment, gel electrophoresis and drying, and quantification of radioactive signals by densitometry or from phosphorimager files. Furthermore, due to the extreme reactivity of RNase, this assay cannot be used to screen for splicing patterns in different samples with genetic variations including single nucleotide polymorphism. Invader RNA assays rely on the detection of a cleaved DNA probe that occurs when hybridized to a complementary RNA. Consequently, using a set of DNA probe corresponding to the sequences of the different isoforms, it is possible to quantify these isoforms (19). However, this is not amenable to analyses of numerous splicing patterns.
The most widely used methods are real-time and end-point RT–PCR. Real-time RT–PCR is nowadays the best method to quantify RNA levels, due to its unequalled sensitivity and the facility to simultaneously analyse several targets. However, in its simplest (and cheapest) design, that is with a fluorescent DNA intercalating dye, only one amplification product can be quantified with a given pair of primers. Consequently, to analyse alternative-splicing patterns, it is necessary to design several primer pairs, each of them being specific for one mRNA isoform (20,21). To accurately measure the isoform ratios, it is therefore necessary to use primer pairs with the same amplification efficiencies, which is often difficult to achieve. In contrast, in end-point PCR, it is generally possible to use only one pair of PCR primers to amplify all the isoforms, and to individually quantify the isoforms. For example, if one of the PCR primers is radiolabelled, the PCR products are easy to detect and quantify after electrophoresis by autoradiography or preferably phosphorimager analysis. On the basis of differential electrophoretic mobilities, it may be possible to directly discriminate between the PCR products derived from the differentially spliced mRNAs. This method can be used for the analysis of splicing of both endogenous mRNAs and recombinant RNAs transcribed from a minigene [e.g. see (18)].
PASP shares with RT–PCR the advantage of being very sensitive. Furthermore, since only one pair of primers is used to amplify all the isoforms, its efficiency does not need to be carefully controlled. Finally, several kinds of splicing event can be detected by radioactive RT–PCR or PASP: the presence of either of two sequences (mutually exclusive exons), the presence or the absence of one sequence (exon skipping, intron retention, alternative 5′ or 3′ sites) or different 3′ mRNA extremities (alternative terminal exons). Pyrosequencing was previously used in a limited number of cases to analyse splicing patterns (12,13), but neither of these articles examined the capacity of RT–PCR-pyrosequencing to measure the ratios of isoforms that originate from this variety of splicing events. The possibility to use two sequencing primers to analyse complex-splicing patterns was also not investigated in these articles. However, PASP has several additional advantages over radioactive RT–PCR. First, no radioactive isotope is required. Second, it is much more rapid, since all steps are performed in 96-wells plates. Even gel electrophoresis is dispensable if the correct quality controls are used. Starting from cDNAs, the time required to generate a raw data file corresponding to one plate is about 4 h. Since three different devices are used (thermocycler, workstation for binding of amplimeres to streptavidin and treatments, pyrosequencer), it is possible to treat several plates within the same working day, allowing several hundreds of splicing events to be analysed, or to fractionate the experiments into different days. A considerable increase in throughput is possible by automating all of these steps.
A challenger to PASP is non-radioactive RT–PCR followed by detection of the amplimeres by capillary electrophoresis. This technology was used at a medium-throughput level to identify splicing signatures of ovarian cancer (22). However, as for radioactive RT–PCR, analysing splicing patterns by RT–PCR capillary electrophoresis requires that all the isoforms have different sizes, a requirement that does not hold for PASP. Furthermore, the isoforms are identified only by their electrophoretic mobilities in RT–PCR capillary electrophoresis, whereas they are identified by sequencing in PASP. This adds an additional level of specificity that is valuable if some aspecific PCR amplification occurred.
A high-throughput method to analyse splicing patterns is with splicing arrays. Initially, the oligonucleotides spotted on the chips were designed so as to hybridize to exon–exon junctions. Consequently, a positive signal was detected for a given spot only if the tested sample included mRNAs that were subject to the corresponding splicing event (23). The determination of probes was based on the characterization of gene exons from known mRNA. Hence, exon boundaries must be known initially and if a mRNA sequence has not been determined, exons will be missed. Furthermore, the differences in melting temperature between probes and the cross-hybridization that are likely to occur at junction probes have to be included in the analysis. Other array designs include the use of probes to target the internal part of constitutive and alternative exons (24–27). A very high-density human ‘exon array’ has been developed by Affymetrix and most global analyses rely on the estimation of the relative abundance of each exon (28). These chip-based technologies provide valuable information on the frequency of alternative-splicing event and exon usage in a quantitative manner and may also lead to the discovery of new splicing events. These approaches allow the simultaneous analysis of thousands of splicing events that may occur within a given mRNA population and that must subsequently be validated. An alternative to microarrays for genome-wide analyses of splicing patterns is deep-sequencing of mRNAs within a population (2,29).
However, due to the cost of either of these two methods, only a limited number of different RNA populations can be tested. In this respect, PASP is complementary to exon-junctions arrays or deep sequencing. These latter methods allow hundreds of thousands of splicing events to be simultaneously analysed in a RNA population, whereas PASP may compare the splicing patterns of several mRNAs from a large number of different origins. For example, with PASP one could compare several organs with different genetic backgrounds, make precise time-course experiments, or rapidly screen for potentially abnormally spliced RNAs in human patients or model animals. PASP provides detailed knowledge about the abundance of individual alternative-splicing transcripts and the possibility to combine this technology with microarray analysis will be of great value in establishing variations in alternative-splicing among different individuals that could have medical implications such as resistance to drug or disease susceptibility.
FUNDING
Association pour la Recherche contre le Cancer (ARC4003 to L.P., doctoral fellowship to V.A.); Agence Nationale de la Recherche (ANR-07-JCJC-0097-01 to L.P.); Région Bretagne (ARED, doctoral fellowships to A.V.). Undergraduate Research Fellowship to A.D. Funding for open access charge: ANR-07-JCJC-0097-01.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank H Beverley Osborne, Serge Hardy, Marie-Dominique Galibert-Anne and members of the EGD and RTO teams for discussions.
REFERENCES
- 1.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 4.Ahmadian A, Ehn M, Hober S. Pyrosequencing: history, biochemistry and future. Clin. Chim. Acta. 2006;363:83–94. doi: 10.1016/j.cccn.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 5.Tost J, Gut IG. Analysis of gene-specific DNA methylation patterns by pyrosequencing technology. Methods Mol. Biol. 2007;373:89–102. doi: 10.1385/1-59745-377-3:89. [DOI] [PubMed] [Google Scholar]
- 6.Shaw RJ, Liloglou T, Rogers SN, Brown JS, Vaughan ED, Lowe D, Field JK, Risk JM. Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br. J. Cancer. 2006;94:561–568. doi: 10.1038/sj.bjc.6602972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhathena A, Mueller T, Grimm DR, Idler K, Tsurutani A, Spear BB, Katz DA. Frequency of the frame-shifting CYP2D7 138delT polymorphism in a large, ethnically diverse sample population. Drug Metab. Dispos. 2007;35:1251–1253. doi: 10.1124/dmd.107.014993. [DOI] [PubMed] [Google Scholar]
- 8.Alderborn A, Kristofferson A, Hammerling U. Determination of single-nucleotide polymorphisms by real-time pyrophosphate DNA sequencing. Genome Res. 2000;10:1249–1258. doi: 10.1101/gr.10.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun A, Ge J, Siffert W, Frey UH. Quantification of allele-specific G-protein beta3 subunit mRNA transcripts in different human cells and tissues by Pyrosequencing. Eur. J. Hum. Genet. 2005;13:361–369. doi: 10.1038/sj.ejhg.5201334. [DOI] [PubMed] [Google Scholar]
- 10.Bentley L, Nakabayashi K, Monk D, Beechey C, Peters J, Birjandi Z, Khayat FE, Patel M, Preece MA, Stanier P, et al. The imprinted region on human chromosome 7q32 extends to the carboxypeptidase A gene cluster: an imprinted candidate for Silver-Russell syndrome. J. Med. Genet. 2003;40:249–256. doi: 10.1136/jmg.40.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Wu H, Chen Z, Zhou G, Kajiyama T, Kambara H. Dye-free gene expression detection by sequence-tagged reverse-transcription polymerase chain reaction coupled with pyrosequencing. Anal. Chem. 2009;81:273–281. doi: 10.1021/ac801964a. [DOI] [PubMed] [Google Scholar]
- 12.Frey UH, Nuckel H, Dobrev D, Manthey I, Sandalcioglu IE, Eisenhardt A, Worm K, Hauner H, Siffert W. Quantification of G protein Gaalphas subunit splice variants in different human tissues and cells using pyrosequencing. Gene Expr. 2005;12:69–81. doi: 10.3727/000000005783992124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szafranski K, Schindler S, Taudien S, Hiller M, Huse K, Jahn N, Schreiber S, Backofen R, Platzer M. Violating the splicing rules: TG dinucleotides function as alternative 3′ splice sites in U2-dependent introns. Genome Biol. 2007;8:R154. doi: 10.1186/gb-2007-8-8-r154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duriez P, Lesimple M, Allo MR, Hardy S. Alternative splicing of Xenopus alphafast-tropomyosin pre-mRNA during development: identification of determining sequences. DNA Cell Biol. 2000;19:365–376. doi: 10.1089/10445490050043335. [DOI] [PubMed] [Google Scholar]
- 15.Hamon S, Le Sommer C, Mereau A, Allo MR, Hardy S. Polypyrimidine tract-binding protein is involved in vivo in repression of a composite internal/3′-terminal exon of the Xenopus alpha-tropomyosin Pre-mRNA. J. Biol. Chem. 2004;279:22166–22175. doi: 10.1074/jbc.M313809200. [DOI] [PubMed] [Google Scholar]
- 16.Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum. Mol. Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 17.Hardy S, Fiszman MY, Osborne HB, Thiebaud P. Characterization of muscle and non muscle Xenopus laevis tropomyosin mRNAs transcribed from the same gene. Eur. J. Biochem. 1991;202:431–440. doi: 10.1111/j.1432-1033.1991.tb16392.x. [DOI] [PubMed] [Google Scholar]
- 18.Le Sommer C, Lesimple M, Mereau A, Menoret S, Allo MR, Hardy S. PTB regulates the processing of a 3′-terminal exon by repressing both splicing and polyadenylation. Mol. Cell Biol. 2005;25:9595–9607. doi: 10.1128/MCB.25.21.9595-9607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner EJ, Curtis ML, Robson ND, Baraniak AP, Eis PS, Garcia-Blanco MA. Quantification of alternatively spliced FGFR2 RNAs using the RNA invasive cleavage assay. RNA. 2003;9:1552–1561. doi: 10.1261/rna.5840803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 21.Kafert S, Krauter J, Ganser A, Eder M. Differential quantitation of alternatively spliced messenger RNAs using isoform-specific real-time RT-PCR. Anal. Biochem. 1999;269:210–213. doi: 10.1006/abio.1999.4016. [DOI] [PubMed] [Google Scholar]
- 22.Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Gervais-Bird J, Madden R, Paquet ER, Koh C, Venables JP, Prinos P, et al. Multiple alternative splicing markers for ovarian cancer. Cancer Res. 2008;68:657–663. doi: 10.1158/0008-5472.CAN-07-2580. [DOI] [PubMed] [Google Scholar]
- 23.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 24.Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Sugnet CW, Srinivasan K, Clark TA, O'Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanchette M, Green RE, Brenner SE, Rio DC. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev. 2005;19:1306–1314. doi: 10.1101/gad.1314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark TA, Sugnet CW, Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 28.Yeo GW, Xu X, Liang TY, Muotri AR, Carson CT, Coufal NG, Gage FH. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput. Biol. 2007;3:1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Dov C, Hartmann B, Lundgren J, Valcarcel J. Genome-wide analysis of alternative pre-mRNA splicing. J. Biol. Chem. 2008;283:1229–1233. doi: 10.1074/jbc.R700033200. [DOI] [PubMed] [Google Scholar]