Abstract
Viruses use alternative splicing to produce a broad series of proteins from small genomes by utilizing the cellular splicing machinery. Since viruses use cellular RNA binding proteins for viral RNA processing, it is presumable that the splicing of cellular pre-mRNAs is affected by viral infection. Here, we showed that herpes simplex virus type 2 (HSV-2) modifies the expression of promyelocytic leukemia (PML) isoforms by altering pre-mRNA splicing. Using a newly developed virus-sensitive splicing reporter, we identified the viral protein ICP27 as an alternative splicing regulator of PML isoforms. ICP27 was found to bind preferentially to PML pre-mRNA and directly inhibit the removal of PML intron 7a in vitro. Moreover, we demonstrated that ICP27 functions as a splicing silencer at the 3′ splice site of the PML intron 7a. The switching of PML isoform from PML-II to PML-V as induced by ICP27 affected HSV-2 replication, suggesting that the viral protein modulates the splicing code of cellular pre-mRNA(s) governing virus propagation.
INTRODUCTION
As many as two-third of human genes produce two or more isoforms from one gene by alternative pre-mRNA splicing (1,2). Alternative splicing is strictly regulated across cell and tissue types, sex determinations, signal-regulated changes and developmental stages to provide a variety of gene functions depending on the situation (3,4). Alternative splicing also contributes to viral proteomic diversity (5). In the case of human immunodeficiency virus type 1, primary RNA transcripts are alternatively spliced to generate more than 40 different mRNAs (6). As viral RNA processing is often catalyzed by cellular RNA-binding proteins such as serine–arginine rich (SR) proteins (7,8), virus infection sometimes affects the host RNA processing factor (9,10). However, the effects of viral proteins on cellular mRNA splicing have poorly been investigated.
Herpes simplex virus type 2 (HSV-2) is a nuclear replicating DNA virus and a highly adapted human pathogen with rapid lytic replication cycle. When the viral capsid makes its entry into the host cell nucleus, HSV-2 genome DNA localizes to discrete nuclear foci called promyelocytic leukemia nuclear bodies (PML-NBs), also known as nuclear domain 10 (ND10) or PML oncogenic domain (POD) (11). PML was originally characterized as part of a fusion protein with RARα cloned from acute PML patients (12–14). PML is expressed in all normal tissues as well as tumor cell lines; however, its expression is reduced in some progressed tumors (15). The size of PML-NBs varies from 0.2 to 1 μm, and their frequency depends on cell type, cycle, and status (16–19). PML-NBs consist of many kinds of proteins involved in various functions (20,21), and are implicated in various cell processes, including apoptosis, DNA repair, transcription, senescence, cell proliferation, signal transduction and viral pathogenicity (19,20,22–29). PML-NBs have been thought to contribute to intrinsic antiviral defense on the interferon pathway (30). However, recent reports have indicated that PML-NBs provide scaffolds for DNA viruses and promote efficient viral propagation (11,24,31). Thus, a simple model is not sufficient to accommodate all accumulated evidence.
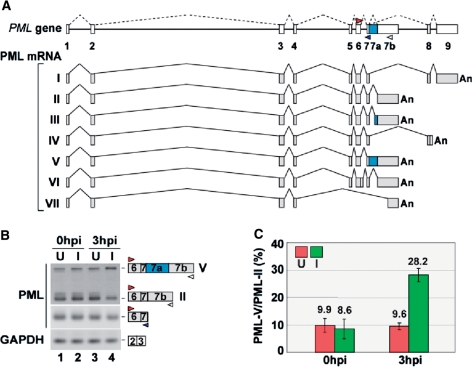
The human PML gene consists of nine major exons, and several alternatively spliced PML transcripts lead to the expression of a multitude of different PML isoforms (32,33), as shown in Figure 1A. PML exon 1 to exon 4, which are common to all isoforms, are translated into the tripartite motif (TRIM) including the RING finger, B-box and coiled-coil domain. On the other hand, PML exon 5 to exon 9 can be alternatively spliced, generating multiple PML isoforms such as PML-I containing the putative exonuclease III domain (34). Furthermore, PML exon 6 contains the nuclear localization signal, and can be excluded for the expression of the cytoplasmic PML-VII isoform, which is essential for TGF-β signaling (27,33). Thus, the PML gene utilizes alternative pre-mRNA splicing for the functional diversity of its own protein products.
Figure 1.
Modulation of PML expression by HSV-2 infection. (A) Schematic representation of the PML gene and mRNA species generated by alternative splicing. The positions of different primer sets used for RT-PCR are indicated by colored arrow heads. (B) RT–PCR analysis (28 cycles) of uninfected (U: lanes 1 and 3) and HSV-2-infected (I: lanes 2 and 4) HeLa cells at MOI 1 (0 hpi and 3 hpi). GAPDH primers were used as a control. (C) Graphic representation of the splicing ratios PML-V/PML-II. Green and red box indicate the splicing ratio in HSV-2-infected (I) and uninfected (U) cells, respectively (n = 3).
In this study, we hypothesized that the conflicting host–virus interactions at PML-NBs may reflect the differential functions of PML isoforms. Consequently, we found that the expression of PML splicing isoforms was switched during HSV-2 infection by alternative splicing. Our group has recently developed a splicing reporter capable of visualization of alternative splicing events in vivo and has also identified novel trans-acting factors (35,36). Here, we newly developed a virus-sensitive splicing reporter whose fluorescent protein expression is changed in HSV-2-infected cells, and we identified infected cell protein 27 (ICP27) as an alternative splicing regulator. ICP27 preferentially interacted with PML pre-mRNA and suppressed intron 7a removal presumably by modulating 3′ splice site (ss) recognition of the cellular trans-acting factor.
MATERIALS AND METHODS
Construction of plasmids
We constructed the reporter minigene PML E6-7b by amplifying the PML genomic DNA fragments spanning from exon 6 to exon 7b and cloning to a pcDNA3 vector (Invitrogen). Constructs expressing myc-tagged HSV-2 cDNAs and Flag-tagged ICP27 were prepared by inserting PCR products from the cDNA of HSV-2-infected HEK293 cells into the pcDNA3 vector. A construct for the preparation of the T-REx293/Flag-ICP27 stable cell line was prepared by inserting PCR products from the cDNA of HSV-2-infected HEK293 cells into the pcDNA5/FRT vector in accordance with the manufacturer's protocol (Invitrogen). Constructs expressing RFP-PML-II and RFP-PML-V were prepared by inserting PCR products from the cDNA of HEK293 cells into the pmRFP-C1 vector (Clontech). The constructs of ICP27 mutant M15, PML-small interference (siRNA)-resistant mutants, PML intron 7a-deletion mutant d1 and PML 3′ ss mutants m1-m4 were made using a QuikChange II XL kit (Stratagene). The cloning primers are shown in Supplementary Table S1.
RT–PCR
RNA was isolated from intact, HSV-2-infected cells, and transfected cells with sepasol RNA I (Nacalai). For reverse transcription, 500 ng of total RNA from each sample was incubated with oligo (dT)20 and Superscript II reverse transcriptase (Invitrogen). PCR products were analyzed by 2% agarose gel electrophoresis, followed by ethidium bromide staining. As shown in Figure 1C, semi-quantitative PCR products were analyzed using the 2100 Bioanalyzer (Agilent Technologies) following the protocol stated in the manuals. The PCR primers are shown in Supplementary Table S2.
Viruses and antibodies
HSV-2 strain G [HSV-2 (G)] and Venus-HSV-2 strain YK381 were used at multiplicities of infection (MOI) based on their plaque-forming unit titers in Vero cells. Anti-Flag M2 antibody, anti-c-myc antibody, anti-ICP27 (8.F.137B) and Pan-PML antibody (H-238) were purchased from Sigma, Nacalai, Abcam and Santa Cruz, respectively. PML-II- and PML-V-specific sera were a kind gift from H. de The (18).
Construction of YK381 expressing Venus fluorescent protein
In pRB5198 (37), a region containing the bidirectional polyadenylation [poly(A)] signals of HSV-1(F) UL21 and UL22 was cloned into pBluescript II KS(+) (Stratagene). To construct p26.5-Venus, a SacI-BstEII fragment of pRB4090 (a kind gift from Dr Bernard Roizman) containing the promoter region of HSV-1(F) UL26.5 and a BamHI–EcoRI fragment of Venus/pCS2 (38) containing the entire open reading frame of Venus were subsequently cloned into pRB5198. The resultant plasmid contains a Venus expression cassette driven by the UL26.5 promoter. The BamHI fragment, 8.2 kb, encoding UL1 to a part of UL5 of the HSV-2 186 viral genome was cloned into pBluescript II KS(+) to yield p2UL3-4. p2UL3-4pac, in which the PacI site was introduced into the region between poly (A) signals for HSV-2 186 UL3 and UL4 genes, was generated by site-specific mutagenesis. p26.5-Venus in 2UL3-4 was constructed by cloning the SacI–KpnI fragment of p26.5-Venus containing the Venus expression cassette into the PacI site of p2UL3-4pac and used as a transfer plasmid for the generation of a recombinant virus YK381 expressing Venus fluorescent protein driven by the UL26.5 promoter, as described previously (38). YK381 exhibits an identical phenotype to wild-type HSV-2 186 in cell cultures and mouse models (T.M. and Y.K., unpublished observation).
Virus infection
HeLa and HEK293 cells were seeded into 6-well plates, and cells reaching 100% confluence were infected with HSV-2(G), as stated in the relevant figure legend. To examine the role of PML in HSV-2 replication, HeLa cells were transfected with PML siRNA for 48 h before the viral infection. For the PML splicing isoform rescue experiments shown in Figure 8F, HeLa cells were transfected with PML siRNA and then with plasmids containing either the siRNA-resistant RFP-PML-II mutant or RFP-PML-V mutant on the next day. HeLa cells were infected with HSV-2(G) for 24 h after plasmid transfection. The production of infectious HSV-2 was assessed by plaque assay in Vero cells (39). Vero cells grown in 6-well plates to nearly 100% confluency were infected with a diluted whole-cell extract of HSV-2(G)-infected cells. The whole cell extract was prepared by three repeats of freeze-thaw disruption of HSV-2(G)-infected cells. After 1 h adsorption, the inoculum was removed and the cell monolayer was overlaid with Dulbecco's modified Eagle medium (DMEM) containing 1% fetal calf serum (FCS) and 0.16 mg/ml pooled human immunoglobulin (Sigma). The overlaid medium was removed after 2 days of infection, and the infected cell monolayer was fixed and stained with methanol and 0.1% crystal violet, respectively.
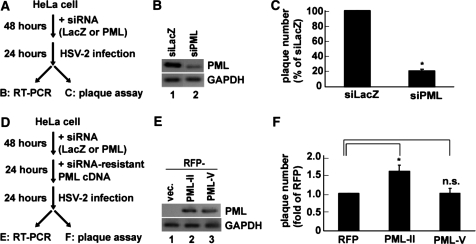
Figure 8.
Differential function of PML isoforms in relation to HSV-2 infection. (A) Experimental procedure for examining the role of PML in HSV-2 viral production. Knockdown of all endogenous PML splicing variants in HeLa cells was performed using siRNA against PML (siPML4). After 48 h of incubation, HeLa cells were infected with HSV-2 at MOI 0.01 for 24 h. RT–PCR analysis (B) and plaque assay (C) were performed to determine knockdown efficiency and viral titer, respectively. (B) RT–PCR analysis, using PML ex3-ex4 primers, of siLacZ- or siPML4-treated HeLa cells. (C) Production of infectious HSV-2 in either siLacZ- or siPML4-treated HeLa cells as assessed by plaque assay (n = 4). (D) Experimental procedure for the rescue assay of PML splicing variants. After 48 h of siRNA treatment, the expression plasmids of either RFP or siRNA-resistant PML II or PML-V were transfected for 24 h. Following 24 h of infection with HSV-2 at MOI 0.01, RT–PCR analysis (E) and plaque assay (F) were performed. (E) RT–PCR analysis of siPML4-treated HeLa cells transfected with the RFP vector, RFP-PML-II siRNA resistant (siR), or RFP-PML-V siR expression plasmids. Details are described in ‘Materials and Methods’ section. (F) Production of infectious HSV-2 in HeLa cells siPML4-treated and then transfected with each PML plasmid as assessed by plaque assay (n = 4). *P < 0.001; n.s., not significant.
Confocal immunofluorescence microscopy
Twenty-four hours after the transfection, cells on 15-mm glass coverslips were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.5% Triton X-100 for 20 min, blocked with the blocking solution (0.2% Gelatin, 1% BSA, 0.05% Tween 20 in PBS) for 30 min and reacted with a diluted primary antibody in the blocking solution for 12 h at 4°C. After incubation, cells were washed extensively with a washing solution [0.05% Tween 20 in PBS (pH 8.0)], incubated for 2 h at room temperature with the appropriate secondary antibody diluted in the blocking solution, and then washed three times with the washing solution. The cells were analyzed under a confocal microscope (Olympus; FV1000; confocal aperture, 300 μm) through a PLAPO 60× NA:1.40 objective lens. ICP27 and PML-NB signals were collected sequentially by excitation with a 488 nm laser and a 543 nm laser, respectively.
Transfection of plasmids and siRNAs
HeLa, HEK293, and Vero cells were grown in DMEM (Nacalai) supplemented with 10% heat-inactivated FCS, and then maintained by the standard protocol. HeLa and HEK293 cells were transfected with GeneJuice (Roche) or Lipofectamine 2000 (Invitrogen) and TransIT293 (Mirus), respectively. All plasmids used for transfection were prepared using a Maxiprep kit (Biogene). HEK293 cells were grown in a monolayer in 6-well plates, and then co-transfected with 500 ng per well of a reporter plasmid and 500 ng of either myc-vector or plasmids expressing myc-tagged HSV-2 proteins.
siRNA transfection was performed in 12-well plates by transfecting HeLa cells with 20 pmol per well of siRNAs against either the LacZ reporter (Invitrogen) or PML (Invitrogen, UCUUGGAUACAGCUGCAUCUUUCCC), using Lipofectamine RNAiMAX (Invitrogen).
In vitro splicing reaction
The PCR products of human T7-PML wt (exon 7, 53 nt; intron 7a, 641 nt; and exon 7b, 213 nt) and T7-PML d1 (exon 7, 53 nt; intron 7a, 257 nt; and exon 7b, 213 nt) were used as the DNA template for T7 transcription. Pre-mRNA substrates were m7GpppG-capped and 32P-labeled by in vitro transcription. In vitro splicing reactions were performed in 20 μl volumes at 30°C under the conditions described by Krainer et al. (40). As shown in Figure 6E, highly purified Flag-ICP27 was added in the reaction system. After the reaction, RNA was subjected to denaturing PAGE analysis and autoradiography.
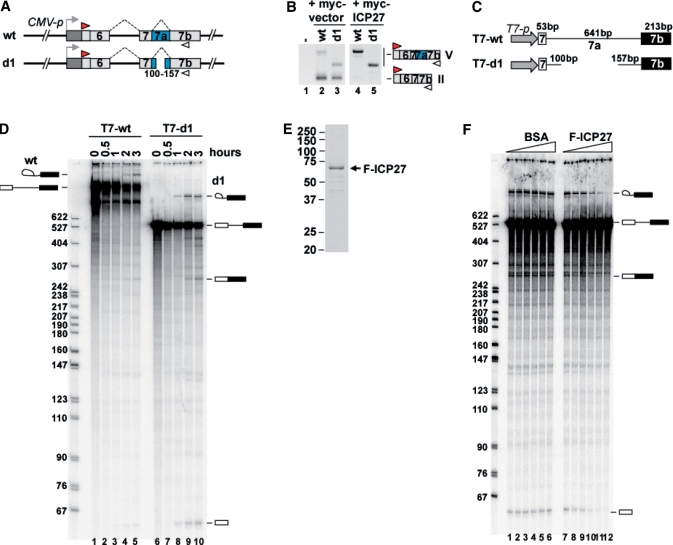
Figure 6.
Inhibition of PML pre-mRNA splicing by ICP27. (A) Schematic representation of E6-7b wt and deletion-mutant splicing reporters, d1. The reporter E6-7b d1 contains a 100-bp fragment downstream of the 5′ ss and a 157-bp fragment upstream of the 3′ ss of intron 7a. (B) RT–PCR analysis of HEK293 cells transfected with Flag-vector (lane 1), and the combination of either E6-7b wt (lanes 2 and 4) or d1 (lanes 3 and 5) with either myc-vector (lanes 2 and 3) or myc-ICP27 (lanes 4 and 5). (C) Schematic representation of three E7-7b DNA templates, T7-wt (exon 7: 53 bp; intron 7a: 641 bp; exon 7b: 213 bp) and T7-d1 (exon 7: 53 bp; intron 7a: 257 bp; exon 7b: 213 bp), which are driven by the T7 promoter. (D) In vitro splicing of T7-wt (lanes 1–5) and T7-d1 (lanes 6–10) for the indicated time. The size of the pBR322 MspI-digested DNA marker is indicated on the right side of the panel. The pre-mRNA, mature mRNA and splicing intermediates are described on both sides of the panel. (E) A 10% SDS–PAGE and CBB staining of the purified Flag-ICP27 from the nuclear extract of T-Rex 293 cells expressing Flag-ICP27. (F) Effects of ICP27 on T7-d1 splicing in vitro. Two-fold dilution series (0, 25, 50, 100, 200 and 400 ng) of BSA (lanes 1–6) and highly purified Flag-ICP27 (lanes 7–12) were added to the in vitro splicing reaction.
RNA immunoprecipitation
Nuclear extracts were prepared from HEK293 cells transfected with plasmids expressing Flag alone, Flag-ICP27 wild-type and M15. 32P-labeled PML exon 7-7b, β-globin and δ-crystallin RNA were prepared by in vitro transcription with [α-32P]UTP and T7 RNA polymerase (Takara). Gel purified RNA was incubated for 30 min at 30°C with each nuclear extract in 20 μl of RNA binding buffer [20 mM HEPES–KOH (pH 7.9), 100 mM KCl, 5% Glycerol, 1% Triton X-100, 2 mM DTT and 0.2 mM PMSF] supplemented with 0.4 U of an RNase inhibitor (Promega), and immunoprecipitation was then performed with Flag-M2 antibody. Each RNA was purified by the protein removal and ethanol precipitation. The purified RNAs were analyzed by denaturing PAGE and imaging using a phosphoimage analyzer (FLA-3000G; FUJIFILM).
Preparation of stable transfected T-REx293 cells
T-REx293 cells (the HEK293 cell line expressing a tetracycline repressor obtained from Invitrogen) were grown following the manufacturer's protocol in DMEM (Nacalai) containing 10% heat-inactivated FCS and 10 mg/ml blasticidin (Invitrogen). To generate stably transfected tetracycline inducible cell lines, T-REx293 cells were co-transfected with either pcDNA5-Flag or pcDNA5-Flag-ICP27. After 48 h post-transfection, these transfected cells were grown in the presence of 0.1 mg/ml hygromycin B following the manufacturer's protocol to select stable transfected clones.
RESULTS
HSV-2 infection switches the expression pattern of PML isoforms
We first compared the splicing patterns of PML transcripts from HeLa cells, with and without HSV-2 infection. Total RNAs of HeLa cells were collected immediately (0 h post-infection, 0 hpi) and 3 h post-infection (3 hpi). The RT–PCR analysis, using the primer sets indicated in Figure 1A, showed that the amounts of mRNAs containing PML-I-specific exon 8-9 and PML-II-specific exon 6-7b were reduced in the HSV-2-infected HeLa cells (Supplementary Figure S1A and Figure 1B, top, lane 4), whereas the amounts of PML mRNA containing constitutive exon 1-4 (Supplementary Figure S1A) and exon 6-7 (Figure 1B. middle, lane 4), and GAPDH mRNA were much less reduced (Figure 1B, bottom, lane 4 and Supplementary Figure S1A).
Moreover, we examined other cellular mRNAs containing constitutive introns (Aly/REF intron 4, Lamin A/C intron 6), alternative introns (Lamin A/C introns 9 and 10) and minor introns (P120 intron 6, HPS1 intron 16) by RT–PCR analysis. The expression levels and splicing patterns of the mRNAs containing these introns were not significantly changed (Supplementary Figure S1B). Interestingly, the amount of mRNA containing PML-V-specific exon 7-7a-7b was increased in the HSV-2-infected cells (Figure 1B, top). The quantitative analysis also showed that the ratio of PML splicing isoforms (PML-V/PML-II) was enhanced by HSV-2 infection (Figure 1C, 2.9-fold in 3 hpi). Consistent with the reduction in PML-II-specific exon 7-7b, PML-II foci recognized by PML-II-specific antibody were reduced in the HSV-2-infected cells (Supplementary Figure S2). These observations indicate that HSV-2 infection changes the alternative splicing of PML pre-mRNA.
The splicing reporter of PML gene is sensitive to HSV-2 infection
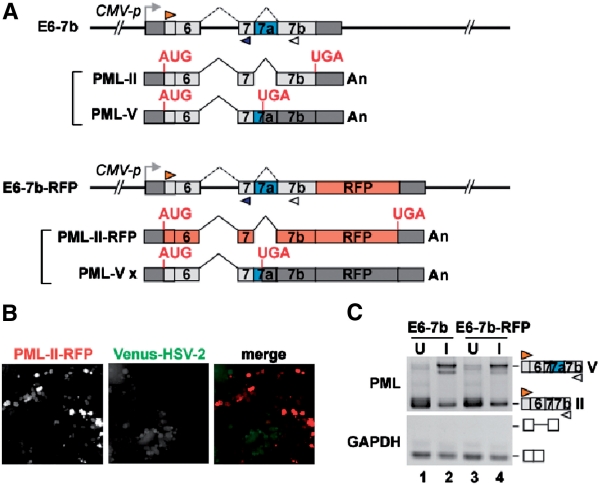
We have recently succeeded in visualizing the tissue-specific regulation of alternative pre-mRNA splicing in live nematodes by developing a transgenic reporter system with fluorescent proteins (35). Using this system, we have identified three genes as alternative splicing regulators (35,36). To clarify the HSV-2-mediated regulatory mechanism of alternative splicing of the PML pre-mRNA, we prepared splicing reporter minigene constructs containing the region from exon 6 to exon 7b of the PML gene without or with the red fluorescent protein (RFP) coding region, and named them E6-7b and E6-7b-RFP, respectively (Figure 2A). The splicing reporter E6-7b-RFP can express RFP only when intron 7a is spliced out and the PML-II form is produced. Notably, when we transfected E6-7b-RFP into HeLa cells, RFP was detected in the transfected cells. To examine whether or not we can use E6-7b-RFP as a screening tool of viral splicing regulators, we observed RFP expression in Venus-HSV-2-infected cells. As we initially assumed, the RFP expression was reduced in the Venus-HSV-2-infected cells in a viral titer-dependent manner (Figure 2B and Supplementary Figure S3), suggesting that the minigene reporter reflects the splicing alteration induced by HSV-2 infection. To examine whether or not the reduction of RFP expression was caused by HSV-2 regulated-pre-mRNA splicing, we performed RT–PCR analysis of HeLa cells transfected with the E6-7b (Figure 2C, lanes 1 and 2) or E6-7b-RFP (Figure 2C, lanes 3 and 4) reporter and then infected with HSV-2 at MOI 10 for 3 hours (Figure 2C, lanes 2 and 4). Regardless of the additional RFP coding region, the promotion of intron 7a retention in the HSV-2-infected HEK293 cells was confirmed by RT–PCR (Figure 2C, lanes 3 and 4). The incomplete retention of intron 7a by HSV-2 infection might be caused by the remaining uninfected cells under the experimental condition, as shown in Figure 1B, lanes 4. These observations indicate that the E6-7b-RFP reporter can be used as a screening tool of viral splicing regulators.
Figure 2.
Visualization of virus-sensitive splicing reporter of the PML gene. (A) Schematic representation of two splicing reporters. The splicing reporter involving the PML gene from exon 6 to exon 7b with (E6-7b-RFP) or without (E6-7b) the RFP coding region, and mRNAs derived from the reporter. Predicted ORFs are indicated in red (PML-II-RFP) or gray (PML-V x), and UTRs in dark gray. The positions of different primer sets used for RT–PCR are indicated. This reporter is driven by the CMV promoter. (B) Microscopy analysis of Venus-HSV-2-infected cells. HeLa cells were infected with Venus-HSV-2 at MOI 0.01. After 3 h infection, E6-7b-RFP was transfected into the cells. After 24 h infection, RFP expression was reduced particularly in Venus-HSV-2 infected cells, although RFP was well detected in uninfected cells. Venus-HSV-2 is indicated in green. (C) RT–PCR analysis of uninfected (U) and infected (I) HeLa cells at MOI 10 (3 hpi), into which either E6-7b or E6-7b-RFP constructs were transfected.
The viral protein ICP27 switches the splicing pattern of PML pre-mRNA
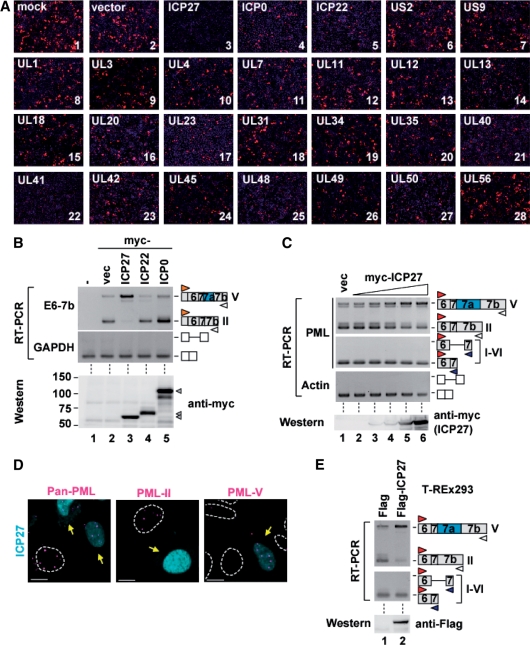
To identify viral splicing regulator(s), HSV-2 cDNAs coding 26 viral nuclear proteins of HSV-2 were co-transfected with PML E6-7b-RFP into HeLa cells, and it was demonstrated that RFP expression was lost only in the cells transfected with the ICP27 or UL41 expression vector (Figure 3A, panels 3 and 22). Furthermore, the reduction of RFP expression was also checked by western blot (Supplementary Figure S4). ICP27 overexpession dramatically switched the mRNA expression pattern of the PML E6-7b reporter from the PML-II-specific exon 7-7b form to the PML-V-specific exon 7-7a-7b form (Figure 3B, top, lane 3), whereas other viral nuclear proteins such as ICP22 or ICP0 did not affect the splicing (Figure 3B, top, lanes 4 and 5). Also, the amounts of expressed viral proteins were almost the same (Figure 3B, bottom). In contrast to ICP27, the overexpression of UL41 encoding a viral nuclease (41) suppressed PML-II-RFP expression owing to the promotion of PML-II-RFP RNA degradation (data not shown). To investigate whether ICP27 can induce switching of the endogenous PML splicing isoform, we transfected myc-tagged ICP27 cDNA and then checked for endogenous PML transcripts by RT–PCR analysis using a combination of exon 6-forward and exon 7b-reverse primers. The results showed that the intron 7a retention of endogenous PML transcripts was promoted in proportion to the amount of transfected ICP27 cDNA (Figure 3C, top and bottom); however, ICP27 did not inhibit the removal of other introns in PML pre-mRNA (Figure 3C, upper middle) or actin pre-mRNA (Figure 3C, lower middle). The foci in myc-tagged ICP27-expressing cells that were recognized by anti-PML-II antibody were decreased (Figure 3D, center), whereas those recognized by anti-PML-V sera were increased (Figure 3D, right) compared with untransfected HeLa cells. Moreover, the endogenous PML-II mRNA was switched to PML-V mRNA in a Flag-tagged ICP27 stable expressing cell line (T-REx293/Flag-ICP27), whose transcription was regulated by tetracycline addition (Figure 3E). However, as far as we have checked, the removal of the other introns containing the constitutive introns, alternative intron and minor introns was not affected by Flag-ICP27 (Supplementary Figure S1C). These observations indicate that ICP27 preferentially switches the expression of PML isoforms from PML-II to PML-V.
Figure 3.
Retention of PML intron 7a by ICP27. (A) Microscopy analysis of HeLa cells transfected with the combinations of the E6-7b-RFP construct and each HSV-2 cDNA performed by detecting RFP signaling (red). DNAs were stained with DAPI (blue). (B) RT–PCR analysis of HEK293 cells transfected with the combinations of E6-7b constructs and plasmids containing myc-tagged HSV-2 cDNA (B, upper panels). Whole-cell extracts of HEK293 cells transfected with these plasmids were subjected to western blot analysis for myc-tag (B, bottom). (C) RT–PCR, using the primers shown in Figure 1A, and western blot analysis of HEK293 cells transfected with 2-fold dilution series of myc-tagged ICP27. (D) Immunofluorescence of HeLa cells transfected with myc-tagged ICP27. PML-NBs were stained with anti-Pan-PML (left), PML-II specific (center) and PML-V specific (right) sera. Scale bar, 10 μm. Yellow arrows indicate ICP27-transfected cells. (E) RT–PCR analysis, using the primers shown in Figure 1A, and western blot analysis for Flag-tag of T-REx293 expressing Flag-tag peptides and Flag-ICP27.
Interestingly, the foci recognized by anti-Pan-PML antibody were unchanged or only slightly decreased (Figure 3D, left). Although PML-I-specific exon 8-9 transcripts were decreased in HSV-2-infected cells, they were not changed in ICP27-expressing cells (Supplementary Figure S1A). These results suggest that PML-I mRNA expression is regulated by other viral factors other than ICP27 or cellular signal induced proteins (e.g. interferon stimulating factors) in HSV-2-infected cells.
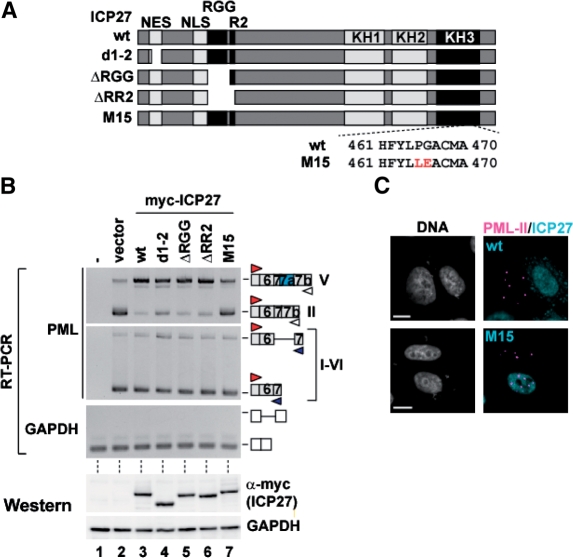
KH3 domain of ICP27 is required to switch the PML isoform
As shown in Figure 4A, ICP27 contains defined functional domains, including a nuclear export signal (NES), the arginine- and glycine-rich motif (RGG box), the RGG box with nearby sequence (R2) and 3 KH domains. To identify the ICP27 region required to promote retention of intron 7a of PML, we prepared a series of ICP27 mutants lacking NES (d1-2), the RGG box (ΔRGG), and the RGG box plus R2 (ΔRR2) (Figure 4A). Overexpression of the deletion mutants of ICP27 retained the promotion activity for intron 7a retention (Figure 4B, top, lanes 4–6), whereas ICP27 mutant M15 containing altered residues at 465 and 466 (P465L/G466E) in the KH3 domain (42) failed to switch the alternative splicing of PML pre-mRNA (Figure 4B, top, lane 7). Wild-type ICP27 and the mutants were equally expressed in individual transfected cells (Figure 4B, bottom), and the foci of PML-II were unaffected by M15 expression (Figure 4C, lower panels). These results indicate that the KH3 domain of ICP27 is critical for the switching of the PML isoform from PML-II to PML-V.
Figure 4.
Effects of ICP27 mutants on PML RNA splicing. (A) Schematic representation of the structure of ICP27 mutants. (B) RT–PCR and western blot analysis of HEK293 cells transfected with either myc-tagged ICP27 wild-type or mutant plasmids. (C) Immunofluorescence of HeLa cells transfected with either myc-tagged ICP27 wild-type or the M15 plasmid, stained with anti-myc antibody and PML-II specific sera.
Since the ICP27 protein of HSV-1 is highly homologous to that of HSV-2, which we used, we next tested whether HSV-1 ICP27 protein could also affect the PML splicing or not. The ICP27 of HSV-1 also promoted the intron retention of PML splicing (Supplementary Figure S5A, lane 4). The ICP27 gene is conserved in all members of the herpesvirus family. The UL69 gene (cytomegalovirus; CMV), EB2 (or SM) gene (Epstein–Barr virus; EBV) and ORF57 gene (Kaposi's sarcoma-associated herpesvirus; KSHV and the herpesvirus saimiri; HVS) are homologues of the ICP27 gene of each virus. In addition, the amino acids PG at 465 and 466 in the KH3 domain of ICP27, which are essential for splicing regulation of PML pre-mRNA (Figure 4B), are conserved within herpesviridae (Supplementary Figure S5B) (43). Furthermore, ICP27 and ORF57 reportedly promote intron retention of viral RNA (44,45). Taken together, these observations strongly suggest that ICP27-mediated alternative splicing of PML pre-mRNA is a common feature in the herpesvirus family.
ICP27 is specifically associated with PML pre-mRNA
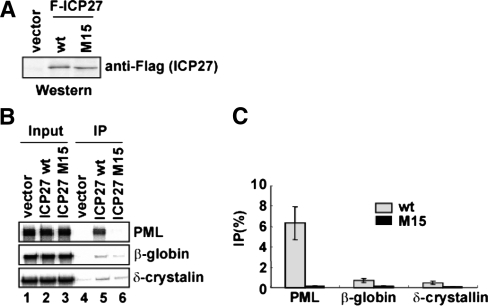
To examine whether or not ICP27 preferentially associates with PML pre-mRNA, we performed RNA immunoprecipitation assay. Three pre-mRNAs containing human PML exon 7 to exon 7b, human β-globin exon 1 to exon 2 and chicken δ-crystallin exon 14 to exon 15, which were synthesized by in vitro transcription and labeled with [α-32P] UTP, were mixed with the nuclear extract from HEK293 cells expressing Flag-tagged ICP27 (Figure 5A). Following incubation, RNAs were immunoprecipitated with anti-Flag antibody. The results showed that Flag-ICP27 was co-immunoprecipitated with PML pre-mRNA more efficiently (9.3- to 14.0-fold) than β-globin and δ-crystallin pre-mRNAs (Figure 5B, lane 5 and 5C, gray bars). Further, RNA immunoprecipitation experiment using the nuclear extract from HEK293 cells expressing Flag-ICP27 mutant M15 demonstrated that Flag-ICP27 mutant M15 was not co-immunoprecipitated with these pre-mRNAs (Figure 5B, lane 6 and 5C, black bars). Moreover, the amounts of Flag-ICP27 wild-type and M15 proteins were almost equally expressed (Figure 5A), indicating that ICP27 specifically associated with PML pre-mRNA, at least among cellular pre-mRNAs, and that the associations depended on the KH domains, which were presumably the RNA binding sites.
Figure 5.
RNA binding specificity of ICP27. (A) Western blot analysis, using anti-Flag antibody, of nuclear extract from HEK293 cells transfected with Flag-vector and Flag-ICP27 wild-type (wt) and M15 expression plasmids. (B) RNA immunoprecipitation. Each 32P-labeled in vitro transcript was incubated with the nuclear extracts from HEK293 cells expressing Flag-tag or Flag-tagged ICP27, and then immunoprecipitated with anti-Flag antibody. (C) Quantitation of three independent experiments is shown. Error bars represent SDs (n = 3).
ICP27 directly inhibits the removal of PML intron 7a
To examine whether or not ICP27 directly inhibits PML pre-mRNA splicing, we performed an in vitro splicing reaction. The DNA template T7-wt containing a T7 promoter and PML exon 7 to exon 7b was prepared by PCR and used for T7 transcription (Figure 6C). The in vitro splicing reaction resulted in the T7-wt transcript slightly generating splicing intermediates (Figure 6D, lanes 1–5), which led us to modify the T7-wt DNA template. We prepared the mutant d1 which has deletion in intron 7a, as described in Figure 6A. Transfection of the d1 construct showed that the PML d1 transcript was efficiently removed from intron 7a and that intron 7a of d1 RNA was retained by myc-ICP27, as well as the PML wt transcript (Figure 6B, lanes 3 and 5). These results indicate that the PML d1 mutant can be useful for in vitro splicing reactions. The in vitro splicing of the T7-d1 transcript showed that it was more efficiently spliced than the T7-wt transcript (Figure 6D, lanes 6–10). Next, we examined the effects of ICP27 on PML splicing. Flag-ICP27 was purified from T-REx293 cells expressing Flag-ICP27, and then the highly purified Flag-ICP27 was added to the in vitro splicing reaction using d1 mutant RNA. The results showed that the productions of T7-d1 spliced mRNA and intermediates were inhibited by the addition of purified Flag-ICP27 in a dose-dependent manner (Figure 6F, lanes 7–12), but not by BSA (Figure 6F, lanes 1–6). These results indicate that ICP27 directly inhibits PML pre-mRNA splicing.
3′ Splice site of intron 7a of PML pre-mRNA is critical for ICP27-mediated switching
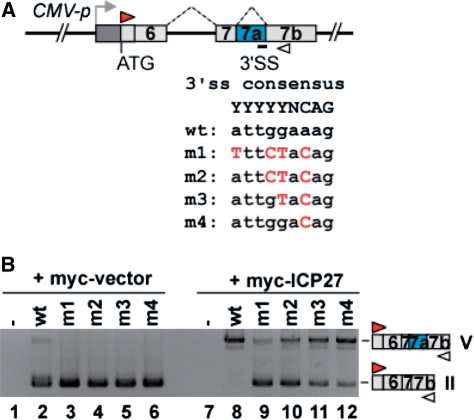
The alternative splicing depends on the utilization of the 5′ ss and 3′ ss at the end of introns (3). The 5′ ss includes a GU dinucleotide at the intron end encompassed within a less conserved consensus sequence. At the other end of the intron, the 3′ ss region has conserved a polypyrimidine tract capable of associating with U2AF65/35, followed by a terminal AG at the extreme 3′-end of the intron (YYYYYNCAG) (46). In contrast, the 3′ ss sequence of PML intron 7a is a purine-rich sequence (ATTGGAAAG). To evaluate the involvement of the atypical 3′ ss of intron 7a for the ICP27-mediated switching from PML-II to PML-V, we mutated the 3′ ss region of intron 7a in the PML E6-7b reporter to match with the 3′ ss consensus sequence (Figure 7A, m1-m4). RT–PCR analysis showed that the ICP27-induced retention of PML intron 7a was diminished in proportion to the number of mutated nucleotides at the 3′ ss (Figure 7B, lanes 8–12).
Figure 7.
The weak 3′ ss essential for ICP27-mediated alternative splicing of PML pre-mRNA. (A) Scheme of the E6-7b construct wild-type and a series of point mutations at the 3′ ss in PML intron 7a. (B) Results of RT–PCR analysis of HeLa cells transfected with each E6-7b cis-mutant and myc-tagged ICP27.
PML splicing isoform II contributes to production of infectious HSV-2
To examine the differential roles of PML isoforms in host-virus interactions, we attempted rescue experiments with a siRNA-resistant PML isoform in PML pre-knocked down HeLa cells. We first efficiently knocked down all PML isoforms from HeLa cells with siRNA (siPML4) against PML exon 4. Following 48 h of siRNA transfection, HSV-2 was infected and then HSV-2 titer was measured by plaque assay (Figure 8A). siPML4 efficiently knocked down endogenous PML isoforms (Figure 8B, lane 2), whereas control siRNA (siLacZ) did not knock down any of them (Figure 8B, lane 1). Plaque assay revealed that HSV-2 replication was suppressed by ∼80% in the PML-knocked-down HeLa cells (Figure 8C), consistent with the results of the immunofluorescence analysis (Supplementary Figure S6). We then transfected these cells with a plasmid encoding either RFP, RFP-PML-II or RFP-PML-V with a silent mutation that makes the PML isoform resistant to siPML4, and thereafter evaluated the plaque-forming activity of the virus (Figure 8D). The rescue experiment showed that RFP-PML-II increased the HSV-2 plaque number (∼1.6-fold RFP), but RFP-PML-V did not (Figure 8F), although the expression levels of the RFP-PML-II and RFP-PML-V mRNAs were similar (Figure 8E, lanes 2 and 3). The result showed that the difference in the rescue experiments between only RFP and RFP-PML-II appeared smaller than the reduction of HSV-2 replication by the knock-down of all PML isoforms. One possible explanation is the transfection efficiencies of RFP (43%), RFP-PML-II (33%) and RFP-PML-V (34%) (Supplementary Figure S7). These observations suggest that the difference in HSV-2 plaque number may become larger if all cells expressed siRNA-resistant PML isoforms, suggesting that PML-II plays a specific role in efficient HSV-2 replication.
DISCUSSION
In this study, we identified ICP27 as a splicing regulator of PML pre-mRNA, showing the possibility that the viral protein switches alternative splicing of specific cellular RNA in association with functional alteration.
Selective splicing regulation by ICP27
HSV ICP27 is one of the immediate early (IE) proteins of HSV and plays an essential role in the expression of viral late genes (47,48). ICP27 interacts and co-localizes with cellular splicing factors, such as small nuclear ribonucleoproteins (snRNPs) (49), SR proteins, SR protein kinase 1 (SRPK1) (50) and spliceosome-associated protein 145 (SAP145) (51), and is suggested to inhibit the splicing of cellular pre-mRNAs by impairing spliceosomal assembly (50). However, considering that some essential viral pre-mRNAs such as ICP0 have introns and require splicing, it is less likely that HSV suppresses constitutive pre-mRNA splicing. Interestingly, we observed that ICP27 specifically bound to PML pre-mRNA (Figure 5B, lane 5) and selectively switched the isoform from PML-II to PML-V by promoting the retention of PML intron 7a (Figure 3E). Our results indicate that ICP27 is an alternative splicing factor, although it does not inhibit all constitutive splicing. Moreover, the observed in vitro splicing suggested that ICP27 directly suppressed the early stage of spliceosome assembly on PML intron 7a, because splicing intermediates (5′ exon and lariat-3′ exon) were reduced by ICP27 addition (Figure 6F).
The selectivity of ICP27-dependent regulation may be related to the relative weakness of the 3′ ss of PML intron 7a. PML intron 7a was not retained even in the presence of ICP27 when we mutated the 3′ ss to the consensus sequence for U2AF65/35 (Figure 7B). Therefore, it is also relevant to examine whether ICP27 selectively binds to the 3′ ss of PML intron 7a by competing with U2AF65/35. Sokolowski et al. (52) showed that ICP27 binds to many different viral sequences by yeast three-hybrid screening of an HSV-1 genomic library; however, the target RNA sequence of ICP27 remains to be elucidated. Since the ICP27 homolog contains more cellular targets than PML pre-mRNA (53), in vitro systematic evolution of ligands by exponential enrichment (SELEX) and in vivo UV cross-linking immunoprecipitation (CLIP) are currently under way in our laboratory to determine the consensus target sequence of ICP27.
A possible role of ICP27-mediated mRNA export on alternative splicing
Transfection experiments revealed that ICP27 promoted the intron 7a retention (Figure 3B, lane 3). Although our results showed that ICP27 directly inhibited the splicing of PML intron 7a in vitro (Figure 6F), ICP27-mediated alternative splicing modulation is still possible to be explained by another mechanism, a promotion of pre-mRNA export by ICP27. It has been shown that ICP27 shuttles between the nucleus and the cytoplasm in host cells (54,55), and that ICP27 homologues stimulate the export of viral mRNAs by interacting with the mRNA export adaptor REF (56–61). The splicing reaction at the weak ss of PML intron 7a may take longer time than that at the consensus ss, and the accelerated mRNA export induced by ICP27 does not provide sufficient time for PML pre-mRNA to be spliced in the nucleus. These two possibilities are not mutually exclusive. It will be of interest to investigate if ICP27 promotes export of pre-mRNAs that harbor introns with weak 3′ ss, such as PML.
The role of ICP27 KH domain on PML alternative splicing
ICP27 M15 mutant failed to alter the expression of PML splicing isoform (Figure 4B, lane 7), suggesting that the KH3 domain is critical for the regulation of splicing. Therefore, we checked whether other KH domains are also essential for splicing regulation or not. RT–PCR analyses indicate that the mutants lacking either the KH2 domain or the KH3 domain were not able to completely switch PML splicing isoforms (data not shown). These results suggest that both the KH2 and the KH3 domains are required for altering the splicing. Unfortunately, the mutant lacking the KH1 domain was not expressed enough for the splicing experiment. It is highly likely that all of three KH domains are required for the regulation of the splicing.
Differential roles of PML isoform
PML isoforms have differential localizations and functions (32–34). For example, p53 interacts with PML-IV, but not with PML-III, and thus it was speculated that p53-PML-IV interaction was required for p53 recruitment into PML-NBs to modulate cell survival (62). Moreover, retinoblastoma protein (pRB) interacts more efficiently with PML-IV than with PML-II, suggesting that pRB–PML-IV interaction plays an important role in the regulation of cell differentiation and proliferation (63). In the present study, we showed that PML-II can enhance HSV-2 proliferation in HeLa cells (Figure 8F). In addition to HSV-2, adenovirus type 5 E4-ORF3 protein reportedly rearranges PML-NBs to a track-like structure by specifically interacting with PML-II (64,65), suggesting that PML-II may function as a modulator of the viral environment. However, as shown in the present study, HSV-2 infection switches the PML isoform from PML-II to PML-V. Apparently, this switching reduces the efficiency of viral replication (Figure 8F) and is thus not favorable to the virus, although it may also contribute to persistent viral infection by controlling viral replication rate through a negative feedback mechanism. Another possibility is that this switching is relevant to host defense to virus production. Because ICP27 is expressed at early stage of virus infection, the infected cell may utilize ICP27 as virus antigen to monitor virus infection and then the cells may try to reduce virus replication rate by producing PML-V, which results in delay of the expansion of virus infection to surrounding cells.
The expression of PML isoforms differs in various cell lines or tissues. For example, PML-I is the dominant form expressed in the brain but not in the liver (18). However, the regulation mechanisms of the cell type- or tissue-specific alternative splicing of PML pre-mRNA have not been clarified to date. In this study, using our original splicing reporter with RFP, we demonstrated that the viral protein ICP27 regulates the switching of the PML isoform from PML-II to PML-V by promoting the retention of PML intron 7a; PML-I, however, was not affected by ICP27 (Supplementary Figure S1A). The mechanism underlying the HSV-induced decrease in PML-I remains to be elucidated. The splicing reporter system used in the present study may pave the way for identifying the cellular regulators of PML alternative splicing, along with viral regulatory proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid (to M.H.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan; the National Institute of Biomedical Innovation (NIBI); Japanese Ministry of Education, Global Center of Excellence (GCOE) Program; International Research Center for Molecular Science in Tooth and Bone Diseases; Program for Improvement of Research Environment for Young Researchers from Special Coordination Funds for Promoting Science and Technology (SCF) commissioned by MEXT of Japan (to N.K.). Funding for open access charge: [Ministry of Education, Culture, Sports, Science, and Technology of Japan (to T.N.)].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to H. de Thé for providing the anti-PML-II and PML-V specific sera. We thank A. Hiraishi and C. Parlayan for critically reading the article; and members of M.H.'s Laboratory for helpful comments.
REFERENCES
- 1.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 4.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz S, Felber BK, Fenyo EM, Pavlakis GN. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J. Virol. 1990;64:5448–5456. doi: 10.1128/jvi.64.11.5448-5456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoltzfus CM, Madsen JM. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr. HIV Res. 2006;4:43–55. doi: 10.2174/157016206775197655. [DOI] [PubMed] [Google Scholar]
- 7.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 8.Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv. Exp. Med. Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- 9.Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature. 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 10.Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y, Suzuki M, Yamamoto N, Herzenberg LA, Herzenberg LA, et al. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl Acad. Sci. USA. 2006;103:11329–11333. doi: 10.1073/pnas.0604616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishov AM, Maul GG. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 13.Kakizuka A, Miller WH, Jr, Umesono K, Warrell RP, Jr, Frankel SR, Murty VV, Dmitrovsky E, Evans RM. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 14.Goddard AD, Borrow J, Freemont PS, Solomon E. Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science. 1991;254:1371–1374. doi: 10.1126/science.1720570. [DOI] [PubMed] [Google Scholar]
- 15.Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, Verbel DA, Cordon-Cardo C, Pandolfi PP. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J. Natl Cancer Inst. 2004;96:269–279. doi: 10.1093/jnci/djh043. [DOI] [PubMed] [Google Scholar]
- 16.Terris B, Baldin V, Dubois S, Degott C, Flejou JF, Henin D, Dejean A. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res. 1995;55:1590–1597. [PubMed] [Google Scholar]
- 17.Koken MH, Linares-Cruz G, Quignon F, Viron A, Chelbi-Alix MK, Sobczak-Thepot J, Juhlin L, Degos L, Calvo F, de The H. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene. 1995;10:1315–1324. [PubMed] [Google Scholar]
- 18.Condemine W, Takahashi Y, Zhu J, Puvion-Dutilleul F, Guegan S, Janin A, de The H. Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res. 2006;66:6192–6198. doi: 10.1158/0008-5472.CAN-05-3792. [DOI] [PubMed] [Google Scholar]
- 19.Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:165–170. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- 20.Zhong S, Salomoni P, Pandolfi PP. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2000;2:E85–E90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann TG, Will H. Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ. 2003;10:1290–1299. doi: 10.1038/sj.cdd.4401313. [DOI] [PubMed] [Google Scholar]
- 22.Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W, Pandolfi PP. The function of PML in p53-dependent apoptosis. Nat. Cell Biol. 2000;2:730–736. doi: 10.1038/35036365. [DOI] [PubMed] [Google Scholar]
- 23.Dellaire G, Bazett-Jones DP. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays. 2004;26:963–977. doi: 10.1002/bies.20089. [DOI] [PubMed] [Google Scholar]
- 24.Ching RW, Dellaire G, Eskiw CH, Bazett-Jones DP. PML bodies: a meeting place for genomic loci? J. Cell Sci. 2005;118:847–854. doi: 10.1242/jcs.01700. [DOI] [PubMed] [Google Scholar]
- 25.Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 26.Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi PP. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 27.Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431:205–211. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- 28.Everett RD. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene. 2001;20:7266–7273. doi: 10.1038/sj.onc.1204759. [DOI] [PubMed] [Google Scholar]
- 29.Regad T, Chelbi-Alix MK. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene. 2001;20:7274–7286. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- 30.Chee AV, Lopez P, Pandolfi PP, Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 2003;77:7101–7105. doi: 10.1128/JVI.77.12.7101-7105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maul GG, Ishov AM, Everett RD. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 32.Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- 33.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 34.Condemine W, Takahashi Y, Le Bras M, de The H. A nucleolar targeting signal in PML-I addresses PML to nucleolar caps in stressed or senescent cells. J. Cell Sci. 2007;120:3219–3227. doi: 10.1242/jcs.007492. [DOI] [PubMed] [Google Scholar]
- 35.Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat. Methods. 2006;3:909–915. doi: 10.1038/nmeth944. [DOI] [PubMed] [Google Scholar]
- 36.Ohno G, Hagiwara M, Kuroyanagi H. STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes Dev. 2008;22:360–374. doi: 10.1101/gad.1620608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 2003;77:1382–1391. doi: 10.1128/JVI.77.2.1382-1391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugimoto K, Uema M, Sagara H, Tanaka M, Sata T, Hashimoto Y, Kawaguchi Y. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 2008;82:5198–5211. doi: 10.1128/JVI.02681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka M, Kodaira H, Nishiyama Y, Sata T, Kawaguchi Y. Construction of recombinant herpes simplex virus type I expressing green fluorescent protein without loss of any viral genes. Microbes Infect./Institut Pasteur. 2004;6:485–493. doi: 10.1016/j.micinf.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 40.Krainer AR, Maniatis T, Ruskin B, Green MR. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 41.Strom T, Frenkel N. Effects of herpes simplex virus on mRNA stability. J. Virol. 1987;61:2198–2207. doi: 10.1128/jvi.61.7.2198-2207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice SA, Lam V. Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. J. Virol. 1994;68:823–833. doi: 10.1128/jvi.68.2.823-833.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soliman TM, Silverstein SJ. Herpesvirus mRNAs are sorted for export via Crm1-dependent and -independent pathways. J. Virol. 2000;74:2814–2825. doi: 10.1128/jvi.74.6.2814-2825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sedlackova L, Perkins KD, Lengyel J, Strain AK, van Santen VL, Rice SA. Herpes simplex virus type 1 ICP27 regulates expression of a variant, secreted form of glycoprotein C by an intron retention mechanism. J. Virol. 2008;82:7443–7455. doi: 10.1128/JVI.00388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majerciak V, Yamanegi K, Allemand E, Kruhlak M, Krainer AR, Zheng ZM. Kaposi's sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J. Virol. 2008;82:2792–2801. doi: 10.1128/JVI.01856-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3' splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 47.Sacks WR, Greene CC, Aschman DP, Schaffer PA. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 1985;55:796–805. doi: 10.1128/jvi.55.3.796-805.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice SA, Knipe DM. Gene-specific transactivation by herpes simplex virus type 1 alpha protein ICP27. J. Virol. 1988;62:3814–3823. doi: 10.1128/jvi.62.10.3814-3823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandri-Goldin RM, Hibbard MK. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum, and the C terminus appears to be required for this interaction. J. Virol. 1996;70:108–118. doi: 10.1128/jvi.70.1.108-118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sciabica KS, Dai QJ, Sandri-Goldin RM. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 2003;22:1608–1619. doi: 10.1093/emboj/cdg166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bryant HE, Wadd SE, Lamond AI, Silverstein SJ, Clements JB. Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J. Virol. 2001;75:4376–4385. doi: 10.1128/JVI.75.9.4376-4385.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sokolowski M, Scott JE, Heaney RP, Patel AH, Clements JB. Identification of herpes simplex virus RNAs that interact specifically with regulatory protein ICP27 in vivo. J. Biol. Chem. 2003;278:33540–33549. doi: 10.1074/jbc.M302063200. [DOI] [PubMed] [Google Scholar]
- 53.Verma D, Swaminathan S. Epstein-Barr virus SM protein functions as an alternative splicing factor. J. Virol. 2008;82:7180–7188. doi: 10.1128/JVI.00344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phelan A, Clements JB. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J. Gen. Virol. 1997;78(Pt 12):3327–3331. doi: 10.1099/0022-1317-78-12-3327. [DOI] [PubMed] [Google Scholar]
- 55.Mears WE, Rice SA. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology. 1998;242:128–137. doi: 10.1006/viro.1997.9006. [DOI] [PubMed] [Google Scholar]
- 56.Hiriart E, Farjot G, Gruffat H, Nguyen MV, Sergeant A, Manet E. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 2003;278:335–342. doi: 10.1074/jbc.M208656200. [DOI] [PubMed] [Google Scholar]
- 57.Malik P, Blackbourn DJ, Clements JB. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 2004;279:33001–33011. doi: 10.1074/jbc.M313008200. [DOI] [PubMed] [Google Scholar]
- 58.Williams BJ, Boyne JR, Goodwin DJ, Roaden L, Hautbergue GM, Wilson SA, Whitehouse A. The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem. J. 2005;387:295–308. doi: 10.1042/BJ20041223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lischka P, Toth Z, Thomas M, Mueller R, Stamminger T. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol. Cell Biol. 2006;26:1631–1643. doi: 10.1128/MCB.26.5.1631-1643.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soliman TM, Sandri-Goldin RM, Silverstein SJ. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 1997;71:9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koffa MD, Clements JB, Izaurralde E, Wadd S, Wilson SA, Mattaj IW, Kuersten S. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 2001;20:5769–5778. doi: 10.1093/emboj/20.20.5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fogal V, Gostissa M, Sandy P, Zacchi P, Sternsdorf T, Jensen K, Pandolfi PP, Will H, Schneider C, Del Sal G. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 2000;19:6185–6195. doi: 10.1093/emboj/19.22.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alcalay M, Tomassoni L, Colombo E, Stoldt S, Grignani F, Fagioli M, Szekely L, Helin K, Pelicci PG. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol. Cell Biol. 1998;18:1084–1093. doi: 10.1128/mcb.18.2.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doucas V, Ishov AM, Romo A, Juguilon H, Weitzman MD, Evans RM, Maul GG. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 65.Hoppe A, Beech SJ, Dimmock J, Leppard KN. Interaction of the adenovirus type 5 E4 Orf3 protein with promyelocytic leukemia protein isoform II is required for ND10 disruption. J. Virol. 2006;80:3042–3049. doi: 10.1128/JVI.80.6.3042-3049.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.