Abstract
Promoterless gene trap vectors have been widely used for high-efficiency gene targeting and random mutagenesis in embryonic stem (ES) cells. Unfortunately, such vectors are only effective for genes expressed in ES cells and this has prompted the development of expression-independent vectors. These polyadenylation (poly A) trap vectors employ a splice donor to capture an endogenous gene's polyadenylation sequence and provide transcript stability. However, the spectrum of mutations generated by these vectors appears largely restricted to the last intron of target loci due to nonsense-mediated mRNA decay (NMD) making them unsuitable for gene targeting applications. Here, we present novel poly A trap vectors that overcome the effect of NMD and also employ RNA instability sequences to improve splicing efficiency. The set of random insertions generated with these vectors show a significantly reduced insertional bias and the vectors can be targeted directly to a 5′ intron. We also show that this relative positional independence is linked to the human β-actin promoter and is most likely a result of its transcriptional activity in ES cells. Taken together our data indicate that these vectors are an effective tool for insertional mutagenesis that can be used for either gene trapping or gene targeting.
INTRODUCTION
Since the advent of homologous recombination and the development of embryonic stem (ES) cell technologies, mouse genetics has become the principal approach for elucidating molecular mechanism(s) in mammalian biology. In the wake of a complete genome sequence, a major focus of the mouse genetics community is to generate mutations in every identifiable gene in the genome (‘genome saturation’). Attempts to reach genome saturation have involved multiple technologies including high-throughput targeting via BAC recombineering and gene trapping. Gene trapping is an attractive insertional mutagenesis strategy as it relies on the random introduction of DNA constructs into ES cells and does not involve the generation of targeting vectors for homologous recombination. In addition to generating a bank of mutations in already annotated genes, gene trap vectors also continue to aid in gene identification, generating insertions into novel and previously uncharacterized transcripts. To fully exploit gene trapping as a resource for genome scale mutagenesis, the International Gene Trap Consortium (IGTC) was established to coordinate screening efforts, produce a searchable database and establish a public repository of mouse ES cell lines harboring gene trap insertions in every, or most genes of the mouse genome (1).
The most widely used gene trap vectors are promoterless and contain a splice acceptor (SA) sequence upstream of a selectable marker or reporter gene (‘SA-type’ or ‘promoter trap vectors’) (2–4). When this type of vector integrates into a gene transcribed in ES cells, the gene trap cassette's selectable marker is expressed under the control of the endogenous gene's promoter. Because the selectable marker in these vectors lacks a promoter, they can also be particularly effective when combined with homology arms and used for gene targeting (‘targeted trapping’) (5). However, these vectors have the caveat that they depend on the expression of the disrupted gene. To circumvent this problem, vectors have been designed that include a heterologous promoter driving expression of a selectable marker that lacks a poly A sequence, but include a splice donor (SD). Integration of this type of vector upstream of a functional poly A sequence then generates a stable transcript and drug resistance (6–8). The uncoupling of antibiotic resistance from the requirement for endogenous gene expression implies that poly A trap vectors can theoretically disrupt a wider range of genes including those that are not expressed in ES cells as well as non-protein coding transcripts.
To date, based on data compiled by the IGTC, gene trap insertions have been identified in approximately 40% of the genome (http://www.sanger.ac.uk/PostGenomics/genetrap/). These have been generated predominantly through the use of various SA-type gene trap vectors, both plasmid- and retroviral-based (1), but also include some poly A trap vector data. While, this is a significant accomplishment, the rate of trapping new genes is progressively diminishing and is currently ∼10% (i.e. one new gene is trapped for every 10 gene trap clones isolated) (9). This trend has also been observed in a privately funded high-throughput gene trap initiative (10), where the occurrence of new insertion events appears to have plateaued at 60% genome coverage.
Based on the rate of accumulation of new mutations, it appears that ∼60–70% of all mouse genes are predicted to be accessible to SA-type vectors (9,11). The accessibility of a locus to trapping (‘trappability’) correlates with both gene size and expression levels (12). Furthermore, different gene trap vectors appear to each have their own insertional ‘hot spots’ (12) and it is now widely accepted that genome saturation can be achieved only through the use of a wider range of vector designs and approaches targeting this ‘untrappable’ 30–40% of the mouse genome (9). Towards this goal, a number of public high-throughput initiatives (KOMP, EUCOMM, NorCOMM) focusing mainly on the conditional disruption of currently untrapped loci by gene targeting have emerged (13,14).
While the inclusion of poly A trap vectors in the IGTC data set has been limited, the uncoupling of antibiotic resistance from the requirement for endogenous gene expression should enhance the accessibility of the genome to trapping. However, these vectors have generally not performed up to expectations, as the combination of a strong internal promoter with an inefficient SD can produce antibiotic resistance even in the absence of splicing onto an endogenous transcript (10,15). Moreover, despite attempts to resolve these problems with different promoter and SD combinations (10,16–18) or through the insertion of a synthetic intron within the selectable marker gene (19), these vectors are limited because of the action of an mRNA surveillance mechanism called nonsense-mediated mRNA decay (NMD) (17,20). NMD promotes the selection of insertional events in the 3′-most intron of target sequences as it triggers the degradation of the selectable marker's transcript based on the presence of a premature termination codon (17–19). Although this bias is still compatible with the engineering of 3′ insertions that generate non-null hypomorphic alleles, it clearly jeopardises the mutagenic potential of these vectors and limits their application in homologous recombination. While, the generation of poly A trap vectors that include an IRES downstream of the selectable marker appears to resolve this problem (17), this cassette has not been shown to function in a targeting context and the majority of poly A trap vectors still suffer from an NMD-based handicap.
Here, we describe a set of novel, expression-independent trapping vectors that we show to be effective for both gene trapping and gene targeting. These constructs contain a novel poly A trap cassette that includes a previously uncharacterized SD sequence derived from the rabbit β-globin gene and a cis-acting mRNA destabilizing AU-rich element (ARE) from the human GM-CSF gene (21). We show that our vectors function efficiently as poly A traps and that the ARE improves the performance of the rabbit β-globin SD sequence by reducing the incidence of background SD read-through events. Interestingly, these vectors showed little 3′ most intron bias in random integration, and could also be targeted to the first intron of the Oct4 locus. Importantly, we show that this ability to overcome the extreme 3′ bias generated by NMD is dependent on the human β-actin promoter, and can be transferred to other gene trap vectors via promoter swaps. We also demonstrate the successful targeting through the use of one of our poly A trap vectors of Protocadherin 21 (Pcdh21), a gene that is expressed at very low levels in ES cells and has been shown to be inaccessible to targeted trapping approaches employing expression-dependent SA-type vectors. Taken together, we show that these expression-independent vectors are efficient tools for gene identification and disruption of previously uncharacterized novel transcripts.
MATERIALS AND METHODS
Vector construction
Vector pEHygroSD2ARE was constructed in two parts using a PacI and AscI linker for assembly of the entire vector. The construction of the 5′ side of this vector (pEGFPβhygro) was described in Tsakiridis et al. (22), and the 3′ side was engineered from pBKnSD (a kind gift from Bill Skarnes), by inserting a synthetic oligonucleotide containing the ARE sequence into the NcoI of the second intron of the rabbit β-globin gene. This entire cassette was flanked on the 5′ side with PacI and 3′ side with AscI and combined with pEGFPβhygro.
Vector pGTIV3 was constructed by permutation of the 5′ side of vector pGTIV2 (a kind gift from Bill Stanford) to the 3′ side of pEHygroSD2ARE. The HindIII restriction site was destroyed and replaced with an AgeI site upstream of pEHygroSD2ARE (SA). An AgeI and an AscI restriction sites were introduced by PCR flanking the 5′ cassette of pGTIV2 vector. The 5′ AgeI/AscI side of pEHygroSD2ARE was then replaced with the 5′ AgeI/AscI fragment of pGTIV2 to construct pGTIV3.
The retroviral gene trap vector GEP-IV3 was constructed according to the following steps: (i) the retroviral cassette pGEP− (23) was digested with ClaI/XhoI and ligated to a XhoI/ClaI polylinker containing FseI and SgrAI restriction sites: (5′-TCGAGCGCCGGTGATTTAAATCACGTCACTGCCCAAAGTTTAAACGGCCGGCCAT-3′) (Vector pGEP-MCS). (ii) The plasmid vector pGTIV3 was digested using NdeI/AgeI and ligated to a NdeI–FseI–loxP–AgeI oligo (5′-TATGTCACGGCCGGCCATAACTTCGTATAGCATACATTATACGAAGTTATA-3′). The resulting vector was then digested with MfeI/PacI and ligated to a MfeI–SgrAI–loxP–PacI oligo (5′-AATTGCGCCGGTGATAACTTCGTATAGCATACATTATACGAAGTTATTTAAT-3′) (Vector pGTIV3-FseI-SgrAI). (iii) Vector pGTIV3-FseI-SgrAI was digested with FseI/SgrAI and ligated to FseI/SgrAI-digested pGEP-MCS to generate the pGEP-IV3 vector.
The Oct4 targeting vector pGTIV3-Oct4 was assembled according to the following steps: (i) pOct4-5′HindIII/NarI plasmid which contains the Oct4 genomic DNA region starting from a HindIII site 104 nt 3′ of the Oct4 AUG in exon 1 to a NarI site in exon 5, was digested with SphI and ligated to a SphI–AgeI–NheI–FseI–NcoI–MfeI–PacI–SgrAI–SphI polylinker (5′-CACCGGTATGCTAGCGGCCGGCCCCATGGCAATTGTTAATTAACGCCGGTGGCATG-3′) (Vector p246-MCS). (ii) Vector pGTIV3 was digested with AgeI/MfeI and ligated to AgeI/MfeI-digested p246-MCS plasmid to generate pGTIV3-Oct4. The control Oct4 targeting vector pGTIV2-Oct4 was constructed during the following cloning steps: (i) Vector pGTIV2 was digested with NotI and ligated to a NotI–FseI–loxP–NotI oligo (5′-GGCCGCATTTAAATCGTACTGCCCAAAGGCCGGCCGTTTAAACGTTAACGC-3′) (Vector pGTIV2-FseI). (ii) Vector pGTIV2-FseI was digested with KpnI/AflIII and ligated to a KpnI–SgrAI–AflIII oligo (5′-CCGTACTGCCCAAACGCCGGTGCGTACGCACGTGGTTTAAACGTTAACA-3′) (Vector pGTIV2-FseI-SgrAI). (iii) Vector pGTIV2-FseI-SgrAI was digested with FseI/SgrAI and ligated to FseI/SgrAI digested plasmid p246-MCS to generate pGTIV2-Oct4. For the generation of the pGTIV3-Oct4-PGK and pGTIV2-Oct4-β-actin Oct4-targeting vectors, both pGTIV3-Oct4 and pGTIV2-Oct4 constructs were digested using NsiI/RsrII and the resulting NsiI-PGK-RsrII fragment was ligated to the NsiI/RsrII-digested pGTIV3-Oct4 vector while the resulting NsiI-β-actin P-RsrII fragment was joined to the NsiI/RsrII-digested pGTIV2-Oct4 plasmid.
The Pcdh21 targeting vector pGTIV3-Pcdh21 was constructed according to the following steps: (i) a targeting vector plasmid containing the Pcdh21 5′ and 3′ homology arms (5 kb and 3 kb, respectively) flanking via AscI sites the gene trap vector pTT0,1,2 (5) (a kind gift from Roland Friedel) was digested using AscI to release the gene trap construct and (ii) subsequently ligated to a AscI–PacI–FseI–SwaI–AflII–EcoRI–SgrAI–MfeI–AscI polylinker (5′-CGCGCCACGGCCACAAGTTCAGCGTGTCTTAATTAAGGC CGGCCATTTAAATCTTAAGGAATTCCGTACTGCCCATCCGCCGGTGCAATTGGG-3′). (iii) The resulting Pcdh21-MCS plasmid was then digested with FseI/SgrAI and ligated to the FseI/SgrAI digested pGTIV3-FseI-SgrAI plasmid described above (see GEP-IV3 construction) to generate the pGTIV3-Pcdh21 vector.
All restriction enzymes were supplied by New England Biolabs. All DNA fragment extractions were performed using the Zymoclean Gel DNA Recovery Kit (Zymo Research). All ligations were performed using the Quick Ligation kit (New England Biolabs) according to the manufacturer's instructions.
Retrovirology
For virus production, the Phoenix ecotropic packaging cell line (24) was transiently transfected with the pGEP-IV3 proviral plasmid either by Lipofectamine2000 (Invitrogen) or by calcium phosphate precipitation and the viral particle-containing supernatant was harvested as described previously (24,25).
Cell culture and manipulation
E14TG2a cells were maintained as described previously (22,26–28). Linearized plasmid vectors were introduced into cells by electroporation using a GenePulserTM (Bio-Rad) as described previously (4). After ∼7–10 days of G418 selection (Sigma) (200–250 μg/ml) resistant colonies were picked, replicated and expanded. For infection with the pGEP-IV3 vector, E14TG2a cells were plated at a density of 3 × 105 cells on 10 cm2 dishes and after overnight culture they were incubated with ES medium containing 1 : 10 dilution of the viral supernatant and 4 μg/ml polybrene (Chemicon International). After ∼20 h, the medium was replaced and cells were subjected to G418 selection (250 μg/ml) for ∼10 days. The NMD inhibition experiment involved the plating of the selected ES cell lines in 12-well plates and their treatment with 100 µg/ml of cycloheximide (CHX) for 4 h. This was followed by RNA extraction using the RNeasy mini kit (Qiagen).
The cell line carrying the EGFP reporter at the Nanog locus (TNG) has been previously described (29). In these cells, an EGFP-loxP-frt-IRES-puro-frt-SPA-MAZ-loxP cassette (30) was placed in the first exon of Nanog such that EGFP was initiated by the Nanog AUG codon. Cre-deleted derivative cell lines were established by transient transfection of CAG Cre-IRES PuroR-pA using Fugene (Roche) according to the manufacturer's instructions. The following day ES cells were trypsinized, re-plated at clonal density and expanded. Cre-mediated deletion of the loxP-frt-IRES-pac-frt-SPA-MAZ-loxP cassette was verified by preparing DNA from individual clones and Southern analysis for the expected alteration in restriction enzyme digestion pattern.
PCR analysis and sequencing
For 3′RACE PCR total RNA was extracted from individual trapped ES cell clones grown in 6-well plates using TRIZOL (Invitrogen). RNA samples were then poly A enriched using the Oligotex mRNA kit (Qiagen) and 3′RACE PCR was performed by employing the GeneRacer kit (Invitrogen) according to the manufacturer's instructions. The sequences of the gene trap vector specific primers (NeoA and NeoB) that were used in conjunction with the kit's primers are shown in Supplementary Table 1. PCR reactions were performed using the Peltier Thermal Cycler PTC-200 (MJ Research). The thermal cycling parameters employed were: 94°C for 2 min; 94°C for 30 s, 72°C for 2 min (X5); 94°C for 30 s, 70°C for 2 min (X5); 94°C for 30 s, 68°C for 30 s and 72°C for 2 min (X20). The resulting PCR products were cleaned up using the PCR Product Pre-Sequencing kit (USB Corporation) and sequenced directly using primers NeoC or T2 (for sequences see Supplementary Table 1). The sequencing reactions were carried out using the BigDye Terminator Cycle Sequencing ready Reaction Kit (Perkin Elmer) by the School of Biological Sciences Sequencing Service (Ashworth Laboratories, University of Edinburgh).
For semi-quantitative RT–PCR analysis, RNA was isolated from wild-type E14TG2a cells as described above and cDNA synthesis was performed using an oligodT primer and Superscript III Reverse Transcriptase (Invitrogen) following the manufacturer's instructions. Primer sequences are shown in Supplementary Table 1. The thermal cycling parameters employed were: 94°C for 2 min; 94°C for 30 s, 55°C for 30 s and 72°C for 1 min (X25–30).
For quantitative real-time PCR analysis, total RNA from CHX-treated cells and controls (1 µg) was used for cDNA synthesis employing SuperScript III RT (Invitrogen) according to the manufacturer's instructions. Real-time PCR was then performed with the LightCycler 480 using the Universal Probe Library System (Roche). The sequences of the primers used are shown in Supplementary Table 1. Primers for EGFP were used with UPL probe no. 67, for Neo with probe no. 51 and for TATA-binding protein (TBP) with probe no. 97.
Bioinformatics
Vector integration sites were determined by performing BLAST analysis of the RACE tags using the NCBI (blastn, http://www.ncbi.nlm.nih.gov/BLAST/), Ensembl Mouse (http://www.ensembl.org/Mus_musculus/blastview) and UCSC (http://genome.ucsc.edu/index.html?org=Mouse) databases. Conservation analysis of the RACE tags was performed using the UCSC web genome server (http://genome.ucsc.edu/index.html?org=Mouse).
Southern blotting
For Southern blot analysis of the Oct4-targeted clones a 324 bp sequence located ∼1141 bp upstream of the first exon of Oct4 was used as a probe. For Southern blot analysis of Pcdh21-targeted clones, a 733 bp sequence located directly downstream of Pcdh21 exon 10 was used. Probes were prepared by PCR using genomic DNA from wild-type E14TG2a ES cells and a high-fidelity Phusion DNA polymerase (Finnzymes). Genomic DNA was extracted from targeted ES cell clones using the DNeasy kit (Qiagen). DNA samples were digested with EcoRI (Oct4) or EcoRV (Pcdh21) (New England Biolabs). Southern blotting was performed according to ref. (31). Radioactive signals were detected using Hyperfilm MP (Amersham) or a Fuji Scanner FLA-3000 (analysed with Aida Image Analyser v.400 software).
Flow cytometry and fluorescence microscopy analysis
Oct4-targeted clones were analyzed for Venus expression by flow cytometry which was performed using a FACSCalibur bench top cytometer equipped with a 488 nm laser (Becton Dickinson) without compensation. Data were analyzed using Cellquest software (Becton Dickinson). Controls were mock transfected cells and non-electroporated wild-type cells. EGFP expression was visualized using an Olympus IX51 inverted microscope (Olympus). Image acquisition and processing were carried out using the Volocity (Improvision) software package.
RESULTS
The inclusion of an ARE improves poly A trap SD function
To design an efficient, expression-independent, gene trap vector we first sought to address the background problems associated with poly A trap vectors. These vectors are known to generate drug-resistant ES cell clones from non-genic vector integrations where antibiotic resistance is a consequence of transcriptional readthrough rather than proper SD usage. We used an RNA instability element (ARE) from GM-CSF (21) to destabilize transcripts generated from the vector in the absence of true splicing events to a downstream endogenous SA sequence. The ARE sequence was placed into the second intron of the β-globin gene (Figure 1) so that when splicing occurs via the strong constitutive SD from exon 2 of β-globin, this instability element is removed from the gene trap transcript. This SD cassette was placed downstream of the human β-actin promoter driving expression of the neomycin resistance gene (neo).
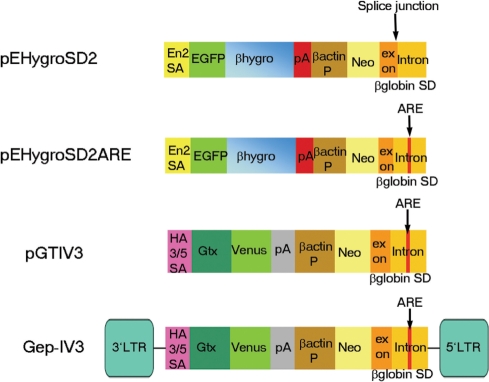
Figure 1.
Schematic representation of the poly A trap constructs employed. SA, splice acceptor; pA, polyadenylation signal; P, promoter; neo, neomycin phosphotransferase gene; SD, splice donor; ARE, AU-rich element; Gtx, synthetic sequence containing Gtx motifs; En2, engrailed-2; HA 3/5, Human adenovirus type 3/5; βhygro, fusion between β-galactosidase and hygromycin resistance genes; LTR, long terminal repeat; EGFP, enhanced green fluorescent protein.
As our goal was to generate insertions that would both produce a mutation and provide a readout of the endogenous gene's expression pattern, these SD cassettes were combined with two different promoterless expression modules. pEHygroSD2ARE combines the SD cassette with a SA-module employing the En-2 SA to drive the expression of a triple fusion between egfp, lacZ and the hygromycin resistance gene (egfpβhygro) (22) (Figure 1). In addition, to generate a particularly sensitive readout of endogenous gene expression we employed a second SA-module (32) that includes a translational amplifier from the 5′ UTR of the homeobox gene Gtx (33) and the enhanced YFP, Venus (34) (plasmid vector pGTIV3 and retroviral vector Gep-IV3) (Figure 1).
To assess the ability of the ARE to enhance gene trap efficiency and reduce background, we compared pEHygroSD2ARE with an identical vector lacking the ARE (termed pEHygroSD2). Both vectors were electroporated into E14TG2a ES cells and colonies selected in G418. Interestingly, the vector containing the ARE produced consistently 1.7-fold fewer G418 resistant colonies (40 ± 3 per plate) than did the vector without the ARE (67 ± 6.5 per plate) (Table 1) suggesting that a significant proportion of the neo resistant colonies obtained in the absence of the ARE contain unspliced neo message and are therefore not true gene trap events. This finding is consistent with a previous study reporting a similar level of reduction in the number of drug-resistant colonies as a result of the inclusion of an ARE (35). To determine whether the ARE had a direct effect on the nature of integration, we expanded a subset of these colonies and used 3′RACE to sequence the gene trap transcript generated by each integrant. In the vast majority of pEHygro2SD2ARE clones (77%), we were able to detect correct SD usage and no evidence of a read-through transcript (Table 1). This contrasts significantly with RACE sequences obtained from clones derived from pEHygro2SD2 insertions, where we observed only 11% correctly spliced transcripts (Table 1). Thus, the presence of the ARE sequence led to a 7-fold improvement in the number of properly spliced integrants and hence enhanced this vector's performance.
Table 1.
The ARE enhances the performance of the β-globin splice donor
| Vector | Average (SD) G418R clonesa | Number G418R clones analyzed | Correct SD usageb (%) | SD read- throughb (%) |
|---|---|---|---|---|
| pEHygroSD2 | 67 (±3) | 45 | 11 | 69 |
| pEHygroSD2ARE | 40 (±6.5) | 43 | 77 | 16 |
aPer electroporation plate.
bPercentages representing RACE products indicative of vector degradation or concatemer formation are not included.
Transcripts disrupted by this new set of vectors
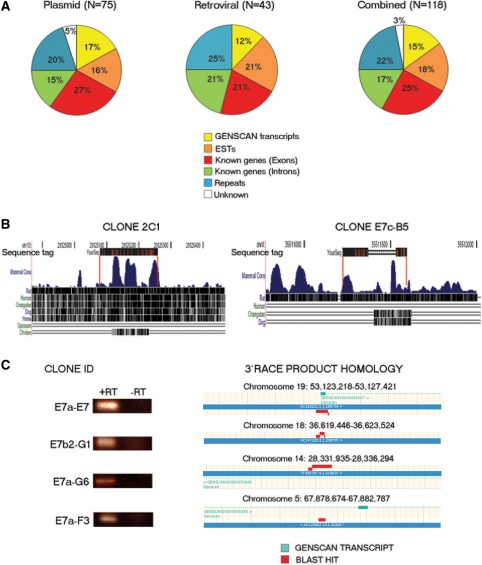
To assess the performance of our vectors in insertional mutagenesis, we introduced them into E14TG2a ES cells either by electroporation (pEHygroSD2ARE and pGTIV3) or retroviral infection (Gep-IV3) and analyzed the gene trap insertions within the resulting G418 resistant clones by 3′RACE PCR. We generated 118 unique sequence tags which were used for all subsequent analysis. The majority of the resulting properly spliced 3′RACE products contained a poly A signal sequence and a poly A tail (data not shown). 3′RACE sequences were analyzed using the BLAST (or BLAT) algorithm and the NCBI, Ensembl and UCSC mouse databases (NCBI m37 mouse assembly). An overview of the resulting 3′RACE products is shown in Supplementary Table 2. Of the 118 sequence tags we obtained, 43% matched reported transcribed sequences (Figure 2A), a similar level to that obtained with other poly A trap vectors (16,18). Of these, 25% matched exons of known genes, and 18% were found to be homologous to EST transcripts (Figure 2A). In addition to these sequences, we also identified tags that appeared spliced to intronic sequences of known genes, probably representing previously undiscovered splice variants (17%), exons or introns in GENSCAN-predicted transcripts (15%) and sequences within regions of the genome that do not appear to map to any known transcript (3%). In a number of cases, we observed that these RACE tags contained sequences corresponding to multiple downstream exons and some examples of this are shown in Supplementary Figure S1. We obtained a significant fraction (22%) of insertions where the RACE sequence was homologous to repeats and retrotransposon sequences (Figure 2A). Because it has been reported that retroviral vectors have a different insertional bias than plasmid based vectors (12), we examined the distribution of integrants for each type of vector independently (Figure 2A) and found little difference in the spectrum of sequence tags, although the plasmid-based vectors appear more efficient at identifying previously unannotated transcripts. We also found that a fraction of the sequence tags (∼20%) corresponding to regions of known transcripts were in the opposite transcriptional orientation from the trapped gene (Supplementary Table 2), a phenomenon that has been observed previously with other gene trap vectors (19,23).
Figure 2.
Nature of poly A trap insertions. (A) BLAST hit distribution of sequence tags derived from the plasmid vectors pEHygro2SD2ARE and pGTIV3 (left) (N = 75), the retroviral vector Gep-IV3 (middle) (N = 43) and their combination (right) (N = 118). (B) Conservation analysis of two indicative sequence tags that were not homologous to any known or predicted transcripts. Genome mapping and conservation analysis were performed using the UCSC genome browser. Red lines represent the boundaries of homology between the tag and the genome. (C) RT–PCR analysis of E14TG2a ES cells using primers designed on RACE products that are not homologous to known exons/ESTs. The trapped ES cell clone IDs that gave rise to the analyzed 3′RACE products are shown on the left. The BLAST-based genome mapping (ENSEMBL genome browser) of the RACE transcripts is also shown. RT, reverse transcriptase.
While, some of the sequence tags corresponding to GENSCAN-predicted, intronic or unknown sequences could represent cryptic or orphan 3′ exons, we believe the majority of them correspond to novel transcripts and alternative/additional exons. Thus, many of these 3′RACE sequences were found to be conserved between different species (Figure 2B). Moreover, part of this conservation appears in distinct clusters with the intervening sequence missing from the RACE product, indicating that these sequences represent conserved intron/exon structures (Figure 2B, clone E7c-B5). Similar conserved and apparently multi-exonic RACE tags have been observed with other gene trap vectors (23). To confirm that these novel sequences represented true spliced transcripts, we performed a further round of 3′RACE PCR on selected clones belonging to this category. Using nested primers based on the initial RACE sequences we obtained further downstream sequence indicating the presence of multiple downstream exons and putative splicing patterns (Supplementary Figure S2). Thus in the cases we have been able to examine, these uncharacterized sequences appear to represent genuine genes rather than individual cryptic 3′ exons. To further test whether these sequence tags that did not map to known exons/ESTs do in fact represent de novo transcripts, we examined the expression of a set of them in wild-type ES cells. Transcripts were selected for analysis based on the expression of the 5′ reporter in our vectors, indicating that they should be expressed in wild-type undifferentiated ES cells. Primers were designed to detect the expression of representative trapped sequences of each class; exons of a GENSCAN-predicted transcript (clone E7a-E7, Figure 2C), transcripts derived from intronic sequence of a curated predicted GENSCAN gene (clones E7a-G6 and E7a-F3, Figure 2C), and a sequence that did not map to any known or predicted transcript (clone E7b2-G1, Figure 2C). Figure 2C shows that all these trapped sequences are indeed expressed in unmodified ES cells indicating that this series of poly A trap vectors is effective at identifying previously uncharacterized or predicted transcripts. A closer inspection of the GENSCAN transcripts contained within our sequence tag set revealed that a significant proportion (8/17, 47%) of them have not been previously trapped by other poly A or SA-type gene trap vectors (Supplementary Table S3) suggesting that our vectors can potentially aid the identification and disruption of previously uncharacterized, novel transcripts.
We also compared the sequence tag set described in Figure 2A to the complete set of tags available on the IGTC database (www.igtc.org). We found that the majority (46/49) of the known genes trapped by our vectors have been previously disrupted by SA-type (promoterless) vectors (Supplementary Table S4) although four of these genes (D2Wsu81e, Bank1, Dpp10, Pitpnm2, Supplementary Table S4) have only been trapped in a screen employing C57BL/6N ES cells (36). Interestingly, two of the sequence tags generated through the use of our vectors (Shd, Grin2b, Supplementary Table S4) represent novel integrations indicating that the β-actinP-neo-β-globin-SDARE poly A trap vectors have the potential to ‘enrich’ for insertions within previously untrapped loci. Both genes are not expressed in early embryonic development, are tissue-specific (37–39) and examination of EST/expression databases (Mouse Genome Informatics: http://www.informatics.jax.org/ and Gene Expression Omnibus repository: http://www.ncbi.nlm.nih.gov/sites/entrez?db=geo&cmd=search&term=;) suggests they are not expressed in undifferentiated ES cells. Targeted trapping of Grin2b has thus far been unsuccessful (www.eucomm.org).
Expression-independent vectors that do not exhibit a severe 3′ bias
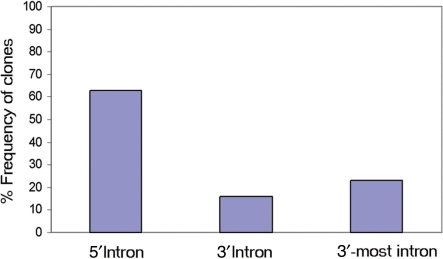
One of the major limitations associated with poly A trap vectors is their inability to generate stable, selectable integrations outside of the last intron of their target genes due to the action of NMD (17). As a result, we were surprised to find that integrations generated by our vectors do not exhibit the severe 3′ bias observed with the widely used poly A trap vectors (17–19) (Table 2). Interestingly, as the stop codon in our vector sequence was located 333 nucleotides upstream of the β-globin splice junction (Supplementary Figure S3), all transcripts generated by this vector regardless of the integration site should have been susceptible to NMD and we should not have been able to obtain any selectable integrants (40). However, not only did we obtain integrations, but also the distribution of these integrations did not show the typical 3′ bias exhibited by standard poly A trap vectors. Thus, as shown in Figure 3, the majority of the clones analyzed (63%) contained vector insertions within the 5′-end of their target genes, while only 23% of integrations occurred in the 3′-most intron. This result indicates that poly A trapping using the β-actinP-neo-β-globinSDARE vectors may somehow overcome the effect of NMD.
Table 2.
Overview of known disrupted genes and their trapped exons
| Gene identity |
Total number of exons | Number of trapped exonsa | |
|---|---|---|---|
| Gene symbol | Accession number | ||
| Ylpm1 | NM_178363 | 22 | 2 |
| Rfx4 | AY342003 | 18 | 8 |
| Q8VE10-2 | AK033896 | 8 | 7 |
| Pnrc2 | AK077403 | 3 | 2 |
| Zmat4 | NM_177086 | 7 | 1 |
| Larp7 | NM_138593 | 13 | 1 |
| Esrrb | NM_011934 | 7 | 6 |
| Tmem57 | BC037192 | 11 | 10 |
| ENSMUSESTT00003795807 | AK140938 | 5 | 4 |
| ENSMUSESTT00000017789 | BF019192 | 4 | 3 |
| Tcerg1 | BC040284 | 22 | 8 |
| Cds2 | AK170888 | 13 | 12 |
| ENSMUSESTG00000015152 | AK013487 | 5 | 1 |
| Dnahc8 | NM_013811 | 95 | 94 |
| Gna13 | NM_010303 | 4 | 1 |
| Flvcr2 | NM_145447 | 10 | 9 |
| Slc39a14 | NM_144808 | 10 | 9 |
| D2Wsu81e | NM_172660 | 12 | 11 |
| Skap2 | NM_018773 | 13 | 7 |
| D2Ertd750e | NM_026412 | 9 | 1 |
| Shd | NM_009168 | 7 | 3 |
| Zbtb24 | NM_153398 | 7 | 6 |
| Ccnd2 | NM_009829 | 5 | 1 |
| Gas6 | NM_019521 | 15 | 13 |
| Ppfibp1 | NM_026221 | 28 | 14 |
| Pde6a | NM_146086 | 22 | 4 |
| Oxa1l | NM_026936 | 10 | 9 |
| Bank1 | NM_001033350 | 17 | 9 |
| Trappc6 | NM_030057 | 6 | 5 |
| Dpp10 | NM_199021 | 26 | 20 |
| Gm672 | NM_201354 | 13 | 2 |
| Pitpnm2 | NM_011256 | 25 | 24 |
| Nvl | NM_026171 | 23 | 9 |
| F730014I05Rik | NM_146129 | 17 | 16 |
| Ube2k | NM_016786 | 7 | 6 |
| ENSMUSESTG00000017061 | BB642399 | 3 | 2 |
| CD59b | NM_181858 | 5 | 1 |
| Grin2b | NM_008171 | 13 | 8 |
| Fgf14 | NM_207667 | 5 | 3 |
| Fnbp1 | NM_001038700 | 14 | 8 |
| Grik4 | NM_175481 | 19 | 18 |
| Tdgf1 | NM_011562 | 7 | 1 |
| RP23-14F5.7-001 | BE630360 | 3 | 1 |
| Lemd1 | NM_001033250 | 4 | 1 |
| Gm525 | NM_001033266 | 4 | 1 |
| Gusb | NM_010368 | 12 | 8 |
| Nob1 | NM_026277 | 9 | 8 |
| ENSMUSESTP00000030261 | LOC100048565 | 3 | 2 |
| Q5F2E7-2 | NM_001024205 | 4 | 1 |
aNumber of exons downstream of vector integration site. Genes consisting of two exons were excluded.
Figure 3.
Distribution of vector insertion sites within trapped genes. The analysis includes clones containing pEHygro2SD2ARE, pGTIV3 and Gep-IV3 vector integrations. Only integrations within genes with well-defined exon–intron structure were included. Insertions within two-exon genes were excluded from the analysis. The 5′- and 3′-introns were defined as being 5′ and 3′ to the middle exon or intron of a gene respectively, according to ref. (17). Vector insertions defined as ‘3’-most intron’ were independently counted and excluded from the 3′ intron group. N = 49.
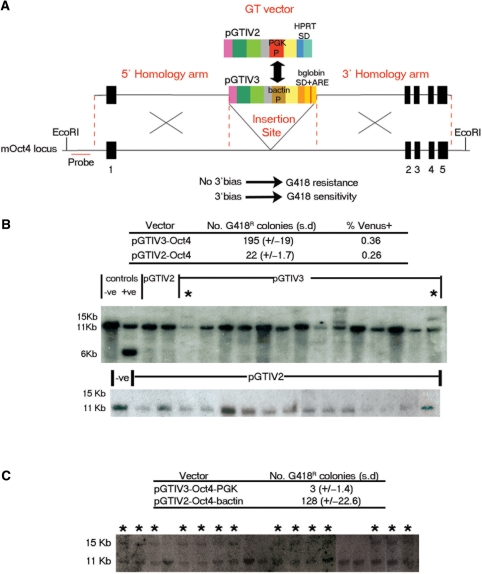
To test whether these particular poly A trap vectors were indeed effectively able to overcome the inhibitory activity of NMD, we asked whether they could function as a selection cassette, when targeted directly to a 5′ intron of a gene by homologous recombination. We compared the ability of pGTIV3 to function as a selection cassette when placed in the first intron of the ES cell regulator Oct4 (41) to the established poly A trap vector pGTIV2 (32). We selected pGTIV2 as it exhibits a 3′ integration bias (W. Stanford and T. Tanaka, personal communication) and is identical to pGTIV3 apart from the SD-selection cassette (Figure 4A). Both gene trap vectors were flanked by sequence homologous to the Oct4 locus and introduced into E14TG2a ES cells. Based on the number of antibiotic resistant colonies produced with both versions of this targeting vector (Figure 4B), targeting of the non-3′ biased pGTIV3 vector appeared 9-fold more efficient than pGTIV2 (Figure 4B). As these vectors contain a promoterless reporter (Venus), we were able to measure the percentage of cells expressing Venus immediately after electroporation and determine that a similar percentage of the population was able to express the 5′ promoterless reporter in each of these constructs prior to selection (0.36% for pGTIV3 and 0.26% for pGTIV2) (Figure 4B). Since the Venus reporter is dependent on endogenous Oct4 expression through a SA, this transcript is not NMD sensitive and therefore we can conclude from the comparable number of Venus positive cells in both electroporations that there were no significant differences in the efficiency with which these vectors integrate and that differences in the numbers of G418 positive colonies represent the ability of the neo SD cassette to function in a 5′ intron. The location of this cassette within the 5′-most intron of Oct4 was confirmed by Southern blot indicating that 2/14 (15%) of G418-resistant, Venus positive, pGTIV3-electroporated clones were correctly targeted (Figure 4B), whereas analysis of a comparable number of pGTIV2-containing clones revealed the absence of any targeting events (Figure 4B).
Figure 4.
Targeted poly A trapping of the Oct4 locus. (A) Schematic representation of the targeted insertion of vectors pGTIV3 and pGTIV2 into the first intron of the mouse Oct4 locus. The location of the probe used for Southern blot analysis of the targeted clones is shown in red. The genomic organization of Oct4 is not drawn to scale. Unbiased insertional preference should theoretically give rise to neomycin resistant clones while tendency to insertion into the 3′ most intron should be associated with loss of neomycin resistance. (B) Top: number of G418 resistant colonies obtained after ES cell electroporation with the pGTIV3 and pGTIV2 Oct4 targeting vectors. The fractions of electroporated cells expressing Venus prior to G418 selection are indicated. Numbers and percentages are an average of three electroporation experiments. Bottom: Southern blot analysis of G418 resistant, Venus positive, pGTIV3 and pGTIV2-Oct4 targeted clones. Genomic DNA was digested using EcoRI (restriction sites are shown in A). Correctly targeted clones should yield an 11 kb (wild-type) and a 15 kb (targeted) band and are indicated by an asterisk. DNA from wild-type E14TG2a ES cells and an independently Oct4 targeted clone (expected bands of 6 and 11 kb) were also included as negative and positive controls, respectively (first two lanes from the left). The analysis of 15 pGTIV2-Oct4 targeted clones is also shown separately (bottom). (C) Targeted poly A trapping of Oct4 after promoter swap between vectors pGTIV3-Oct4 and pGTIV2-Oct4. The human β-actin promoter of the insertionally unbiased pGTIV3-Oct4 vector was exchanged for the PGK promoter present in the 3′ biased pGTIV2-Oct4 vector (represented by the arrow in A). Top: number of G418 resistant colonies obtained after electroporation with the pGTIV3-Oct4-PGK and pGTIV2-Oct4-β-actin modified constructs. Bottom: Southern blot analysis of G418 resistant, Venus positive clones electroporated with the pGTIV2-Oct4-β-actin vector. Genomic DNA was digested and probed as in B. Correctly targeted clones should yield an 11 kb (wild-type) and a 15 kb (targeted) band and are indicated by an asterisk; (n = 2).
The human β-actin promoter is linked to NMD-independent integration
As our poly A trap vectors are able to generate antibiotic resistant clones irrespective of the site of integration and therefore appear to overcome the effect of NMD, we wanted to know if this property was linked to the presence of particular sequences. It has been previously shown that certain promoter sequences can affect the post-transcriptional fate of mRNAs and rescue otherwise NMD-sensitive transcripts from NMD (42). Furthermore, NMD efficiency is variable (43,44) and transcripts expressed at high levels are often not completely eliminated by NMD probably due to saturation of the NMD machinery (45–47) (Oliver Mühlemann, personal communication). We therefore sought to investigate whether the human β-actin promoter used in the selection cassette of all our vectors mediates relatively unbiased gene trapping and targeting. To test this possibility, we swapped the human β-actin promoter present in our NMD-independent pGTIV3-Oct4 targeting vector with the PGK promoter present in the NMD biased pGTIV2-Oct4 vector (depicted by the arrow in Figure 4A). We found that the β-actin containing, pGTIV2-Oct4 vector yielded about 34-fold more G418-resistant colonies (average of 128 colonies/electroporation) than pGTIV3-Oct4-PGK (3 colonies/per electroporation) (Figure 4C). Correct targeting of this cassette to the first intron of Oct4 was confirmed by Southern blot in the majority of the neo resistant, Venus-positive clones (14/19, 73%) electroporated with vector pGTIV2-Oct4-β-actin (Figure 4C). However, in all six neo resistant clones obtained with pGTIV3-Oct4-PGK no targeting was detected (data not shown). From this we conclude that the ability of our SD vectors to generate antibiotic resistance outside of the last intron of a gene is linked to the specific nature of the promoter employed.
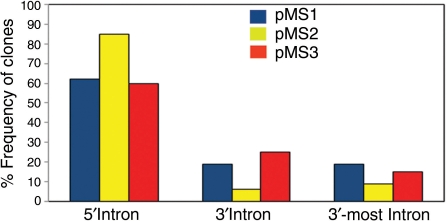
Interestingly, we noticed that a series of older poly A trap vectors (pMS1-3) used by the IGTC also contain the human β-actin promoter. These vectors, (pMS1, pMS2 and pMS3, http://www.cmhd.ca/genetrap/vectors.html) (8) have not been used extensively, but we were able to identify 114 insertions in genes with known intron-exon structure. This data set is summarized in Figure 5 (a detailed list of the pMS vector insertions analyzed is shown in Supplementary Table S5) and we found that, as with the vectors reported here, pMS vectors also appear to overcome the NMD mediated 3′ bias, supporting the notion that the β-actin promoter can confer positional independence to poly A trap vectors.
Figure 5.
Distribution of vector insertion sites within genes trapped by the β-actin promoter-containing pMS1 (blue), pMS2 (yellow) and pMS3 (red) gene trap vectors. The classification of the insertions is the same as in Figure 3. A detailed list of the pMS insertions used for the analysis is shown in Supplementary Table S5. N = 40 for pMS1 and pMS3 and N = 34 for pMS2.
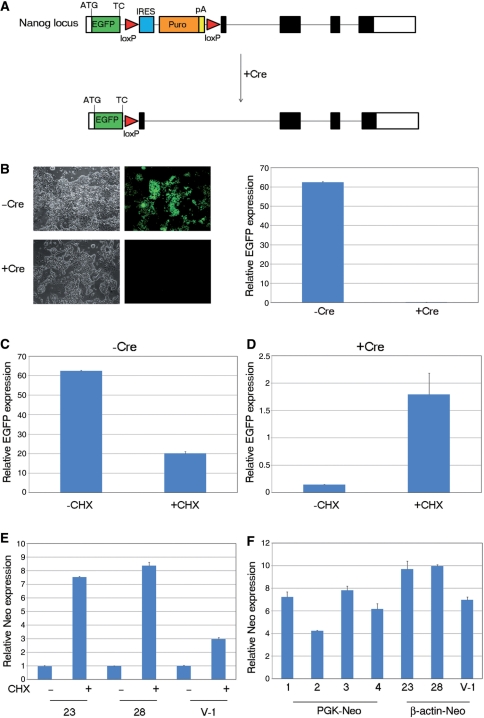
While these data indicate that our vectors are immune to the positional bias generated by NMD, it had been reported previously that an Ig-μ minigene driven by the human β-actin promoter is susceptible to NMD (48). To examine whether poly A trapping/targeting with our human β-actin promoter-containing constructs is subject to NMD we treated selected cell lines with the NMD inhibitor CHX (49). Thus in instances where a targeted transcript is subject to NMD, limited treatment of this cell line with CHX should result in enhanced transcript levels detectable by quantitative RT–PCR. To confirm that we could detect NMD in this fashion we exploited an EGFP reporter cell line for the ES cell regulator, Nanog (50) that could be rendered sensitive to NMD by transfection with the Cre recombinase. This strategy is shown in Figure 6A and features an EGFP cassette inserted into the first exon of Nanog directly at the Nanog translation initiation codon followed by the EGFP stop codon and a LoxP flanked IRES-puro poly A cassette. The unrecombined cell line expresses EGFP from the Nanog locus as the EGFP coding sequence reads directly into the IRES-puro poly A without splicing. However, following Cre mediated recombination, the IRES-puro poly A is removed, placing the EGFP stop codon into exon 1 of Nanog and generating a spliced transcript downstream of EGFP, which is now sensitive to NMD. Consequently, following Cre transfection, EGFP expression is no longer detected (Figure 6B). When these cells and the unrecombined parental cell line were treated with CHX, levels of EGFP transcript were significantly increased only in the recombined NMD sensitive cell line (Figure 6C and D). As CHX treatment under these conditions appeared an effective antagonist of NMD, we asked whether it stimulated levels of neo mRNA produced by gene trap integrations located in the first intron of target genes. Surprisingly, we found that CHX treatment led to enhanced neo mRNA levels when neo expression was driven from either the first intron of Oct4 (clones 23 and 28, Figure 6E) or a 5′ gene trap insertion (V-1, Figure 6E).
Figure 6.
Assessing the effect of NMD inhibition. (A) Schematic representation of the strategy employed for the generation of Nanog-GFP ES cells. An EGFP-loxP-IRES-Puro-pA-loxP cassette was placed in the first exon of the ES cell pluripotency factor Nanog directly at the Nanog translation initiation codon. Cre recombinase removes the loxP flanked polyadenylation signal (and preceding IRES-PuroR) causing the EGFP termination codon to be placed in the first exon of the 4-exon Nanog transcript. The Nanog genomic locus was not drawn in scale. ATG, Nanog translation initiation codon; TC, termination codon; Puro, puromycin resistance gene. White boxes represent untranslated exonic regions and solid black boxes coding regions (B) Left: fluorescence microscopy of Nanog-GFP ES cells before and after expression of Cre recombinase. Brightfield images of the same cell populations are also shown. Right: real-time RT–PCR expression analysis of EGFP in Nanog-GFP ES cells before and after expression of Cre recombinase. (C and D) Real-time RT–PCR analysis of relative EGFP expression levels in Nanog-GFP ES cells before (C) and after (D) removal of the loxP flanked cassette in the presence or absence of CHX. Values were normalized to the expression levels of the housekeeping gene TBP. Error bars represent SD, CHX. (E) Relative neo expression levels in two Oct4-targeted ES cell clones (23 and 28) and one gene trap clone (V-1) carrying a vector insertion within the first intron of the F730014I05Rik gene with or without CHX treatment. Data are shown as relative expression to the CHX untreated controls. Values were normalized to the expression levels of the housekeeping gene tbp. The graph is representative of three independent experiments and error bars correspond to SD, CHX. (F) Comparison of relative neo expression levels between four ES cell clones (1–4) carrying insertions with the PGK-neo-containing vector pGTIV2 and clones 23, 28 and V-1 which have been targeted (23,28) or trapped (V-1) using β-actin-neo vectors. Values were normalized to the expression levels of the housekeeping gene TBP.
As our β-actin driven SD cassettes are subject to NMD, some other property must make them immune to the extreme 3′ bias observed with other poly A trap vectors. Although our vectors contain a minimal promoter fragment of 468 bp upstream of the human β-actin transcription start site, we considered whether this promoter might be able to generate sufficient levels of neo transcript even in the presence of NMD to allow for antibiotic selection. Thus, we compared the level of neo transcript in the three NMD sensitive clones described in Figure 6E to the level generated by selectable random integrants containing the 3′ biased PGK promoter-driven neo poly A trap cassette (pGTIV2). In all cases, we observed that the β-actin promoter driven levels of neo expression were higher than those generated by the PGK promoter, despite the destabilizing activity of NMD on the β-actin driven transcripts (Figure 6F), We presume that the four PGK neo clones used here contained integrations in the last intron of their target locus as these vectors typically exhibit a severe 3′ bias (80–90%) (17–19) (W. Stanford, T. Tanaka, personal communication) and neo expression in all four clones failed to respond to CHX (Supplementary Figure S4). Taken together these observations support the notion that as a stable chromosomal integrant, the particular β-actin promoter fragment employed here is effective at generating sufficient transcript levels in ES cells to guarantee success in selection despite mRNA surveillance mechanisms such as NMD.
Gene targeting of a locus expressed at low levels in ES cells
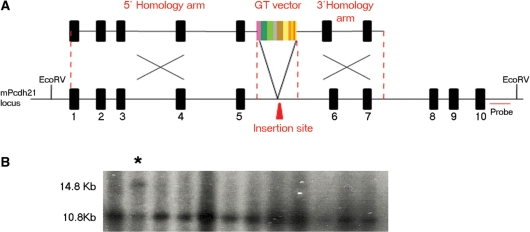
The accessibility of a set of 127 genes to gene targeting with high efficiency promoterless vectors was recently tested and found to be directly proportional to the level of expression of the target locus in ES cells (5). The ability to target our SD vectors to a 5′ intron suggests that they might be a new and effective means for the targeting of those genes inaccessible to promoterless vectors. To test this possibility, we selected the gene Pcdh21 that was both expressed at low levels in ES cells and found by Friedel et al. (5) to be untargetable. We replaced the SAβgeo cassette in their original targeting vector with pGTIV3 (Figure 7) so that we could assay the effectiveness of our SD cassette with identical homology arms. Upon introduction of this vector into ES cells we expanded 20 colonies and found a correctly targeted clone (Figure 7B). We conclude from this our NMD-independent poly A trap cassettes may be effective tools for use in targeted trapping of genes that are not expressed in ES cells.
Figure 7.
Targeted poly A trapping of the Pcdh21 locus. (A) Schematic representation of the targeted insertion of vector pGTIV3 into the fifth intron of the mouse Pcdh21 locus. The location of the probe used for Southern blot analysis of the targeted clones is shown in red. The genomic organization of Pcdh21 is not drawn to scale. Only 10 of the 17 Pchd21 exons are shown. (B) Southern blot analysis of representative G418 resistant ES cell clones electroporated with the pGTIV3-Pcdh21 vector. Genomic DNA was digested using EcoRV (restriction sites are shown in A). Correctly targeted clones should yield an 10.8 kb (wild-type) and a 14.8 kb (targeted) band and a correctly targeted clone is indicated by an asterisk.
DISCUSSION
In this article, we have described a series of expression-independent gene trap vectors that do not suffer from the extreme 3′ insertional bias observed for other poly A trap vectors. We found that the incorporation of an ARE in the design of these vectors reduced significantly the occurrence of ‘background’ drug resistant ES cell clones arising from SD readthrough events. The use of these vectors also appears to enrich for insertions within unknown and predicted novel transcripts. Importantly, we also show that our poly A trap vectors can be introduced via homologous recombination into the 5′ side of two different genes and this suggests that our expression and NMD-independent poly A trap vectors may be effective tools for disrupting genes expressed at low levels in ES cells.
Our initial vector design incorporated an RNA instability element from the 3′ UTR of the human GM-CSF gene. This element was designed to ensure that viable integrations are only achieved when splicing removes this sequence from the transcript generated by the vector's internal promoter, thereby promoting the selection of ‘true’ poly A trap events. The combination of this element with a SD encompassing exon 2 and the adjacent intronic sequence from rabbit β-globin, appears to make a particularly effective selection unit. While RNA destabilizing sequences have been incorporated into other poly A trap vectors (16,18,35), their effectiveness had not been tested at the molecular level. Here, we report the first evidence that these elements can be introduced into the intronic sequence downstream of a SD to achieve a 7-fold enrichment in correctly spliced gene trap transcripts.
Analysis of the set of sequence tags generated through the use of this new class of SD vectors suggests that these function efficiently as poly A trap vectors. The percentage of sequence tags that were homologous to either predicted open reading frames or ESTs (43%) was similar to that achieved with other poly A trap vectors (16,18). We also identified at least two well-characterized late neural markers (Shd and Grin2b) that are unequivocally not expressed in ES cells and have not been trapped by promoterless vectors. However, in addition to these sequences, we identified an additional set of sequence tags (18%) that were not homologous to any known exonic or EST sequences. While similar sequences have been identified with other poly A vectors they were generally dismissed as ‘non-genic’ (12). However, we show that some of these transcripts are expressed in wild-type ES cells and thus represent genuine transcripts that are not yet present in the EST databases. While our analysis suggests at least some of these transcripts are expressed in undifferentiated ES cells, their identification exclusively by poly A traps suggests that their level of expression cannot be selected for using promoterless vectors. The presence of similar sequences in the gene trap databases has been commented on (23,51) and some of these sequences have been shown to be expressed during development and in a tissue-specific manner (16,23,51). The level of conservation observed in some of these transcription units, suggests that we may have identified a number of previously uncharacterized exons, novel alternatively spliced variants or non-coding RNAs.
From the sequence tags we identified in known genes, it became apparent that our poly A trap vectors do not exhibit the severe NMD mediated 3′-most intron bias observed in other vectors of this class. However, while we observed that 63% of our vector insertions localized to the 5′ half of a gene, these vectors cannot be considered completely unbiased as we also observed a considerable set of insertions (23%) in the last intron of their target locus. Does this represent a bias? According to Shigeoka et al. (17), the addition of an IRES to a poly A trap vector shifted the frequency of 3′ most insertions from 88% to 6%, but they also reported that 70% of their insertions were found in the 5′ half of target genes. Thus, while the IRES appears to have eliminated the influence of NMD, their use of a retrovirus may have introduced a slight 5′ bias (11,52). As even plasmid based promoterless vectors exhibit some 5′ bias (52), it is difficult to establish criteria for unbiased integration. Thus, while it is impossible to say whether our vectors are truly unbiased, they represent a considerable improvement over the majority of poly A trap vectors.
To demonstrate explicitly that these vectors could function at the 5′-end of a gene, we targeted our SD cassette to the 5′-end of the Oct4 gene by homologous recombination, an achievement not possible with an equivalent 3′ biased poly A trap vector. As SD vectors have been in use for some time now and the majority are susceptible to NMD, we thought it important to determine the sequences responsible for this surprising finding. Based on promoter swaps between a 3′ biased poly A trap cassette and our vectors we established that this NMD independence was due to the β-actin promoter. Although neo expression from our vectors was still subject to NMD we found that neo transcript levels generated from the β-actin promoter were greater than that achieved by NMD-independent selectable integrants employing the PGK promoter. Thus, the strength of the human β-actin promoter fragment used here is sufficient to overcome the inhibitory effect of NMD and establish drug resistance in the majority of insertions regardless of intronic position. This was particularly surprising as it has been previously reported that the PGK promoter is stronger than β-actin in human T cells (53). While we were unable to determine the β-actin promoter fragment used in that study, it is likely that the human β-actin fragment used in our gene trap vectors is particularly effective in ES cells. Although, our data demonstrates that promoter strength is responsible for the effective NMD independence of our vectors, they may also accrue an additional advantage from the particular neo gene used here. As far as we can determine, all poly A trap vectors that exhibit a severe 3′ bias also employ a mutant version of the neomycin phosphotransferase gene, which has been shown to be associated with a reduction in the enzyme's activity without affecting the stability of neo mRNA or protein levels (4,54).
We believe the major determinants of the efficient and relatively unbiased performance of our vector were promoter strength and message stability. A strong promoter, like β-actin, would normally produce high levels of splice donor read-through and in most cases require an RNA instability element (ARE) to ensure these transcripts were not expressed at levels sufficient for selection. While a weak promoter would not generate significant levels of splice donor read-through, it would be sensitive to NMD. Thus, the highly effective vectors generated here were the result of a fortuitous and inseparable combination of a strong promoter with a means to target unspliced message for degradation.
Our observation that the β-actin promoter can produce sufficient transcript in the presence of NMD to generate viable antibiotic resistance suggests that in cell types that express high levels of a target gene, NMD may not be sufficient to generate a null phenotype. As a result, phenotypes that rely on the introduction of premature stop codons (e.g. through Cre-lox mediated exon removal) as a means to target degradation of a transcript may not always represent true nulls, but in fact produce truncated protein products and this suggests some care should be taken in the design of conditional targeting vectors.
For the first time, we have shown that a poly A trap vector can be employed for gene targeting applications. We found that our vectors could be used for the successful targeting of both Oct4 and Pcdh21. Previous attempts to target Pcdh21 using a SA-type gene trap construct have been unsuccessful (5). Consistent with this observation, Pcdh21, unlike Oct4, is expressed at low to negligible levels in ES cells. Interestingly, there is a clear difference in our observed targeting frequencies for Oct4 and Pcdh21 and perhaps this reflects some influence of the endogenous gene's expression levels on the efficiency of selection for SD vectors. Global analysis of available gene trap insertions suggests that poly A trapping efficiency is modestly influenced by gene expression levels with high expression enhancing the trappability of a locus ∼2-fold as compared to 75-fold for promoterless vectors (12). So is targeted poly A trapping a more efficient means to target low level expressed genes? Promoterless vectors are unable to access these genes and Pcdh21 is listed as a failed project by EUCOMM (http://www.eucomm.org/) based on attempts with traditional promoter containing vectors. Thus our vectors may represent an improvement in targeting over existing technology, although this is yet to be rigorously tested. Despite this caveat, this work represents the first demonstration that poly trap vectors may be effective tools for gene targeting.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust (062965), Medical Research Council (G0701428 and G120/215), Engineering and Physical Sciences Research Council (EP/D038197/1) and Leukemia Research Fund (0279 and 6036). J.M.B. is a senior fellow of the Medical Research Council. Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank William Skarnes William Stanford, Tetsuya Tanaka, Marc Tessier-Lavigne and Roland Friedel for plasmids and advice. We are also grateful to Jan Vrana and Martin Waterfall for assistance with flow cytometry. Finally, we are grateful to Mark Bradley, Keisuke Kaji, Alessandra Livigni, Andrew Smith, Maurice Canham, Violetta Karwacki-Neisius, Oliver Mühlemann and Dasa Longman for critical comments and technical advice.
Footnotes
Present address: Elena Tzouanacou, Biologie Moleculaire du Developpement, Institute Pasteur 25, rue du Dr. Roux, 75724 Paris cedex 15, France.
REFERENCES
- 1.Skarnes WC, von Melchner H, Wurst W, Hicks G, Nord AS, Cox T, Young SG, Ruiz P, Soriano P, Tessier-Lavigne M, et al. A public gene trap resource for mouse functional genomics. Nat. Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gossler A, Joyner AL, Rossant J, Skarnes WC. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science. 1989;244:463–465. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- 3.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 4.Skarnes WC, Moss JE, Hurtley SM, Beddington RS. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc. Natl Acad. Sci. USA. 1995;92:6592–6596. doi: 10.1073/pnas.92.14.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedel RH, Plump A, Lu X, Spilker K, Jolicoeur C, Wong K, Venkatesh TR, Yaron A, Hynes M, Chen B, et al. From The Cover: gene targeting using a promoterless gene trap vector (‘targeted trapping’) is an efficient method to mutate a large fraction of genes. Proc. Natl Acad. Sci. USA. 2005;102:13188–13193. doi: 10.1073/pnas.0505474102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niwa H, Araki K, Kimura S, Taniguchi S, Wakasugi S, Yamamura K. An efficient gene-trap method using poly A trap vectors and characterization of gene-trap events. J. Biochem. 1993;113:343–349. doi: 10.1093/oxfordjournals.jbchem.a124049. [DOI] [PubMed] [Google Scholar]
- 7.Zambrowicz BP, Friedrich GA, Buxton EC, Lilleberg SL, Person C, Sands AT. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature. 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 8.Salminen M, Meyer BI, Gruss P. Efficient poly A trap approach allows the capture of genes specifically active in differentiated embryonic stem cells and in mouse embryos. Dev. Dyn. 1998;212:326–333. doi: 10.1002/(SICI)1097-0177(199806)212:2<326::AID-AJA17>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 9.Skarnes WC. Two ways to trap a gene in mice. Proc. Natl Acad. Sci. USA. 2005;102:13001–13002. doi: 10.1073/pnas.0506279102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zambrowicz BP, Abuin A, Ramirez-Solis R, Richter LJ, Piggott J, BeltrandelRio H, Buxton EC, Edwards J, Finch RA, Friddle CJ, et al. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc. Natl Acad. Sci. USA. 2003;100:14109–14114. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnutgen F, De-Zolt S, Van Sloun P, Hollatz M, Floss T, Hansen J, Altschmied J, Seisenberger C, Ghyselinck NB, Ruiz P, et al. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc. Natl Acad. Sci. USA. 2005;102:7221–7226. doi: 10.1073/pnas.0502273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nord AS, Vranizan K, Tingley W, Zambon AC, Hanspers K, Fong LG, Hu Y, Bacchetti P, Ferrin TE, Babbitt PC, et al. Modeling insertional mutagenesis using gene length and expression in murine embryonic stem cells. PLoS ONE. 2007;2:e617. doi: 10.1371/journal.pone.0000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedel RH, Seisenberger C, Kaloff C, Wurst W. EUCOMM–the European conditional mouse mutagenesis program. Brief Funct. Genomic Proteomic. 2007;6:180–185. doi: 10.1093/bfgp/elm022. [DOI] [PubMed] [Google Scholar]
- 14.Floss T, Schnutgen F. Conditional gene trapping using the FLEx system. Methods Mol. Biol. 2008;435:127–138. doi: 10.1007/978-1-59745-232-8_9. [DOI] [PubMed] [Google Scholar]
- 15.Hirashima M, Bernstein A, Stanford WL, Rossant J. Gene-trap expression screening to identify endothelial-specific genes. Blood. 2004;104:711–718. doi: 10.1182/blood-2004-01-0254. [DOI] [PubMed] [Google Scholar]
- 16.Matsuda E, Shigeoka T, Iida R, Yamanaka S, Kawaichi M, Ishida Y. Expression profiling with arrays of randomly disrupted genes in mouse embryonic stem cells leads to in vivo functional analysis. Proc. Natl Acad. Sci. USA. 2004;101:4170–4174. doi: 10.1073/pnas.0400604101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shigeoka T, Kawaichi M, Ishida Y. Suppression of nonsense-mediated mRNA decay permits unbiased gene trapping in mouse embryonic stem cells. Nucleic Acids Res. 2005;33:e20. doi: 10.1093/nar/gni022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osipovich AB, Singh A, Ruley HE. Post-entrapment genome engineering: first exon size does not affect the expression of fusion transcripts generated by gene entrapment. Genome Res. 2005;15:428–435. doi: 10.1101/gr.3258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Q, Donahue SL, Moore-Jarrett T, Cao S, Osipovich AB, Ruley HE. Mutagenesis of diploid mammalian genes by gene entrapment. Nucleic Acids Res. 2006;34:e139. doi: 10.1093/nar/gkl728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nature Rev. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 21.Xu N, Chen CY, Shyu AB. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsakiridis A, Tzouanacou E, Larralde O, Watts TM, Wilson V, Forrester L, Brickman JM. A novel triple fusion reporter system for use in gene trap mutagenesis. Genesis. 2007;45:353–360. doi: 10.1002/dvg.20301. [DOI] [PubMed] [Google Scholar]
- 23.Chen WV, Delrow J, Corrin PD, Frazier JP, Soriano P. Identification and validation of PDGF transcriptional targets by microarray-coupled gene-trap mutagenesis. Nat. Genet. 2004;36:304–312. doi: 10.1038/ng1306. [DOI] [PubMed] [Google Scholar]
- 24.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl Acad. Sci. USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao S, Osborne CS, Bharadwaj RR, Pasceri P, Sukonnik T, Pannell D, Recillas-Targa F, West AG, Ellis J. Retrovirus silencer blocking by the cHS4 insulator is CTCF independent. Nucleic Acids Res. 2003;31:5317–5323. doi: 10.1093/nar/gkg742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison GM, Brickman JM. Conserved roles for Oct4 homologues in maintaining multipotency during early vertebrate development. Development. 2006;133:2011–2022. doi: 10.1242/dev.02362. [DOI] [PubMed] [Google Scholar]
- 27.Morrison GM, Oikonomopoulou I, Migueles RP, Soneji S, Livigni A, Enver T, Brickman JM. Anterior definitive endoderm from ES cells reveals a novel role for FGF signaling. Cell Stem Cell. 2008;3:402–415. doi: 10.1016/j.stem.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 28.Zamparini AL, Watts T, Gardner CE, Tomlinson SR, Johnston GI, Brickman JM. Hex acts with beta-catenin to regulate anteroposterior patterning via a Groucho-related co-repressor and Nodal. Development. 2006;133:3709–3722. doi: 10.1242/dev.02516. [DOI] [PubMed] [Google Scholar]
- 29.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 30.Ashfield R, Patel AJ, Bossone SA, Brown H, Campbell RD, Marcu KB, Proudfoot NJ. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 1994;13:5656–5667. doi: 10.1002/j.1460-2075.1994.tb06904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 32.Tanaka TS, Davey RE, Lan Q, Zandstra PW, Stanford WL. Development of a gene-trap vector with a highly sensitive fluorescent protein reporter system for expression profiling. Genesis. 2008;46:347–356. doi: 10.1002/dvg.20404. [DOI] [PubMed] [Google Scholar]
- 33.Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl Acad. Sci. USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 35.Ishida Y, Leder P. RET: a poly A-trap retrovirus vector for reversible disruption and expression monitoring of genes in living cells. Nucleic Acids Res. 1999;27:e35. doi: 10.1093/nar/27.24.e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen GM, Markesich DC, Burnett MB, Zhu Q, Dionne KM, Richter LJ, Finnell RH, Sands AT, Zambrowicz BP, Abuin A. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 2008;18:1670–1679. doi: 10.1101/gr.078352.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oda T, Kujovich J, Reis M, Newman B, Druker BJ. Identification and characterization of two novel SH2 domain-containing proteins from a yeast two hybrid screen with the ABL tyrosine kinase. Oncogene. 1997;15:1255–1262. doi: 10.1038/sj.onc.1201299. [DOI] [PubMed] [Google Scholar]
- 38.Sasner M, Buonanno A. Distinct N-methyl-D-aspartate receptor 2B subunit gene sequences confer neural and developmental specific expression. J. Biol. Chem. 1996;271:21316–21322. doi: 10.1074/jbc.271.35.21316. [DOI] [PubMed] [Google Scholar]
- 39.Sato A, Sekine Y, Saruta C, Nishibe H, Morita N, Sato Y, Sadakata T, Shinoda Y, Kojima T, Furuichi T. Cerebellar development transcriptome database (CDT-DB): profiling of spatio-temporal gene expression during the postnatal development of mouse cerebellum. Neural Netw. 2008;21:1056–1069. doi: 10.1016/j.neunet.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 40.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 41.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 42.Enssle J, Kugler W, Hentze MW, Kulozik AE. Determination of mRNA fate by different RNA polymerase II promoters. Proc. Natl Acad. Sci. USA. 1993;90:10091–10095. doi: 10.1073/pnas.90.21.10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zetoune AB, Fontaniere S, Magnin D, Anczukow O, Buisson M, Zhang CX, Mazoyer S. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet. 2008;9:83. doi: 10.1186/1471-2156-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the nonsense mediated decay pathway. Nucleic Acids Res. 2007;35:4542–4551. doi: 10.1093/nar/gkm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohn F, Buhler M, Muhlemann O. Nonsense-associated alternative splicing of T-cell receptor beta genes: no evidence for frame dependence. RNA. 2005;11:147–156. doi: 10.1261/rna.7182905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paillusson A, Hirschi N, Vallan C, Azzalin CM, Muhlemann O. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res. 2005;33:e54. doi: 10.1093/nar/gni052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 48.Buhler M, Paillusson A, Muhlemann O. Efficient downregulation of immunoglobulin mu mRNA with premature translation-termination codons requires the 5′-half of the VDJ exon. Nucleic Acids Res. 2004;32:3304–3315. doi: 10.1093/nar/gkh651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 50.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 51.Roma G, Cobellis G, Claudiani P, Maione F, Cruz P, Tripoli G, Sardiello M, Peluso I, Stupka E. A novel view of the transcriptome revealed from gene trapping in mouse embryonic stem cells. Genome Res. 2007;17:1051–1060. doi: 10.1101/gr.5720807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansen J, Floss T, Van Sloun P, Fuchtbauer EM, Vauti F, Arnold HH, Schnutgen F, Wurst W, von Melchner H, Ruiz P. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc. Natl Acad. Sci. USA. 2003;100:9918–9922. doi: 10.1073/pnas.1633296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper LJ, Topp MS, Pinzon C, Plavec I, Jensen MC, Riddell SR, Greenberg PD. Enhanced transgene expression in quiescent and activated human CD8+ T cells. Hum. Gene Ther. 2004;15:648–658. doi: 10.1089/1043034041361217. [DOI] [PubMed] [Google Scholar]
- 54.Yenofsky RL, Fine M, Pellow JW. A mutant neomycin phosphotransferase II gene reduces the resistance of transformants to antibiotic selection pressure. Proc. Natl Acad. Sci. USA. 1990;87:3435–3439. doi: 10.1073/pnas.87.9.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.