Abstract
Most nucleic acid-based technologies rely upon sequence recognition between an oligonucleotide and its nucleic acid target. With the aim of improving hybridization by decreasing electrostatic repulsions between the negatively charged strands, novel modified oligonucleotides named Zip nucleic acids (ZNAs) were recently developed. ZNAs are oligonucleotide–oligocation conjugates whose global charge is modulated by the number of cationic spermine moieties grafted on the oligonucleotide. It was demonstrated that the melting temperature of a hybridized ZNA is easily predictable and increases linearly with the length of the oligocation. Furthermore, ZNAs retain the ability to discriminate between a perfect match and a single base-pair-mismatched complementary sequence. Using quantitative PCR, we show here that ZNAs are specific and efficient primers displaying an outstanding affinity toward their genomic target. ZNAs are particularly efficient at low magnesium concentration, low primer concentrations and high annealing temperatures, allowing to improve the amplification in AT-rich sequences and potentially multiplex PCR applications. In reverse transcription experiments, ZNA gene-specific primers improve the yield of cDNA synthesis, thus increasing the accuracy of detection, especially for genes expressed at low levels. Our data suggest that ZNAs exhibit faster binding kinetics than standard and locked nucleic acid-containing primers, which could explain why their target recognition is better for rare targets.
INTRODUCTION
Nucleic acid-based technologies are widely used in research and diagnostics. Such techniques rely upon specific sequence recognition between a synthetic oligonucleotide and its complementary sequence within a nucleic acid strand. PCR-based assays, and particularly qPCR or RT-qPCR are the most commonly used methods for detecting and quantifying genes or their expression (1–3). These powerful techniques need to be carefully implemented to be specific, accurate and sensitive. For several years, scientists in both academia and industry have been devoting their efforts towards facilitating assay design and optimization, decreasing the time of analysis or detecting multiples targets simultaneously in a single reaction or in parallel in a run. In practice, numerous improvements have been made in PCR platforms, laboratory consumables or PCR reagents for this common time-saving goal. Chemistry has also made significant contributions, with the development of probes and modified synthetic oligonucleotides such as minor groove binder (MGB) (4,5) or locked nucleic acid (LNA) modified analogues (6–8). Both types of modifications enhance hybridization, most probably through non-ionic interaction mechanisms (9). Another approach for improving nucleic acid hybridization is to decrease the electrostatic repulsion between negatively charged nucleic acid strands. Peptide nucleic acids (PNAs) are uncharged molecules that have shown a large improvement in hybridization. However, because the backbone is modified, PNA oligomers are not recognized by polymerases (10) and cannot be used as primers in PCR. Based on the same principle of reducing the negative charge of the oligonucleotide, the phosphoramidite-based conjugation of spermine residues to oligonucleotides as cationic units has been recently described (11,12). Oligocation conjugation was shown to favour hybridization with a complementary sequence by clipping the strands together like a zipper (11), hence their name Zip nucleic acids (ZNAs). This stepwise spermine conjugation was indeed shown to raise the Tm in a smooth and linear way, while preserving mismatch discrimination. Base sequence and conjugation site (3′ or 5′) did not influence the effect on Tm. Rather, it appeared that the melting temperature increase of ZNA brought about by spermine conjugation only depends on the length of the oligonucleotide and on the number of grafted spermines, providing the possibility to predict and finely tune the Tm of any given sequence (13). However, whether the oligospermine moiety does interfere with processing by a DNA polymerase, or whether ZNAs are capable of specific recognition of a target in a whole genome were essential questions to be addressed before considering their use in nucleic acid-based techniques.
In the present study, we show that ZNAs are potent primers for PCR and RT-qPCR, with some distinct advantages over other chemically modified oligonucleotides. Moreover, a comparative evaluation of ZNAs in qPCR provides a better understanding of the mechanism by which ZNAs improve the hybridization.
MATERIALS AND METHODS
Oligonucleotides
ZNA oligonucleotides were synthesized on a standard oligonucleotide synthesizer as described (11,12), DMT-ON purified on reverse phase cartridge (Poly-Pak II, Glen Research) and characterized on anion-exchange HPLC and ESI-TOF mass spectrometry.
Standard and LNA-containing primers were synthesized and DMT-ON purified by Eurogentec (Seraing, Belgium).
Nucleic acid extraction
Genomic DNA was isolated from SiHa cells (cervical carcinoma, ATCC HTB-35) containing 1–2 integrated copies of HPV16 or HPV-negative A549 cells (lung carcinoma, ATCC CCL-185) using DNeasy Blood and Tissue kit (Qiagen), according to the manufacturer's instructions. Purity and concentration were assessed by agarose gel electrophoresis and UV spectrophotometry. Aliquots were stored at −20°C and kept at +4°C for short-term use. Total RNA was isolated from HeLa cells (human cervical cancer cell, ATCC CCL-2) or A549 cells using RNA NOW reagent (Biogentex) following manufacturer's instructions. After isopropanol precipitation, RNA was resuspended in RNase-free water and stored at −80°C. RNA quality was assessed by agarose gel electrophoresis and A260/280 ratio.
qPCR
All qPCR reactions were performed in a final volume of 10 µl in a Rotor-Gene 6000 instrument (Corbett Lifescience) under the conditions specified in the figure legends. For PCR inter-run comparison, threshold settings were constant. Amplification of expected product was assessed by melting curve analysis (14).
RT-qPCR
cDNA synthesis was performed using Superscript III Reverse transcriptase (Invitrogen). Total RNA, dNTPs and gene-specific primers were incubated in a 10 µl volume at 65°C for 5 min to denature the RNA and then chilled on ice for at least 1 min. cDNA synthesis mix containing 5× First strand buffer (Invitrogen), dithiothreitol (DTT) and Superscript III was added to RNA/primer mix to a final volume of 20 µl containing: RNA 200 ng, gene-specific primers as indicated, dNTPs 500 µM, Tris–HCl 50 mM pH 8.3, KCl 75 mM, MgCl2 3 mM, DTT 5 mM and SuperScript III RT (200 U). The reaction was performed at 50°C for 30 min, and heat inactivated at 85°C for 5 min then chilled on ice. In experiments investigating the role of MgCl2, magnesium-free 10× RT buffer supplied with SuperScript III First Strand Synthesis System (Invitrogen) was used for the reverse transcription (RT) reaction, inducing slight modifications as follows: Tris–HCl 20 mM (pH 8.4), KCl 50 mM, DTT 10 mM, MgCl2 1.5, 3 or 5 mM. Incubation time was 50 min at 50°C as recommended by the manufacturer.
Subsequent qPCR reactions for HPRT1 and HMGA2 quantifications were performed in Sensimix (Quantace) following cycling conditions: 95°C (15 s) and 60°C (60 s). For probe-based detection, a single-tube format (custom TaqMan gene expression assay, Applied Biosystems) was used (see Table 1 for primers and probes sequence details). For Sybr Green-based qPCR reactions, identical primer pairs (HPRT1F/HPRT1R or HMGA2F/HMGA2R) were used at the concentration of 200 nM each.
Table 1.
Sequence information
| Gene | Primer | 5′-3′ sequences | Position | NCBI Accession number | ||
|---|---|---|---|---|---|---|
| HPV16-E7 | % GC | Tma | NM001526 | |||
| DNA-E7F | GAG GAG GAG GAT GAA ATA GAT GGT | 45.8 | 62.1°C | 658–681 | ||
| DNA-E7R | GCC CAT TAA CAG GTC TTC CAA | 47.8 | 61.3°C | 816–796 | ||
| ZNA-E7F | Z4-GAG GAG GAG GAT GAA ATA GAT GGT | |||||
| ZNA-E7R | Z4-GCC CAT TAA CAG GTC TTC CAA | |||||
| LNA-E7F | G+AG GA+G GAG GAT GAA ATA GAT GGT | |||||
| LNA-E7R | GC+C CAT +TAA CAG GTC TTC CAA | |||||
| HPV16-L1 | ||||||
| DNA-L1F | TTT GTT ACT GTT GTT GAT ACT AC | 30.4 | 56.4°C | 6624–6646 | ||
| DNA-L1R | GAA AAA TAA ACT GTA AAT CAT ATT C | 20.0 | 53.9°C | 6741–6765 | ||
| ZNA-L1F | Zn-TTT GTT ACT GTT GTT GAT ACT AC | |||||
| ZNA-L1R | Zn-GAA AAA TAA ACT GTA AAT CAT ATT C | |||||
| LNA-L1F | TT+T GTT +ACT GTT GTT GAT ACT AC | |||||
| LNA-L1R | GA+A AAA +TAA ACT GTA AAT CAT ATT C | |||||
| HPRT1 | Localization | NM000194 | ||||
| HPRT1F | TCCTTGGTCAGGCAGTATAATCCA | Exon6 | 527–550 | |||
| HPRT1R | GTCTGGCTTATATCCAACACTTCGT | Exon7 | 580–566 | |||
| HPRT1probe | FAM-CACCAGCAAGCTTGC-MGBnfq | Junction 6–7 | 613–592 | |||
| ZNA-HPRT1R | Z4-GTCTGGCTTATATCCAACACTTCGT | |||||
| HMGA2 (variant 1) | NM003483 | |||||
| HMGA2F | CTAGGAAATGGCCACAACAAGTTG | Exon4 | 1050–1073 | |||
| HMGA2R | GGCAGACTCTTGTGAGGATGT | Exon5 | 1129–1109 | |||
| HMGA2probe | FAM-CTGCTCAGGAGGAAAC-MGBnfq | Junction 4–5 | 1086–1101 | |||
| ZNA-HMGA2R | Z4-CTAGGAAATGGCCACAACAAGTTG | |||||
Zn denotes n spermine units, +N denotes LNA nucleotide.
aTm prediction from IDT SciTools web server for 100 nM oligonucleotide and 3 mM MgCl2. At 1.5 mM MgCl2, the Tm is predicted to be reduced by 2°C.
All the studies were performed in accordance with the MIQE guidelines (Minimum Information for Publication of Quantitative real-time PCR Experiments) (15).
RESULTS
In order to evaluate the specificity and the performances of ZNAs as PCR primers, we implemented a diagnostic model consisting of detecting the high-risk human papillomavirus type 16 (HPV 16) genome integrated into human genomic DNA. In this model, genomic DNA was extracted from SiHa cells, containing 1–2 copies of viral genome per cell (target genomic DNA); DNA from uninfected human A549 cells was used as negative control. We used two previously described primer pairs (16,17) to amplify the viral E7 oncogene and L1 structural gene. Each primer pair had either 46–48% or 20–30% GC content (Table 1) and thus represented medium- and low-GC content primers, respectively. We synthesized the ZNA primers by conjugating four or five spermines at the 5′-end of the oligonucleotide sequence and compared their performances with those of their standard counterparts (unconjugated DNA primers) and LNA primers containing two LNA nucleotides incorporated near the 5′-end (7,8).
Gradient PCR
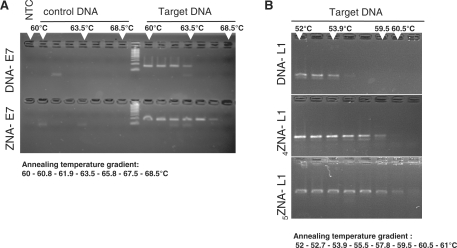
We first evaluated ZNA primers by conventional PCR using a gradient PCR procedure. ZNA oligonucleotides were able to prime DNA synthesis and drive PCR amplification indicating that the oligospermine tail located at the 5′-end does not impede the polymerase (Figure 1). PCR products of the expected size (Figure 1A, 159 bp for E7 gene; Figure 1B, 142 bp for L1 target) were exclusively amplified from target template (genomic DNA from SiHa cells versus non-infected A549 cells), showing that amplifications using ZNAs are specific. We confirmed amplicons identity by DNA sequencing. ZNAs enable the annealing step to be carried out at higher temperature compared with standard DNA primers, up to +4°C for both E7 and L1 primers conjugated to four spermines. This stabilization was further increased with five spermine moieties (Figure 1B) confirming that Tm is modulated by the number of spermine units grafted onto the oligonucleotide (11,13). No PCR product was detected using control genomic DNA even at the lowest temperature, except for some primer–dimers also occurring using unconjugated primers. This indicates that ZNA primers stabilize hybridization without affecting specificity. Moreover, the addition of spermine units stabilizes both E7 and L1 sequences in the same extent, while variability is observed for LNA modifications (Figure 1 and Supplementary figure).
Figure 1.
Conventional gradient PCR. Target or control genomic DNA was amplified following an annealing temperature gradient procedure (Bio-Rad iCycler) with DNA-E7 (A, upper lanes) or ZNA-E7 (A, lower lanes) primer pairs (100 nM each); with DNA-L1 (B, upper lanes) or ZNA-L1 primer pair (500 nM each) containing four spermines (B, medium lanes) or five spermines (B, lower lanes). Each sample was amplified in the presence of 10 ng genomic DNA, 10 mM Tris–HCl (pH 9), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 200 µM dNTP (each) and 0.04 U/µl of EconoTaq DNA Polymerase (Lucigen). Cycling protocol was 94°C (20 s), annealing (20 s), 72°C (15 s) × 35 cycles (A) or 30 cycles (B). Final reactions were analysed on 4% agarose gel (Seakem LE, Cambrex) stained with ethidium bromide.
PCR optimization, efficiency and sensitivity
To further characterize ZNAs as PCR primers, qPCR using SYBR Green detection was implemented to evaluate both the specificity and the efficiency of PCR. PCR efficiency depends on the binding efficiency of both primers to their target sequence. As magnesium concentration, annealing temperature and primer concentration are known to critically impact primer–template duplex formation, we focused on those three parameters.
Magnesium concentration and annealing temperature
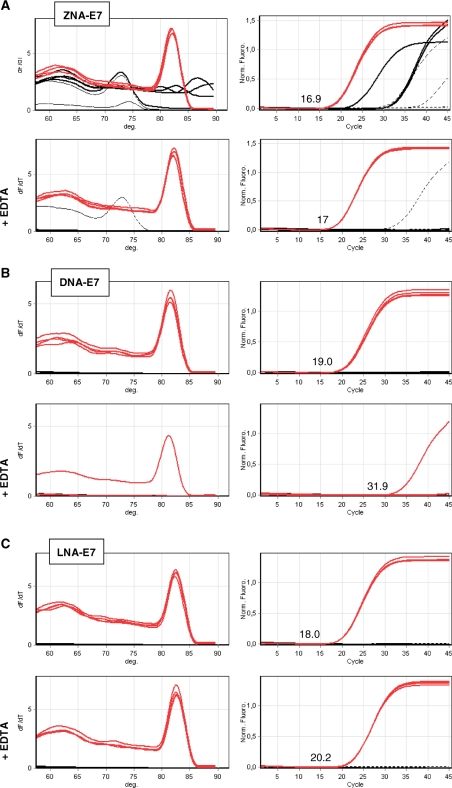
We first carried out standard two-step PCR reactions using ZNA, DNA and LNA primers specific for E7. As shown in the upper panels in Figure 2A–C, ZNA primers gave rise to a single amplification product in target samples with a quantification cycle value (Cq) (15) of 16.9, achieved one and two cycles earlier than LNA and DNA, respectively. However, ZNA primers also drove non-specific amplifications and primer–dimer artefacts in both control samples (genomic DNA and no template control), which were not observed with DNA and LNA primers. Since ZNA oligonucleotides display intrinsically reduced electrostatic repulsions, we hypothesized that the off-target amplifications observed in control samples might result from inappropriate salt content of the commercial qPCR mix. Indeed, cations and especially magnesium play a central role in PCR. They increase the stability of the primer–template hybrids by reducing electrostatic repulsions between strands, but excess reduces specificity by tolerating elongation of mismatched primers or promoting primer–dimers artefacts. The catalytic activity of the polymerase is sensitive to these environmental factors as well. Postulating an excess of cations in qPCR mixes optimized for standard oligonucleotides, we then added 1 mM EDTA to the 3 mM magnesium-containing reaction mix in order to decrease Mg2+ (lower panels in Figure 2A–C). PCR reactions driven by LNA primers were impeded (Cq shift of +2 cycles) and reactions primed by standard DNA primers were dramatically inhibited (only one out of four target replicates detected, Cq shift of 13 cycles). In contrast, EDTA addition did not affect ZNA oligonucleotides for target amplifications. Moreover, EDTA also prevented background signal in control samples, clearly showing that ZNAs require lower magnesium content.
Figure 2.
SYBR Green-based qPCR of HPV16-E7 using ZNA, DNA and LNA primers. Target genomic DNA (10 ng, red lines), control DNA (10 ng, black thin lines) and no template control (black dotted lines) were amplified in quadruplicate with 100 nM of ZNA-E7 (A), DNA-E7 (B), LNA-E7 (C) primer pairs. All reactions were performed using Sensimix NoRef DNA kit (Quantace) without (upper panels in A–C) and with 1 mM EDTA (lower panels in A–C) as indicated. Melting curves (left panels) and amplification plots (right panels) are represented. Cq values for target amplification are indicated. Cycling profile was 95°C (10 s), 60°C (1 min).
In order to confirm this result, we diluted the qPCR mix twice in order to decrease the salt content. MgCl2 requirement was then investigated by performing reactions at final concentrations of 1.5, 3 and 4 mM. Cq values of on- and off-target amplifications driven by ZNA, DNA or LNA primers at annealing temperatures of 57°C, 60°C and 63°C are given in Table 2. Target amplifications performed with ZNA were efficient at 1.5 mM MgCl2 and Cq values remained unchanged over the studied MgCl2 concentrations and annealing temperatures. Higher MgCl2 decreased specificity as shown by earlier Cq values displayed in control DNA samples. DNA and LNA primers performed poorly at 1.5 mM MgCl2. For those oligonucleotides, the original 3 mM magnesium concentration was the best condition to improve on-target amplification, while maintaining low non-specific signals. At 57°C, target amplifications yielded later Cq (4 and 3.5 additional cycles for DNA and LNA primers, respectively) than those primed with ZNA. Further increase of MgCl2 concentration to 4 mM had little effect. The higher Cq value observed with DNA and LNA primers does not result from too high annealing temperatures because Cq values remained unchanged at 60°C for DNA and even decreased from 57°C to 63°C for LNA primers. This latter observation might rather be due to the destabilization of secondary structures or non-specific binding due to LNA moieties that compete with proper binding of LNA primers. Similar Cq values were finally achieved by increasing the DNA and LNA primer concentrations (data not shown), suggesting that ZNA oligonucleotides exhibit an intrinsically higher affinity for their target.
Table 2.
Cq values obtained with ZNA, DNA and LNA primers under various MgCl2 concentrations and annealing temperatures
| Primer type | MgCl2 (mM) |
Cq target DNA |
Cq control DNA |
||||
|---|---|---|---|---|---|---|---|
| 57°C | 60°C | 63°C | 57°C | 60°C | 63°C | ||
| ZNA | 1.5 | 16.5 ∓ 0.10 | 16.4 ∓ 0.03 | 16.2 ∓ 0.06 | 27.4 ∓ 0.57 | 28.1 ∓ 0.04 | 34.4 ∓ 0.48 |
| 3 | 16.7 ∓ 0.21 | 16.9 ∓ 0.08 | n.d | 19.6 ∓ 0.03 | 22.3 ∓ 0.07 | n.d | |
| 4 | 16.5 ∓ 0.05 | n.d | n.d | 17.6 ∓ 2.10 | n.d | n.d | |
| DNA | 1.5 | 27.5 ∓ 0.10 | 30.7 ∓ 0.52 | 33.2 ∓ 0.32 | >45 | >45 | >45 |
| 3 | 20.4 ∓ 0.03 | 20.2 ∓ 0.11 | 21.8 ∓ 0.11 | 34.5 ∓ 1.09 | 35.4 ∓ 1.7 | 38.5 ∓ 1.23 | |
| 4 | 19.6 ∓ 0.00 | n.d | n.d | 31.0 ∓ 0.18 | n.d | n.d | |
| LNA | 1.5 | 24.6 ∓ 0.38 | 24.1 ∓ 0.04 | 24.0 ∓ 0.23 | 38.0 ∓ 1.41 | >45 | >45 |
| 3 | 20.0 ∓ 0.13 | 19.4 ∓ 0.12 | 19.1 ∓ 0.06 | 29.6 ∓ 0.6 | 32.6 ∓ 2.79 | 35.2 ∓ 1.51 | |
| 4 | 19.4 ∓ 0.06 | n.d | n.d | 28.5 ∓ 0.04 | n.d | n.d | |
Target genomic DNA (10 ng) or control DNA (10 ng) were amplified in duplicate with 100 nM of each E7 primer type as specified. All reactions were performed in duplicate (replicate standard deviation provided by the Rotor-Gene 6000 analysis software is indicated) using Sensimix NoRef kit 0.5×. MgCl2 was adjusted to the indicated concentration. Cycling conditions were 94°C for 20 s, annealing for 20 s, 72°C for 15 s.
n.d., not determined.
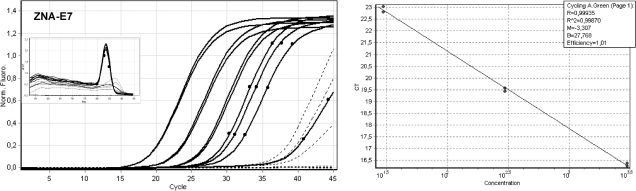
We assessed each primer pair's specificity at its optimal magnesium concentration (1.5 mM for ZNA, 3 mM for LNA and DNA) by calculating the Cq difference between control genomic DNA (non-specific amplification) and target genomic DNA (specific amplification) (Table 3). ZNA and LNA exhibited comparable specificities at all annealing temperatures. Due to higher Tm, both modified primers were less specific than DNA primers at 57°C (delta Cq ∼ 10 versus 14.1). However, primer specificity was restored while maintaining target amplification efficiency by increasing the annealing temperature. This is further demonstrated in Figure 3 by showing amplification reactions of serial dilutions of target genomic DNA spiked in control DNA. In this experiment, the annealing temperature was 67°C, that is, 7°C above the expected Tm of standard DNA primers at 1.5 mM MgCl2. ZNA primers drove highly efficient (E = 1.01, R2 = 0.999) and sensitive PCR, as three out of four replicates containing only 3–6 copies of viral target were detected quantitatively. Non-specific amplification products in control DNA samples occurring 10 cycles after the last dilution do not affect specificity or sensitivity of detection. It is noteworthy that while spike-in experiments were intentionally performed for evaluating ZNA specificity, we observed that highly diluted target samples are only detectable under these conditions, where control DNA behaves as a carrier. Thus, the ability of ZNA to detect low amounts of target depends on the total amount of nucleic acid present in the sample and may require addition of a nucleic acid carrier, most probably to avoid adsorption artefacts.
Table 3.
Primer specificity
| Primer type | MgCl2 (mM) |
Cq (control)–Cq (target) |
||
|---|---|---|---|---|
| 57°C | 60°C | 63°C | ||
| ZNA | 1.5 | 10.9 | 11.7 | 18.2 |
| DNA | 3 | 14.1 | 15.2 | 16.7 |
| LNA | 3 | 9.6 | 13.2 | 16.1 |
The difference between off- and on-target amplification was calculated from data in Table 1 for each E7 primer pair type at their respective optimal MgCl2 concentration as indicated.
Figure 3.
Amplification of HPV16-E7 using ZNA primers at high annealing temperature and low salt. Serial dilutions of target genomic DNA (10 ng to 10 pg) spiked in control DNA were amplified with 100 nM of each ZNA-E7 primers. All reactions were performed in duplicate (black thin) except 10 pg (3–6 copies of HPV16 genome), which was performed in quadruplicate (black circles). Control DNA (10 ng) and no template control are represented in dotted lines. Melting curves are shown in left top corner. The 10 pg sample was not taken into account for PCR efficiency calculation (standard curve, right panel). All reactions were performed using Sensimix NoRef DNA kit 0.5×. Cycling conditions were 94°C (20 s), 67°C (20 s), 72°C (15 s).
Taken together, our data show that when comparing the three primer types under their optimal PCR conditions, ZNA primers exhibit the highest sensitivity. Even though reaction conditions may need to be adapted, high sensitivity is achieved without compromising specificity.
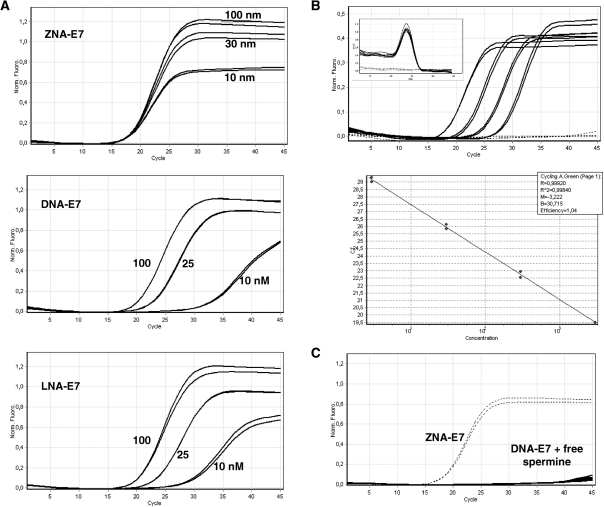
Primer concentration
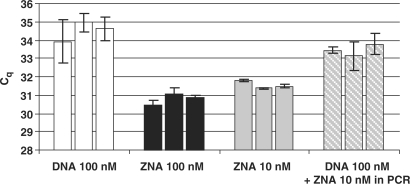
To evaluate the respective affinity of DNA, LNA and ZNA oligonucleotides for the target, we carried out PCR reactions using low amounts of primers. Magnesium was optimally adjusted for each primer type and amplifications were performed at the annealing temperature of 60°C which is compatible with all the primers. As shown in Figure 4A, the PCR performance was dramatically affected when DNA and LNA primers were used at low concentration. A gradual decrease in yield was observed with decreasing primer concentration and both primer types were almost unproductive at 10 nM. In contrast, decreasing ZNA concentration as low as 10 nM did not impact the Cq value. Moreover, for ZNA, the PCR reaction remained highly efficient as shown by the standard curve in Figure 4B. Sensitivity was not altered as samples containing 3–6 copies of target were quantitatively detected. Finally, decreasing ZNA primer concentration restored full specificity as amplifications only occurred in target samples (Figure 4B). Adding increasing amounts of free spermine to the reaction did not allow standard DNA primers to drive amplification when used at the concentration of 10 nM (Figure 4C). This observation demonstrates that a covalent linkage of spermines to the oligonucleotide is required to confer ZNA oligonucleotides an exceptionally high affinity towards their complementary sequence. Interestingly, the level of the PCR plateau is decreasing with ZNA primers concentration, indicating a reduction in the final amount of product at the reaction end-point (Figure 4A, upper panel). The ability of ZNAs to stop production of amplicon molecules early in the reaction without affecting other PCR reaction features is of interest for multiplex PCR applications where simultaneous target amplifications in a single reaction compete for finite amounts of reagents.
Figure 4.
Effect of primer concentration on PCR. (A) Variable amounts of E7-specific primers were used to amplify 10 ng of target genomic DNA. Primers were ZNA-E7 (upper panel), DNA (middle panel) or LNA primers (lower panel). (B) Serial dilutions of target genomic DNA (10 ng to 10 pg) spiked in control DNA were amplified with 10 nM each of ZNA-E7 primers. Control DNA (10 ng) and no template control are shown (dotted lines). Melting curves (left top corner in upper panel in B) and standard curve (lower panel in B) are shown. (C) DNA-E7 primers were used at 10 nM with increasing amounts of free spermine (0, 10, 40 and 80 nM). Amplifications using 10 nM ZNA-E7 are shown (dotted lines). All reactions were carried out in duplicates using Sensimix NoRef DNA kit (Quantace) 0.5×. MgCl2 was generally 1.5 mM except 3 mM for LNA and DNA primers in (A). Cycling conditions were 95°C (10 s), 60°C (1 min).
AT-rich sequences and standardization
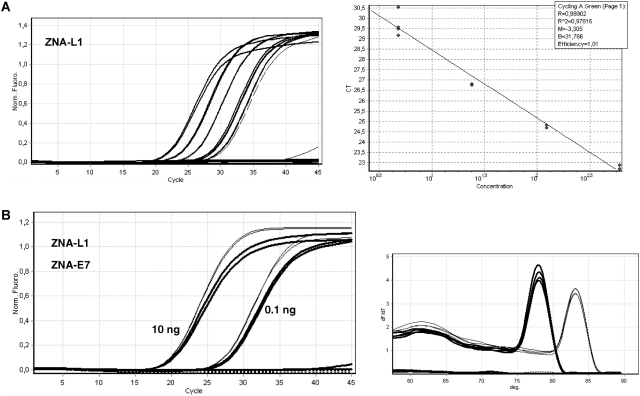
In order to evaluate ZNAs performance in various GC contexts, we assessed viral genome detection using AT-rich L1 primer pairs containing 20–30% GC (Table 1) and compared PCR performances with those obtained with E7 primers exhibiting 46–48% GC. As expected, primer base sequence had a large impact on PCR performances and optimal reaction conditions for standard DNA oligonucleotides. At appropriate annealing temperature (50°C), a 5-fold larger amount of DNA-L1 primers or a combination of more primers and higher MgCl2 concentrations is required to achieve the PCR performance obtained with DNA-E7 primers (data not shown). In contrast, ZNA-L1 primers containing four spermines operated optimally (Figure 5A; E = 1.01, R2 = 0.989) at the same primer and magnesium concentrations used for ZNA-E7 oligonucleotides (100 nM primers, 1.5 mM MgCl2). The reaction sensitivity remained unaffected as all replicate samples containing 5–10 target copies and half replicate samples containing 1–2 target copies were detected. Strikingly, both types of ZNA primers led to similar Cq's [19.7 for 2 ng (L1 primers, Figure 5A) versus 19.5 for 1 ng (E7 primers, Figure 3)]. Thus, while DNA primer pairs with different base sequences exhibit different intrinsic affinities for their targets requiring different reaction conditions, their ZNA counterparts yield constant performances under standardized conditions; the only variable being the annealing temperature. ZNA-L1 primers indeed performed best at an annealing temperature of 55°C [i.e. 2°C above the estimated Tm of standard DNA-L1 primers (Figure 5A)], and increasing it to 60°C induced a Cq shift of three cycles to the right (data not shown). However, using such AT-rich primers at a standard annealing temperature of 60°C could be of interest for hydrolysis probe-based detection, multiplex PCR or high-throughput experiments. We, therefore, grafted an additional spermine residue to the ZNA primers, which led to optimal amplification efficiency and sensitivity at 60°C (data not shown). We confirmed this result using a PCR reagent dedicated to fast PCR (Quantifast Sybr Green kit, Qiagen). According to the manufacturer, no magnesium adjustment was required; thus, all reactions were carried out under exactly the same conditions. As shown in Figure 5B, neither DNA-L1 nor primers containing two LNA moieties were capable of amplifying the L1 target at 60°C, while ZNA primers containing five spermines performed efficiently with Cq's similar to ZNA-E7 oligonucleotides.
Figure 5.
DNA amplification with AT-rich ZNA primers. (A) Serial dilutions (2 ng to 3.2 pg) of target genomic DNA spiked in control DNA were amplified with ZNA-L1 primers containing four spermine units (100 nM each). Samples containing 2, 0.4 and 0.08 ng were amplified in duplicates and those with 16 and 3.2 pg corresponding to 5–10 copies and 1–2 copies, respectively were amplified in quadruplicate. The 3.2 pg sample (dashed lines) was not taken into account for PCR efficiency calculation (A, right panel). No amplification was detected in the controls. All reactions were performed using Sensimix NoRef DNA kit 0.5× concentrated. Cycling conditions were 94°C (20 s), 55°C (20 s), 72°C (15 s). (B) Target genomic DNA (10 or 0.1 ng spiked in control DNA) or control DNA (10 ng) was amplified with 100 nM ZNA-L1 primers containing five spermines (thin lines), DNA-L1 or LNA-L1 primers (dotted lines) using the QuantiFast SYBR Green PCR kit (Qiagen). Solid curves show amplifications using 100 nM ZNA-E7 primers. Cycling conditions were 95°C (10 s) and 60°C (30 s). Melting curves are represented (B, right panel).
Taken together, our results show that ZNAs drive efficient, sensitive and specific PCR at high annealing temperature and low magnesium concentration. In particular, ZNAs improve primers performing poorly such as AT-rich sequences, allowing their use under standardized conditions and circumventing magnesium and primer concentration adjustments. The ZNA Tm can be tuned by varying the number of spermines, thus giving the possibility of easily designing efficient primers operating at any given annealing temperature.
Fast cycling, high sensitivity
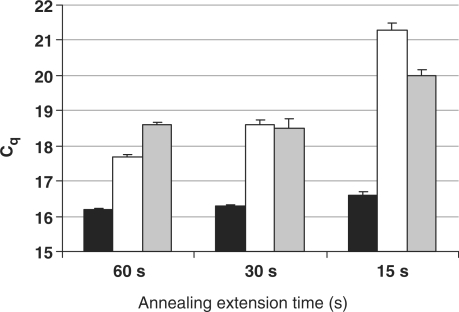
In order to explore the impact of spermine conjugation on hybridization kinetics, we compared ZNA, LNA and DNA priming activities following a universal, a fast and a very fast cycling protocol with combined annealing/elongation steps of 60, 30 and 15 s, respectively. Target DNA (10 ng) was amplified in quadruplicates using each type of E7 primer pair. Mean Cq values and standard deviations are represented in Figure 6. ZNA primers yielded a constant Cq value irrespective of PCR conditions, which is overall the lowest Cq measured for the three primer species at any cycling condition. Repeatability was not affected by fast cycling protocols as demonstrated by SDs < 0.1 and confidential intervals ranging below 0.30 cycles (data not shown). However, fast cycling had a dramatic impact on DNA primers efficiency and to a lower extent on LNA primers as shown by the increase in Cq's. The same results were obtained using a mix dedicated to fast cycling (data not shown), which suggested that ZNA oligonucleotides exhibit faster binding kinetics.
Figure 6.
Fast cycling PCR. Mean Cq values and standard deviations (error bars) from quadruplicate reactions carried out with ZNA-E7 (black bars), DNA-E7 (white bars) or LNA-E7 (grey bars) primers as a function of annealing/extension time. Reactions were carried out on 10 ng of target genomic DNA with 100 nM each primer using Sensimix NoRef DNA kit (Quantace) 0.5× concentrated. MgCl2 was 1.5 mM (ZNA primers) and 3 mM (LNA and DNA primers). Cycling conditions were 95°C (10 s), 60°C as indicated.
In keeping with the above results, we observed that ZNA-primed amplifications were detected 2–3 cycles earlier than LNA- and DNA-primed reactions (Figure 6), whatever the length of the annealing extension step. In order to better understand the underlying mechanism of the improved sensitivity of ZNAs, we performed serial dilutions amplifications of target genomic DNA using each primer pairs and compared PCR efficiencies (Table 4). Interestingly, earlier Cq's provided by ZNA primers were not associated with a higher PCR efficiency (E = 0.94 for ZNA and 0.96 for DNA primers). This indicates that the binding of ZNA, DNA or LNA primers to the template are comparable in the central-cycle window of the reaction where PCR efficiency is measured, amplicon concentration being sufficient to emit detectable fluorescence. Our data, therefore, suggest that ZNA oligonucleotides are capable of binding their target with a greater efficiency during the early stages of PCR, when the template is present in low amounts. This result gives further support to the exceptionally high affinity of ZNA primers for their target.
Table 4.
PCR efficiencies (E) calculated from the slope of standard curves using ZNA-E7, DNA-E7 and LNA-E7 primers
| Primers | Cqa | E | R2 b |
|---|---|---|---|
| ZNA-E7 | 17.7 ∓ 0.01 | 0.94 | 0.999 |
| DNA-E7 | 20.7 ∓ 0.09 | 0.96 | 0.993 |
| LNA-E7 | 19.3 ∓ 0.00 | 0.89 | 0.999 |
Target genomic DNA (10 ng) and 10-fold serial dilutions (up to 0.01 ng) spiked in 10 ng of control DNA were amplified in duplicate with 100 nM each primer using QuantiFast SYBR Green PCR kit (Qiagen). Cycling conditions were 95°C (10 s), 60°C (30 s).
aCq obtained from amplification of 10 ng target DNA.
bFrom the linear regression.
Reverse transcription using ZNAs
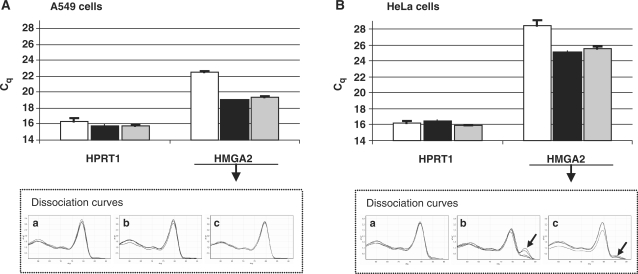
The fast and efficient binding of ZNA primers to low amounts of template during the initial PCR cycles prompted us to investigate whether a ZNA primer could improve Reverse transcription (RT), the initial and the most crucial step in RT-qPCR. In particular, we addressed the RNA to cDNA conversion of low-expressed messengers as they are prone to higher variability as well as reliability issues (18). For these experiments, we quantified the HMGA2 oncogene, a member of the high mobility group AT-hook family of non-histone chromatin proteins which is a low-expressed gene in HeLa cervix carcinoma cells and is over-expressed in A549 lung carcinoma cells (19). Hypoxanthine phosphoribosylltransferase 1 gene (HPRT1) was chosen as calibration reference. The reverse PCR primers HMGA2R and HPRT1R were used as gene-specific primers for the RT step (Table 1). The ZNA-HMGA2R primer was synthesized with four spermines at the 5′-end, and used for priming a pool of total RNA from A549 or HeLa cells using Superscript III (Invitrogen). The same RNA pool was reverse transcribed using a standard DNA-HMGA2R primer under identical conditions. The standard DNA-HPRT1 primer was simultaneously added to all samples to allow HPRT1 expression measurement as an internal control. After the RT step, reactions were subsequently subjected to qPCR with SYBR Green I detection using the standard DNA primer pair. As shown in Figure 7A, the HMGA2 mRNA from A549 cells was detected three cycles earlier when the RT reaction was primed with the ZNA primer (Cq = 19.08 and 19.42, respectively, for 100 and 10 nM) instead of the DNA primer (Cq = 22.59). HPRT1 calibrations (Cq = 16.3, 15.8 and 15.8) confirmed identical amounts of total RNA in the samples. The PCR efficiency for both HPRT1 and HMGA2 targets were 100% as assessed by amplifying serial dilutions of poly(dT)-primed cDNA from A549 cells (data not shown). Thus, a three-cycle difference in Cq corresponds to a 8-fold difference in starting cDNA material, indicating that the ZNA primer is most likely to drive the RNA to cDNA conversion with a 8-fold higher efficiency than its standard counterpart. However, an alternative explanation was that DNA and ZNA primers both generate cDNA with the same efficiency, but that the material is amplified with a greater sensitivity in the subsequent PCR reaction due to residual ZNA-HMGA2R primer from the RT reaction (respectively, 10 and 1 nM in Figure 7). Addition of 1 nM ZNA-HMGA2R to the PCR reaction had no impact on amplification of cDNA primed with the standard DNA primer (data not shown), ruling out the latter hypothesis. Primer–dimers occurred in RT negative controls lacking the reverse transcriptase or lacking RNA when ‘primed’ with ZNA-HMGA2R. However, their Cq values were >32 which corresponds to less than one molecule under our conditions and the use of the specific hydrolysis probe avoided their detection (data not shown). In HeLa cells, HMGA2 is ∼60-fold less abundant than in A549 cells as shown by the six-cycle Cq increase (Figure 7B). We confirmed that the ZNA RT primer is providing more cDNA than the standard DNA primer (Cq = 25.1 and 28.4, respectively, for 100 nM). However, careful analysis of melting curves also showed that off-target products were generated in addition to the specific amplicon. Although absent in A549 cDNA (Figure 7A, plots b and c), ZNA-induced mispriming occurred when RT was performed with RNA extracted from HeLa cells, i.e. when the target was present at very low concentration (Figure 7B, plot b). Mispriming, however, decreased upon reduction of the ZNA primer concentration to 10 nM (Figure 7B, plot c). We then performed the same experiment using the specific hydrolysis probe to detect exclusively the specific amplicon (Figure 8). RT reactions were carried out in triplicate and each cDNA was quantified by qPCR in duplicates. We confirmed that the ZNA primer allows earlier detection of the HMGA2 mRNA, thus, demonstrating that the presence of non-specific amplification material did not interfere with quantification of the HMGA2 target using SYBR Green for detection in the previous experiment. More importantly, lower Cq values were also associated with less variability as shown by the much lower standard deviations between qPCR duplicates. Thus, when used as primers in RT, ZNAs allow a more accurate quantification of weakly expressed genes by generating larger amounts of cDNA molecules. Finally, ZNA-HMGA2R primer used at 100 nM provided lower Cq values than at 10 nM (mean Cq = 30.8 and 31.6, respectively). Given that the same difference in Cq's was observed when DNA-primed cDNA was amplified in the presence or absence of additional 10 nM ZNA primer (mean Cq = 34.5 and 33.5, respectively), we conclude that ZNAs improve RT-qPCR at both steps: RNA to cDNA conversion and subsequent qPCR.
Figure 7.
ZNA primers improve RNA to cDNA conversion. Mean Cq values for HPRT1 and HMGA2 were determined by RT-qPCR on 200 ng of total RNA extracted from A549 (A) and HeLa (B) cells. Dissociation curves of HMGA2 amplification product are represented in lower panels (a–c). RT reactions were performed in duplicate with 100 nM HPRT1R and 100 nM DNA-HMGA2R (white bars, plot a), 100 nM ZNA-HMGA2R (black bars, plots b) or 10 nM ZNA-HMGA2R (grey bars, plot c). Each cDNA corresponding to 10 ng of total RNA was subjected to SYBR Green-based qPCR in duplicate. Error bars indicate standard deviation.
Figure 8.
ZNA primers improve the accuracy of low-abundant gene expression measurement. Quantification of HMGA2 target in three independent RT-qPCR reactions. Bars represent the Cq value for each RT reaction and error bars indicate qPCR duplicate standard deviations. cDNA was obtained by RT of 200 ng of total RNA extracted from HeLa cells using 100 nM HPRT1R and 100 nM DNA-HMGA2R (white bars, dashed bars), 100 nM ZNA-HMGA2R (black bars) or 10 nM ZNA-HMGA2R (grey bars). Each cDNA corresponding to 10 ng of total RNA was subjected to qPCR using a specific hydrolysis probe for detection. White and dashed bars represent qPCR reactions on the same cDNA samples but 10 nM of ZNA-HMGA2R primer was added in qPCR reaction represented in dashed bars.
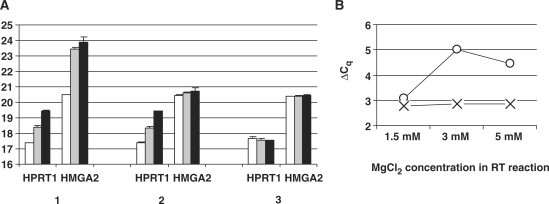
We finally addressed the role of magnesium in RT. Instead of using the 5× First-Stand Buffer supplied with the reverse transcriptase and containing 3 mM final MgCl2 concentration as in previous experiments, we used the magnesium-free 10× RT buffer (Invitrogen). RT reactions were carried out with 1.5, 3 or 5 mM MgCl2 and 100 nM of HPRT1- and HMGA2-specific primers. Three RT priming conditions were compared: both primers being standard DNA sequences, or their ZNA counterparts or the combination DNA-HPRT1 and ZNA-HMGA2 (Figure 9A, conditions 1, 3 and 2, respectively). Interestingly, standard DNA primers operated optimally at 1.5 mM MgCl2, with RT efficiency similar to ZNA primers. However, RT reactions primed with DNA primers were inhibited by increasing magnesium concentrations as shown by the increase in Cq values. Moreover, magnesium variations did not affect HMGA2 and HPRT1 quantification to the same extent. Measuring HMGA2 gene expression relative to HPRT1 was, thus, subjected to large variations up to 66% depending on experimental conditions (Figure 9B). In contrast, when ZNA primers were used for cDNA synthesis, Cq's remained constant and provided a robust and precise quantification of the target gene.
Figure 9.
Magnesium induces RT efficiency variability. (A) Mean Cq values for HPRT1 and HMGA2 determined by RT-qPCR using 100 nM of each gene-specific primer DNA-HPRT1R/DNA-HMGA2R (1), DNA-HPRT1R/ZNA-HMGA2R (2) or ZNA-HPRT1R/ZNA-HMGA2R (3) for cDNA synthesis. RT reactions were performed using 200 ng of total RNA extracted from A549 cells with MgCl2 1.5 mM (white bars), 3 mM (grey bars) or 5 mM (black bars). Each cDNA corresponding to 2 ng of total RNA was subjected to SYBR Green-based qPCR in duplicate. Error bars indicate the standard deviation. (B) ΔCq = Cq(HPRT1) − Cq(HMGA2) as a function of MgCl2 concentration in the RT reaction. ΔCq were calculated from DNA priming conditions [circles, from data (1) in A] and ZNA priming conditions [cross, from data (3) in A].
DISCUSSION
In the present study, we have addressed the specificity of ZNA oligonucleotides by using them as primers for PCR detection of a genomic target. We have shown that ZNAs exhibit a very high affinity for their target that requires adapting experimental conditions, but by doing so, the ZNA displays strict recognition selectivity. Similarly to standard oligonucleotides, ZNA specificity indeed relies on optimized conditions including salt concentration, annealing temperature or target and primer concentrations that may be different from those optimized for standard molecules.
First, we have shown that ZNAs require low magnesium concentrations. At higher MgCl2 concentrations needed for unmodified primers, ZNAs may generate off-target amplifications and primer–dimer artefacts. Beside its catalytical role for polymerase activity, magnesium increases the stability of primer–template hybrids by reducing electrostatic repulsions between strands. When present in excess, divalent cations decrease specificity by stabilizing mismatched duplexes that can be extended. For ZNA, the oligospermine tail covalently attached to the oligonucleotide plays to some extent the role of magnesium for shielding negative charges repulsion between strands. In this context, magnesium is no longer required to facilitate specific base pairing. Yet, qPCR reactions are generally performed with ready-to-use mixes including all needed components except primers and target. There is a plethora of kits available in the market exhibiting various performances and shifting from one or another often requires adjustment of experimental conditions. Usually the complete reagent composition is not disclosed but magnesium concentration, when provided, is within 3–6 mM. Thus, to counteract the presence of excess magnesium, we have chosen to add EDTA, a known inhibitor of PCR shown to affect differentially PCR reactions, partly depending on amplicon GC content and primer Tm's (20). We have tested seven different PCR mixes from various manufacturers, and all performed well with ZNAs when complemented with EDTA (data not shown). We also observed that once the optimal EDTA concentration was determined for the ZNA pair with ∼50% GC, no further optimization was required even for the ZNA pair with 20–30% GC content. Conversely, no magnesium adjustment was required for those ZNA primer pairs with different GC contents. Thus, ZNA may circumvent fastidious salt optimizations generally required for each conventional primer pairs to achieve efficient and specific PCR reactions, thereby presenting an advantage for setting-up PCR reactions. It may be of interest in future work to investigate whether ZNAs are more resistant to PCR inhibitors depleting free Mg2+ that may be potentially co-purified during nucleic acid extraction (21,22).
We have also demonstrated that ZNA primers allow PCR to be run at higher annealing temperature than using unmodified primers. Interestingly, the highest annealing temperature which did not affect PCR efficiency was in agreement with Tm predictions. Given that electrostatic forces are non-directive, the Tm increase per spermine is independent of the base sequence (13). The ZNA Tm was found to be easily predictable, using a simple mathematical relation depending on the intrinsic DNA oligonucleotide Tm, on the length N of the oligonucleotide and on the number z of spermines: Tm(ZNA) = Tm(DNA) + 36z/(N − 3.2). This straightforward linear dependence contrasts with LNAs, where Tm increments (and sometimes decrements) are highly sequence and context dependent (23). However, the constants of the above mathematical relation depend on the buffer. The Tm increment per spermine is dependent on the ionic strength (the lower the salt concentration, the higher the impact of spermine) and on the pH, with less stable binding of ZNAs to their target sequences at alkaline pH where spermines are less protonated. We estimate that PCR conditions at annealing steps using a classical reagent containing 10 mM Tris buffer, 50 mM NaCl and 3 mM MgCl2 are not significantly different from the HEPES 10 mM, pH7.4, NaCl 150 mM buffer used by Behr and collaborators (13), given that the pH value of Tris decreases with increasing temperatures and is close to neutrality between 60°C and 72°C (24). According to this postulate, the Tm increase per spermine for a 22-mer oligonucleotide is predicted to be 36/(22 − 3.2) = 1.9°C per spermine. In the present study, ZNA containing four and five spermines were, thus, expected to exhibit a stabilization of 7.6°C and 9.5°C over DNA oligonucleotides. Interestingly, PCR were successfully conducted using ZNAs at annealing temperatures approximately 6°C and 8°C above the maximum annealing temperatures implemented for unmodified sequences, suggesting a direct correlation between calculated Tm and annealing temperatures. More data will be generated to develop a prediction tool capable of accurately calculating the Tm in various buffers. Still ZNAs already provide means of finely and accurately manipulating the Tm of a given oligonucleotide sequence. The possibility of easily standardizing the Tm is of prime interest for numerous PCR applications, such as multiplex PCR or high-throughput experiments. For example, we showed the potential of ZNAs for improving poorly performing AT-rich DNA primers. Their ZNA counterparts enabled us to conduct PCR under standardized conditions of magnesium and primer concentrations at an annealing temperature of 60°C, yielding the same high performance as a ZNA primer pair with a standard 50% GC content under the same conditions. ZNAs will, thus, help to design efficient assays within AT-rich regions to be run under universal conditions. In particular, this will provide the ability to implement 5′-nuclease assays in such sequences where standard primers exhibit low Tm.
In addition, we found that ZNAs are able to optimally detect a genomic target at very low concentration (10-fold less than DNA- or LNA-substituted primers in the present study). Interestingly, reducing ZNA primers concentration did not affect any PCR reaction feature (Cq values, dynamic range, sensitivity and efficiency), but decreased the level of the plateau. Several factors have been proposed to explain the amplification arrest in the late phase of PCR, including competitive binding of DNA polymerase to its amplification product or depletion of dNTPs (25). Our data support the existence of a competition between primer–template and amplicon–amplicon reannealing (26). Furthermore, the possibility to stop the reaction early while maintaining PCR performances, which is only observed with ZNAs, is of interest for multiplex PCR applications. When one of the targets is much more abundant than the others at the beginning of the reaction, its early amplification may potentially inhibit amplification of the less abundant targets in the reaction by depleting reaction components. To avoid such a competition, the primers concentration for the most abundant target should be limited in order to stop the reaction soon after the signal detection. This limitation, however, often affects PCR efficiency (27). Instead, the exceptional high affinity of ZNAs allows their use at considerably lower concentration without impacting PCR efficiency. Moreover, low ZNA concentrations also ensure lower background amplifications.
Another strong advantage of ZNA primers demonstrated here is their ability to improve the sensitivity of detection in the early stage of PCR reaction when the target is present in low abundance. Later during the reaction, when the amplicon is detectable, no difference in PCR efficiency was observed compared to reactions primed with DNA or LNA primers. The greater binding of ZNA primers at the beginning of the reaction results in an improvement of the PCR sensitivity corresponding to earlier Cq values. Mechanistically, formation of the primer–template complex is a second-order reaction proportional of both primer concentration and target amount. The finding that ZNAs are prone to trigger productive amplification at low concentrations together with the ability of improving recognition of rare targets suggests that ZNAs exhibit a high primer–template association rate. It is likely that spermine conjugation increases the local concentration of ZNA in the template vicinity, resulting in a significant increase in effective concentration of primer near the target sequence. Our result supports a previous model suggesting that spermine binds DNA with a high affinity, crawls along the molecule within the minor groove until the oligonucleotide finds its target sequence (28,29).
In order to further understand the mechanism underlying the high affinity of ZNAs, we performed qPCR amplification using fast cycling protocols. We demonstrated that ZNAs are not affected by a drastic reduction of the annealing time, compared with DNA or LNA primers, suggesting that primer–template formation occurs with faster kinetics than with DNA or LNA primers. Such an acceleration of hybridization has been observed previously for uncharged PNAs or oligonucleotides conjugated with cationic peptides (30,31). Consistent with the previous finding that LNA-increased stability results from a slower dissociation rather than faster association of the complexes (32), our data support a distinct mechanism to stabilize hybridization of ZNAs, explaining the different behaviours observed in the present study for both modified oligonucleotides. In particular, we have demonstrated here that ZNAs exhibit improved performances using rapid PCR, and particularly an earlier detection relative to DNA and LNA oligonucleotides. Having emerged in order to reduce the PCR analysis time, fast cycling protocols have nevertheless been shown to be associated with loss of sensitivity or increase in variability (33). Whether this is also the case for ZNAs remains to be explored in detail.
The greater ability to bind rare targets prompted us to investigate whether ZNA could improve RT which is used in most techniques to quantify gene expression. RT-qPCR is the most widely used method to determine expression levels, but it depends on numerous critical considerations about assay design, technical and analytical aspects to provide reliable data (34). The first molecular step in RT-qPCR is the conversion of RNA to cDNA which has been shown to be the source of most of variability (18). One of the reasons leading to a lack of reproducibility is the low efficiency of reverse transcriptase that has been estimated to be ∼20%, dropping to 6% for low-abundance RNA templates (35). Interestingly, we have shown here that ZNA primers improve the yield of RNA to cDNA conversion under standard conditions. For a given sample, higher amount of cDNA templates are obtained and used as substrate for subsequent qPCR reaction leading to earlier detection. This result is of particular interest in the context of low-abundant targets, since accurate measurements are compromised by the inherent variability of PCR amplification in the presence of small amount of template (36,37). We have indeed demonstrated that the use of the ZNA RT primer significantly increases the reliability of HMGA2 mRNA measurement in HeLa total RNA. Moreover, achieving a higher yield of cDNA provides the possibility to dilute samples, thereby offering the possibility to overcome inhibition due to the presence of contaminants from biological samples.
Finally, we have performed RT reactions with various concentrations of magnesium and observed that the increase in magnesium concentration induces a decrease in priming efficiency using standard DNA primers, leading to high variations in target quantification. This suggests that a slight variation of salt content in the RNA preparation may interfere with RT efficiency, leading to extensive variations in Cq determinations and finally to inaccurate data. This also may lead to inconsistent results between studies or laboratories. In contrast, RT reactions primed with ZNAs were not affected by magnesium variations, providing a more accurate measurement of the gene expression. Furthermore, this observation possibly explains in part the mechanism by which ZNAs improve RNA to cDNA conversion. Indeed, salt stabilizes intramolecular interactions of RNA molecules and potentially the primer accessibility to its binding site if the latter is prone to be folded. Due to its high affinity, and particularly fast binding kinetics, ZNA may be capable of binding the primer site even when the single-stranded conformation has short lifetimes. This would explain the greater priming activity of ZNAs under standard magnesium concentrations that is not observed when MgCl2 was lower. In agreement with this hypothesis, the relative improvement of ZNA RT priming seems to be dependent on the nature of target and not on its amount. Indeed, HMGA2 mRNA to cDNA conversion yield was shown to be 8-fold increased in both HeLa and A549 extracts in which the level of expression of RNA messenger differs significantly, while HPRT1 RT appeared to be only 2-fold more efficient using ZNA primer compared with its standard counterpart.
In conclusion, ZNAs are easy-to-design modified oligonucleotides that exhibit a very high affinity for the target sequence without affecting the specificity of recognition. Extensive use of ZNAs will provide a detailed evaluation of the potency of these new modified oligonucleotides. However, this study demonstrates that used as primers, ZNAs offer the ability to work at higher annealing temperature, low primer concentrations, low magnesium concentrations and under fast protocols. Thus, they may be ideal for applications such as multiplex PCR or amplification in AT-rich regions. Our data also show that ZNAs are particularly promising in RT, since they improve the yield of cDNA synthesis. Their evaluation as qPCR probes is in progress. Finally, this work has given us a deeper understanding of ZNA hybridization and the improvement provided which will also benefit to other nucleic acid-based technologies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Competitivity cluster Alsace Biovalley; the Ministère de l’Industrie et the Direction Générale des Entreprises (project number 2005-2319-06A-045); the Région Alsace and the Communauté Urbaine de Strasbourg. Funding for open access charge: Polyplus-transfection SA.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We warmly thank Jean-Marc Strub (CNRS, France) for mass spectrometry analysis of ZNA oligonucleotides as well as Pierre Muller and Vincent Cirimèle (ChemTox, France) for their implication and support.
REFERENCES
- 1.Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR—a perspective. J. Mol. Endocrinol. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- 2.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonak J, Lind K, Sindelka R, Sjoback R, Sjogreen B, Strombom L, et al. The real-time polymerase chain reaction. Mol. Aspects Med. 2006;27:95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 4.Afonina I, Zivarts M, Kutyavin I, Lukhtanov E, Gamper H, Meyer RB. Efficient priming of PCR with short oligonucleotides conjugated to a minor groove binder. Nucleic Acids Res. 1997;25:2657–2660. doi: 10.1093/nar/25.13.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, et al. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa JM, Ernault P, Olivi M, Gaillon T, Arar K. Chimeric LNA/DNA probes as a detection system for real-time PCR. Clin. Biochem. 2004;37:930–932. doi: 10.1016/j.clinbiochem.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 7.Latorra D, Arar K, Hurley JM. Design considerations and effects of LNA in PCR primers. Mol. Cell Probes. 2003;17:253–259. doi: 10.1016/s0890-8508(03)00062-8. [DOI] [PubMed] [Google Scholar]
- 8.Levin JD, Fiala D, Samala MF, Kahn JD, Peterson RJ. Position-dependent effects of locked nucleic acid (LNA) on DNA sequencing and PCR primers. Nucleic Acids Res. 2006;34:e142. doi: 10.1093/nar/gkl756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arora A, Kaur H, Wengel J, Maiti S. Effect of locked nucleic acid (LNA) modification on hybridization kinetics of DNA duplex. Nucleic Acids Symp. Ser. 2008;52:417–418. doi: 10.1093/nass/nrn212. [DOI] [PubMed] [Google Scholar]
- 10.Pellestor F, Paulasova P. The peptide nucleic acids, efficient tools for molecular diagnosis (Review) Int. J. Mol. Med. 2004;13:521–525. [PubMed] [Google Scholar]
- 11.Pons B, Kotera M, Zuber G, Behr JP. Online synthesis of diblock cationic oligonucleotides for enhanced hybridization to their complementary sequence. Chembiochem. 2006;7:1173–1176. doi: 10.1002/cbic.200600178. [DOI] [PubMed] [Google Scholar]
- 12.Voirin E, Behr JP, Kotera M. Versatile synthesis of oligodeoxyribonucleotide-oligospermine conjugates. Nat. Protoc. 2007;2:1360–1367. doi: 10.1038/nprot.2007.177. [DOI] [PubMed] [Google Scholar]
- 13.Noir R, Kotera M, Pons B, Remy JS, Behr JP. Oligonucleotide-oligospermine conjugates (zip nucleic acids): a convenient means of finely tuning hybridization temperatures. J. Am. Chem. Soc. 2008;130:13500–13505. doi: 10.1021/ja804727a. [DOI] [PubMed] [Google Scholar]
- 14.Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 15.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 16.de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 1995;76(Pt 4):1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 17.Hesselink AT, van den Brule AJ, Groothuismink ZM, Molano M, Berkhof J, Meijer CJ, Snijders PJ. Comparison of three different PCR methods for quantifying human papillomavirus type 16 DNA in cervical scrape specimens. J. Clin. Microbiol. 2005;43:4868–4871. doi: 10.1128/JCM.43.9.4868-4871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stahlberg A, Hakansson J, Xian X, Semb H, Kubista M. Properties of the reverse transcription reaction in mRNA quantification. Clin. Chem. 2004;50:509–515. doi: 10.1373/clinchem.2003.026161. [DOI] [PubMed] [Google Scholar]
- 19.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huggett JF, Novak T, Garson JA, Green C, Morris-Jones SD, Miller RF, Zumla A. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC Res. Notes. 2008;1:70. doi: 10.1186/1756-0500-1-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Soud WA, Radstrom P. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 2001;39:485–493. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radstrom P, Knutsson R, Wolffs P, Lovenklev M, Lofstrom C. Pre-PCR processing: strategies to generate PCR-compatible samples. Mol. Biotechnol. 2004;26:133–146. doi: 10.1385/MB:26:2:133. [DOI] [PubMed] [Google Scholar]
- 23.McTigue PM, Peterson RJ, Kahn JD. Sequence-dependent thermodynamic parameters for locked nucleic acid (LNA)-DNA duplex formation. Biochemistry. 2004;43:5388–5405. doi: 10.1021/bi035976d. [DOI] [PubMed] [Google Scholar]
- 24.Owczarzy R, Moreira BG, You Y, Behlke MA, Walder JA. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry. 2008;47:5336–5353. doi: 10.1021/bi702363u. [DOI] [PubMed] [Google Scholar]
- 25.Kainz P. The PCR plateau phase – towards an understanding of its limitations. Biochim. Biophys. Acta. 2000;1494:23–27. doi: 10.1016/s0167-4781(00)00200-1. [DOI] [PubMed] [Google Scholar]
- 26.Gevertz JL, Dunn SM, Roth CM. Mathematical model of real-time PCR kinetics. Biotechnol. Bioeng. 2005;92:346–355. doi: 10.1002/bit.20617. [DOI] [PubMed] [Google Scholar]
- 27.Raja S, El-Hefnawy T, Kelly LA, Chestney ML, Luketich JD, Godfrey TE. Temperature-controlled primer limit for multiplexing of rapid, quantitative reverse transcription-PCR assays: application to intraoperative cancer diagnostics. Clin. Chem. 2002;48:1329–1337. [PubMed] [Google Scholar]
- 28.Schmid N, Behr JP. Location of spermine and other polyamines on DNA as revealed by photoaffinity cleavage with polyaminobenzenediazonium salts. Biochemistry. 1991;30:4357–4361. doi: 10.1021/bi00231a035. [DOI] [PubMed] [Google Scholar]
- 29.Schmid N, Behr JP. Recognition of DNA sequences by strand replacement with polyamino-oligonucleotides. Tetrahedron Lett. 1995;36:1447–1450. [Google Scholar]
- 30.Corey DR. 48000-fold acceleration of hybridization by chemically modified oligonucleotides. J. Am. Chem. Soc. 1995;117:9373–9374. [Google Scholar]
- 31.Iyer M, Norton JC, Corey DR. Accelerated hybridization of oligonucleotides to duplex DNA. J. Biol. Chem. 1995;270:14712–14717. doi: 10.1074/jbc.270.24.14712. [DOI] [PubMed] [Google Scholar]
- 32.Christensen U, Jacobsen N, Rajwanshi VK, Wengel J, Koch T. Stopped-flow kinetics of locked nucleic acid (LNA)-oligonucleotide duplex formation: studies of LNA-DNA and DNA-DNA interactions. Biochem. J. 2001;354:481–484. doi: 10.1042/0264-6021:3540481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hilscher C, Vahrson W, Dittmer DP. Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability. Nucleic Acids Res. 2005;33:e182. doi: 10.1093/nar/gni181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J. Biomol. Tech. 2004;15:155–166. [PMC free article] [PubMed] [Google Scholar]
- 35.Curry J, McHale C, Smith MT. Low efficiency of the Moloney murine leukemia virus reverse transcriptase during reverse transcription of rare t(8;21) fusion gene transcripts. Biotechniques. 2002;32:768. doi: 10.2144/02324st02. 770, 772, 754–765. [DOI] [PubMed] [Google Scholar]
- 36.Karrer EE, Lincoln JE, Hogenhout S, Bennett AB, Bostock RM, Martineau B, Lucas WJ, Gilchrist DG, Alexander D. In situ isolation of mRNA from individual plant cells: creation of cell-specific cDNA libraries. Proc. Natl Acad. Sci. USA. 1995;92:3814–3818. doi: 10.1073/pnas.92.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levesque-Sergerie JP, Duquette M, Thibault C, Delbecchi L, Bissonnette N. Detection limits of several commercial reverse transcriptase enzymes: impact on the low- and high-abundance transcript levels assessed by quantitative RT-PCR. BMC Mol. Biol. 2007;8:93. doi: 10.1186/1471-2199-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.