Abstract
Pif-1 proteins are 5′→3′ superfamily 1 (SF1) helicases that in yeast have roles in the maintenance of mitochondrial and nuclear genome stability. The functions and activities of the human enzyme (hPif1) are unclear, but here we describe its DNA binding and DNA remodeling activities. We demonstrate that hPif1 specifically recognizes and unwinds DNA structures resembling putative stalled replication forks. Notably, the enzyme requires both arms of the replication fork-like structure to initiate efficient unwinding of the putative leading replication strand of such substrates. This DNA structure-specific mode of initiation of unwinding is intrinsic to the conserved core helicase domain (hPifHD) that also possesses a strand annealing activity as has been demonstrated for the RecQ family of helicases. The result of hPif1 helicase action at stalled DNA replication forks would generate free 3′ ends and ssDNA that could potentially be used to assist replication restart in conjunction with its strand annealing activity.
INTRODUCTION
Helicases are motor proteins that unwind duplex nucleic acids (1). Processive helicases initiate unwinding at origins of replication and are responsible for providing the single-stranded template for DNA replication. However, under conditions of stress when replication forks stall or collapse at sites remote from origins of replication specialized helicases are required to re-start replication. These processes are best understood in bacteria (2), but homologues of prokaryotic DNA repair helicases are also known in eukaryotic cells. This is exemplified by the RecQ family of helicases that unwind DNA in the 3′→5′ direction (3) and are involved in homologous recombination, the re-start of stalled replication forks and the implementation or transduction of signals that enforce an S-phase checkpoint. Several hereditary cancer predisposition syndromes resulting from mutations of RecQ genes are known demonstrating the importance of this helicase family in the maintenance of genetic stability (4).
The Pif1 protein has also been identified as a helicase required for genome stability. Saccharomyces cerevisiae Pif1 (S.c.Pif1) was first recognised as a non-essential nuclear gene required for the repair of mitochondrial DNA after UV or chemical damage (5), and for mitochondrial genome maintenance at elevated temperature (6). S.c.Pif1 was later re-identified as a gene with a function in telomere length maintenance (7) and a Schizosaccharomyces pombe gene with a function in genomic DNA repair, revealed by the sensitivity of mutants to DNA-alkylating agents (8). Pif proteins are non-processive 5′→3′ helicases and member of helicase superfamily 1 [SF1; (9–11)]. S.c.Pif-1 acts as an inhibitor of telomere addition in the telomere maintenance process (12). Other studies implicate S.c.Pif1 in ribosomal DNA (rDNA) replication (13) and Okazaki fragment processing (14), where it functions in conjunction with the Dna2 helicase/nuclease. Baker’s yeast also has a second Pif-like gene, RRM3 [40% identical and 60% similar; (15)]. The role of Rrm3 appears to be to promote replication processes by displacing DNA bound proteins or resolving DNA secondary structures (13,16). Schizosaccharomyces pombe encodes only one Pif protein that is essential for completion of chromosomal DNA replication and hence viability (8,11,12).
Pif1 is conserved in eukaryotes and also shares significant homology with the Escherichia coli helicase RecD, principally in the seven core SF1 helicase motifs. RecBCD is a bipolar helicase/nuclease complex that is required for Okazaki fragment processing and recombination-mediated rescue of stalled replication forks, suggesting this as another possible replication function of eukaryotic Pif1 (17). Little is known about the replication pathways in which the human enzyme (hPif1) functions. Like S.cPif1, the hPif1 gene is not essential (18) and there are nuclear and mitochondrial splice variants (19). The protein associates with telomeres in vivo, inhibits telomerase in vitro (17) and siRNA-mediated depletion results in cell-cycle delay at S-phase, suggesting a role in chromosome maintenance associated with DNA replication (19). We have characterized the DNA binding and unwinding properties of purified recombinant hPif1 helicase domain (hPifHD) and the full-length nuclear form of the enzyme. Figure 1A details the hPifHD fragment employed and the organization of the conserved motifs. In the N-terminal 1–200 amino acids there is only limited sequence conservation and it has been suggested recently that this domain may have strand-annealing activity (20). We show here that the hPif helicase core domain interacts preferentially with ssDNA molecules greater than 35 bases and that ssDNA interactions promote its DNA remodeling activities that include dsDNA unwinding and also ssDNA annealing. However, hPifHD and the full-length enzyme also act on synthetic stalled DNA replication fork-like structures in vitro, where the ds- and ssDNA components of the substrate contribute to structure-specific recognition. The net result of hPif action on such substrates is to displace an oligonucleotide corresponding to the putative leading replication strand. These data suggest that hPif may act on stalled replication forks and this activity may be important for genome stability.
Figure 1.
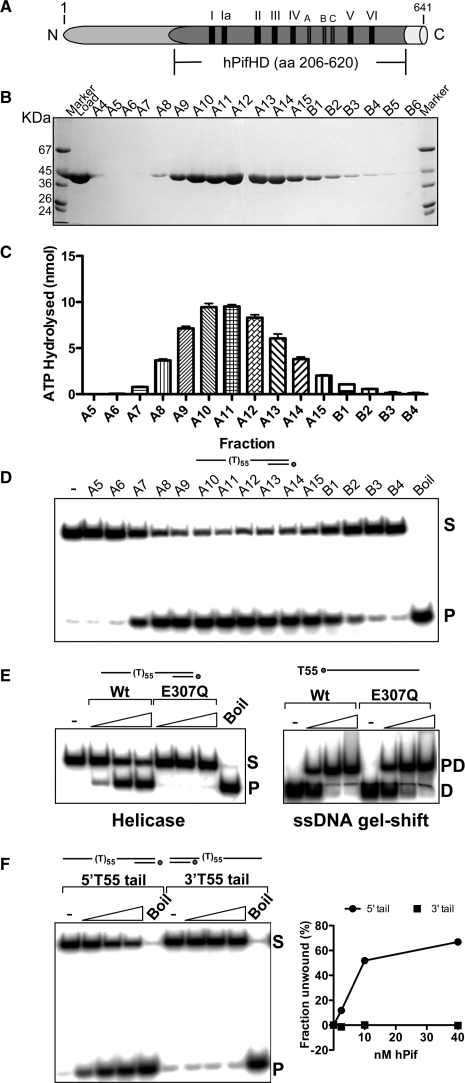
Purification and enzymatic activities of hPifHD. (A) Domain diagram of hPif. Amino acids 206–620 correspond to the conserved helicase domain. Relative positions of the seven conserved SF1 helicase motifs (I–VI) are indicated, and also motifs A, B and C (unknown function) characteristic of the same helicase family. (B) hPifHD fractions from a Superdex 75 column analysed on a 10% SDS-PAGE gel. The molecular weight of the purified protein corresponded to that predicted for hPifHD (45.5 kDa). The peak protein concentration was in fraction A11. (C) ATPase activity of fractions was determined in the presence of a 55-base poly T oligonucleotide and correlated with protein concentration. (D) Helicase activity of the peak fractions was determined using a 32P labeled substrate with a 55 base T tail and a 20 bp duplex portion (PST55). S, native substrate; P, ssDNA product as determined by boiling the substrate. (E) Helicase activity was completely abolished by a mutation, E307Q, in the Walker B ATPase motif (S, substrate; P, single-stranded product), but the mutant retained wild-type ssDNA-binding activity (PD, protein–DNA complex; D, free DNA). (F) hPifHD unwinds DNA in the 5′→3′ direction; statistical data for 3 repeats.
MATERIALS AND METHODS
Expression and purification of hPifHD and full length hPif1 protein
Human hPif1 (nuclear form amino acids 1–641) and the hPif helicase domain (hPifHD amino acid residues 206–620) were cloned as a fusion protein with glutathione S-transferase in pET11c, as described previously (21). GST-hPifHD was expressed in E. coli BL21(DE3) cells for 8 h at 22°C, and GST-hPif1 at 20°C after the cultures had reached OD600 ∼0.8. All purification steps were at 4°C. Cells were lysed in 50 mM Tris–HCl pH 7.5, 400 mM NaCl, 10 mM EDTA, 10% v/v glycerol, 10 mM DTT and 1 mM PMSF (‘lysis buffer’) after treatment with lysozyme. The lysate was cleared (40 000 × g, 30 min) and the supernatant mixed with 0.125 volumes of 5 M NaCl before the addition of polyethyleinamine P (pH 8) to 0.5% w/v. After 5 min the solution was cleared (25 000 × g, 10 min) and proteins precipitated by the addition of 0.291 g of (NH4)2SO4/ml of supernatant (25 000 × g, 20 min). The pellet was dissolved in lysis buffer/0.4 M NaCl/1 mM PMSF and incubated with glutathione sepharose (GE Healthcare; 16 h, 1 ml of beads per 30 g of E. coli). Beads were washed sequentially in batch (2 × 10 min with 10 bead volumes) in lysis buffer/1 M NaCl/1 mM PMSF, lysis buffer/0.2 M NaCl/1 mM PMSF/0.5% v/v NP-40 and finally lysis buffer/0.2 M NaCl/1 mM PMSF/0.1% v/v NP-40. The beads were loaded into a column and washed with 40 bed volumes of lysis/1 M NaCl/1 mM PMSF and lysis/0.2 M NaCl/no PMSF. Protein was eluted from the column with GST elution buffer at 300 mM NaCl, as described previously (21), and digested with thrombin (∼10 units per 30 g starting wet weight E. coli) for ∼16 h to cleave off the GST moiety. hPifHD was purified further by ion exchange chromatography (Source-Q), on heparin sepharose (20 mM NaPhosphate buffer pH 7.2, 2.5 mM DTT, 10% glycerol, 0.5 mM EDTA, 0.5 mM PMSF, 0.1–0.6 M NaCl gradient) and finally a Superdex 75 column (GE Healthcare) equilibrated in 20 mM NaPhosphate pH 7.5, 300 mM NaCl, 10% v/v glycerol, 2.5 mM DTT, 0.1 mM EDTA. Peak fractions were pooled, concentrated and stored at −80°C. hPif1 was further purified by ion exchange chromatography (Source-S) and gel filtration (Superdex 75). hPifHD protein concentrations were determined from Abs280 readings obtained in the presence of 7.5 M guanidinium hydrochloride using a molar absorption coefficient of 47 555 cm−1 M−1. hPif concentration was determined by BioRad assay.
Helicase and strand annealing assays
Test substrates were generated by annealing synthetic oligonucleotides (SigmaGenosys) to yield the substrates described and illustrated in Supplementary Figure S1. For each substrate, one or all of the oligonucleotides, as indicated, was end-labeled with 32P using polynucleotide kinase (pnk) and (γ-32P)ATP (6000 Ci/mmol). Substrates were gel purified from 8% (19 : 1) poly-acrylamide gels (1× TBE running buffer). The helicase and strand annealing reaction buffer was 20 mM HEPES-NaOH pH 7.5, 20 mM NaCl, 5 mM MgCl2, 2 mM ATP, 1 mM DTT, 0.1 mg/ml BSA and 0.1% v/v NP-40. Reactions were performed at 20°C for 30 min and terminated by the addition of 0.2 volumes of 120 mM EDTA, 0.6% w/v SDS, 60% v/v glycerol and 0.1% w/v bromophenol blue. Products were resolved on 8% (19 : 1) poly-acrylamide gels and visualized and quantified following exposure of dried gels to a phosphorimaging plate (Fujifilm).
The substrate for the helicase processivity assay was generated from 4 oligonucleotides: 5′(T)55-CGAATTCGAGCTCGGTACCC (oligo PS1), 5′GTACCGAGCTCGAATTCG (oligo PS2), 5′CGAATTCGAGCTCGGTACCC (oligo PS3) and 5′GTACCGAGCTCGAATTCGGG (oligo PS4). Oligonucleotide PS2 was phosphorylated using (γ-32P)ATP and oligonucleotides PS3 and PS4 with cold ATP using pnk. After phosphorylation, oligonucleotides PS1 and PS2 were combined at equimolar concentrations, as were oligonucleotides PS3 and PS4, and annealed. Oligonucleotide PS1+2 was combined with a 5-fold molar excess of oligonucleotide PS3+4, fresh ATP added to 10 mM and ligated for 1 h at room temperature with 400 cohesive end units of T4 DNA ligase (NEB). The substrate was extracted with phenol/chloroform and ethanol precipitated. Free primer (PS2) was removed by electrophoresing the DNA briefly in an 8% polyacrylamide gel and recovering the mature products by soak elution following identification by autoradiography.
DNA-binding reactions and gel-shift assays
DNA-binding reactions were performed with helicase substrates and single-stranded oligonucleotides (poly T), end-labeled and purified as described above. The reaction buffer for binding was 20 mM HEPES-NaOH pH 7.5, 75 mM NaCl, 5% v/v glycerol, 1 mM DTT, 1 mg/ml BSA and 0.1% v/v NP-40. Reactions were incubated for 20 min at 20°C before resolving complexes on 5% poly-acrylamide gels (29 : 1) using 0.25× TBE running buffer and pre-run for 2–3 h before loading. Visualization and quantification was as described above.
ATPase assays
ATPase activity was determined in 20 mM HEPES-NaOH pH 7.5, 75 mM NaCl, 5 mM MgCl2, 5 mM ATP, 0.0125 µM (γ-32P)ATP (6000 Ci/mmol), 1 mM DTT, 0.1 mg/ml BSA and 0.1% v/v NP-40. Reactions were incubated at 20°C and phosphate released determined by the charcoal binding method of Iggo and Lane (22).
RESULTS
Purification of the hPifHD core helicase domain (hPifHD)
The hPif1 ORF (641 amino acids) was amplified by PCR from HEK 293 cells following reverse transcription of mRNA. The sequence assembled from multiple amplification products matched the existing sequence in the data-base except for a single conservative substitution at codon 69. The sequences outside this core helicase motif are not required for helicase activity (17,23) and we expressed and purified to near homogeneity the helicase domain of hPif1 (hPifHD, amino acids 206–620, 45.5 kDa predicted, see Figure 1A), free from the GST tag. Figure 1B shows an SDS-PAGE analysis of the peak fractions from the final gel filtration step of purification. Extents of ssDNA-dependent ATPase and helicase activity corresponded exactly with the protein concentration of the peak fractions (Figure 1C and D). Furthermore, a mutation of the conserved ATPase Walker B motif (E307Q) completely abolished helicase activity (and ATPase activity, not shown), but not the ssDNA-binding activity of hPifHD assayed with a 32P-labeled 55 base poly T oligonucleotide (Figure 1E). We also performed a helicase polarity assay with helicase substrates consisting of a 20 bp dsDNA segment and either a 3′ or 5′ ssDNA tail of 55 T residues. Like other Pif1 proteins, hPifHD only unwound the substrate with a 5′ single-stranded DNA (ssDNA) overhang and is thus by convention a 5′→3′ helicase (Figure 1F).
hPifHD binds and initiates unwinding on substrates with an extended 5′ ssDNA tail
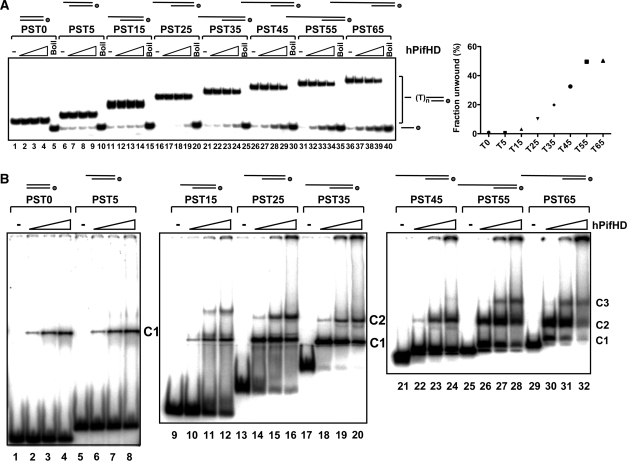
To determine how the length of the 5′ overhang effects the efficiency of initiation of unwinding, substrates were generated with a 20 bp dsDNA portion and 5′ poly T ssDNA tails of increasing length (0–65 residues, substrates PST0-65, see Supplementary Figure S1 for a detailed description of the substrates used). Substrates with 5′ T tails up to 15 residues (PST0, PST5 and PST15, lanes 5–15) are unwound poorly or not at all by the hPifHD enzyme (Figure 2A, lanes 1–15). As the length of the 5′ tail increases from 25 to 55 residues (PST25 to PST55, lanes 16–35), the initiation of unwinding was progressively more efficient. No significant increase in the extent of unwinding was observed when the 5′ tail length was increased to 65 residues (lanes 36–40 and graphed data).
Figure 2.
hPifHD DNA unwinding and DNA binding as a function of 5′ tail length. (A) 32P-end-labeled substrates were generated with a 20-bp dsDNA portion and 5′ T tails of increasing length. The position of the label is indicated with a circle on the depiction of the substrate as in all subsequent figures. Reaction products (0.1 nM substrate; 2.5, 10 and 40 nM hPifHD) were resolved on poly-acrylamide gels and the extents of unwinding determined from quantified phosphorimages. Boil indicates the heat-denatured substrates. The graph on the right shows that unwinding increases with tail lengths up to 55 bases (data for 40 nM hPifHD, average of three repeats). (B) Binding of hPifHD to partially single-stranded substrates, as in (A) in the absence of ATP/Mg2+ Binding reactions (0.1 nM probe; 0.1, 2.5 and 5 nM hPifHD) were resolved on native poly-acrylamide gels. Two predominant complexes (C1 and C2) formed on probes with ssDNA tails of 15 bases or more, and a third formed at higher concentrations (C3). As the size of the probes increased, complexes became more difficult to resolve, particularly C1 from free probe. However, careful comparison of the lanes with no protein with those containing protein reveal distinct shifts, for example, lane 21 compared with lane 22, PST45.
To complement the unwinding assays we also examined hPifHD binding, without ATP, to the helicase substrates using a gel-shift assay (Figure 2B). hPifHD bound to all the substrates tested (PST0 to PST65, 0.1–5 nM hPifHD protein), but the extent of binding increased with increasing 5′ ssDNA tail length. Substrates PST0, PST5 and PST15 that were poorly unwound all bound hPifHD, but at lower extents compared with the other substrates (lanes 1–12). Long dsDNA sequences (60 bp) were also bound poorly by hPifHD (see below). There was a significant increase in binding to substrate PST25 (lanes 13–16) compared with PST15 and two predominant complexes, C1 and C2, were observed. For all other substrates (PST35–PST65, lanes 17–32), there were also two main complexes formed and probe binding activity was similar in each case. However, a third complex (C3) also appeared at high protein concentrations for substrates PST45–PST65, and there was increased retention of the substrate in the wells. Complex C2 was the predominant complex to form on the substrates that were most efficiently unwound (PST55 and PST65). The binding affinity of hPifHD and the ability to form higher order complexes on the substrates therefore correlates well with the ability to initiate unwinding. Furthermore, similar binding patterns were observed in the presence of nucleotide cofactors, although the binding affinities of all substrates tested were significantly reduced (Supplementary Figure 2A).
Binding of hPifHD to ssDNA
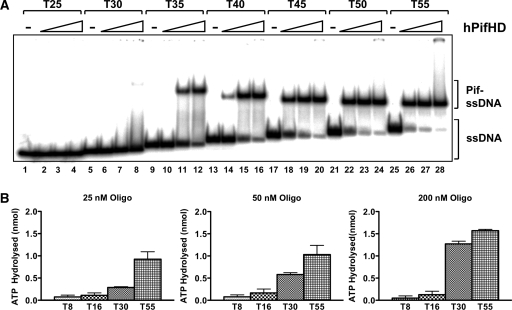
We also tested hPifHD binding to poly T oligonucleotides of increasing length (Figure 3A). The protein did not bind oligonucleotides of 25 bases (lanes 1–4) or less (not shown) over the protein concentration range tested (0.1–2.5 nM). Poly T oligonucleotides from 30 up to 50 residues bound with increasing affinity, complementing the observations for the partially single-stranded test substrates described above. However, only a single discrete shifted complex was observed for all oligonucleotides and, curiously, the mobility of the shifted complex increased with the increasing length of the oligonucleotide. Furthermore, our protein–DNA titration indicate a one to one stoichiometry for hPifHD ssDNA binding, suggesting that the enzyme does not multimerise on ssDNA like the hexameric helicases.
Figure 3.
hPifHD interactions with ssDNA. (A) Binding to 32P-end-labeled poly T probes of increasing length (0.1 nM probe; 0.1, 0.5 and 2.5 nM hPifHD) was assesed by gel-shift assay. Significant binding was detected only with probes of 35 bases or greater (lanes 9–28). (B) Stimulation of hPifHD ATPase activity as a function of probe length and concentration. hPifHD (50 nM) was incubated with oligonucleotides T8, T16, T30 and T55 at three concentrations, 25, 50 and 200 nM, and product release (from 5 µl of reaction) determined after 10 min. During this time period ATPase activity was in the linear range.
To complement the ssDNA binding studies we also tested how oligonucleotide length stimulated the ATPase activity of hPifHD (Figure 3B). In the absence of ssDNA or with dsDNA the ATPase activity of the enzyme is low (Supplementary Figure 2B). With the concentration of hPifHD fixed at 50 nM we measured ATPase activity at different ssDNA concentrations (25, 50 and 200 nM) of four polynucleotides of different length, T8, T16, T30 and T55. ATPase activity in the presence of oligonucleotides T8 and T16 was low compared with T30 and T55 and did not rise significantly as the ratio of DNA to protein increased. The stimulation of ATPase activity by T55 was greater than that for T30 at each ratio of DNA to protein and each increased with the ssDNA concentration. However, the stimulation of ATP hydrolysis promoted by T30 relative to T55 increased with higher DNA to protein ratios. The ability of oligonucleotides of increasing length to stimulate hPifHD ATPase activity therefore parallels their relative DNA binding affinities observed in the gel-shift assay.
Unwinding of replication fork-like substrates by hPifHD
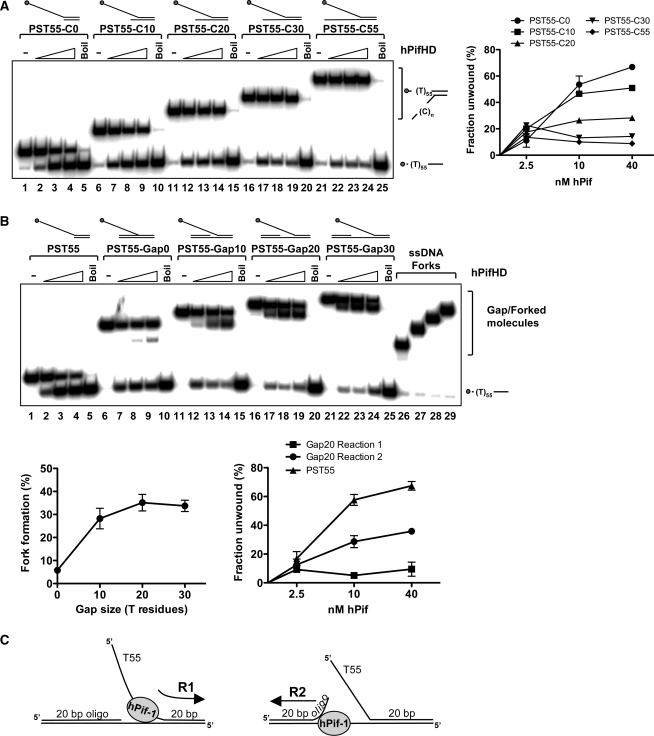
Duplex DNA with ssDNA extensions represent the simplest DNA substrates for hPifHD unwinding, so we therefore tested the unwinding of a more complex series of fork-like DNA structures. First, we generated fork-like structures with two ssDNA tails, by adding a 3′ poly C-tail to the substrate PST55 (55 base 5′ T tail, 20 bp duplex), as illustrated in Figure 4. As shown in Figure 4A, for fork-like molecules with a 3′ tail as short as 10 residues the proportion of substrate unwound by hPifHD increased slightly compared with PST55 at the lowest concentration of protein tested (lanes 7, 12, 17 and 22 compared with lane 2 and graph on the right). However, as the protein concentration was increased, the 3′ tails of increasing length were increasingly inhibitory for unwinding.
Figure 4.
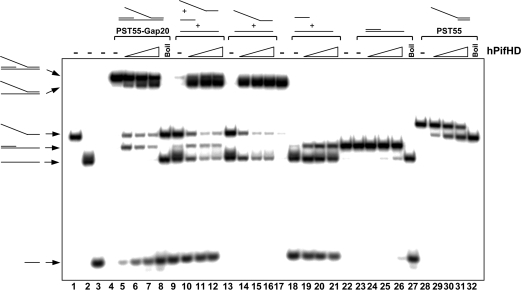
Unwinding of fork substrates by hPifHD. (A) Fork-like substrates were generated comprising of a 20 bp duplex and a 55 base 5′ T tail (PST55/PST55-C0) and a 3′ poly C tail of increasing length (10, 20, 30 and 55 residues, from PST55-C10 to PST55-C55 respectively). Assays were performed as in Figure 1A. (B) Unwinding of gapped substrates by hPifHD. Unwinding substrates were designed with two 20 bp duplex regions of equivalent Tm, one with a 55 base 5′ T-tail, annealed to a complementary strand; an increasing length of unpaired T residues was introduced between the duplex portions on the bottom strand to generate gapped molecules. Assays were performed and analyzed as described for Figure 1A. Lanes 26–29 are markers for the single-stranded forked products produced. Statistical data show fork formation as a function of gap size (10 nM hPifHD, graph on the left) and a comparison of products formed for PST55-Gap20 (graph on the right). (C) The apparent unwinding reactions on the fork substrate are described as R1 (engagement of the free T55 5′ tail) and R2, engagement of the ssDNA of the gapped duplex.
We next tested fork-like molecules with a dsDNA component instead of the 3′ poly C extensions. We designed the test substrates so that both portions of the duplex were 20 bp and the same Tm, and increased the spacing between the duplex portions of the substrate stepwise from a nick to a gap of up to 30 unpaired T residues on the bottom strand. These are therefore flap-like substrates. The oligonucleotide with the 55-base T tail was displaced from the substrate with no unpaired T residues on the bottom strand (20 bp duplex arm) to a similar extent to the substrate with a 20 C residue 3′ tail (Figure 4A, lanes 11–15 compared with Figure 4B, lanes 6–10). Surprisingly however, the unwinding products of this and all other flap substrates consisted of fork molecules without the 20 bp 5′ oligonucleotide, as well as the ssDNA oligonucleotide with the T55 ssDNA tail (Figure 4B). Increasing the spacing between the duplex portions of the molecule from a nick to 10 unpaired T residues resulted in significant stimulation of formation of the ssDNA-forked molecules (∼6-fold, Figure 4B, lanes 11–15 compared with 6–10). Increasing the length of the unpaired T residues between the duplex portions of the molecule to 20 (PST55-Gap20) promoted fork formation further, but there was no further increase with 30 unpaired T residues (PST55-Gap30; Figure 4B, lanes 16–24). The graph below on the left shows fork formation as a function of unpaired T residues for reactions at 10 nM hPifHD. Quantification of the assays performed with the PST55-Gap20 substrate demonstrated that the balance of product formation was biased significantly toward formation of the ssDNA-fork [i.e. displacement of 20 base oligonucleotide; reaction 2 (R2)], rather than displacement of the 5′ T55 tailed molecule [reaction 1 (R1)], as shown in the graph on the right. In Figure 4C we have summarized the actions of hPifHD on these substrates (reactions R1 and R2).
hPifHD possesses an intrinsic strand annealing activity
One interpretation of the data in Figure 4B is that hPifHD can engage gapped dsDNA molecules with an ssDNA flap and unwind either of the duplex portions of such molecules as summarized in Figure 4C. Another possibility is that the enzyme first engages and unwinds the flap duplex part (R1), reengages the substrate to displace the 20 bp oligonucleotide, and then promotes re-annealing of the molecules that comprise the ssDNA-fork. To test the latter, we determined whether hPifHD had strand annealing activity using the component oligonucleotides of substrate PST55-Gap20, each end-labeled with 32P (Figure 5, lanes 1–3). For the substrate PST55-Gap20 (lanes 4–8), the products of hPifHD action included the products of reactions 1 and 2 but not the free bottom strand of the template. Displacement of the 20 base oligonucleotide from this substrate was significantly more efficient than its displacement from the substrate lacking the extended free 5′ T flap (lanes 23–27), suggesting that generation of the ssDNA fork-like molecules predominantly occurs by direct displacement of the 20-mer from PST55-Gap20. Surprisingly, when all three component-strands of PST55-Gap20 were mixed and incubated under the same helicase reaction conditions we detected a robust strand annealing activity (lanes 9–12). Annealing of the long oligonucleotides to generate the single-stranded fork-like molecule was more efficient compared with annealing of the 20 base oligonucleotide to its complementary strand (lanes 13–22 and 9–12). Strand annealing activity did not require ATP binding and hydrolysis (data not shown). The conserved hPif1 helicase domain therefore possesses an intrinsic strand annealing activity, complicating a quantitative assessment of hPifHD helicase activity on ssDNA fork and flap substrates.
Figure 5.
Strand annealing activity of hPifHD. Lanes 1–3, labeled component strands of the flap substrate PST55-Gap20. Lanes 4–8, reaction products for hPifHD action on the substrate PST55-Gap20. Lanes 9–12, annealing of the component oligonucleotides of substrate PST55-Gap20 catalysed by hPifHD. The long strands of the substrate are preferentially annealed. Lanes 13–17 and 18–22, annealing of individual pairs of the component oligonucleotides of PST55-Gap20. Lanes 23–27 and 28–32, hPifHD action on the substrate lacking the ssDNA flap oligonucleotide and the reference substrate PST55, respectively.
hPifHD unwinds short stretches of dsDNA
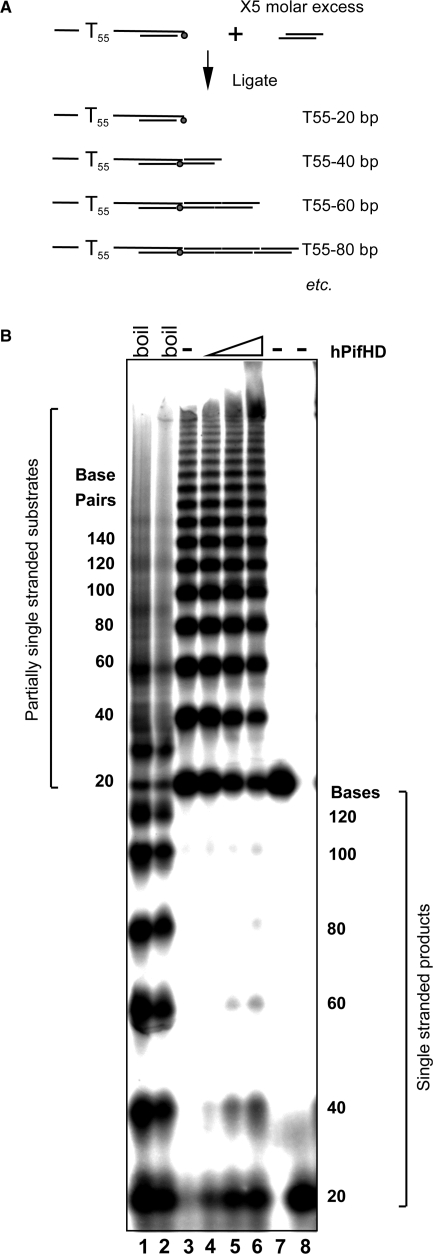
The results described above indicate that the strand displacement and strand annealing activities of hPifHD compete. The apparent proficiency with which the substrate PST55 is unwound is likely a result of the inefficiency with which the enzyme catalyses the re-annealing of short duplex (20 bp) sequences. Given this, we sought to determine the effective strand displacement activity of hPifHD on extended duplex substrates. Substrates were generated with a 55 T 5′ ssDNA tail and a variable double stranded portion increasing in 20 base increments. The method that we used to generate the mix was based on the ligation of linkers to substrate PST55, modified to include a cohesive end that was phosphorylated with [γ-32P]ATP. The substrate mix thus generated contains a population of molecules with an increasing duplex portion for hPifHD unwinding (Figure 6A). Figure 6B, lanes 1 and 2 show the substrate boiled and analysed on a native polyacrylamide gel, with or without formamide loading buffer. Lane 3 shows the native substrate and lanes 4–6 unwinding of the substrate in the presence of increasing concentrations of hPifHD. Only the partially single-stranded substrate molecules with a 20 bp duplex were effectively displaced by the enzyme. The efficiency of unwinding of the 40 bp substrate was less than 50% of that of the 20 bp species and the maximum effective length of unwinding was less than 100 bp. We also note that the intensity of very high substrate bands appeared to increase with increasing hPifHD concentration in the reactions. It is unclear how these products form but one distinct possibility is that they arise due to the strand annealing activity of the enzyme.
Figure 6.
Action of hPifHD on substrates with an extended duplex. The ability of hPifHD to unwind progressively longer stretches of duplex DNA was assessed using a population of substrate molecules with a 55-base 5′ T tail and a duplex portion increasing in 20 base increments. (A) Schematic diagram of substrate generation. Two pairs of phosphorylated oligonucleotides were annealed as illustrated. The oligonucleotide annealed to the strand with a 55 base T extension was phosphorylated with (γ-32P)ATP. Double stranded oligonucleotides were ligated with a 5-fold molar excess of the non-tailed duplex oligo generating a series of products with a 55 base 5′ T tail and duplex portions increasing in 20 bp increments. (B) Unwinding products were resolved on a 6% (19 : 1) native poly-acrylamide gel. Lanes 1 and 2 show the substrates heat-denatured with and without formamide present. Lane 3 is the native substrate and lanes 4–6 unwinding product generated by hPifHD (2.5, 10 and 40 nM hPifHD). Lanes 7 and 8 are markers for the minimal substrate (55 bases 5′ ssDNA, 20 bp duplex), native and denatured respectively. The fractions (%) unwound for the 20, 40, 60 and 80 bp substrate molecules were 18, 8, 1.3 and 0% at 2.5 nM hPifHD; 41, 27, 8.5 and 1.8% at 10 nM hPifHD and 47, 43, 12.4 and 5% at 40 nM hPifHD.
hPifHD binds and unwinds DNA resembling stalled replication forks
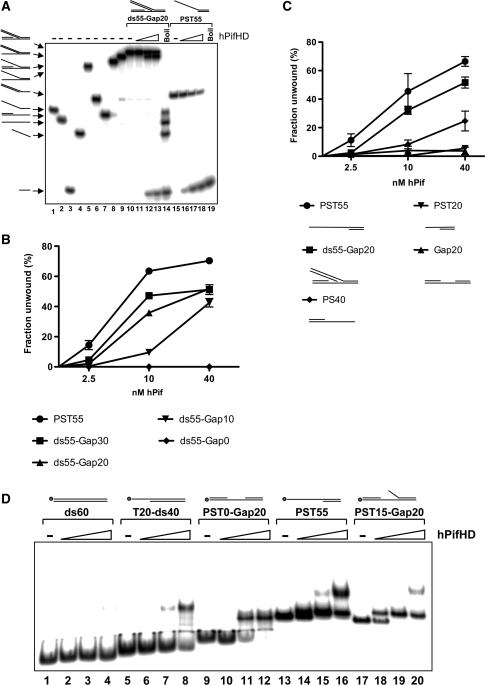
We also generated fork substrates, like those described in Figure 4B, where the free 5′ ssDNA tail was replaced by a 55 bp duplex portion (ds55), and varied the ssDNA component of the molecule by introducing unpaired T residues between the fork junction and the 20 bp duplexes on the bottom strand, the only ssDNA for hPifHD binding. We first tested the substrate with 20 unpaired T residues on the bottom strand (ds55-Gap20, Figure 7A), with all component oligonucleotides end-labeled with 32P. The only detectable reaction products for this substrate appeared to result from direct displacement of the 20 base oligonucleotide from the bottom strand (lanes 10−14), and the reaction efficiency approached that of our reference substrate PST55 (lanes 15−19 and see below).
Figure 7.
Binding and unwinding of stalled replication fork-like substrates by hPifHD. (A) A substrate (ds55-Gap20) was generated with two 20 bp duplex portions separated by 20 unpaired T residues with one extended to form a 55 bp dsDNA flap as illustrated. Each component oligonucleotide was end-labeled with 32P (lanes 1–4), and all possible combinations of annealed oligonucleotide generated (lanes 5–10). The only products of hPifHD action on the substrate ds55-Gap20 resulted from displacement of the 20 base oligonucleotide (lanes 10–14). Lanes 15–19, substrate PST55. (B) Fork-like substrates were generated and analysed, as described in (A), except that the number of unpaired T residues on the bottom strand was varied from 0 (a nick) to 30 residues. Substrates with 20 and 30 unpaired T residues (ds55-Gap20 and ds55-Gap30) were unwound to similar extents. The substrate with 10 unpaired T residues was less efficiently unwound at lower protein concentrations, while the substrate with a nick was not unwound by the enzyme. (C) Direct comparison of ds55-Gap20 with the substrate lacking the 55 bp flap (Gap20), or the substrate with a 20 base T overhang (PST20), reveals the dependence of the dsDNA arm of the substrate for efficient unwinding. (D) Binding of hPifHD to gapped DNA molecules and comparison to other substrates. Binding of hPifHD (0.1, 0.5 and 2.5 nM) to labeled DNA probes (0.1 nM) was analyzed by gel-shift. hPifHD bound the probe PST0-Gap20 with 20 unpaired T residue gap at high protein concentrations more effectively than the 60 bp probe (ds60) or the 40 bp duplex with a 20 base 5′ T tail (T20-ds40; lanes 9–12 compared with 1–4 and 5–8). The addition of 15 5′ T residues to the gapped substrate to generate a fork-like molecule (PST15-Gap20) further increased hPifHD-binding activity to levels comparable with PST55 (lanes 17–20 compared with lanes 9–16). Addition of larger ss- or dsDNA flaps resulted in complexes that were difficult to resolve.
We next compared substrates, where the number of unpaired T residues on the bottom strand was varied from 0 (a nick) to 30 bases in 10 base increments (ds55-Gap0-30, Figure 7B). As above, all the strands of the substrates were labeled and all possible combinations of annealed oligonucleotides generated so the products could be unambiguously identified by electrophoresis. As before, the only unwinding products of these substrates were for displacement of the 20 base oligonucleotides (R2). Displacement of the 20 mer from ds55-Gap10 (10 unpaired T residues) was only slightly less efficient than from the substrates with 20 and 30 unpaired T residues at the higher protein concentrations, and the nicked substrate was not unwound at all. In Figure 7C we show that without the 55 bp duplex arm (substrate Gap20), only minimal unwinding is observed, as with the substrate with a 20 base T overhang (PST20).
DNA structure and hPifHD DNA binding
Since a gapped duplex component of the substrate appeared necessary but not sufficient for unwinding (Figure 7B and C), we next asked if hPifHD had significant binding affinity for a simple gapped duplex molecule (PST0-Gap20; 0.1–2.5 nM protein). As before, DNA binding was assessed using the gel-shift assay (Figure 7D). Minimal hPifHD binding was observed with a 60 bp duplex molecule based on the lower strand of PST0-Gap 20 (lanes 1–4). This same 60 bp sequence also stimulated ATPase activity poorly compared with T55 ssDNA (Supplementary Figure 2). A probe with a 40 bp duplex and a 20 T residue 5′ tail (T20-ds40) was bound by hPifHD at the higher protein concentrations (lanes 5–8), consistent with previous observations (Figure 2). When the 20 unpaired T residues were positioned between the two 20 bp duplex portions of the molecule (PST0-Gap20), there was a significant increase in binding affinity compared with T20-ds40: Approximately 50% of the probe was bound by a 5-fold lower concentration of hPifHD (lanes 5–8 compared with lanes 9–12). Addition of a short 15 base 5′ T tail to the gapped substrate (PST-15-Gap20, lanes 17–20) resulted in a further increase in binding affinity, to an extent similar to the substrate PST55 (lanes 13–16). Similar binding affinities were observed with other double or ssDNA tails of greater length but complexes became difficult to resolve (not shown). These data therefore complement the results of the unwinding data (Figure 7B and C) and suggest that the gapped duplex portion of the replication fork structures tested contributes significantly to substrate recognition by hPifHD.
Unwinding of stalled replication forks by full-length hPif
We have also over-expressed and purified recombinant full-length hPif1, although the enzyme is more difficult to obtain in quantities of similar purity to hPifHD. In Supplementary Figure 3, we demonstrate that the enzyme has DNA strand displacement, DNA annealing and size dependent ssDNA binding activities similar to the core helicase module. Our data therefore indicate that hPif1 is a structure specific DNA helicase with specificity for synthetic DNA structures resembling putative stalled replication forks and that the initiation of unwinding activity is dependent on both arms of the fork. These DNA recognition and processing activities are intrinsic to the hPif1 helicase module.
DISCUSSION
Genetic and biochemical studies in yeast and mammalian cells indicate a role for the Pif1 proteins in telomere stability and Okazaki fragment processing (7,14,24,25), although it appears that the enzymes do not have identical functions even in related yeast species (8,11). Work with yeast also implicates these enzymes in the maintenance of mitochondrial and nuclear genome stability (5,6). The non-equivalence of function in related yeast species highlights the need for careful studies in mammalian cells. This assertion is strengthened in the light of recent, although contentious, data for other replication proteins. Zheng et al. (26) have proposed that the human DNA2 helicase/nuclease, a key enzyme in Okazaki fragment processing, is a mitochondrial enzyme while the yeast homologue has a nuclear function in DNA replication. However, more recently Duxin et al. (27) have presented evidence that human DNA2 participates in both nuclear and mitochondrial DNA maintenance. The Pif1 protein is envisioned to act upon specific nucleic acid structures that form during replication and require resolution before it can be completed. These include, in yeast, long flaps that escape FEN1 cleavage on the lagging replication strand (25). At the biochemical level, the helicase activity of yeast Pif1 resides solely in the conserved C-terminal two-thirds of the protein [∼400 amino acids; (17,23)]. Here we show that the corresponding human enzyme specifically recognizes and unwinds substrates envisioned to form when replication forks stall, implying a role for the human enzyme in processing such structures.
Using linear partially single-stranded substrates we first established that initiation of 5′→3′ helicase activity by hPifHD is dependent on an extended 5′ ssDNA tail of 55 bases (Figure 2). Complementing this, our ssDNA binding studies demonstrate high affinity binding to a minimal ssDNA sequence of 35 residues, with increasing affinity up to 45–50 residues. Furthermore, the binding of multiple proteins to the longest oligonucleotide tested (55 T residues) was not observed and the protein–DNA titrations suggest a 1 : 1 stoichiometry of binding. ATP hydrolysis by hPifHD requires ssDNA and the ssDNA length dependence for hPifHD binding apparent by gel-shift analysis is also observed for the stimulation of ATPase activity required for strand displacement (Figure 3). This ssDNA length is large in comparison with other known helicases (28) and the data also hint at an unusual mode of binding as the mobility of the protein–DNA complex appears to increase with DNA length (Figure 3A). One distinct possibility is that the ssDNA wraps around hPifHD, contacting one or more extended or remote binding sites on the protein surface. hPifHD binding affinity for the partially single-stranded helicase substrates also increased progressively with ssDNA tail length (Figure 2B). hPifHD bound predominant as two complexes, C1 and C2, of which the higher-order species (C2) forms more readily on the substrates that are more efficiently unwound. The related SF1 and SF2 helicases generally act as monomers or dimers (29–31). Although the composition of the hPifHD unwinding complex is currently uncertain, the data indicate that the cellular DNA substrates of hPif1 may recruit multiple monomers and that functional co-operativity of monomers may lead to improved helicase action, as with other SF1 helicases such as the E. coli Rep protein (31).
Helicases are viewed to translocate along one nucleic acid strand when unwinding (1). During the productive phase of unwinding the nucleic acid contacts made by the helicase would not necessarily resemble those for the initial structure-specific recognition of a preferred substrate. Thus, although partially single-stranded test substrates may be efficiently bound and unwound by a helicase they can give an incomplete view of substrate specificity. When we tested hPifHD unwinding of ssDNA fork molecules we observed only a modest stimulation of unwinding at low protein concentrations. However, higher protein concentrations, that otherwise promoted unwinding of the linear substrate PST55, appeared inhibitory for unwinding (Figure 4A). Similar results were obtained when the 3′ arm of the fork was converted to duplex DNA and a variable number of T residues introduced between this duplex portion and the flap junction (Figure 4B): Displacement of the 5′ flap oligonucleotide (reaction 1) appeared to be inhibited in favor of displacement of the 20 base oligonucleotide (reaction 2), even when very short stretches (10 bases or less) of unpaired T residues were available on the putative loading strand. The exact mechanism of reaction 2 on these substrates is difficult to determine. Our subsequent analysis demonstrated that although direct displacement of the 20 base oligonucleotides from its complementary strand is inefficient compared with the parent flap structure (Figure 5, lanes 4–8 compared with lanes 23–27), a concerted mechanism of displacement coupled with the strand annealing activity of hPifHD could prevail. Several SF1 and SF2 helicases have been shown also to have ssDNA strand annealing activity (32–34). We were surprised to observe this annealing activity since Gu et al. (20) recently reported that N-terminal sequences outside the core hPif1 helicase domain alone have strand-annealing activity. In the case of human RecQ5β protein, the core helicase domain (analogous to hPifHD we have analyzed) has a strong strand annealing but not strand displacement activity in the absence of an adjoining regulatory domain (35). Our data demonstrate that full-length hPif1 has very similar DNA unwinding and strand-annealing (and also DNA binding) properties as those described for hPifHD (Supplementary Figure 3), and hence the two catalytic activities appear to compete freely.
Although interpretation of the precise action of hPIf1 on ssDNA flap structures is problematic due to the strand annealing activity of the enzyme, the action of hPif1 on putative stalled replication fork structures is clearer. R2, potentially resembling leading strand reversal of such forks, is robust. This activity is significant even on substrates that contain only small regions of ssDNA on the loading strand of the fork (as little as 10 and as much as 20 unpaired T residues, Figure 7), much smaller than those required for efficient ssDNA binding and stimulation of ATPase activity (Figure 3), or initiation of unwinding on simple partially single-stranded test substrates (Figures 2 and 7). Furthermore, initiation of unwinding is also dependent on structural features of the second arm of the synthetic replication fork. The dsDNA component is essential, although dsDNA itself does not bind or stimulate hPifHD ATPase activity efficiently (Figures 7 and S2). On the basis of these data we speculate that hPif1 may recognize and process stalled DNA replication forks, reversing the leading replication strand to generate a free 3′ end and ssDNA. With hPif1 alone, this displaced DNA sequence may be small (Figure 6). However, this could be extended if hPif1 unwinding were increased by a processivity factor as is the case with other SF-1 helicases such as RecD and PcrA (36,37), or may be sufficient if further processing involves the strand annealing activity of the enzyme (Figure 5). We also speculate that the structure specific mode of DNA recognition may restrict hPif1 activity in vivo. Although substrates with extended 5′ ssDNA tails are good substrates for unwinding in vitro, competition with RPA binding may prevent unwinding of such substrates in vivo. The ssDNA component of the stalled replication-fork like substrates we have tested is essential for the initiation of unwinding, but small in comparison to the described high affinity binding site size for RPA [>30 nt; (38,39)], thus such a competition for binding may favour hPif1 and this should be testable in vitro using the systems we have described. In summary, we suggest that hPif may have a role in replication control and replication fork restart mechanisms as has been described for other helicases (40). In the light of recent yeast genetics studies implicating S.c.Pif1 in processing G-quadraplex structures in vivo (41), it is also possible that hPif1 may rescue stalled forks resulting specifically from non-B form DNA structure formation. Our data will also facilitate the biochemical testing of this possibility in the human system.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Yorkshire Cancer Research grants (to C.M.S. and M.M.). Funding for open access charge: Yorkshire Cancer Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Tomas Lindahl for critically reading the manuscript.
REFERENCES
- 1.Singleton M, Dillingham M, Wigley D. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 2.Cox M, Goodman M, Kreuzer K, Sherratt D, Sandler S, Marians K. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 3.Bachrati C, Hickson I. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 4.Karow J, Wu L, Hickson I. RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev. 2000;10:32–38. doi: 10.1016/s0959-437x(99)00039-8. [DOI] [PubMed] [Google Scholar]
- 5.Foury F. These d’agregation. Louvain, Belgium: Universite Catholique de Louvain; 1990. The metabolism of mitochondrial DNA. [Google Scholar]
- 6.Foury F, Kolodynski J. Pif mutation blocks recombination between mitochondrial ρ+and ρ− genomes having tandemly arrayed repeats units in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1983;80:5345–5349. doi: 10.1073/pnas.80.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz V, Zakian V. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka H, Ryu GH, Seo YS, Tanaka K, Okayama H, MacNeill S, Yuasa Y. The fission yeast pfh1+ gene encodes an essential 5′ to 3′ DNA helicase required for the completion of S-phase. Nucleic Acids Res. 2002;30:4728–4739. doi: 10.1093/nar/gkf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lahaye A, Leterme S, Foury F. PIF1 DNA helicase from Sacchromyces cerevisiae. Biochemical characterization of the enzyme. J. Biol. Chem. 1993;268:26155–26161. [PubMed] [Google Scholar]
- 10.Lahaye A, Stahl H, Thines-Sempoux D, Foury F. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryu GH, Tanaka H, Kim DH, Kim JH, Bae SH, Kwon YN, Rhee JS, MacNeil SA, Seo YS. Genetic and biochemical analyses of Pfh1 DNA helicase function in fission yeast. Nucleic Acids Res. 2004;32:4205–4216. doi: 10.1093/nar/gkh720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou JQ, Monson E, Teng SC, Schulz V, Zakian V. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 13.Ivessa A, Zhou J-Q, Zakian V. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 14.Budd M, Reis C, Smith S, Myung K, Campbell J. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase δ. Mol. Cell Biol. 2006;26:2490–2500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bessler J, Torres J, Zakian V. The Pif1 subfamily of helicases: region-specific DNA helicases. Trends Cell Biol. 2001;11:60–65. doi: 10.1016/s0962-8924(00)01877-8. [DOI] [PubMed] [Google Scholar]
- 16.Ivessa A, Zhou J-Q, Schulz V, Monson E, Zakian V. Sacchromyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang DH, Zhou B, Huang Y, Xu LX, Zhou JQ. PIf1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res. 2006;34:1393–1404. doi: 10.1093/nar/gkl029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snow E, Mateyak M, Paderova J, Wakeham A, Iorio C, Zakian V, Squire J, Harrington L. Murine Pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol. Cell Biol. 2007;27:1017–1026. doi: 10.1128/MCB.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Futami K, Shimamoto A, Furuichi Y. Mitochondrial and nuclear localization of human Pif1 helicase. Biol. Pharm. Bull. 2007;30:1685–1692. doi: 10.1248/bpb.30.1685. [DOI] [PubMed] [Google Scholar]
- 20.Gu Y, Masuda Y, Kamiya K. Biochemical analysis of human Pif helicase and functions of its N-terminal domain. Nucleic Acids Res. 2008;36:6295–6308. doi: 10.1093/nar/gkn609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castella S, Bingham G, Sanders C. Common determinants in DNA melting and helicase-catalysed DNA unwinding by papillomavirus replication protein E1. Nucleic Acids Res. 2006;34:3008–3019. doi: 10.1093/nar/gkl384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iggo R, Lane D. Nuclear protein p68 is an RNA-dependent ATPase. EMBO J. 1989;8:1827–1831. doi: 10.1002/j.1460-2075.1989.tb03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boule JB, Zakian V. Roles of Pif1-like helicases in the maintenance of genomic stability. Nucleic Acids Res. 2006;32:4205–4216. doi: 10.1093/nar/gkl561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mateyak M, Zakian V. Human Pif helicase is cell cycle regulated and associates with telomerase. Cell Cycle. 2006;5:2796–2804. doi: 10.4161/cc.5.23.3524. [DOI] [PubMed] [Google Scholar]
- 25.Rossi M, Pike J, Wang W, Burgers P, Campbell J, Bambara R. Pif1 helicase directs eukaryotic Okazaki fragments towards the two-nuclease cleavage pathway for primer removal. J. Biol. Chem. 2008;283:27483–27493. doi: 10.1074/jbc.M804550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng L, Zhou M, Guo Z, Lu H, Dai H, Qiu J, Yakubovskaya E, Bogenhagen D, Demple B, Shen B. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of replication and repair intermediates. Mol. Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duxin J, Dao B, Martinsson P, Rajala N, Guittat L, Campbell J, Spelbrink J, Stewart S. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell Biol. 2009;15:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel S, Picha K. Structure and function of hexameric helicases. Ann. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 29.Korolev S, Hsieh J, Gauss G, Lohman T, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 30.Velankar S, Soultana P, Dillingham M, Subramanya H, Wigley D. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 31.Wong I, Chao K, Bujalowski W, Lohman T. DNA-induced dimerisation of the Escherichia coli Rep helicase. J. Biol. Chem. 1992;267:7596–7610. [PubMed] [Google Scholar]
- 32.Garcia P, Liu Y, Jiricny J, West S, Janscak P. Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanagaraj R, Saydam N, Garcia P, Zheng L, Janscak P. Human RECQ5β helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Felice M, Aria V, Esposito L, De Falco M, Pucci B, Rossi M, Pisani FM. A novel DNA helicase with strand-annealing activity from the crenarchaeon Sulfolobus solfataricus. Biochem. J. 2007;408:87–95. doi: 10.1042/BJ20070134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren H, Dou SX, Zhang XD, Wang PY, Liu JL, Janscak P, Hu JS, Xi XG. The zinc-binding motif of human RECQ5beta suppresses the intrinsic strand-annealing activity of its DExH helicase domain and is essential for the helicase activity of the enzyme. Biochem. J. 2008;412:425–433. doi: 10.1042/BJ20071150. [DOI] [PubMed] [Google Scholar]
- 36.Soultanas P, Dillingham M, Papadopoulos F, Phillips S, Thomas C, Wigley D. Plasmid replication initiator RepD increases the processivity of PcrA DNA helicase. Nucleic Acids Res. 1999;27:1421–1428. doi: 10.1093/nar/27.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dillingham M, Kowalczykowski S. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim C, Paulus B, Wold M. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 39.Mitas M, Chock J, Christy M. The binding-site size of Escherichia coli single-stranded-DNA-binding protein and mammalian replication protein A are 65 and ≥54 nucleotides respectively. Biochem. J. 1997;324:957–961. doi: 10.1042/bj3240957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu L, Hickson I. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- 41.Ribeyre C, Lopes J, Boule J, Piazza A, Guedin A, Zakian V, Mergny J, Nicholas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadraplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.