Abstract
Correction of a defective gene is a promising approach for both basic research and clinical gene therapy. However, the absence of site-specific targeting and the low efficiency of homologous recombination in human cells present barriers to successful gene targeting. In an effort to overcome these barriers, we utilized triplex-forming oligonucleotides (TFOs) conjugated to a DNA interstrand crosslinking (ICL) agent, psoralen (pTFO-ICLs), to improve the gene targeting efficiency at a specific site in DNA. Gene targeting events were monitored by the correction of a deletion on a recipient plasmid with the homologous sequence from a donor plasmid in human cells. The mechanism underlying this event is stimulation of homologous recombination by the pTFO-ICL. We found that pTFO-ICLs are efficient in inducing targeted gene conversion (GC) events in human cells. The deletion size in the recipient plasmid influenced both the recombination frequency and spectrum of recombinants; i.e. plasmids with smaller deletions had a higher frequency and proportion of GC events. The polarity of the pTFO-ICL also had a prominent effect on recombination. Our results suggest that pTFO-ICL induced intermolecular recombination provides an efficient method for targeted gene correction in mammalian cells.
INTRODUCTION
One approach to gene therapy is to replace defective genes with wild-type functional genes at a targeted site in the genome (1). This approach has the advantage of ensuring gene expression at endogenous levels under the physiological control of normal gene regulatory elements. To achieve site-specific gene correction, an homologous sequence containing the wild-type gene serving as a template is required. Another requirement is the use of a strategy to efficiently direct the homologous sequence to the targeted site in the genome. Enhancing homologous recombination (HR) provides one such strategy; however, the frequency of targeted HR in mammalian cells is roughly two to three orders of magnitude lower than random integration (2). To address this issue, we have employed triplex technology to enhance the frequency of HR in mammalian cells. Triplex-forming oligonucleotides (TFOs) are small single-stranded oligonucleotides that can bind specifically to the purine-rich strand of a target duplex DNA (3,4). TFOs have been used to increase the level of HR at specific sites by forming a triplex structure at the targeted site (5–7). However, conjugation of a DNA damaging reagent such as psoralen to the TFO can further enhance its ability to stimulate HR (5,7,8).
Psoralens are naturally occurring planar tri-heterocyclic compounds consisting of a furan ring and a pyrone ring. Psoralen can intercalate in DNA, and upon irradiation by UVA light (365 nm), psoralen can form a monoadduct with a pyrimidine base on either the furan side or the pyrone side (9); furan-sided monoadducts are readily converted into interstrand crosslinks (ICLs) by absorption of a second photon at 365 nm. The psoralen ICL is asymmetric, and the repair of psoralen ICLs is also asymmetric with a preference for incision on the furan side of the ICL (10,11). The preferred psoralen crosslinking site is a 5′TpA-3′ sequence; however psoralen binds/intercalates DNA with little sequence specificity. TFOs conjugated to psoralen can be used to target psoralen to specific sites in the genome, thereby directing the formation of site-specific ICLs. Previous studies have demonstrated that TFO-directed psoralen ICLs (pTFO-ICLs) can induce HR between two direct repeats, which results from an HR sub-pathway, single-strand annealing (SSA) (5,6,8,12). However, in typical gene therapy applications, intermolecular HR (HR between two DNA molecules) is required. Thus, in this study we characterized the ability of pTFO-ICLs to enhance intermolecular HR. We measured the HR frequencies induced by pTFO-ICLs at targeted sites, and characterized the factors involved, such as the effect of polarity of the pTFO-ICL and the degree of homology between the two DNA molecules on HR frequencies and spectra. We constructed a series of plasmid substrates with different deletion sizes in the supF reporter gene with the TFO binding site in both orientations relative to the targeted supF gene. Our results demonstrate that the pTFO-ICL induced significantly higher levels of targeted intermolecular HR in a plasmid substrate containing a small deletion (4 bp), but not in plasmids containing a larger deletion (24 bp). This effect depended on the polarity of the TFO relative to the targeted supF gene, with a higher frequency of HR on those plasmids with the ICL located further, as opposed to nearer, the supF gene target. Analysis of recombinant spectra revealed that targeted gene correction was predominant only with substrates containing the short (4 bp) deletion.
MATERIALS AND METHODS
Plasmid constructions
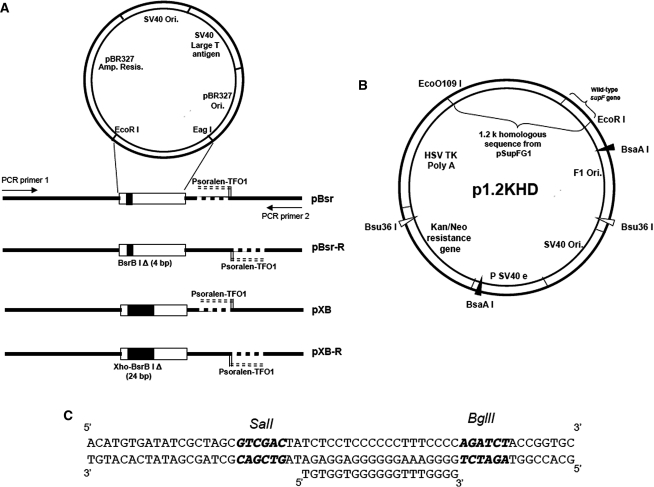
The shuttle vector, pSupFG1, was modified to generate two types of recipient plasmids serving as target substrates, as shown in Figure 1A: one set of plasmids contained a small deletion (4 bp) within the supF target gene at the BsrBI restriction site (original position 37–41 bp); the second group of plasmids contained a larger (24 bp) deletion within the targeted supF gene, from the XhoI restriction site to the BsrBI restriction site (original position 17–41 bp). In all plasmids, a 60 bp DNA linker sequence containing a pTFO1 (psoralen-5′-TGTGGTGGGGGGTTTGGGG-3′) binding site was inserted after an EagI restriction site (original position 124 bp; Figure 1C). The pTFO1 binding site was inserted in both orientations relative to the targeted supF gene, i.e. such that the TFO-directed ICL is at the same distance from the supF gene, but the TFO target duplex sequence is either closer to or further from the supF gene (Figure 1A). The plasmid substrates were confirmed by DNA sequencing. All these substrate plasmids produce white colonies in MBM7070 indicator Escherichia coli when grown on X-Gal, IPTG, ampicillin (XIA) plates.
Figure 1.
Schematic of recipient (A) and donor (B) plasmids. (A) Psoralen ICL sites are at the same position in all plasmids, the dashed line indicates the purine-rich strand bound by pTFO1. This TFO binding sequence was inserted in the plasmids via an insertion of 73 bp shown in (C). (B) The donor plasmid, p1.2kHD, has a 1.2 kb fragment homologous to the recipient plasmids, but contains the wild-type supF gene. p1.2kHD has two restriction enzyme sites for BsaAI and Bsu36I, which are absent from the recipient plasmids. (C) The insertion sequence containing the TFO1 binding sequence. The unique SalI site is used to confirm the presence of the ICL, and the unique BglII site is used to confirm the presence of the triplex structure formed by TFO binding.
The donor plasmid, p1.2kHD, contains a 1.2 kb fragment homologous to a region in the pSupFG1 plasmid containing the wild-type supF gene. This plasmid was constructed by digesting pSupFG1 with EcoRI and EcoO109I restriction enzymes. The 2.5 kb donor plasmid backbone was obtained from the pEGFP-C1 plasmid (Clontech) by digestion with EcoO109I and EcoRI. Ligation of these two fragments yielded the donor plasmid, p1.2kHD. This plasmid contains two unique restriction enzyme sites, BsaAI and Bsu36I (Figure 1B), which are absent from the substrate/recipient plasmids. These sites are used to digest and eliminate the donor plasmid; following co-transfection of both substrate and donor plasmids into HeLa cells, the recovered plasmids were digested with these two restriction enzymes prior to plasmid transformation of the indicator bacterial strain MBM7070 to eliminate the donor plasmid, thereby avoiding any HR events generated in the indicator bacteria.
Substrate preparation
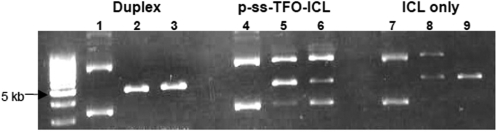
Duplex plasmid substrates (‘Dup’) are the original plasmids described above, in the absence of any treatment. UVA-treated plasmids (‘UVA’) were the duplex plasmid substrates irradiated with 1.8 J/cm2 of UVA (at 365 nm). Triplex plasmids (‘Triplex’) are the duplex plasmids (1.5 μg) incubated with pTFO1 (5′-psoralen-TGTGGTGGGGGGTTTGGGG-3′) (1 μl of TFO at 1.2 × 10⊟4 M) in triplex binding buffer [TBB: 10 mM Tris–HCl, pH 7.6, 10 mM MgCl2 and 10% (vol/vol) glycerol] in a total volume of 50 μl at 37°C overnight, without UVA irradiation. The TFO-directed psoralen ICL-containing plasmids (‘pTFO-ICL’) are the duplex plasmid substrates incubated with pTFO1 under the same conditions as the ‘Triplex’ plasmids except that these plasmids were irradiated with 1.8 J/cm2 UVA light (365 nm) to generate the ICL. The ICL-only plasmid (‘ICL’) is the duplex plasmid treated in a similar fashion as ‘pTFO-ICL’ plasmid, but with the TFO removed, leaving only the site-specific ICL. To prepare this plasmid substrate, we used a TFO, p-ss-TFO1, which has the psoralen tethered to the 5′-end of TFO1 via a disulphide linker. After overnight incubation and UVA irradiation to crosslink the psoralen to the duplex target, 10 mM DTT was added to the reaction, and the reaction was further incubated at 65°C for 6 h to detach the TFO, as we have previously described (13). BglII and SalI digestion can be used to verify the removal of the TFO and the presence of the ICL, respectively (Figures 1C and 2). Samples were covered with Mylar film during UVA irradiation, which effectively eliminates UV with wavelengths <320 nm. The pBsr-R ‘DSB’ plasmid substrate was prepared by co-digesting the plasmid with EcoRV and SalI enzymes to produce a DNA double-strand break (DSB) with no homology at the ends.
Figure 2.
Enzyme digestion to confirm the plasmid substrate containing an ICL. Lanes 1, 4, 7: supercoiled plasmids with no treatment; lanes 2, 5, 8: plasmids treated with SalI; lanes 3, 6, 9: plasmids treated with BglII. Duplex plasmid can be cut by both SalI and BglII (lanes 2 and 3), but the plasmid with the p-ss-TFO-ICL is resistant to both SalI and BglII digestion (lanes 5 and 6). However, if a plasmid containing a p-ss-TFO-ICL is treated with DTT, the plasmid can be cut by BglII (linearized band in Lane 9), but not by the SalI enzyme (Lane 8). This indicated that TFO1 had been removed by DTT treatment and the psoralen ICL was still present at the SalI restriction site.
Recombination assays
Approximately 5 × 105 HeLa cells were seeded in T-25 flasks 24 h prior to transfection with reporter plasmids. The cells were transfected with a plasmid mixture containing 1.5 μg substrate plasmid and 4.5 μg donor plasmid (∼1 : 4 molar ratio) using Geneporter transfection reagent, according to the manufacturer’s instructions (Genlantis, San Diego, CA, USA). The next day, the cells were washed twice with PBS, and incubated in 5 ml EMEM culture medium containing 10% FBS, 2 mM l-glutamine, and 1% Pen-strep antibiotics. The cells were cultured in 5% CO2 at 37°C for another 48 h. Seventy-two hours after transfection, cells were harvested with trypsin–EDTA followed by centrifugation at 1000 r.p.m. at 4°C for 5 min. After one wash with PBS, the cell pellets were lysed and the plasmid DNA was isolated with Qiagen Miniprep kit (Qiagen, Valencia, CA, USA), extracted twice with phenol/chloroform/isoamyl alcohol (25 : 24 : 1), and ethanol precipitated. The dried plasmid DNA was dissolved in 60 μl ddH2O and passed through a G25 column by spinning at 3500 r.p.m. for 2 min to remove any residual salt. Finally, 7 μl of NEB buffer 3, 0.7 μl BSA and 1 μl each of DpnI, BsaAI and Bsu36I were added to the flow-through (final volume 70 μl) and incubated at 37°C for 6 h. The plasmid DNA was then re-extracted from the digestion reaction with phenol/chloroform/isoamyl alcohol (25 : 24 : 1). After washing twice with 75% ethanol and drying the plasmid, the DNA was dissolved in 10 μl ddH2O. Next, 3 μl of plasmid DNA was used to transform competent MBM7070 indicator bacteria with the Gene Pulser apparatus (BioRad, Hercules, CA, USA), followed by suspension in 500 μl SOC media and incubation at 37°C on a shaker for 45 min. Finally, the bacterial cells were spread on LB plates containing XIA medium for blue-white colony screening as we have previously described (8).
Characterization of recombinants
The recombinant (blue) colonies were picked and streaked onto another LB plate prior to performing colony PCR for analysis of recombinant structures. The primer sequences used for DNA structure analysis of recombinant plasmids were: 5′-ATAATACCGCGCCACATAGC-3′ (upstream), and 5′-GCCTACATACCTCGCTCTGC-3′ (downstream). PCR reaction conditions consisted of denaturing at 90°C, annealing at 54.7°C and elongation at 72°C. After 35 cycles, the PCR products were digested with the BsrBI restriction enzyme (0.5 μl of BsrBI in a 15 μl reaction volume) for 3 h at 37°C. Then the samples were loaded onto a 1% agarose gel and electrophoresed at 80 V for 2 h. Finally, the gel was visualized by EtBr staining. The colonies showing an abnormal restriction digestion profile compared to wild-type pSupFG1 plasmid were re-streaked and a second confirmative PCR and BsrBI digestion was performed. In addition, these complex recombinant plasmids were sequenced by standard DNA sequencing methods.
Statistical analysis
The significance of the differences in homologous recombination frequencies between groups was conducted by using Student's t-test. To analyze the differences in recombinant spectra, we utilized the pair-wise proportional test.
RESULTS
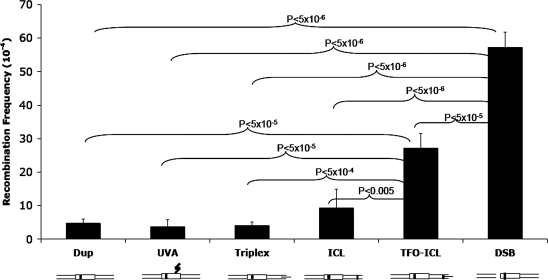
TFO-directed psoralen ICLs stimulate intermolecular HR
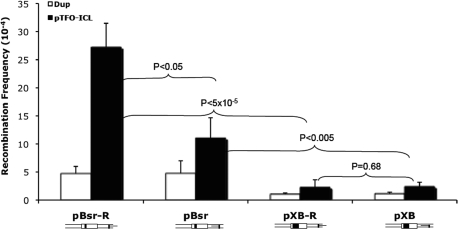
The recipient plasmid, pBsr-R, was subjected to treatment with a psoralen-conjugated TFO (pTFO) and UVA irradiation to measure enhancement of HR by site-specific pTFO-ICL damage. In addition, other treatments representing various controls included UVA irradiation only, pTFO treatment only (no ICL), removal of the TFO from the pTFO-psoralen/UVA-irradiated substrate (ICL only), and restriction enzyme digestion to generate a DSB, all as described in ‘Materials and Methods’ section. The recombination frequency was determined as the number of blue colonies/(total number of white + blue colonies). Blue colonies can result from both homologous recombination and illegitimate recombination (as discussed below). The duplex plasmid (Dup) without any treatment served as a negative control. As shown in Figure 3, the pTFO-ICL induced a significantly higher recombination frequency (27.2 × 10⊟4, ∼6-fold above background) than background (Dup, 4.7 × 10⊟4), UVA irradiated plasmid only (UVA, 3.7 × 10⊟4, near background levels), triplex structure only (Triplex, 4.2 × 10⊟4, near background levels) or the ICL only (ICL, 9.4 × 10⊟4, ∼2-fold above background). The plasmid containing the DSB served as a positive control, and as expected, stimulated recombination (57.2 × 10⊟4, ∼12-fold above background) more than any other treatment. Although the ICL alone induced a higher HR frequency (9.4 × 10⊟4) than Dup (4.7 × 10⊟4), UVA (3.7 × 10⊟4) and Triplex (4.2 × 10⊟4) plasmid substrates, there were no significant differences among these treatments (Figure 3).
Figure 3.
pTFO-ICL treatment can stimulate intermolecular recombination. All results are from three independent experiments (error bars represent the SD), P-values were derived from Student's t-test. Different treatments are illustrated below the bars.
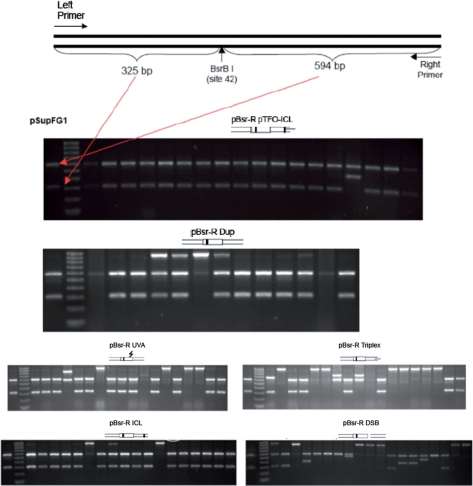
Colony PCR and BsrBI digestion confirm pTFO-ICL-induced targeted gene conversions
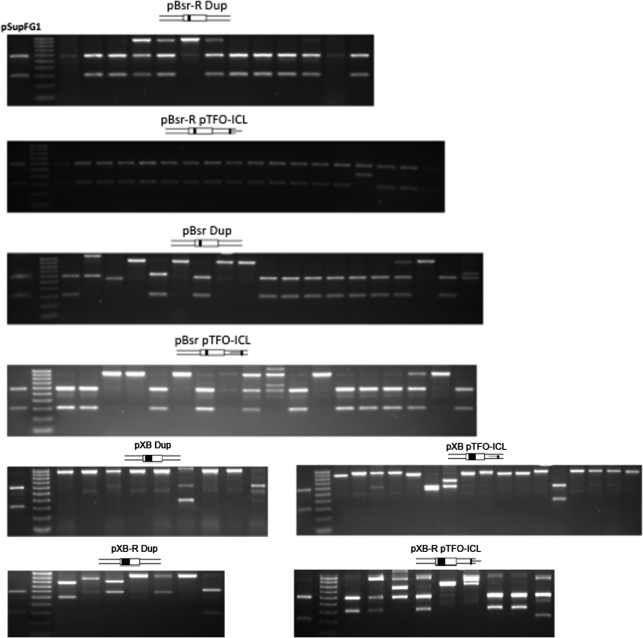
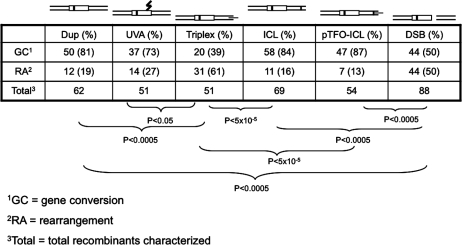
The recombinant spectra were analyzed by colony PCR plus BsrBI digestion. The BsrBI restriction site is unique in the parental pSupFG1 plasmid. This site was eliminated during the construction of recipient plasmids. If the recipient plasmid underwent targeted gene conversion within the mutant supF gene, then the restored wild-type supF gene should have a BsrBI site such that a PCR product could be cut into two fragments with lengths of 325 bp and 594 bp (Figure 4). Figure 4 shows representative examples of colony PCR plus BsrBI digestion from pBsr-R plasmids with different treatments. The summarized data are shown in Table 1. The major recombination products of Dup, UVA, ICL and pTFO-ICL plasmid substrates were gene conversions (at least 72%), but the pTFO-ICL plasmid showed the highest GC frequency among all these treatments (Figure 3), indicating that the pTFO-ICL is effective in inducing both high GC frequency and high GC proportion. In contrast, most recombination products from Triplex and DSB plasmid substrates resulted from illegitimate recombination events (at least 50%).
Figure 4.
Recombination spectrum analysis by colony PCR plus BsrBI digestion shows the different effects of the various treatments on the recombination spectrum.
Table 1.
Intermolecular recombination events in plasmid pBsr-R
 |
The size of the deletion in the supF gene affects pTFO-ICL-induced gene conversion frequency and spectrum
To investigate the effect of deletion size on pTFO-ICL-induced HR, we used plasmids with different sizes of deletions in the supF gene. In plasmids with the same polarity of the pTFO-ICL, the pTFO-ICL induced significantly higher HR frequencies in the plasmids containing the smaller deletion (Figure 5). pTFO-ICL-induced HR frequency in pBsr-R was 27.2 × 10⊟4, which was ∼11-fold higher than that in pXB-R (2.3 × 10⊟4). Statistical analysis demonstrated a significant difference between them (P < 5 × 10⊟5). Similarly, pTFO-ICL-induced HR frequency in the pBsr plasmid (11 × 10⊟4) was ∼4-fold higher than that of the pXB plasmid (2.4 × 10⊟4), which is also significant (P < 0.005).
Figure 5.
The effects of target gene deletion size on pTFO-ICL-induced intermolecular recombination frequency. All results are from three independent experiments (error bars represent the SD), P-values were derived from Student's t-test. Different treatments are illustrated below the bars.
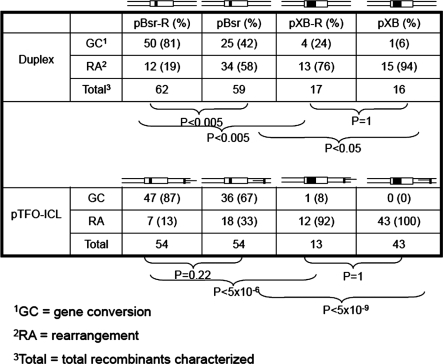
In addition to the impact on HR frequency, deletion size also affected the HR spectrum induced by a pTFO-ICL. Figure 6 and Table 2 demonstrate that the major products in plasmids with the smaller 4 bp deletion (pBsr-R and pBsr) were targeted GC events (∼87% in pBsr-R and ∼67% in pBsr). In contrast, illegitimate recombinants were the major product (>90%) in plasmids containing the larger 24 bp deletions (pXB-R and pXB). Statistical analysis showed that there was a significant difference (P < 5 × 10⊟6) between plasmids with smaller deletions (4 bp) versus larger deletions (24 bp) in pTFO-ICL treatment groups, regardless of the polarity of the pTFO-ICL.
Figure 6.
The effects of target gene deletion size on pTFO-ICL-induced intermolecular recombination spectrum. Representative samples of colony PCR plus BsrBI digestion demonstrate the effects of deletion sizes on the recombination spectrum.
Table 2.
Effects of deletion size and ICL polarity on HR
 |
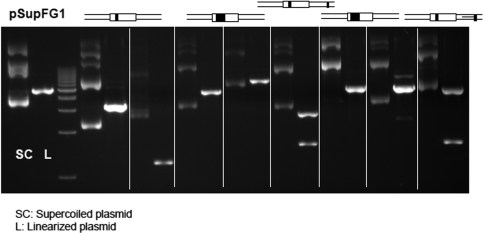
To confirm recombinant spectra, we re-streaked those blue colonies with abnormal patterns of colony PCR and BsrBI digestion. The results confirmed the presence of an abnormal profile from these blue colonies. Furthermore, we isolated the plasmid DNA from these abnormal colonies and subjected them to DNA sequencing and BsrBI digestion. Results of sequencing indicated the presence of a wild-type supF gene in some recombinants (in addition to inserted sequences). Although some of the recombinants did not contain a wild-type supF gene in the sequenced regions, BsrBI digestion linearized all of these the plasmids, indicating the presence of wild-type supF gene within the plasmids, but beyond the sequenced region (Figure 7).
Figure 7.
Confirmation of the presence of a wild-type supF gene in plasmids with aberrant colony PCR plus BsrBI digestion profiles. SC: supercoiled plasmid. L, BsrBI linearized plasmid. The plasmids with (L) or without (SC) BsrB1 treatment are shown.
pTFO-ICL polarity affects the recombination frequency
In plasmid substrates with small deletions (4 bp), the recombination frequency in pBsr-R with pTFO-ICL treatment was higher than that of pBsr (P < 0.05) (Figure 5). The psoralen ICLs in these two plasmids were located the same distance from the supF gene. The only difference was the polarity of triplex structure relative to the supF gene. The position of the pTFO-ICL in pBsr-R had the TFO binding site located further from the supF gene, while the TFO binding site was nearer to the supF gene in plasmid pBsr, as shown in Figure 1A. Although the pTFO-ICL induced a higher frequency of GC in pBsr-R (Figure 5), the proportion of GC events between these two plasmids was not significantly different (Table 2; P = 0.22). For those plasmids containing the larger deletions (24 bp), pXB-R and pXB, both recombination frequencies and spectra were very similar and no significant differences were detected regardless of the pTFO-ICL polarity. Therefore, the effect of pTFO-ICL polarity on GC frequency and spectrum depends on the deletion size in the homologous region of the recipient plasmid.
DISCUSSION
This study was undertaken to examine the effects of the positioning of a pTFO-ICL lesion on induced intermolecular homologous recombination in mammalian cells; in addition, we wished to determine how other parameters such as whether the removal of the TFO from the pTFO-ICL and the size of deletions in the homologous target influenced HR incited by site-specific DNA damage. Results presented here support our previous conclusion that efficient site-specific induction of homologous recombination requires both the TFO-induced triplex structure and the psoralen ICL (8). Although plasmids containing only the ICL or the pTFO-ICL have the psoralen ICL structure in common, the presence of the triplex structure in the pTFO-ICL was observed to significantly stimulate HR. However, the triplex structure alone (in the absence of the psoralen ICL) induced HR only slightly above background levels (Figure 3), similar to the function of the triplex structure in SSA (6,8). There is the possibility that the TFO (in the absence of the covalent psoralen–DNA linkage) might be displaced from its target site during recombination in these substrates. However, if this were indeed the case, then the TFO only plasmid should have the same recombination frequency and spectrum as the untreated (Dup) plasmid. While the recombination frequencies between Dup and Triplex plasmids are very similar, the recombination spectra are significantly different (Figure 4 and Table 1, P < 0.0005). Therefore, the triplex structure is likely present on the substrate in the cells, but the processing of the triplex structure in the absence of the ICL may differ from that of the pTFO-ICL. In support of this interpretation, it has been shown that triplex-induced HR requires functional XPA, and the triplex alone is not as efficient as the pTFO-ICL in inducing SSA, suggesting different pathway utilization in processing these two lesions (6,14). Psoralen ICLs in the absence of the triplex structure may be primarily repaired by a dual incisions around the ICL on the purine-rich strand, which would displace the strand containing the psoralen adduct, producing a single-strand gap that could be bypassed by translesion synthesis polymerases (6,12,15–17). However, some of the intermediate products with single-stranded gaps may be repaired via an HR pathway to yield targeted GC products.
The triplex structure may also recruit certain repair proteins more efficiently than the psoralen ICL alone, such as the MMR repair protein complex MSH2–MSH3 (MutSβ) (18,19). MSH2 is required for the efficient processing of TFO-directed psoralen ICLs, but not for pTFO-ICL-induced mutagenesis (20), suggesting that MSH2 might channel the psoralen ICL into a more error-free pathway (i.e. HR). In fact, MutSβ deficiency can lead to reduced levels of HR (21).
Our analysis of recombinant structures confirms that targeted gene conversions are efficiently induced by TFO-ICLs. The significantly lower proportion of targeted GC events in the Triplex plasmid substrate compared with the pTFO-ICL containing plasmids might also be explained by different repair proteins or pathways being involved. As noted above, MMR proteins could play a role in pTFO-ICL repair (18–20,22). Since MMR can inhibit non-homologous recombination (23–26), the recombination events in the plasmids containing the pTFO-ICL may shift toward targeted homologous GC.
As a positive control for inducing HR, we included recipient plasmids containing a DSB in our studies. Although, as expected, the DSB containing recipient plasmid induced the highest recombination frequency among all the treatment groups (Figure 3), ∼50% of the recombinants were the result of illegitimate recombination (Table 1). The GC proportion from DSBs is significantly lower than that from pTFO-ICL treatment (P < 0.0005). This result is consistent with a previous report that ∼70% of recombination products are generated by illegitimate recombination (27), indicating that DSB repair in mammalian cells occurs predominantly via a non-homologous end joining mechanism (28,29). The recombination spectra differed between the plasmids containing a DSB and those containing a pTFO-ICL, which also implies that each is subject to different repair processing. On the other hand, although previous reports have demonstrated that DSB formation occurs during psoralen ICL processing (30–35), all the plasmid substrates containing psoralen ICLs may not be converted into DSB intermediates. Several proposed ICL repair models imply the presence of intermediate products with a single-stranded gap and an oligonucleotide flap containing the psoralen ICL (12,36–39). If a homologous sequence is available, the 3′-OH end of the single-stranded gap may be able to initiate HR via a synthesis-dependent strand annealing (SDSA) mechanism (40–42).
The data presented in this study are the first to demonstrate a significant enhancement of induced HR between two DNA plasmids in mammalian cells by pTFO-ICLs, when the donor sequence is presented on a replicating plasmid. Previous studies on pTFO-ICLs used TFOs tethered to donor oligonucleotides (43–45) or non-replicating donor plasmids (27), which induced only low levels of HR. In this latter study, although the donor sequence was presented in an SV40-derived shuttle vector, the lymphoblastoid cells used could not support plasmid replication and the length of homology was only about 180 bp (27). In our study, the donor plasmid and cell line (HeLa) supported replication, which could generate more donor plasmids for HR. Another important factor is the length of the homologous sequence between the donor and recipient plasmids. In our donor plasmid, the homologous sequence length between recipient and donor plasmids is about 1.2 kb, whereas in these previously reported experiments, the homologous sequences were <300 bp (27,43–45). Longer homologous sequences could be important in stimulating HR, since there is a minimum requirement of homology for stable heteroduplex formation (46–49).
In our previously reported results (8) we observed that for intramolecular recombination, gene conversions induced by TFO-directed psoralen crosslinks occurred preferentially at smaller (8 bp) as opposed to larger (24 bp) deletions in the supF gene. In the results reported here for intermolecular recombination, we varied the size of the deletion (4 bp versus 24 bp) in the target plasmid to assess what effect target deletion size has on pTFO-ICL induced gene conversion in this system. As shown in Figure 5, target plasmids with the smaller, 4 bp deletion (pBsr-R and pBsr) exhibited significantly higher levels of induced recombination than those with the larger, 24 bp deletion (pXB-R and pXB). Results presented in Table 2 clearly indicates that plasmids pBsr-R and pBsr are more readily converted than pXB-R and pXB plasmids, and that this is the case regardless of whether or not recombination is induced by TFO-directed psoralen ICLs. Thus, pTFO-ICL induced frequencies, but not the recombinant spectra, are apparently influenced by the size of the target deletion. There is significant sequence heterology in the immediate vicinity of the supF target in all these plasmids. This heterology is the result of the differences in the sizes of the deletions, as well as to an insertion of 60 bp containing the pTFO binding site which is present in the target, but not the donor plasmid. This may result in less stable heteroduplex formation than in the completely homologous sequences downstream from the supF target, and could explain why shorter conversion tracts may be more productive in generating functional recombinants.
The recombinants resulting from pTFO-ICL treatment of the 4 bp deletion target plasmids, pBsr-R or pBsr, may have been generated from DSB intermediates (30–35,50,51), from single-stranded gap intermediates (12,36–39), or from both. The triplex structure is resistant to DNA endonuclease digestion in vitro (52); thus, we postulate that DSBs might form in vivo only on the side of the psoralen ICL away from the triplex structure. In pBsr-R containing the pTFO-ICL, a ‘clean’ DSB may form downstream, but proximal, to the supF gene with the pTFO-ICL remaining on the side of the DSB distal to supF. On the other hand, for the pBsr plasmid containing a pTFO-ICL, the ‘dirty’ end (containing the pTFO-ICL) is located proximal to the supF gene, and the DSB is downstream. The ‘clean’ DSB can be used to initiate HR directly, but the ‘dirty’ end must be further processed to initiate HR, because only the DSB end without an ICL could be used to invade the homologous sequence on the donor DNA template (37). Therefore, in plasmid pBsr containing a pTFO-ICL, HR may be mainly initiated from a single-stranded gap intermediate after unhooking the psoralen from the purine-rich strand of the duplex (6,12,13,15–17). The 3′-OH end of the single-stranded gap could then initiate a recombination event via a SDSA mechanism (40–42). Homology-dependent DNA synthesis could then lead to the formation of a functional supF gene. Thus, we speculate that pTFO-ICL-induced GC may result predominantly from SDSA in these substrates. Further studies are warranted to more completely elucidate the specific mechanism involved, since increased understanding of this process will lead to more efficient gene targeting approaches.
FUNDING
National Institutes of Health/NCI (CA097175, CA093729 to K.M.V.); NIEHS Center grant ES007784 for Facility Core services. Funding for open access charge: CA093729.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Ms Sarah Henninger for article preparation, and Kevin Lin for assistance with statistical analyses.
REFERENCES
- 1.Friedmann T. A brief history of gene therapy. Nat. Genet. 1992;2:93–98. doi: 10.1038/ng1092-93. [DOI] [PubMed] [Google Scholar]
- 2.Roth D, Wilson J. Illegitimate Recombination in Mammalian Cells. Washington, DC: American Society for Microbiology; 1988. [Google Scholar]
- 3.Cooney M, Czernuszewicz G, Postel EH, Flint SJ, Hogan ME. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988;241:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 4.Beal PA, Dervan PB. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 5.Luo Z, Macris MA, Faruqi AF, Glazer PM. High-frequency intrachromosomal gene conversion induced by triplex-forming oligonucleotides microinjected into mouse cells. Proc. Natl Acad. Sci. USA. 2000;97:9003–9008. doi: 10.1073/pnas.160004997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faruqi AF, Datta HJ, Carroll D, Seidman MM, Glazer PM. Triple-helix formation induces recombination in mammalian cells via a nucleotide excision repair-dependent pathway. Mol. Cell Biol. 2000;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Nairn RS, Vasquez KM. Processing of triplex-directed psoralen DNA interstrand crosslinks by recombination mechanisms. Nucleic Acids Res. 2008;36:4680–4688. doi: 10.1093/nar/gkn438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- 10.Van Houten B, Gamper H, Holbrook SR, Hearst JE, Sancar A. Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a defined position. Proc. Natl Acad. Sci. USA. 1986;83:8077–8081. doi: 10.1073/pnas.83.21.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barre FX, Asseline U, Harel-Bellan A. Asymmetric recognition of psoralen interstrand crosslinks by the nucleotide excision repair and the error-prone repair pathways. J. Mol. Biol. 1999;286:1379–1387. doi: 10.1006/jmbi.1999.2550. [DOI] [PubMed] [Google Scholar]
- 12.Faruqi AF, Seidman MM, Segal DJ, Carroll D, Glazer PM. Recombination induced by triple-helix-targeted DNA damage in mammalian cells. Mol. Cell Biol. 1996;16:6820–6828. doi: 10.1128/mcb.16.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen LA, Wang H, Van Houten B, Vasquez KM. Efficient processing of TFO-directed psoralen DNA interstrand crosslinks by the UvrABC nuclease. Nucleic Acids Res. 2008;36:7136–7145. doi: 10.1093/nar/gkn880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datta HJ, Glazer PM. Intracellular generation of single-stranded DNA for chromosomal triplex formation and induced recombination. Nucleic Acids Res. 2001;29:5140–5147. doi: 10.1093/nar/29.24.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 16.Zheng H, Wang X, Legerski RJ, Glazer PM, Li L. Repair of DNA interstrand cross-links: interactions between homology-dependent and homology-independent pathways. DNA Repair. 2006;5:566–574. doi: 10.1016/j.dnarep.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Svoboda DL, Taylor JS, Hearst JE, Sancar A. DNA repair by eukaryotic nucleotide excision nuclease. Removal of thymine dimer and psoralen monoadduct by HeLa cell-free extract and of thymine dimer by Xenopus laevis oocytes. J. Biol. Chem. 1993;268:1931–1936. [PubMed] [Google Scholar]
- 18.Zhang N, Lu X, Zhang X, Peterson CA, Legerski RJ. hMutSbeta is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol. Cell Biol. 2002;22:2388–2397. doi: 10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J, Jain A, Iyer RR, Modrich PL, Vasquez KM. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic Acids Res. 2009;37:4420–4429. doi: 10.1093/nar/gkp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6:551–557. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang N, Liu X, Li L, Legerski R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair. 2007;6:1670–1678. doi: 10.1016/j.dnarep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Q, Vasquez KM. Human MLH1 protein participates in genomic damage checkpoint signaling in response to DNA interstrand crosslinks, while MSH2 functions in DNA repair. PLoS Genet. 2008;4:e1000189. doi: 10.1371/journal.pgen.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabisiewicz A, Worth L., Jr Escherichia coli MutS,L modulate RuvAB-dependent branch migration between diverged DNA. J. Biol. Chem. 2001;276:9413–9420. doi: 10.1074/jbc.M005176200. [DOI] [PubMed] [Google Scholar]
- 24.Calmann MA, Marinus MG. MutS inhibits RecA-mediated strand exchange with platinated DNA substrates. Proc. Natl Acad. Sci. USA. 2004;101:14174–14179. doi: 10.1073/pnas.0406104101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Worth L, Jr, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc. Natl Acad. Sci. USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rayssiguier C, Thaler DS, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 27.Sandor Z, Bredberg A. Triple helix directed psoralen adducts induce a low frequency of recombination in an SV40 shuttle vector. Biochim. Biophys. Acta. 1995;1263:235–240. doi: 10.1016/0167-4781(95)00109-t. [DOI] [PubMed] [Google Scholar]
- 28.Jackson SP, Jeggo PA. DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem. Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 29.Sargent RG, Brenneman MA, Wilson JH. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dardalhon M, Averbeck D. Pulsed-field gel electrophoresis analysis of the repair of psoralen plus UVA induced DNA photoadducts in Saccharomyces cerevisiae. Mutat Res. 1995;336:49–60. doi: 10.1016/0921-8777(94)00037-7. [DOI] [PubMed] [Google Scholar]
- 31.Meniel V, Magana-Schwencke N, Averbeck D. Preferential repair in Saccharomyces cerevisiae rad mutants after induction of interstrand cross-links by 8-methoxypsoralen plus UVA. Mutagenesis. 1995;10:543–548. doi: 10.1093/mutage/10.6.543. [DOI] [PubMed] [Google Scholar]
- 32.Miller RD, Prakash L, Prakash S. Genetic control of excision of Saccharomyces cerevisiae interstrand DNA cross-links induced by psoralen plus near-UV light. Mol. Cell Biol. 1982;2:939–948. doi: 10.1128/mcb.2.8.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jachymczyk WJ, von Borstel RC, Mowat MR, Hastings PJ. Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems for DNA repair: the RAD3 system and the RAD51 system. Mol. Gen. Genet. 1981;182:196–205. doi: 10.1007/BF00269658. [DOI] [PubMed] [Google Scholar]
- 34.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 35.Dardalhon M, de Massy B, Nicolas A, Averbeck D. Mitotic recombination and localized DNA double-strand breaks are induced after 8-methoxypsoralen and UVA irradiation in Saccharomyces cerevisiae. Curr. Genet. 1998;34:30–42. doi: 10.1007/s002940050363. [DOI] [PubMed] [Google Scholar]
- 36.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 37.Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 38.Sladek FM, Munn MM, Rupp WD, Howard-Flanders P. In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5′-exonuclease of DNA polymerase I. J. Biol. Chem. 1989;264:6755–6765. [PubMed] [Google Scholar]
- 39.Cole RS. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc. Natl Acad. Sci. USA. 1973;70:1064–1068. doi: 10.1073/pnas.70.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strathern JN, Klar AJ, Hicks JB, Abraham JA, Ivy JM, Nasmyth KA, McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982;31:183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- 41.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson DO, Holloman WK. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc. Natl Acad. Sci. USA. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan PP, Lin M, Faruqi AF, Powell J, Seidman MM, Glazer PM. Targeted correction of an episomal gene in mammalian cells by a short DNA fragment tethered to a triplex-forming oligonucleotide. J. Biol. Chem. 1999;274:11541–11548. doi: 10.1074/jbc.274.17.11541. [DOI] [PubMed] [Google Scholar]
- 44.Knauert MP, Kalish JM, Hegan DC, Glazer PM. Triplex-stimulated intermolecular recombination at a single-copy genomic target. Mol. Ther. 2006;14:392–400. doi: 10.1016/j.ymthe.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 45.Rogers FA, Vasquez KM, Egholm M, Glazer PM. Site-directed recombination via bifunctional PNA-DNA conjugates. Proc. Natl Acad. Sci. USA. 2002;99:16695–16700. doi: 10.1073/pnas.262556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watt VM, Ingles CJ, Urdea MS, Rutter WJ. Homology requirements for recombination in Escherichia coli. Proc. Natl Acad. Sci. USA. 1985;82:4768–4772. doi: 10.1073/pnas.82.14.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen P, Huang HV. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.King SR, Richardson JP. Role of homology and pathway specificity for recombination between plasmids and bacteriophage lambda. Mol. Gen. Genet. 1986;204:141–147. doi: 10.1007/BF00330201. [DOI] [PubMed] [Google Scholar]
- 49.Ayares D, Chekuri L, Song KY, Kucherlapati R. Sequence homology requirements for intermolecular recombination in mammalian cells. Proc. Natl Acad. Sci. USA. 1986;83:5199–5203. doi: 10.1073/pnas.83.14.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magana-Schwencke N, Henriques JA, Chanet R, Moustacchi E. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc. Natl Acad. Sci. USA. 1982;79:1722–1726. doi: 10.1073/pnas.79.6.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fisher LA, Bessho M, Bessho T. Processing of a psoralen DNA interstrand cross-link by XPF-ERCC1 complex in vitro. J. Biol. Chem. 2008;283:1275–1281. doi: 10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- 52.Cheng AJ, Van Dyke MW. Monovalent cation effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 1993;21:5630–5635. doi: 10.1093/nar/21.24.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]