Abstract
The expression and function of the skeletal muscle chloride channel CLCN1/ClC-1 is regulated by alternative splicing. Inclusion of the CLCN1 exon 7A is aberrantly elevated in myotonic dystrophy (DM), a genetic disorder caused by the expansion of a CTG or CCTG repeat. Increased exon 7A inclusion leads to a reduction in CLCN1 function, which can be causative of myotonia. Two RNA-binding protein families—muscleblind-like (MBNL) and CUG-BP and ETR-3-like factor (CELF) proteins—are thought to mediate the splicing misregulation in DM. Here, we have identified multiple factors that regulate the alternative splicing of a mouse Clcn1 minigene. The inclusion of exon 7A was repressed by MBNL proteins while promoted by an expanded CUG repeat or CELF4, but not by CUG-BP. Mutation analyses suggested that exon 7A and its flanking region mediate the effect of MBNL1, whereas another distinct region in intron 6 mediates that of CELF4. An exonic splicing enhancer essential for the inclusion of exon 7A was identified at the 5′ end of this exon, which might be inhibited by MBNL1. Collectively, these results provide a mechanistic model for the regulation of Clcn1 splicing, and reveal novel regulatory properties of MBNL and CELF proteins.
INTRODUCTION
Myotonic dystrophy (dystrophia myotonica, DM) type 1, or DM1, is a genetic disorder with multi-systemic symptoms, such as myotonia, progressive muscle loss, cataracts, cardiac conduction defects, insulin resistance and cognitive impairments (1). DM1 is caused by the expansion of a CTG trinucleotide repeat in the 3′-untranslated region (UTR) of the DM protein kinase (DMPK) gene (2–4). Evidence suggests that the expanded CUG repeats transcribed from a mutated allele cause RNA gain-of-function effects that affect the function of other cellular factors. This concept is supported by transgenic mice (HSALR) expressing an expanded CUG repeat inserted in an unrelated gene (human skeletal actin, HSA) that manifest myotonia and abnormal muscle histology (5). Recently, a second locus of DM has been identified, and CCTG repeat expansion in intron 1 of the ZNF9 gene was found to be causative of DM type 2 (DM2; 6). Remarkably, in the nuclei of cells of patients with both DM1 and DM2, RNA inclusions containing CUG and CCUG repeats, respectively, have been observed (6–8). In addition, abnormalities in RNA metabolism have been found in the cells of DM patients. Splicing of certain genes is misregulated in DM1. These genes include cardiac troponin T (cTNT/TNNT2), insulin receptor (IR), chloride channel 1 (CLCN1), fast skeletal troponin T (TNNT3), sarcoplasmic/endoplasmic reticulum Ca2+-ATPase (SERCA) 1 and others (9–14). The splicing patterns of some of these genes are also misregulated in DM2 patients and the HSALR mouse (12,15,16). These results suggest that certain RNA-binding proteins that regulate pre-mRNA splicing of these genes are abnormally influenced by the mutant transcripts containing expanded CUG/CCUG repeats (17).
Two protein families—muscleblind-like (MBNL) and CUG-BP and ETR-3-like factor (CELF) proteins—may play major roles in the pathogenesis of DM. MBNL proteins MBNL1/EXP, MBNL2/MBLL/MLP1 and MBNL3/MBXL/CHCR are orthologs of the Drosophila muscleblind protein, which is involved in the terminal differentiation of photoreceptor and muscle cells in the fly (18,19). All three MBNL proteins can colocalize with RNA inclusions of expanded CUG/CCUG repeats in both DM1 and DM2 cells (20). MBNL1 binds directly to both CUG and CCUG repeat RNA in a length-dependent manner in vitro (21,22). Therefore, these proteins are considered to be sequestered by the expanded RNA through direct interactions, and their cellular functions can be disrupted in both types of DM. Remarkably, knockout mice of Mbnl1 manifest some DM-like symptoms, including myotonia, abnormal muscle histology and cataracts (13). More recently, Mbnl2 knockout mice were reported to manifest myotonia (23). Importantly, cellular studies have demonstrated that MBNL proteins can directly regulate the alternative splicing of the cTNT and IR genes, which are misregulated in DM1 patients (24,25). These results strongly support the hypothesis that loss of function of MBNL proteins leads to the misregulation of splicing in DM.
CELF proteins are another protein family involved in the pathogenesis of DM1. CELF proteins CUG-BP/CUGBP1/BRUNOL2, ETR-3/CUGBP2/NAPOR/BRUNOL3, CELF3/TNRC4/BRUNOL1, CELF4/BRUNOL4, CELF5/BRUNOL5 and CELF6/BRUNOL6 are multi-functional proteins that play regulatory roles in translation, RNA editing, mRNA stability, as well as splicing (26–29). CUG-BP regulates the alternative splicing of cTNT exon 5, IR exon 11 and CLCN1 intron 2 (9–11). In DM1 patients, the expression of CUG-BP protein is elevated because of protein stabilization induced by PKC-mediated phosphorylation (10,30,31). Moreover, CUG-BP transgenic mice can reproduce some of the muscular abnormalities observed in DM1 or its congenital form, including aberrant splicing and muscle histology (32,33). CUG-BP acts antagonistically against MBNL proteins in the splicing regulation of cTNT and IR (24,25), suggesting that altered CELF activities, in addition to the loss of MBNL function, can induce aberrant splicing in DM1. However, the extent to which these proteins can account for splicing abnormalities and the pathogenesis of DM remains unclear.
Myotonia is a characteristic symptom of both DM1 and DM2 and has been linked with a loss of function of CLCN1 chloride channel caused by aberrant splicing of its pre-mRNA in DM patients (11,12). The CLCN1 protein is a muscle-enriched voltage-gated chloride channel and is important for stabilizing the resting potential of muscle membrane (34). More than 50 mutations of CLCN1 have been found in myotonia congenita, another genetic disorder with myotonia, directly linking defects of CLCN1 with the pathogenesis of myotonia (35). In DM1 patients, the abnormal inclusion of alternative exons 6B and/or 7A and retention of intron 2 of CLCN1 have been observed (11,12). These aberrant-splicing patterns can lead to CLCN1 transcripts containing premature termination codons, resulting in an enhanced degradation of transcripts through the mechanism of nonsense-mediated mRNA decay (NMD), or the production of truncated proteins having a dominant-negative effect (12,36). Consistently, the levels of CLCN1 mRNA and protein are considerably lower in the muscle of DM patients (11,12). Thus, the misregulated splicing of CLCN1 in DM1 leads to a reduction in CLCN1 activity, which can be causative of myotonia.
In mouse models expressing an expanded CUG repeat (HSALR) or lacking Mbnl1, the inclusion of exon 7A in mouse Clcn1 increased, as in human CLCN1 of DM patients (12,13). It is important to note that the introduction of exogenous MBNL1 into HSALR mice by viral administration reversed the misregulation of Clcn1 splicing as well as the myotonic phenotype (37). Furthermore, antisense oligonucleotide (AON)-induced exon 7A skipping resulted in the upregulation of Clcn1 mRNA and protein levels and eliminated myotonia in both HSALR and Mbnl1 knockout mice (38). This suggests that the misregulation of Clcn1 splicing alone can explain the pathogenesis of myotonia in these mouse models. Therefore, MBNL proteins play an essential role in the splicing regulation of Clcn1 and are probably involved in the pathogenesis of myotonia in DM. Although the loss of function of MBNL was reproduced in these two mouse models, another pathogenic pathway involving CELF proteins might have been underrepresented in these models, which did not show elevated CUG-BP protein levels (13,16,39). Therefore, it is important to ask whether CELF proteins are involved in the regulation of CLCN1/Clcn1 splicing. Indeed, CUG-BP binds directly to an element in intron 2 and promotes the retention of this intron (11). Furthermore, CUG-BP transgenic mice exhibit increased inclusion of Clcn1 exon 7A, even though the manifestation of myotonia is not clear (33). Thus, it is important to characterize the roles of MBNL and CELF proteins in the regulation of Clcn1 splicing to understand the mechanism of myotonia in DM. Although increased exon 7A inclusion is the most frequent abnormality of CLCN1/Clcn1 splicing in DM (12), the mechanism of its regulation is still unclear.
We established a Clcn1 minigene assay system and identified multiple cis- and trans-acting factors that regulate the alternative splicing of Clcn1 exon 7A. The essential role of MBNL proteins in the normal splicing pattern of Clcn1 was verified. Our results also highlight some CELF proteins as antagonistic regulators against MBNL proteins.
MATERIALS AND METHODS
cDNA clones and constructs
MBNL1 and MBNL2 were amplified by polymerase chain reaction (PCR) from a human skeletal muscle cDNA library (BD Marathon-Ready human cDNA; Clontech). MBNL3 was amplified from a human liver cDNA library. Because the amplified MBNL3 cDNA clones contained an extra exon compared to a previous MBNL3 sequence, this exon was deleted by PCR-mediated mutagenesis. CELF proteins ETR-3, CELF3, CELF4, CELF5 and CELF6 were amplified from cDNA libraries of either brain or skeletal muscle of human origin. CUG-BP was amplified from pSRD/CUG-BP (40). Forward primers for the amplification of these cDNAs contained BamHI, BglII, or EcoRI sites, whereas the reverse primers contained either SalI or XhoI sites to add these restriction sites to the PCR products. Fragments of these cDNAs were inserted into the BamHI-XhoI or EcoRI-XhoI site of pSecDK, a mammalian expression vector with a myc-tag that was modified from pSecTagA (Invitrogen) to delete the Igκ chain leader sequence. Constructs encoding GST-MBNL140 has been described previously (22). The Clcn1 minigene fragment covering exons 6 to 7 was amplified from mouse genomic DNA by PCR using the primer pair Clcn1-Fw and Clcn1-Rv, into which a restriction site for BamHI or SalI was added. Similarly, the corresponding fragment of human CLCN1 was amplified using primers CLCN1-Fw and CLCN1-Rv. The minigene fragments were ligated into pGEM-T Easy vector (Promega). The minigene fragments in pGEM-T Easy were cleaved by BamHI and SalI and then subcloned into the BglII-SalI site of pEGFP-C1 (Clontech). A series of deletion mutants of Clcn1 was generated by PCR-mediated mutagenesis. To construct heterologous minigenes, we inserted alternative exons with flanking regions such as Clcn1 451–720 into the BglII-SalI sites of pEGFP-Tpm2-ex1-2 vector (see Supplementary ‘Materials and Methods’ section). Primer sequences are listed in Supplementary Table S1. Detailed information on the sequences and construction of mutant minigenes is available upon request. DM18 and DM480 contain a fragment of the 3′ region of DMPK with a CTG18 and interrupted CTG480 repeats, respectively (Supplementary Figure S4).
R-miR, a vector modified from pcDNATM6.2-GW/EmGFP-miR (Invitrogen), was utilized for RNA interference (RNAi) experiments. R-miR contains a cDNA fragment of monomeric RFP (mRFP) in place of EmGFP and an Esp3I recognition site introduced downstream of mRFP. DNA fragments corresponding to a portion of an artificial microRNA (Supplementary Table S2) were designed using BLOCK-iT RNA Designer (Invitrogen website) and inserted into the Esp3I site of R-miR.
The FANTOM3 clone plasmids encoding murine Mbnl3 (E430034C16), Cugbp1 (4432412L08), Cugbp2/Etr-3 (9530098D08) and Celf4 (C130060B05) used for testing RNAi efficiency in Neuro2a cells were provided by Dr Hayashizaki (41). Murine Mbnl1 and Mbnl2 were amplified from a mouse brain cDNA library. The N-terminal regions of Mbnl1, Mbnl2 and Mbnl3 and the full-length open reading frames of Cugbp1, Cugbp2 and Celf4 were amplified with primers containing restriction sites, digested by the restriction enzymes, and inserted into the BglII-SalI site of the pEGFP-C1 vector. All constructs were confirmed by sequencing.
Cellular splicing assay
Cells transfected with plasmids for the expression of a protein and a minigene were harvested 48 h post-transfection. Typically, cells were cultured in 12-well plates and transfected with 0.5 μg plasmids for protein expression (or cognate empty vector) and 0.01 μg plasmids for the expression of a minigene. Total RNA was extracted and purified using either the acidic guanidine phenol chloroform method or RNeasy Mini kit (Qiagen) including DNase treatment. Typically, 1.0 μg total RNA was reverse-transcribed using the ThermoScript RT-PCR System (Invitrogen) or Revertra Ace -α- (TOYOBO) with a 1 : 1 mixture of oligo dT and random hexamer as primers. Minigene fragments were amplified by PCR using a fluorescein isothiocyanate (FITC)-labeled forward primer for the 3′ region of the EGFP sequence (FITC-GFP-Fw) and a gene-specific reverse primer (Clcn1-Rv for Clcn1 or CLCN1-Rv for CLCN1 Supplementary Table S1). For Tpm2-based minigenes, primers FITC-GFP-Fw and Tpm2-ex2-splicing-Rv2 were used for amplification. PCR products were resolved by 2.0–2.5% agarose gel electrophoresis. By sampling at multiple cycles, the cycle numbers of PCR were adjusted such that the amplification was within the logarithmic phase. The fluorescence of PCR products was captured and visualized by LAS1000 or LAS3000 (FUJIFILM). The intensity of band signals was quantified using Multigauge software (FUJIFILM). The ratio of exon 7A inclusion in Clcn1 and CLCN1 was calculated as (7A inclusion)/(7A inclusion + 7A skipping) × 100.
Quantitative PCR
Gene-specific primers were designed using Primer Express software (Applied BioSystems) and are listed in Supplementary Table S3. These primer sets were mixed with cDNA samples and Power SYBR Green PCR Master Mix (Applied BioSystems). Real-time amplification and quantification were performed using an ABI7700 (Applied BioSystems) following the manufacturer's protocol.
Gel shift analysis
GST and GST-MBNL1 were purified as described previously (22). Oligo DNA templates corresponding to Clcn1(473–518) (CTGCCCAGGCACGGTCTGCAACAGAGAAGCACGACGGGCGAGGCAGCCCTAT AGTGAGTCGTATTACCCC), Clcn1(GAA) (CTGTTCTTCTTCTTCCTGCAACAGAGAAGCACGACGGGCGA GGCAGCCCTATAGTGAGTCGTATTACCCC), and Clcn1(a504c) (CTGCCCAGGCACGGGCTGCAACAGAGAAGCACGACGGGCG AGGCAGCCCTATAGTGAGTCGTATTACCCC) were purchased from Invitrogen and annealed with another DNA fragment for the T7 promoter (GGGGTAATACGACTCACTATAGGG). Using this partial duplex as a template, we transcribed RNA using T7 RNA polymerase (MEGAscript T7 kit; Ambion). The RNA was purified by phenol–chloroform extraction followed by ethanol precipitation. The purified RNA was treated with alkaline phosphatase, then labeled by T4 polynucleotide kinase in the presence of γ-32P-ATP. The labeled RNA was purified using a Nucaway spin column (Ambion). The procedures and reaction mixture used in the gel shift analysis are described in our previous report (22).
Ribonucleoprotein immunoprecipitation
Ribonucleoprotein immunoprecipitation (RIP) was performed as described previously (42), with minor modifications. The amount of co-precipitated minigene RNA was quantified by SYBR green-based Quantitative PCR (qPCR) using the primers listed in Supplementary Table S3. The procedure is described in the Supplementary ‘Materials and Methods’ section.
RESULTS
MBNL and CELF proteins regulate the inclusion of Clcn1 exon 7A
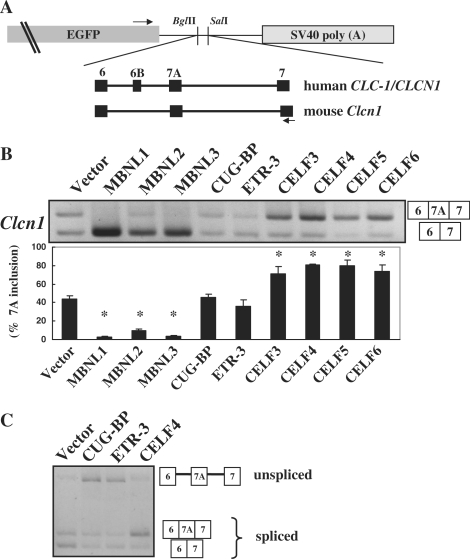
To examine whether the MBNL and CELF family proteins can regulate the splicing of Clcn1, we created a minigene covering exons 6 to 7 of the mouse Clcn1 gene (Figure 1A). It is important to note that because the inclusion of exon 7A does not produce a premature termination codon in the context of our Clcn1 minigene, the spliced products containing exon 7A are not substrates of NMD. Thus, the minigene would provide more faithful splicing patterns compared to the endogenous Clcn1. We utilized non-muscle cell lines to minimize the effect of muscle-dependent backgrounds and focus on the direct effects of transgenes. When the Clcn1 minigene was transfected into COS-7 cells, 45% of the spliced products contained exon 7A (Figure 1B). Next we expressed myc-tagged MBNL or CELF proteins with the Clcn1 minigene and examined the patterns of Clcn1 splicing. The expressions of MBNL and CELF proteins were confirmed by Western blot analysis using an anti-myc antibody (Supplementary Figure S6A). All three MBNL proteins strongly repressed exon 7A inclusion (Figure 1B). In contrast, CELF3, CELF4, CELF5 and CELF6 proteins significantly promoted the inclusion of 7A (Figure 1B). Remarkably, CUG-BP and ETR-3 did not alter the ratio of exon 7A inclusion (Figure 1B). These two proteins increased the unspliced product and reduced the spliced products with or without exon 7A (Figure 1C).
Figure 1.
Splicing regulation of MBNL and CELF proteins. (A) Structure of chloride channel minigenes. Both human CLCN1/ClC-1 and mouse Clcn1 minigenes were subcloned between the BglII and SalI sites of pEGFP-C1. Black boxes represent exons of the minigenes. Arrows indicate the position of primers used in the splicing assays. Exon 6B is a human-specific exon and is absent in Clcn1. (B) Splicing regulation of Clcn1 by MBNL and CELF proteins. Representative results of cellular splicing assays using the Clcn1 minigene in COS-7 cells. The upper bands correspond to a splice product containing exon 7A, whereas lower bands correspond to a splice product lacking exon 7A. Bar chart shows quantified results of exon 7A inclusion (mean ± SD, n = 3). Statistical significance was analyzed by analysis of variance (ANOVA) and Dunnett's multiple comparisons. All MBNL proteins and CELF proteins except for CUG-BP and ETR-3 showed significant differences (*P < 0.0001) compared to the empty vector. (C) CUG-BP and ETR-3 increased an unspliced product of the Clcn1 minigene. Structures of PCR products are indicated.
CELF4 and expanded CUG repeats act antagonistically against MBNL1 in the splicing regulation of Clcn1
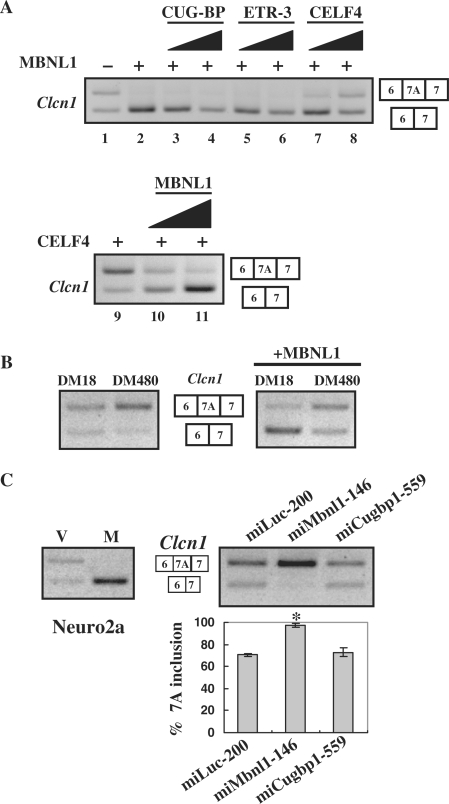
We investigated whether CELF proteins can antagonize the effect of MBNL1. Among CELF proteins that promoted exon 7A inclusion in Clcn1, CELF4 was used for the following analyses because this protein is expressed in muscle (26,43). CELF4 and two other muscle-expressed CELF proteins, CUG-BP and ETR-3, were co-transfected with MBNL1, as well as the Clcn1 minigene. As shown in Figure 2A, CELF4 increased the ratio of exon 7A in a dose-dependent manner (lanes 7 and 8), whereas CUG-BP or ETR-3 did not (lanes 3–6). When MBNL1 was titrated in the presence of CELF4, a decrease in exon 7A inclusion was observed, depending on the dosage of MBNL1 (Figure 2A, lanes 9–11). These results demonstrate that MBNL1 and CELF4, but not CUG-BP or ETR-3, can regulate Clcn1 exon 7A splicing in an antagonistic manner.
Figure 2.
MBNL1 is antagonized by CELF4 and expanded CUG repeats. (A) Antagonistic effects of CELF4 against MBNL1 in the splicing regulation of Clcn1. Clcn1 minigene was co-transfected with MBNL1 or MBNL1 plus CELF protein into COS-7 cells. Upper panel: lane 1, the pattern of empty vector-transfected cells; lanes 2–8, a constant amount of MBNL1-encoding vector and increasing amounts of one of the CELF-encoding vectors were co-transfected at ratios of 1 : 0 (lane 2), 1 : 1 (lanes 3, 5 and 7) and 1:3 (lanes 4, 6 and 8). Lower panel: similarly, a CELF4-encoding vector and MBNL1-encoding vector were transfected at ratios of 1 : 0 (lane 9), 1 : 1 (lane 10) and 1 : 3 (lane 11). The total amount of transfected plasmids was held constant by adjusting the amounts of empty vector. (B) Effects of an expanded CUG repeat on the chloride channel splicing. Clcn1 minigene was transfected with an expression vector of DMPK harboring a normal (DM18) or expanded (DM480) CTG repeat (left panel). Either normal or expanded DMPK vector was co-transfected with MBNL1 and the Clcn1 minigene, and the splicing patterns were analyzed (right panel). (C) Results of Clcn1-splicing assays in Neuro2a cells. MBNL1-mediated repression of exon 7A inclusion in Neuro2a cells (left). ‘V’ and ‘M’ indicate empty vector and MBNL1, respectively. Note that the basal inclusion of exon 7A in Neuro2a cells was higher than that in COS-7 cells. Clcn1-splicing regulation was affected by RNAi of Mbnl1 but not Cugbp1 in Neuro2a cells (right). Bar chart shows the quantified results of exon 7A inclusion (mean ±SD, n = 4). According to ANOVA and Dunnett's test, miMbnl1-146 induced a statistically significant increase of exon 7A compared to miLuc-200 (*P < 0.0001), whereas miCugbp1-559 did not (P = 0.36).
We also examined DMPK constructs harboring either CTG18 (DM18) or interrupted CTG480 (DM480) in the 3′-UTR. When these constructs were expressed with the Clcn1 minigene, DM480 increased exon 7A inclusion, whereas DM18 showed little effect (Figure 2B, left). These constructs were next expressed in the presence of MBNL1. As shown previously, MBNL1 alone strongly repressed the inclusion of exon 7A. DM480 reversed the repression of exon 7A inclusion by MBNL1, whereas DM18 did not (Figure 2B, right). Thus, expanded CUG repeats can antagonize the effect of MBNL1 on Clcn1 splicing.
Finally, we performed knockdown experiments using vector-based RNAi to examine whether the regulation of Clcn1 splicing is dependent on the dose of endogenous MBNL1 or CUG-BP. The RNAi vector used here expresses an artificial microRNA that is exactly complementary to a region of its target gene, leading to degradation of the target mRNA. For RNAi experiments, we utilized Neuro2a and HeLa cells rather than COS-7 because of the availability of cDNA sequence information essential for the design of microRNA. First, the overexpression of MBNL1 in Neuro2a cells strongly repressed the inclusion of exon 7A as in COS-7 cells (M, Figure 2C). An RNAi vector miMbnl1-146 was effective in reducing both Mbnl1 and Mbnl2 and increased the inclusion of Clcn1 exon 7A compared to a control vector miLuc-200 (Figure 2C and Supplementary Figure S1). Another RNAi vector, miMbnl1-236, also increased exon 7A inclusion (data not shown). We also used HeLa cells for RNAi experiments and found that the knockdown of MBNL1 resulted in an increase in exon 7A inclusion (Supplementary Figure S2). Therefore, the splicing of Clcn1 exon 7A is regulated by MBNL in a dose-dependent manner. In contrast, an effective RNAi vector targeting endogenous Cugbp1, miCugbp1-559, did not alter exon 7A splicing (Figure 2C and Supplementary Figure S1). We were not able to confirm the knockdown of endogenous Etr-3/Cugbp2 and Celf4 because of very low or undetectable endogenous expression in the cell lines used in this study (data not shown); thus, the RNAi of Etr-3 and Celf4 was not tested in the splicing analysis.
Involvement of the exon 7A sequence in the splicing regulation by MBNL1
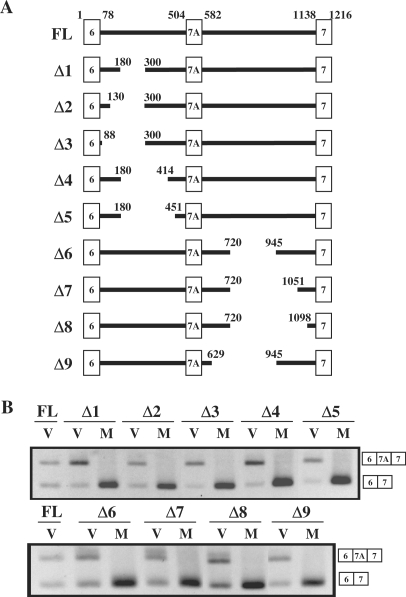
To understand the mechanism by which MBNL1 represses the inclusion of Clcn1 exon 7A, we tried to define the regions of the Clcn1 minigene that are responsive to MBNL1. For this purpose, we examined a series of Clcn1deletion mutants lacking a region in either introns 6 or 7A (Δ1–Δ9, Figure 3A). As shown in Figure 3B, deletions in intron 6 altered the basal splicing pattern. For example, the Δ1 mutant exhibited increased inclusion of exon 7A, suggesting the presence of an element in the deleted region that represses exon 7A inclusion. However, this mutant and the other intron 6 mutants (Δ2–Δ5) were strongly repressed by MBNL1 (Figure 3B, upper panel). Similarly, the series of intron 7A mutants did not lose responsiveness to MBNL1 (Δ6–Δ9, Figure 3B). Thus, large portions of these intronic regions could be excluded from the region critical for responsiveness to MBNL1.
Figure 3.
Intronic deletion analysis of Clcn1. (A) Structure of Clcn1 deletion mutants. Deletion series of the Clcn1 minigene were generated by PCR-mediated mutagenesis. FL corresponds to the full-length Clcn1 minigene covering exons 6–7 and is the same construct as that used in Figures 1 and 2. The positions of nucleotides at the termini of exons and the junction of deleted regions are indicated. (B) Splicing analysis of Clcn1 deletion mutants in COS-7 cells. Cellular-splicing assays were performed using deletion mutants Δ1–Δ9 and MBNL1. Lanes ‘V’ and ‘M’ indicate the co-expression of each deletion minigene with empty vector and MBNL1, respectively. Splicing patterns were detected by RT–PCR, as in Figure 1B.
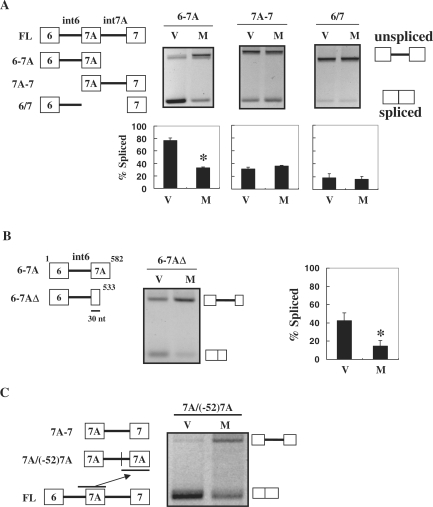
Next we examined another series of deletion mutants (Figure 4A, left). When minigene 6-7A was expressed with the empty vector, both spliced and unspliced products were observed (Figure 4A). Co-expression with MBNL1 resulted in the repression of splicing (Figure 4A). In the case of 7A-7, the expression of MBNL1 did not cause a significant change compared to that of the empty vector (Figure 4A). To examine whether the responsiveness of 6-7A was dependent on the sequence of exon 7A, we analyzed the 6/7 mutant, in which the sequence of exon 7A was virtually replaced with that of exon 7. Interestingly, this replacement completely abolished the responsiveness to MBNL1 (Figure 4A, right panels). The observed loss of response in the 6/7 mutant was not due to inefficient basal splicing of exon 7, because replacement of the first 12 nt of exon 7 in the 6/7 mutant with an exonic-splicing enhancer (ESE) markedly increased the spliced products but did not improve the responsiveness to MBNL1 (Supplementary Figure S3). Thus, exon 7A should contain at least part of the MBNL1-responsive region. In addition, intron 6 alone was insufficient for response to MBNL1. To further define the responsive region, we tested another minigene, 6-7AΔ, a 3′ deletion of the 6-7A fragment containing only 30 nt from the 5′ part of exon 7A. This fragment was still responsive to MBNL1 (Figure 4B). Finally, we tested a minigene derived from 7A-7 that had exon 7 as well as the preceding 52 nt replaced by the corresponding region of exon 7A (Figure 4C). This minigene, 7A/(−52)7A, exhibited increased unspliced products upon MBNL1 expression, suggesting that the substituted fragment could mediate the response to MBNL1 in a heterologous context (Figure 4C). These results suggest the involvement of a 5′ portion of exon 7A (30 nt), as well as its upstream 52-nt region in the MBNL1-mediated-splicing regulation, at least in the context of the truncated minigenes.
Figure 4.
Involvement of exon 7A sequence in splicing regulation by MBNL1. (A) Response of truncated Clcn1 minigenes to MBNL1 is dependent on exon 7A. Structures of 6-7A, 7A-7 and 6/7 deletion mutants are indicated in the upper left. Splicing analysis of these mutants shows both spliced and unspliced products as indicated. The bar chart shows the quantified ratio of spliced products. (B) Deletion in the 3′ region of exon 7A did not abolish the response to MBNL1. The structure of the 6-7AΔ mutant is shown (left). Splicing products of 6-7AΔ and quantification are shown as in (A). (C) Splicing regulation of the 7A/(−52)7A minigene. 7A/(−52)7A was made from 7A-7 by replacing exon 7 and its upstream 52 nt with the corresponding region of exon 7A (left). Splicing of 7A/(−52)7A was responsive to MBNL1 (right). Lanes ‘V’ and ‘M’ indicate co-transfection with the empty vector and MBNL1, respectively. Bars represent the ratio of spliced bands. Statistical analysis was performed by two-tailed t-test in comparison with the empty vector. *P < 0.05 from three independent experiments.
A region around the 3′ splice site of exon 7A mediates the splicing regulation of MBNL1
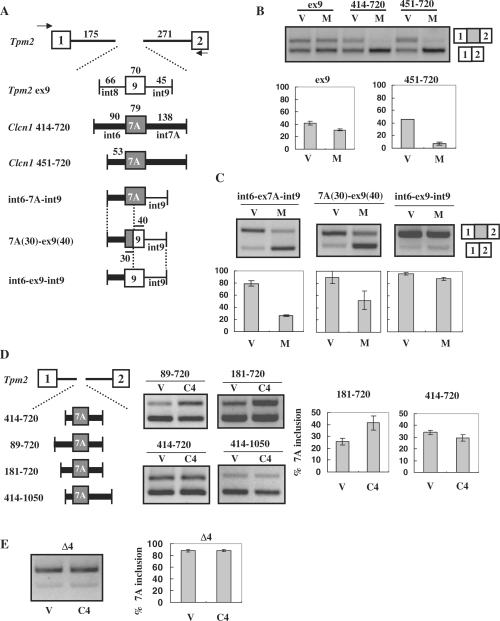
To further analyze the regions critical for exon 7A splicing, we generated heterologous minigenes in which the exon of interest was inserted in the context of constitutive exons of mouse tropomyosin 2 (Tpm2), a gene distinct from Clcn1. Clcn1 fragments covering 414–720 or 451–720, or a reference fragment covering Tpm2 exon 9 and its flanking intronic regions, were inserted into a Tpm2 fragment covering exons 1 to 2 (Figure 5A). MBNL1 repressed Clcn1 exon 7A inclusion of the heterologous minigenes, demonstrating that the inserted fragments of Clcn1 were sufficient for response to MBNL1 (Figure 5B). In contrast, MBNL1 showed little effect on the inclusion of Tpm2 exon 9 (Figure 5B). Next we made an additional chimeric minigene derived from the 451–720 minigene by replacing a region derived from intron 7A with a sequence derived from Tpm2 intron 9 (int6-ex7A-int9; Figure 5A). Although int6-ex7A-int9 was still responsive to MBNL1, another minigene with a further replacement of exon 7A with Tpm2 exon 9 (int6-ex9-int9) exhibited only a slight response to MBNL1 (Figure 5C). Thus, the exon 7A sequence was essential for regulation by MBNL1. Furthermore, a minigene with an intermediate replacement, 7A(30)-ex9(40), in which the first 30 nt of the exon and its upstream intronic region were derived from Clcn1, was responsive to MBNL1 (Figure 5C). These results suggest that a region covering a 3′ portion of intron 6, as well as the first 30 nt of exon 7, can mediate splicing regulation by MBNL1. This is consistent with the results of the truncated minigenes displayed in Figure 4.
Figure 5.
Exon 7A splicing regulation in heterologous minigenes. (A) Structure of the Tpm2-based heterologous minigene. Fragments of Tpm2 covering exons 1 to 2 were inserted downstream of EGFP. Test exons together with their flanking regions were inserted into intron 1 of Tpm2. Intronic fragments derived from Clcn1 are indicated by thick lines, whereas those derived from Tpm2 (regions flanking exon 9) are indicated by thin lines. Exonic sequences of Clcn1 exon 7A and Tpm2 exon 9 are indicated by grey and white boxes, respectively. (B) Splicing assay results using Tpm2-based heterologous minigenes in COS-7 cells. Upper bands correspond to the spliced products containing an exon inserted between Tpm2 exon 1 and 2. ‘V’ and ‘M’ indicate empty vector and MBNL1, respectively. Compared with Tpm2 ex9, Clcn1-derived 414–720 and 451–720 minigenes exhibited evident responses to MBNL1. Bar chart shows quantified results of the splicing assay (n = 3). (C) Splicing regulation of heterologous minigenes containing a portion of Clcn1 intron 6. Results of the splicing assay are shown as in B. The structures of minigenes are shown in A. (D) Determination of the Clcn1 region responsible for CELF4-mediated exon 7A inclusion. Tpm2-based heterologous minigenes covering a Clcn1 region indicated by the numbers were tested for their responsiveness to CELF4 (C4). The splicing assay was performed as in B, except that CELF4 was used in place of MBNL1. In the case of 181–720, CELF4 expression induced a significant increase compared to control (P < 0.05, n = 3, two-tailed t-test). (E) Splicing analysis of the Δ4 mutant minigene and CELF4. The structure of Δ4 is described in Figure 3A. Splicing analysis results are shown as in D. CELF4 did not significantly alter the splicing of Δ4 (P = 0.97, n = 3, two-tailed t-test).
We also tested several minigenes to identify the region of Clcn1 responsive to CELF4. Interestingly, CELF4 did not increase the exon 7A inclusion of the 414–720 construct, which was responsive to MBNL1 (Figure 5B and D). However, minigenes covering 89–720 and 181–720, but not 414–1050, increased the inclusion of exon 7A upon CELF4 overexpression (Figure 5D and data not shown). Therefore, an element responsive to CELF4 was located in the region of 181–413, which was distinct from the regions responsive to MBNL1 identified previously. Lastly, we tested the Δ4 mutant that lacks the putative responsive region for CELF4 (Figure 3A). CELF4 did not promote exon 7A inclusion of Δ4 (Figure 5E). Thus, 181–413 was essential for the regulation of exon 7A by CELF4.
MBNL1 binds directly to the transcript of the Clcn1 minigene
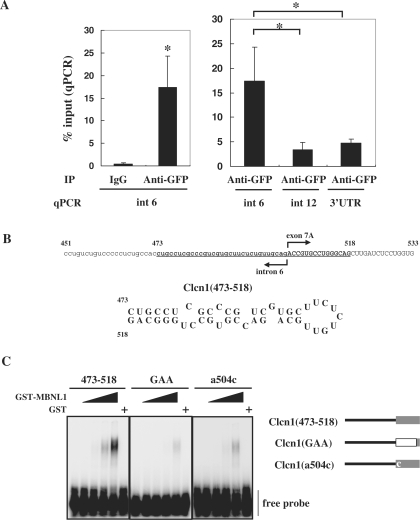
We next examined whether MBNL1 associates with transcripts of Clcn1 in cells. In a ribonucleoprotein immunoprecipitation (RIP) analysis (42), intracellular RNA–protein complexes were reversibly fixed with paraformaldehyde and then co-immunoprecipitated. Fixation allowed for stringent washing of immunoprecipitates to exclude post-lysis association between RNA and proteins. We utilized a Neuro2a cell line stably expressing EGFP-fused MBNL1. This cell line exhibited a lower basal inclusion of exon 7A of endogenous Clcn1 compared to normal Neuro2a cells (Supplementary Figure S5). EGFP-MBNL1 was precipitated by beads pretreated with anti-GFP antibody or rabbit IgG. RNA fragments derived from specific regions of endogenous Clcn1 in the precipitates were detected by quantitative PCR. The Association of Clcn1 intron 6 and EGFP-MBNL1 was detected only when immunoprecipitated by anti-GFP antibody (Figure 6A, left). Importantly, this association was specific because intron 6 was highly enriched compared to other Clcn1 regions (Figure 6A, right).
Figure 6.
MBNL1 associates with Clcn1 RNA. (A) The intracellular association between EGFP-MBNL140 and the transcript of endogenous Clcn1 was analyzed by RIP. The cell lysate of a cell line stably expressing EGFP-MBNL1 was used for immunoprecipitation. The RNA fraction co-precipitated with anti-GFP antibody or IgG was reverse-transcribed. The amount of Clcn1 (pre-)mRNA was quantified by real-time quantitative PCR using a primer set for the indicated regions of the Clcn1 gene (n = 5). Bars represent the amount of Clcn1 RNA co-precipitated with anti-GFP antibody or control IgG normalized by the amount of the Clcn1 RNA in the input fraction (mean ± SD). Left: EGFP-MBNL1 was co-immunoprecipitated with RNA fragments containing Clcn1 intron 6 by GFP antibody but not control IgG (*P < 0.01, two-tailed t-test). Right: RIP analysis of multiple Clcn1 regions. Three regions of Clcn1 (intron 6, intron 12 and 3′-UTR) were amplified from immunoprecipitates of the GFP antibody. Intron 6 was significantly more enriched than the other regions (*P < 0.01, ANOVA and Tukey's test). (B) A hairpin structure predicted in the putative MBNL1-responsive region of Clcn1. Nucleotides 473–518 of Clcn1 are indicated in boldface (upper). Predicted secondary structure of the fragment Clcn1(473–518) (lower). (C) Binding between GST-MBNL1 and Clcn1(473–518) or its mutants was examined by gel shift analysis (left). 32P-labeled probes were incubated with or without GST-MBNL1 (0.225, 0.45, 0.9, 1.8 μM) or GST (1.8 μM). The reaction mixture was separated by native PAGE and visualized by autoradiography. Structure of probes used in gel shift analysis (right). In the Clcn1(GAA) mutant, the first 12 nt of exon 7A are substituted by (GAA)4 repeats. The Clcn1(a504c) mutant contains a point mutation of the first nucleotide of exon 7A but is otherwise the same as Clcn1(473–518).
In the results described previously, a region around the boundary of intron 6 and exon 7A was suggested to play an important role in the response to MBNL1 (Figures 4 and 5C). Previously, it was reported that MBNL1 prefers mismatch-containing double-stranded structures (22,44,45). It is interesting that a region around the 3′ splice site of exon 7A (corresponding to 473–518) was predicted to form a hairpin structure by Mfold (http://www.bioinfo.rpi.edu/applications/mfold; Figure 6B; 46). We examined whether MBNL1 can bind to this fragment, Clcn1(473–518), by gel shift analysis. GST-fused MBNL1, but not GST alone, bound to the Clcn1 fragment in a dose-dependent manner (Figure 6C). GST-MBNL1 exhibited an apparently lower affinity for Clcn1(GAA), a mutant RNA in which the first 12 nt of exon 7A were substituted by a (GAA)4 repeat (Figure 6C). Therefore, the sequence at the 5′ end of exon 7A contributes to the binding of MBNL1.
The 5′ end of exon 7A contains an ESE
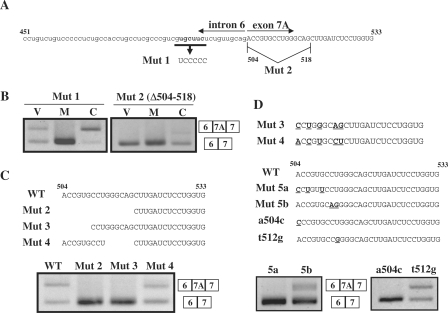
Region 451–533 of Clcn1 consistently accounted for the responsiveness to MBNL1 in multiple contexts (Figures 4B and 5C). We noticed that this region contained a YGCU(U/G)Y motif (Y: C or U; Figure 7A), which has been reported as a binding consensus sequence for MBNL1 (24). A mutant minigene containing a mutation of this motif (but otherwise the same as the full-length minigene) was examined in a splicing assay (Mut 1, Figure 7A). Mut1 was fully responsive to MBNL1 as well as to CELF4 (Figure 7B). Thus, this motif itself was dispensable for the response to MBNL1 in exon 7A splicing.
Figure 7.
Analysis of the 5′ region of exon 7A in the Clcn1 minigene. (A) Sequence of nt 451–533 of the Clcn1 minigene that corresponds to a 3′ portion of intron 6 and a 5′ portion of exon 7A. Substituted nucleotides in Mut 1 are shown in boldface. The first 15 nt of exon 7A were deleted in Mut 2. (B) Splicing assays results for Mut 1 and Mut 2 minigenes in COS-7 cells. Mut 2 exhibited a complete loss of exon 7A inclusion. (C) Effects of deletions in the 5′ region of Clcn1 exon 7A on basal-splicing efficiency. Deleted regions in Mut 2, Mut 3 and Mut 4 are indicated (upper). Splicing assay results of Mut 2/3/4/minigenes transfected into COS-7 cells are shown (lower). WT: wild type. (D) Point mutations at the 5′ end of exon 7A disrupted the basal inclusion of exon 7A. Alignment of part of Mut3 and Mut4 sequences (upper). Different nucleotides between Mut 3 and Mut 4 are indicated by boldface and underlined text. Point mutations that partly mimic the difference between Mut 3 and Mut 4 were introduced in the Mut 5a, Mut 5b, a504c and t512g mutants, as indicated by boldface and underlined text (middle). Splicing assay results of the mutant minigenes in COS-7 cells (lower).
The preceding analyses suggested that the 5′ end of exon 7A is involved in the responsiveness to MBNL1 (Figures 4B and 5C). To characterize the role of this region, we first examined a minigene lacking the first 15 nt of exon 7A (Mut 2, Figure 7A). Unexpectedly, this deletion resulted in the complete exclusion of exon 7A, even without MBNL1 overexpression (Figure 7B), indicating the presence of an ESE in this region that is essential for the basal inclusion of exon 7A. We further tested 6-nt deletions in either the 5′ or 3′ parts of the 15-nt region (Mut 3 and Mut 4, Figure 7C). Only Mut 3 exhibited complete exon 7A skipping like Mut 2 (Figure 7C). Direct comparison of the sequences of Mut 3 and Mut 4 showed that 5 nucleotides differed between these mutants (Figure 7D, upper). We then introduced substitutions of these nucleotides into the full-length minigene (Mut 5a and Mut 5b, Figure 7D). Both Mut 5a and 5b exhibited reduced basal inclusion of exon 7A, with a stronger effect from Mut 5a (Figure 7D). Finally, we tested two point mutants, a504c and t512g, to define the region of the ESE. Whereas the t512g mutation had no effect on the basal splicing of exon 7A, mutant a504c totally disrupted the inclusion of exon 7A (Figure 7D). Because Mut 5b (c511a and t512g), but not t512g alone, showed a reduction in exon 7A inclusion, mutation c511a should be critical for exon 7A inclusion. GST-MBNL1 bound to an RNA containing the a504c mutation with a lower affinity compared to Clcn1(473–518) (Figure 6C), suggesting that the strong repressive effect of a504c was due to the direct disruption of the ESE rather than a hypersensitivity to MBNL1. Thus, we concluded that the 8 nt at the 5′ end of exon 7A, 504–511, contain an ESE essential for the basal inclusion of exon 7A.
DISCUSSION
CLCN1/Clcn1 splicing is a key event in DM. Although the misregulation of splicing has been well established as a characteristic abnormality of DM, few misregulated genes have a clear causal relationship to symptoms of DM. Clcn1 misregulation can account for myotonia in DM model mice (38,47,48). As demonstrated recently, the skipping of exon 7A induced by AON reversed the myotonic phenotype of DM model mice (38), making CLCN1 splicing a promising target for therapeutic approaches. Understanding Clcn1/CLCN1 splicing would aid in the design of rational strategies for correcting CLCN1 expression to perhaps prevent myotonia. In addition, the identification of molecular pathways causative of Clcn1 missplicing is important for the evaluation or interpretation of DM model mice. Besides the aforementioned model mice, myotonic phenotypes are observed in several other mouse models (49–52). It is unclear whether the loss of MBNL function can explain all of these myotonic mice (51). In these contexts, a Clcn1 minigene should be useful for identifying and characterizing the factors that modulate the splicing pattern of Clcn1. Indeed, we have identified multiple determinants for the regulation of exon 7A splicing using a Clcn1 minigene and its derivatives.
MBNL proteins regulate Clcn1 exon 7A
Previous genetic studies have indicated the involvement of MBNL proteins in the regulation of Clcn1 splicing. The loss of either Mbnl1 or Mbnl2 can cause myotonia in mice, whereas overexpression of Mbnl1 rescues the myotonic phenotype in HSALR mice (13,23,37). However, these studies have not excluded the indirect involvement of MBNL proteins such that they regulate muscle differentiation or maintenance, which in turn alters the activity of other splicing factors to result in altered Clcn1 splicing. Indeed, exon 7A of Clcn1 is regulated through muscle development and is preferentially included in neonatal muscle but not in adult muscle (16,47). Here, we have demonstrated that the splicing regulation of Clcn1 exon 7A by MBNL1 was observed in COS-7, HeLa and Neuro2A cell lines (Figures 1 and 2 and Supplementary Figure S2). Thus, the regulation of exon 7A can be determined directly by the expression level of MBNL proteins, even without the context of muscle cells. The inclusion of exon 7A was repressed by the overexpression of MBNL proteins but increased by their knockdown (Figures 1 and 2). In addition, MBNL1 associated with the pre-mRNA of Clcn1 in the cell (Figure 6A), and direct binding between MBNL1 and an RNA fragment of Clcn1 was verified in vitro (Figure 6C). These results are consistent with the model that MBNL proteins directly regulate CLCN1/Clcn1 and that the loss of MBNL function leads to CLCN1/Clcn1 misregulation in DM.
Differential effects of CELF proteins on Clcn1 splicing
In contrast to MBNL proteins, CELF3/4/5/6 promoted increased inclusion of exon 7A of mouse Clcn1 (Figure 1B). Among these CELF proteins, CELF4 is expressed in a wide variety of tissues, including muscle (26,43). Although mice deficient in Celf4 have been reported to manifest a complex seizure phenotype (53), the physiological function of CELF4 is largely unclear. Although an elevation of CUG-BP and ETR-3 proteins was observed in DM1 patients, the other CELF proteins have not been well characterized. The expression level, intracellular localization, and activity of CELF4 (and CELF3/5/6) should be investigated in the context of DM. Although Clcn1 is enriched in muscle, it is expressed in other tissues including brain even at a low level (data not shown). Because some CELF proteins are enriched in the brain (43), they might play a role in keeping Clcn1 expression at a low level in the brain or possibly other tissues through a splicing-mediated regulation of expression.
In contrast, CUG-BP and ETR-3 did not directly change the ratio of exon 7A inclusion in our mouse Clcn1 minigene (Figures 1B and 2C). This was unexpected because the inclusion of exon 7A has been reported to be increased in CUG-BP transgenic mice (33). It is possible that the regulation of exon 7A by CUG-BP is dependent on some specific cellular conditions and/or the genomic context of Clcn1 that is missed in the minigene. Alternatively, the transgenic mouse might involve indirect or secondary effects of long-term CUG-BP overexpression. Nevertheless, our results might be compatible with a role of CUG-BP or ETR-3 in the misregulation of mouse Clcn1, as the overexpression of these proteins could reduce spliced Clcn1 products and increase intron retention (Figure 1C). It should be noted, however, that both CUG-BP and ETR-3 increase neither the inclusion of exon 7A nor the unspliced product in the human CLCN1 minigene in our preliminary results (Kino et al., unpublished data). The determinants of the difference between human and mouse minigenes are currently under investigation. Our results also revealed a functional heterogeneity among CELF proteins. The Clcn1 minigene would allow us to determine the cause of the difference in the effects of CELF proteins on exon 7A inclusion.
Multiple factors involved in the regulation of Clcn1 exon 7A splicing
To clarify the regulatory mechanism of exon 7A splicing, we analyzed a series of Clcn1 mutant minigenes. We found that the regulation of exon 7A by MBNL1, as well as their binding, was at least partly mediated by the sequence of exon 7A itself. Furthermore, the MBNL-responsive regions were mapped roughly to a boundary region of exon 7A and its flanking intronic sequence, or 451–533. In the previous examples, MBNL1 regulated its target exons through intronic sequences of cTNT, TNNT3, and SERCA1 (24,45,54). In contrast, neither intron 6 nor 7A of Clcn1 was sufficient for response to MBNL1 (Figures 4A and 5C). Our results suggest that not only intronic but also exonic sequences should be considered for analyses such as genome-wide identification of the MBNL splicing targets.
We found an ESE located at the 5′ end of exon 7A (504–511) that was essential for the basal inclusion of exon 7A (Figure 7). The nucleotide sequence of this ESE is completely conserved in human CLCN1. In general, ESEs are important motifs for both constitutive and alternative splicing and are bound by SR proteins that enhance the recognition of an exon and promote splicing (55,56). The ESEfinder algorithm (http://rulai.cshl.edu/tools/ESE/; 57) predicted that the candidate region overlaps with potential recognition sites for multiple SR proteins (data not shown). However, the prediction pattern went unchanged even when a critical mutation (a504c) was introduced. Interestingly, the ESE in the 5′ end of exon 7A overlaps with the region targeted by the AON effective against myotonia in mouse models (38). Hybridization of the AON may prevent the functioning of not only the polypyrimidine (PY) tract and 3′ splice site but also the ESE, leading to efficient exon 7A skipping.
Previously, we have suggested double-stranded RNA with mismatches as putative MBNL1-binding targets (22), which was supported by recent analyses directly demonstrating that stem-loop structures with a pyrimidine mismatch are binding sequences of MBNL1 and mediate its splicing regulation (44,45). In contrast, YGCU(U/G)Y motifs have been reported as binding sites for MBNL1 in cTNT and SERCA1 (24,54). In the case of cTNT, however, the YGCU(U/G)Y motifs were later shown to be part of a stem-loop structure (44). Here, we found a stem-loop structure in the junctional region between intron 6 and exon 7A as a candidate region for MBNL1-binding. Indeed, this fragment could bind with recombinant MBNL1 (Figure 6C). Furthermore, substitutions that alter this hairpin region impaired the responsiveness to MBNL1 (Figures 4A, 5C, Supplementary Figure 3) and the binding of RNA to MBNL1 (Figure 6C). Interestingly, this hairpin region overlaps with splicing signals, such as the PY tract, 3′ splice site and the ESE described previously. In particular, the ESE overlaps with an RNA region essential for efficient binding to MBNL1 (Figures 6C and 7). Therefore, one possible explanation of how MBNL1 acts on exon 7A splicing is that MBNL1 binds to the hairpin structure and inhibits the access of splicing factors to these splicing signals, like the AON described previously. The Clcn1(GAA) mutant exhibited residual binding to MBNL1 (Figure 6C). This might be explained by the presence of a YGCU(U/G)Y motif in intron 6.
Lastly, the region of Clcn1 that mediates the effect of CELF4 was located within 181–413 (Figure 5D and E), which was not necessary for the regulation by MBNL1 (Figure 3). Therefore, antagonistic regulation by MBNL1 and CELF4 is mediated by distinct regions of the minigene. This is similar to the case of cTNT exon 5, which is regulated by CUG-BP and MBNL1 through distinct binding motifs (24). Because the Δ4 mutant exhibited a higher basal inclusion of exon 7A (Figure 5E), the deleted region (181–413) may contain an element that represses exon 7A inclusion. Perhaps CELF4 might antagonize this repressive element, which might explain how the effect of CELF4 was lost in the case of the Δ4 mutant.
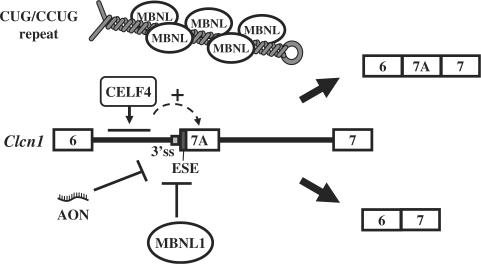
Our current model for Clcn1 regulation is depicted in Figure 8. Alternative splicing of exon 7A can be regulated by MBNL and by certain CELF proteins, such as CELF4. Expanded CUG/CCUG repeats may deplete MBNL and decrease the probability of binding between MBNL and the pre-mRNA of Clcn1. The ESE located at the 5′ end of exon 7A and the flanking splicing signals might be common targets of MBNL and the previous AON. Taken together, our results reveal novel regulatory properties of MBNL and CELF proteins and provide a molecular basis for the mechanism of splicing regulation of Clcn1, which might underlie rational therapeutic strategies targeting myotonia in DM.
Figure 8.
Model of mouse Clcn1 splicing regulation by multiple factors. Cis- and trans-acting factors involved in the splicing regulation of Clcn1 exon 7A are depicted. MBNL1 represses exon 7A inclusion through a region containing the 5′ region of exon 7A as well as its flanking intronic sequence. An exonic splicing enhancer (ESE) is located at the 5′ end of exon 7A. An antisense oligonucleotide (AON) previously reported (38), as well as MBNL1, might act in part through inhibiting this ESE. The facilitation of exon 7A inclusion by CELF4 is mediated by a region located in intron 6. Expanded CUG/CCUG repeat RNA may deplete MBNL proteins, resulting in the facilitation of exon 7A inclusion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Health, Labor and Welfare; Ministry of Education, Culture, Sports, Science, and Technology of Japan; Human Frontier Science Program. JSPS Research Fellowship for Young Scientists and the RIKEN Special Postdoctoral Researchers Program to Y.K. Funding for open access charge: Ministry of Health, Labor and Welfare, Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Yoshihide Hayashizaki for FANTOM3 clones and Dr Gen Matsumoto for the R-miR vector. They are also grateful to the staffs of the Research Resource Center of RIKEN Brain Science Institute for DNA sequencing and cell sorting.
REFERENCES
- 1.Harper PS. Myotonic Dystrophy. 3rd. London: WB Saunders; 2001. [Google Scholar]
- 2.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 3.Fu YH, Friedman DL, Richards S, Pearlman JA, Gibbs RA, Pizzuti A, Ashizawa T, Perryman MB, Scarlato G, Fenwick RG, et al. Decreased expression of myotonin–protein kinase messenger RNA and protein in adult form of myotonic dystrophy. Science. 1993;260:235–238. doi: 10.1126/science.8469976. [DOI] [PubMed] [Google Scholar]
- 4.Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C, Narang M, Barcelo J, O'Hoy K, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 5.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 6.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 7.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl Acad. Sci. USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 10.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 11.Charlet-B N, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 12.Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 13.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 14.Kimura T, Nakamori M, Lueck JD, Pouliquin P, Aoike F, Fujimura H, Dirksen RT, Takahashi MP, Dulhunty AF, Sakoda S. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum. Mol. Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 15.Savkur RS, Philips AV, Cooper TA, Dalton JC, Moseley ML, Ranum LP, Day JW. Insulin receptor splicing alteration in myotonic dystrophy type 2. Am. J. Hum. Genet. 2004;74:1309–1313. doi: 10.1086/421528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 17.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 18.Begemann G, Paricio N, Artero R, Kiss I, Perez-Alonso M, Mlodzik M. muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development. 1997;124:4321–4331. doi: 10.1242/dev.124.21.4321. [DOI] [PubMed] [Google Scholar]
- 19.Artero R, Prokop A, Paricio N, Begemann G, Pueyo I, Mlodzik M, Perez-Alonso M, Baylies MK. The muscleblind gene participates in the organization of Z-bands and epidermal attachments of Drosophila muscles and is regulated by Dmef2. Dev. Biol. 1998;195:131–143. doi: 10.1006/dbio.1997.8833. [DOI] [PubMed] [Google Scholar]
- 20.Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 21.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- 23.Hao M, Akrami K, Wei K, De Diego C, Che N, Ku JH, Tidball J, Graves MC, Shieh PB, Chen F. Muscleblind-like 2 (Mbnl2) -deficient mice as a model for myotonic dystrophy. Dev. Dyn. 2008;237:403–410. doi: 10.1002/dvdy.21428. [DOI] [PubMed] [Google Scholar]
- 24.Ho TH, Charlet-B N, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dansithong W, Paul S, Comai L, Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 26.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 2004;23:406–417. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anant S, Henderson JO, Mukhopadhyay D, Navaratnam N, Kennedy S, Min J, Davidson NO. Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. J. Biol. Chem. 2001;276:47338–47351. doi: 10.1074/jbc.M104911200. [DOI] [PubMed] [Google Scholar]
- 29.Mukhopadhyay D, Houchen CW, Kennedy S, Dieckgraefe BK, Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 30.Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 31.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 33.Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum. Mol. Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 34.Kleopa KA, Barchi RL. Genetic disorders of neuromuscular ion channels. Muscle Nerve. 2002;26:299–325. doi: 10.1002/mus.10164. [DOI] [PubMed] [Google Scholar]
- 35.Pusch M. Myotonia caused by mutations in the muscle chloride channel gene CLCN1. Hum. Mutat. 2002;19:423–434. doi: 10.1002/humu.10063. [DOI] [PubMed] [Google Scholar]
- 36.Berg J, Jiang H, Thornton CA, Cannon SC. Truncated ClC-1 mRNA in myotonic dystrophy exerts a dominant-negative effect on the Cl current. Neurology. 2004;63:2371–2375. doi: 10.1212/01.wnl.0000148482.40683.88. [DOI] [PubMed] [Google Scholar]
- 37.Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orengo JP, Chambon P, Metzger D, Mosier DR, Snipes GJ, Cooper TA. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi N, Sasagawa N, Usuki F, Kino Y, Kawahara H, Sorimachi H, Maeda T, Suzuki K, Ishiura S. Coexpression of the CUG-binding protein reduces DM protein kinase expression in COS cells. J. Biochem. 2001;130:581–587. doi: 10.1093/oxfordjournals.jbchem.a003022. [DOI] [PubMed] [Google Scholar]
- 41.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 42.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 43.Ladd AN, Nguyen NH, Malhotra K, Cooper TA. CELF6, a member of the CELF family of RNA-binding proteins, regulates muscle-specific splicing enhancer-dependent alternative splicing. J. Biol. Chem. 2004;279:17756–17764. doi: 10.1074/jbc.M310687200. [DOI] [PubMed] [Google Scholar]
- 44.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lueck JD, Lungu C, Mankodi A, Osborne RJ, Welle SL, Dirksen RT, Thornton CA. Chloride channelopathy in myotonic dystrophy resulting from loss of posttranscriptional regulation for CLCN1. Am. J. Physiol. Cell Physiol. 2007;292:C1291–C1297. doi: 10.1152/ajpcell.00336.2006. [DOI] [PubMed] [Google Scholar]
- 48.Lueck JD, Mankodi A, Swanson MS, Thornton CA, Dirksen RT. Muscle chloride channel dysfunction in two mouse models of myotonic dystrophy. J. Gen. Physiol. 2007;129:79–94. doi: 10.1085/jgp.200609635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seznec H, Agbulut O, Sergeant N, Savouret C, Ghestem A, Tabti N, Willer JC, Ourth L, Duros C, Brisson E, et al. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum. Mol. Genet. 2001;10:2717–2726. doi: 10.1093/hmg/10.23.2717. [DOI] [PubMed] [Google Scholar]
- 50.O'Cochlain DF, Perez-Terzic C, Reyes S, Kane GC, Behfar A, Hodgson DM, Strommen JA, Liu XK, van den Broek W, Wansink DG, et al. Transgenic overexpression of human DMPK accumulates into hypertrophic cardiomyopathy, myotonic myopathy and hypotension traits of myotonic dystrophy. Hum. Mol. Genet. 2004;13:2505–2518. doi: 10.1093/hmg/ddh266. [DOI] [PubMed] [Google Scholar]
- 51.Mahadevan MS, Yadava RS, Yu Q, Balijepalli S, Frenzel-McCardell CD, Bourne TD, Phillips LH. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat. Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen W, Wang Y, Abe Y, Cheney L, Udd B, Li YP. Haploinsuffciency for Znf9 in Znf9+/- mice is associated with multiorgan abnormalities resembling myotonic dystrophy. J. Mol. Biol. 2007;368:8–17. doi: 10.1016/j.jmb.2007.01.088. [DOI] [PubMed] [Google Scholar]
- 53.Yang Y, Mahaffey CL, Bérubé N, Maddatu TP, Cox GA, Frankel WN. Complex seizure disorder caused by Brunol4 deficiency in mice. PLoS Genet. 2007;3:e124. doi: 10.1371/journal.pgen.0030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hino S, Kondo S, Sekiya H, Saito A, Kanemoto S, Murakami T, Chihara K, Aoki Y, Nakamori M, Takahashi MP, et al. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum. Mol. Genet. 2007;16:2834–2843. doi: 10.1093/hmg/ddm239. [DOI] [PubMed] [Google Scholar]
- 55.Berget SM. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2314. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 56.Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 57.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.