Abstract
Motif overrepresentation analysis of proximal promoters is a common approach to characterize the regulatory properties of co-expressed sets of genes. Here we show that these approaches perform well on mammalian CpG-depleted promoter sets that regulate expression in terminally differentiated tissues such as liver and heart. In contrast, CpG-rich promoters show very little overrepresentation signal, even when associated with genes that display highly constrained spatiotemporal expression. For instance, while ∼50% of heart specific genes possess CpG-rich promoters we find that the frequently observed enrichment of MEF2-binding sites upstream of heart-specific genes is solely due to contributions from CpG-depleted promoters. Similar results are obtained for all sets of tissue-specific genes indicating that CpG-rich and CpG-depleted promoters differ fundamentally in their distribution of regulatory inputs around the transcription start site. In order not to dilute the respective transcription factor binding signals, the two promoter types should thus be treated as separate sets in any motif overrepresentation analysis.
INTRODUCTION
How cells establish and maintain their transcriptome remains one of the fundamental questions in cell biology. Transcription factors together with DNA-methylation, histone modifications and micro RNAs are the key components of the regulatory repertoire of the cell. Detection of transcription factor (TF)-binding site motifs common to a set of co-expressed genes is a central component of the in silico characterization of transcriptional regulation and transcriptional regulatory networks. In the absence of comprehensive genome-wide experimental TF-binding data, the standard bioinformatics procedure starts with the extraction of putative promoter sequences for the co-expressed genes. The sequences are sometimes further refined by phylogenetic footprinting (1,2). Subsequently, algorithms are applied that either try to find new DNA sequence motifs overrepresented in the promoters (3,4), or that search the sequence space for occurrences of known TF-binding motifs (5). The latter approach relies on databases like JASPAR (6) and Transfac (7) to provide motif descriptions for the TFs involved in the regulation of the genes of interest. With the ever growing number of characterized binding motifs such approaches are becoming increasingly popular. For a number of applications, overrepresentation calculations based on the annotation of discrete-binding sites (1) are being complemented with affinity based approaches, which avoid the artificial separation between binding sites and non-binding sites in the prediction of TF target promoters but instead assign continuous binding probabilities to all sites in the sequence based on thermodynamic considerations (2,8,9). Such affinity based methods were shown to emulate the in vivo TF-binding behavior more quantitatively than hit-based approaches (10,11). When applied to sets of tissue-specific genes overrepresentation analyses and affinity based approaches were able to identifying key regulators for a limited number of gene sets derived from e.g. muscle and liver while they largely fail to produce meaningful results for many other tissues such as lung and brain.
To understand the source of the underlying difficulties for enrichment testing more deeply, we need to look at what is known about promoters and their binding site content. The classical textbook depiction of a eukaryotic proximal promoter shows the core promoter flanked by tissue-specific regulatory inputs. The eukaryotic RNA polymerase II core promoter thereby typically includes several sequence elements such as an initiator signal coinciding with the transcription start site (TSS), a TATA box and two or three other motifs such as a CAAT or GC-box [for a review of these elements, see e.g. (12,13)]. Alternatively, the whole promoter can either be partially or completely overlapped by a CpG island. In line with this model, Saxonov (14) made the striking observation that the CpG content of vertebrate promoters shows a distinct bimodal distribution. Using the central dip in this distribution as demarcation line about half of the promoters can be classified as having high CpG content (HCPs) while the others are considered to have low CpG content (LCPs).
Many pioneering vertebrate enrichment analyses used promoters of genes expressed at a high level in a terminally differentiated tissue. Those promoters were typically of the LCP class and had a landmark TATA box about 30-bp upstream of TSSs (15). On the other hand, ubiquitously expressed (‘housekeeping’) genes and developmental regulators, typically lack a TATA box but overlap with a CpG island thus falling in the HCP class. This broad dichotomy is statistically very convincing, but by no means perfect. More recent genome-wide studies revealed that a TATA box is present in only a minority of tissue-specific promoters (16,17) and together with other elements can occur also in CpG-rich promoters (18). In accordance with this, many tissue-specific genes from brain (19) and testis (20) do not have TATA-box containing promoters characteristic of genes expressed specifically in liver or muscle.
In this article we show that, while most sets of tissue-specific genes contain a considerable percentage of CpG-rich promoters, the observable tissue-specific motif overrepresentation information within proximal promoters is coming almost exclusively from CpG-depleted promoters. In contrast, CpG-rich promoters turn out to be of little or no utility for this type of analysis, even when the genes driven by them have clear tissue preference. We show that an a priori separation of the two promoter classes (LCP and HCP) gives a stronger, more robust, and spatially constrained binding affinity signal in the CpG-depleted promoters, and therefore recommend this as a general approach for the analysis of motif enrichment in co-regulated gene sets.
MATERIALS AND METHODS
Expression data and tissue-specificity
The expression of a given gene in one of the 15 mouse tissues (Figure 1) is determined by analyzing corresponding EST clusters from the GeneNest database (21), which includes the annotation of the originating tissue for each EST. To detect EST clusters whose distribution of ESTs derived from various tissues differ significantly from the expected distribution we applied a χ2-test. All clusters with a P-value <10−3 were subjected to a binomial test such that we obtain a P-value describing the likelihood of observing a given number of ESTs from a given tissue in an EST cluster of given size. These EST cluster P-values reflect the degree of over-expression of a given gene in a given tissue and were successfully used previously to predict tissue-specific expression of genes (9,21). Here we use the P-values to rank all genes with respect to a given tissue.
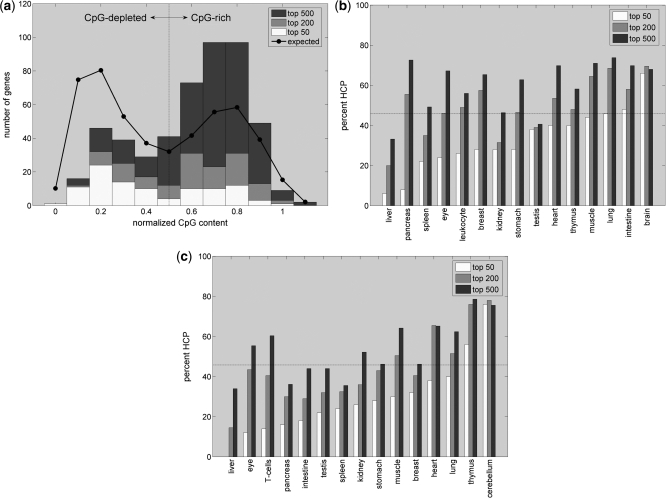
Figure 1.
(a) Bimodal CpG distribution across the promoters of the 50, 200 or 500 most heart-specific genes. The fraction of HCPs in the 200 and 500 gene sets is larger than expected based on the CpG content across all mouse promoters (the black line indicates the expected CpG distribution for random gene sets of size 500). (b) and (c) show the contribution of HCP genes to other tissue sets of indicated size based on EST or microarray data, respectively. The contribution of HCP promoters usually increases with gene set size and is >20% even for most sets of size 50. Sets with more than 200 genes often contain an excess of HCP genes compared to the fraction of 46% of HCPs across all Ensembl promoters (indicated by horizontal lines).
For the analysis of microarray data we refer to the GNF data set from Su et al. (22). After taking the mean expression intensity across replicate microarrays we compute a Z-score for each gene across all tissues. These Z-scores are subsequently used to rank all genes for a given tissue.
Sequence data and promoter CpG content
All mouse promoter sequences as well as the annotation of the corresponding TSSs for 28 205 mouse genes are taken from the Ensembl database version 46 (23). The normalized CpG content of a given promoter measures the ratio of observed over expected CpGs in the promoter and is computed using the following equation:
| 1 |
where all Gs and Cs in the region ranging from −500 to +500 bp around the TSS are being considered. In general, a normalized CpG content <1 indicates that the promoter has less CpGs than expected based on its overall GC content. Here, based on the bimodality of the normalized CpG content in vertebrates, promoters with normalized CpG content <0.5 are classified as CpG-depleted (LCP) while promoters with CpG ≥ 0.5 are considered CpG-rich (HCP). To avoid a strong influence of only predicted Ensembl genes with potentially random promoter composition we restrict the enrichment analysis to those 18 938 mouse genes for which unigene EST clusters have been identified.
Affinity predictions and hit based-binding site annotation
We rely on the collection of 588 vertebrate position frequency matrices (PFM) provided by JASPAR (6) and the Transfac database version 11.1 (7) to describe the binding motif of a given TF. PFMs report the frequency with which a certain base occurs at a given position in alignments of known binding sites of a given TF. To predict the binding strength of a given TF to a promoter sequences we utilize the TRAP method (11). In contrast to motif matching algorithms which make a binary distinction between binding sites and non-binding sites, TRAP avoids this artificial separation by instead computing the occupancy of a TF to each site in the sequence using equation:
| 2 |
where ΔEi(λ) is the energy difference or mismatch energy—scaled by the parameter λ—between the binding energy of the factor to site i and the lowest binding energy possible with the factor bound to its consensus site. The second matrix dependent parameter R0 sets the binding energy of the factor to the consensus site as well as the TF concentration. The nucleotide dependent mismatch energies for each site in the promoter sequence are computed as follows:
| 3 |
where vi,max is the frequency of the consensus base at position i in the PFM and vi,α is the frequency of the observed base at position i in the matrix. Eventually, TRAP obtains the expected number 〈N〉 of TFs bound to the sequence window by summing over the individual probabilities from all sites in the window with length L:
| 4 |
Importantly, aside from avoiding an artificial discretization between bound and unbound states 〈N〉 also allows for a more natural ranking of target promoter sequences with respect to a given TF then do discrete hit counts. As input TRAP requires for each TF a PFM suitable for computing the mismatch energies ΔE, a DNA sequence of interest and the setting of the two parameters λ and R0. As was derived previously, λ is set to a value of 0.7 for all matrices and R0 is derived for each matrix individually using the formula:
| 5 |
where W is the number of informative positions in the TF matrix, which are defined as every column in the PFM with information content exceeding 0.1 bits. The information content of position i in the matrix is computed as the Kullback–Leibler entropy given by:
| 6 |
where νi,α is the frequency of base α at position i. Matrix positions which fall below the entropy cutoff do not contribute to the mismatch energy in equation [Equation (3)].
Discrete binding sites for a given TF are being annotated using a standard approach of shifting a position specific score matrix derived from the PFMs over a promoter sequence. Sites exceeding a score threshold that balances the expected number of false positive hits with the expected number of false negatives are annotated (24).
Enrichment testing using PASTAA and Z-scores
For the enrichment analysis based on continuous TF-binding affinities returned by TRAP we utilize the recently published PASTAA method (9). PASTAA starts by ranking all mouse genes promoters according to their predicted affinity for a given TF. At the same time the genes are also ranked according to their association with a given tissue measured by the EST enrichment P-values. After applying a cutoff to the ranked affinity and tissue lists a hypergeometric test is used to determine the significance of the overlap between the top target genes of the TF and the top ranking genes of the tissue. Cutoffs on the two lists are thereby chosen iteratively in such a way that the obtained hypergeometric P-values are minimized (see Supplementary Figure S1 for details). The negative logarithms of these optimal P-values are subsequently used as affinity enrichment scores.
To test for an enrichment of discrete TF-binding sites obtained from the balanced cutoff method (24) within promoters of tissue-specific genes we utilize a test statistic published previously by Sui et al. (1). Hereby, for each factor and tissue the following Z-score is computed:
| 7 |
where x is the number of binding sites residing in the promoters of the genes assigned to a given tissue, μ is the average number of binding sites residing in the promoters of the background set (here all genes not assigned to the tissue) and σ is the variance of the number annotated hits over all promoters in the background set. For a given tissue the five PFMs with largest Z-score are reported.
Shifting window approach to detect promoter regions with highest TF-binding affinity
For a given TF to assess a preference in the location of maximal affinity with respect to the TSS we shift 200-bp windows in consecutive steps of 100 bp across the promoter regions ranging from −1-kb upstream to +1-kb downstream of the TSSs. For each window start position we compute the binding affinity for the TF to the 200-bp sequence in the window. To detect a preference in the binding location among promoters of tissue-specific genes we retain for each gene the location of the window with highest affinity (Supplementary Figure S2). Subsequently we rank these windows based on their affinity and report the location of the top 50 windows among the 500 genes assigned to a given tissue.
Similarly, to find the location of strongest TF-binding affinity enrichment among tissue-specific genes we shift 200-bp windows across the promoters of all 18 938 genes. PASTAA is then applied to evaluate the significance of the overlap between 500 tissue-specific genes and the target genes predicted based on the affinities from the 200-bp windows starting at a given position.
Matching LCP and HCP genes
In order to avoid a systematic bias in the enrichment analysis towards genes with CpG-depleted promoters, which on average tend to have more significant tissue enrichment, we first select for each tissue the set of 500 CpG-rich genes with highest specificity for the tissue. We then construct a set of 500 genes with CpG-depleted promoters by selecting for each gene in the HCP set the LCP gene with the most closely matching but not more significant tissue P-value (Supplementary Figure S3). The PASTAA enrichment analysis is then performed on both the HCP and matched LCP set separately with background sets consisting of all the other HCP genes (10 996) or LCP genes (∼6942) with less significant tissue enrichment, respectively. It has to be noted that the above procedure of constraining the LCP gene sets causes a considerable reduction in observed enrichment scores when compared to LCP sets constructed by simply taking the 500 most specific LCP genes for each tissue. Alternatively, we therefore also performed enrichment testing on either the top 500 genes for each tissue irrespective of CpG content or the 500 most tissue-specific LCP genes. In the former case, all other mouse genes (18438) are used as background set, in the latter case all other 6942 LCP genes.
Lastly, tissue gene sets may also be defined using a cutoff on the tissue specificity P-values or Z-scores. This procedure offers the advantage that tissue sets do not contain genes with limited or no real specificity for the respective tissue. Enrichment scores can thus be expected to be overall stronger. On the other hand, this procedure results in LCP and HCP sets of sometimes very different size thereby making the subsequently obtained TF enrichment scores less well comparable. To ensure that the inclusion of less tissue specific genes into the gene sets does not result in enrichment artifacts we performed enrichment testing also on tissue specific LCP and HCP gene sets defined by a cutoff of 10−3 on the EST P-values. Results of this analysis closely resemble those obtained from the tissue gene sets of static size 500 and are shown in the Supplementary Data.
An overview of the different ways to define tissue sets and the corresponding enrichment analyses is shown in Supplementary Figure S4.
RESULTS
Typical sets of tissue-specific genes contain a considerable fraction of genes with CpG-rich promoters
The construction of a set of co-expressed genes with tissue-specific expression pattern is a prerequisite for any motif overrepresentation analysis aimed at finding TFs involved in the regulation of the genes. It has become textbook knowledge that promoters of tissue-specific genes tend to be CpG-depleted while housekeeping genes with broad expression patterns have CpG-rich promoters (25). However, when background gene sets are not chosen carefully and controlled for GC content, TFs with GC-rich binding motifs such as SP1 (consensus sequence GGGGCGGGGT) tend to be found as the most overrepresented TF-binding motifs (9) indicating a considerable contributions from genes with CpG-rich promoters to sets of tissue-specific genes. We therefore first analyze to what extent the assumption of tissue-specific genes having only CpG-depleted promoters holds true for comprehensive sets of tissue-specific genes derived from either EST or microarray data. Such gene sets, often containing hundreds of genes, are frequently used in overrepresentation analysis aimed at identifying common regulating TFs (26–31).
To this end we compute for each promoter the CpG content given by the ratio between the numbers of observed versus expected CpG dinucleotides around the TSS (see ‘Materials and Methods’ section). In mouse, the resulting bimodal CpG distribution across all promoters dips at roughly 0.5 (see black line in Figure 1a). We thus set the border for separating LCP versus HCP to this value resulting in about 46% of all Ensembl mouse promoters falling into the HCP category.
For tissue-specific genes we find that the percentage of LCP and HCP promoters depends strongly on the tissue under consideration (Figure 1b). While promoters of liver-specific genes are strongly CpG-depleted, 70–80% of promoters from genes expressed specifically in brain are CpG-rich. Results are hereby comparable between tissue gene sets derived from microarray (22) and EST data (21). As expected, over all tissues there is a clear trend for the most tissue-specific genes to fall into the class of CpG-depleted promoters. However, with the exception of liver, even when restricting the analysis to only the 50 most specific genes in each tissue a considerable proportion of promoters are CpG-rich. As exemplified for differently sized sets of heart-specific genes (Figure 1a), larger gene sets even tend to contain an excess of CpG-rich promoters compared to what is expected based on the CpG distribution across all 28 205 Ensembl mouse promoters (Figure 1b and c). We conclude that gene sets derived based on tissue-specificity typically contain a mixture of genes belonging to the LCP and HCP categories and are by no means only composed of genes with CpG-depleted promoters.
General TFs associate with both, HCP and LCP genes, across all tissues
Having established that most tissue-specific gene sets contain a considerable percentage of genes with CpG-rich promoters, we next investigate whether binding signals for general TFs show a tendency to occur in CpG-rich or CpG-depleted promoters and whether such a preference is tissue-specific. To this end, we compute binding affinities for 200-bp sequence windows that are shifted in steps of 100 bp along all promoters. In order to assess a possible preference of a factor for high or low CpG promoters we split the tissue gene sets into two groups. The first group contains for each tissue the 500 most specifically expressed HCP genes. The second group contains the 500 LCP genes whose expression P-values match most closely those of the genes put into the HCP group but with the restriction of being less tissue-specific than the corresponding HCP gene (Supplementary Figure S3). Subsequently, we report for each factor the locations of windows with highest affinity in each gene set (see ‘Materials and Methods’ section and Figure S2). As shown in Supplementary Figures S5 and S6, we find a weak trend for the general TFs and core promoter motifs to occur preferentially in CpG-rich promoters. The exceptions are YY1, a general TF implicated in pinpointing the transcription start position, whose high affinity sites are found exclusively within 100-bp downstream of CpG-rich promoters, and the TATA box motif which occurs more frequently upstream of CpG-depleted rather than CpG-rich promoters. As might be expected, when performing enrichment testing (see ‘Materials and Methods’ section) we find none of these general motifs to be strongly overrepresented in any of the tissue-specific gene sets (see Supplementary Figure S8 for results from the CAAT box).
Location of sites with maximal affinity for HNF1 and MEF2 demonstrates strong difference in regulatory input to CpG-rich and CpG-depleted promoters
We now turn to TFs with tissue-specific activity and ask whether high affinity regions for such factors occur preferentially in the CpG-rich or CpG-depleted promoters of genes with tissue-specific expression. Two of the best described associations between sets of tissue-specific genes and TFs are that of hepatocyte nuclear factor, HNF1, with sets of liver specific genes, and that of the muscle enhancer factor, MEF2, with sets of muscle and heart specific genes (1,9,28,30,32). We therefore chose these two tissues and factors as a detailed test case before investigating the situation for a wider range of tissues and TFs. To identify regions of high affinity for HNF1 and MEF2 we again report the location of sequence windows with highest affinity with respect to the TSSs (see ‘Materials and Methods’ section).
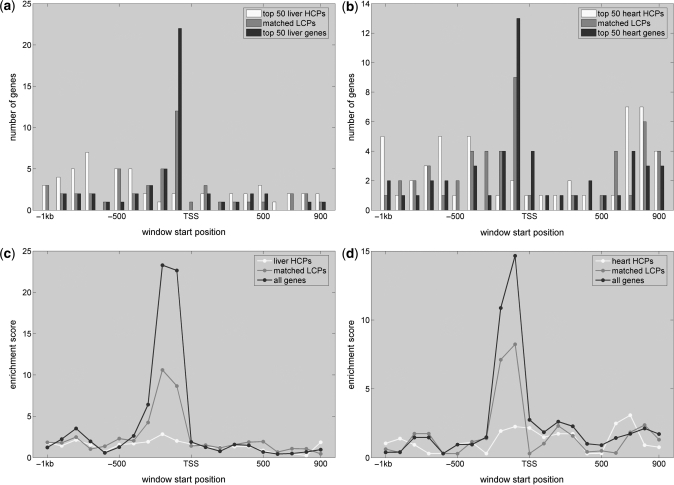
As shown in Figure 2a and b, high affinity windows of HNF1 and MEF2 accumulate in proximal promoters of the 500 most liver and heart specific genes, between 0 and 200 bp upstream of the corresponding TSSs. Evaluating separately the set of 500 most tissue-specific HCP genes and the set of 500 LCP genes whose expression P-values match most closely those of the genes in the HCP group (Figure S3) reveals that high affinity windows accumulate only near the TSS of CpG-depleted promoters (Figure 2a and b) while the strongest affinities observed in HCP genes are scattered randomly across the promoters (for the situation across the other tissues see Supplementary Figure S7).
Figure 2.
Enrichment of high affinity sites for HNF1 and MEF2 near the TSS of liver- and muscle-specific genes with CpG-depleted promoters. (a) and (b) Sequence windows with highest affinity are preferentially located directly upstream of the TSS (red bars). This signal is due to sites in the CpG-depleted promoters as no preferential binding pattern is observed when restricting the analysis to CpG-rich promoters (compare blue and yellow bars for results from CpG-rich and CpG-depleted genes, respectively). (c) and (d) show the corresponding PASTAA enrichment scores (see ‘Materials and Methods’ section) for each sequence window as well as the separate sets of high and low CpG promoters.
In order to quantify to what extent the observed accumulation of high affinity sites is restricted to only the genes specifically expressed in the corresponding tissue, we perform enrichment testing as described in ‘Materials and Methods’ section. As shown in Figure 2c and d when analyzing the 500 most tissue-specific genes we find a clear peak in TF target gene enrichment only when performing the analysis for the sequence windows directly upstream of the TSS. The accumulation of high affinity sites for HNF1 and MEF2 near the TSS is thus restricted to the promoters of genes from the corresponding tissues. Performing the enrichment test separately on the 500 HCP and 500 P-value matched LCP genes shows that the observed associations between HNF1 and liver, and MEF2 and heart, are almost exclusively due to the contributions from CpG-depleted promoters. Similar enrichment scores for HNF1 and MEF2 are obtained also only for CpG-depleted promoters from kidney and muscle, respectively, but not for promoters from other tissues (see Supplementary Figure S8 for the target gene enrichment in other tissues).
Enriched motifs reside in CpG-depleted promoters across all tissue-specific gene sets
We extend the above analysis from liver and heart to all 15 tissues from Figure 1 and first analyze where across the tissue-specific promoters we find the strongest enrichment for high affinity sites from any of the 588 vertebrate TFs from Transfac (7) and JASPAR (6). Performing the enrichment testing on the 500 most specific genes of each tissue, irrespective of the CpG content of their promoters, we find a very strong peak in TF affinity enrichment within 200-bp upstream of the TSS across all tissues except lung and breast (see Supplementary Figure S9a). The TFs corresponding to these strongest enrichments match well to the factors that have previously been implied as potential regulators for these tissues [Table 1; (33–40)]. Following the procedure of separating tissue-specific genes into HCP and matched LCP groups, we next assessed whether the observed enrichment stems from high affinity sites in CpG-rich or depleted promoters. As shown in Figure 3a), when performing the enrichment analysis on the HCP groups we find no clear peak in affinity enrichment near the TSS for any of the tissues except testis. Also, for most tissues the enrichment analysis returns general binding motifs such as TATA and CAAT as the most strongly associated motifs. In contrast, very strong enrichment directly upstream of the TSS is observed when performing the analysis for the groups of 500 P-value matched LCP genes (Figure 3b). In fact, for most tissues a better enrichment is obtained when performing the analysis on all CpG-depleted promoters alone rather than on LCP and HCP genes combined (Table 1 and Supplementary Figure S9b). Together these findings indicate a lack of tissue-specific binding signals in the proximal regions of HCP promoters and a very strong accumulation of binding signals right upstream of the TSS of LCP genes. An interesting exception is observed for the neuron-restrictive silencing factor, NRSF, whose binding signals are enriched much more strongly in brain specific genes of the HCP (enrichment P-value 8.3 × 10−7) rather than the LCP class (P-value 7.8 × 10−2, Table 1).
Table 1.
Top ranking matrices for 200 bp proximal promoters from LCP, HCP and joint gene sets
| ALL | LCP | HCP | ||||
|---|---|---|---|---|---|---|
| Brain | EGR_Q6 | 5.90 | CHCH_01 | 11.32 | NRSF_01 | 6.08 |
| Eye | GATA1_03 | 16.62 | GATA1_03 | 16.93 | LRF_Q2 | 8.44 |
| Liver | HNF1_01 | 30.81 | HNF4_Q6_01 | 38.27 | NFY_Q6_01 | 8.00 |
| Kidney | HNF1_01 | 30.27 | HNF1_01 | 24.02 | NFY_Q6_01 | 5.18 |
| Intestine | HNF4_Q6_01 | 17.02 | HNF4_Q6_01 | 16.87 | MTATA_B | 6.56 |
| Stomach | TATA_01 | 11.17 | LMO2COM_02 | 9.18 | TATA_C | 6.81 |
| Pancreas | TATA_01 | 9.71 | PTF1BETA_Q6 | 9.18 | PEA3_Q6 | 6.93 |
| Muscle | SRF_C | 11.58 | MEF2_Q6_01 | 10.74 | SRF_C | 6.26 |
| Heart | MEF2_Q6_01 | 15.42 | MEF2_Q6_01 | 13.77 | SRF_Q5_02 | 5.97 |
| Leukocyte | NFKAPPAB_01 | 9.99 | ELF1_Q6 | 14.29 | OCT1_B | 5.13 |
| Spleen | ICSBP_Q6 | 16.02 | ICSBP_Q6 | 18.65 | STAT1_01 | 6.62 |
| Thymus | ETS_Q6 | 12.20 | ETS_Q6 | 17.34 | ZF5_01 | 4.73 |
| Lung | NGFIC_01 | 4.90 | ZF5_01 | 7.55 | CAAT_01 | 4.51 |
| Breast | PEBP_Q6 | 6.11 | SMAD4_Q6 | 7.09 | IK2_01 | 3.87 |
| Testis | CREBP1CJUN_01 | 11.26 | CREBP1CJUN_01 | 24.12 | VMYB_02 | 12.06 |
For each tissue the top associated matrix and the corresponding enrichment score from PASTAA is shown (see ‘Materials and Methods’ section) depending on whether the analysis was performed on all 500 tissue-specific promoters indiscriminate of CpG content, on the 500 tissue-specific genes with CpG-rich or on the 500 tissue-specific genes with CpG-depleted promoters. Matrices in bold correspond to well documented TF-tissue associations and are preferentially discovered for sets of LCP genes.
Figure 3.
TF-binding affinity enrichment near the TSS of tissue-specific genes with either CpG-rich (a) or CpG-depleted promoters (b). The height of each bar corresponds to the PASTAA enrichment score of the most significant association that is found for the corresponding tissue. With the exception of testis, no significant enrichment signals are detected when analyzing the tissue sets containing the 500 most specific CpG-rich promoters. In contrast, enrichment peaks strongly for tissue-specific sets of 500 P-value matched LCP genes when computing TF-binding affinities for 200-bp windows directly upstream of the TSSs.
TFs associate preferentially with CpG-depleted promoters
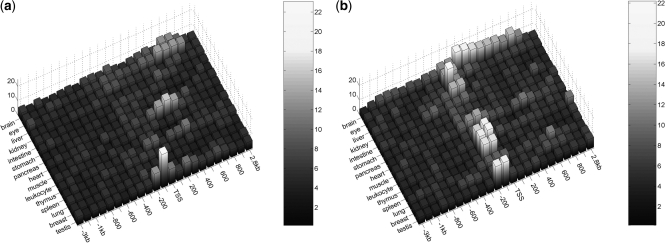
Having tested the enrichment across all tissues, we now switch from the tissue-centric to a TF-centric view and assess with which promoter class each of the 588 vertebrate TF matrices associates preferentially. To this end, we again perform enrichment testing on the 15 tissue sets, this time reporting the promoter location of the most significant association and the average CpG content of its assumed target genes for each of the 588 vertebrate matrices. As shown in Figure 4, about one third of all matrices show the strongest association with any one of the 15 tissues within 200-bp upstream of the TSS. For the vast majority of factors the average CpG content of the target genes is thereby smaller than 0.5, again indicating that high affinity peaks reside preferentially within CpG-depleted promoters. A similar picture is observed not only for the sequence window at the TSS but across the whole promoter region ranging from −3 kb to +3 kb. This finding also strongly underlines a fundamental difference in the regulatory mechanisms of CpG-rich and CpG-depleted promoters of tissue-specific genes.
Figure 4.
TF targets have low average CpG content. Yellow and blue bars indicate the number of matrices whose target genes have an average CpG content ≥0.5 and <0.5, respectively. Red bars indicate the overall propensity to find the most significant association between matrices and any of the tissues at a particular window position. About a third of all matrices show the strongest association with any of the tissues when computing the binding affinities for the window ranging from −200 to 0 bp upstream of the TSS, indicating a strong location preference for the proximal promoter (see red bars). The target genes of the vast majority of matrices thereby have an average CpG content <0.5 (compare yellow and blue bars).
General implications for enrichment testing
Several approaches for detecting overrepresented motifs in promoter sets utilize the annotation of discrete TF-binding sites rather than continuous binding affinities (1,41). To evaluate for such methods the effect of having HCPs included in sets of tissue-specific promoters, we assess the top regulators for the tissues liver, kidney, muscle and eye, as suggested by a Z-score statistic applied to discrete binding site predictions. A similar statistic was used previously to determine an enrichment of discrete binding sites in sets of co-regulated genes (1,9). As shown in Table 2 and in accordance with previous studies, when analyzing the top 500 kidney- and liver-specific genes the approach recovers HNF1 and HNF4 as the top associated regulators. In contrast, for the 500 most muscle- and eye-specific genes we find GC-rich motifs including SP1 and WT1 as top ranking. The situation worsens when performing the enrichment analysis on the 500 most tissue-specific HCP genes with the background gene set consisting of all other 10 996 HCP genes. In this case, GC-rich motifs are found as top ranking in all tissues (indicating that the tissue-specific HCP genes possess particularly CpG-rich promoters). In contrast, when using the 500 P-value matched LCP genes (together with the remaining ∼6942 LCP genes as background) we find well characterized TF–tissue associations for all tissues including MEF2 for muscle and cone rod specific TF CRX for eye. At the same time, general TFs such as SP1 are not found among the top ranking factors in either tissue. This finding indicates that an incorporation of CpG-rich promoters in sets of co-regulated genes hampers not only affinity-based enrichment testing approaches but also methods based on discrete binding site predictions.
Table 2.
Top ranking matrices returned by a hit based z-score statistic
| ALL | HCP | matched LCP |
|---|---|---|
| Kidney | ||
| HNF1_01 | SP1_Q6 | HNF4_01 |
| HNF4_Q6_01 | SP1_Q4_01 | HNF4_Q6_01 |
| HNF4_01 | SP1_Q6_01 | P53_01 |
| HNF1_C | ZF5_01 | HNF1_01 |
| SP1_Q4_01 | GC_01 | COUP_01 |
| Liver | ||
| HNF4_Q6_01 | SP1_Q4_01 | HNF4_Q6_01 |
| HNF4_01 | SP1_Q6 | HNF4_01 |
| DR1_Q3 | GC_01 | HNF1_C |
| HNF4_01_B | SP1_Q6_01 | COUP_01 |
| COUP_01 | SP1_Q2_01 | DR1_Q3 |
| Muscle | ||
| GC_01 | ZF5_01 | MEF2_02 |
| SP1_Q6 | AP2_Q6_01 | MEF2_03 |
| SP1_Q4_01 | SP1_Q6 | SRF_Q5_02 |
| ZF5_01 | WT1_Q6 | DR1_Q3 |
| WT1_Q6 | E2F_Q2 | SRF_Q6 |
| Eye | ||
| AP2_Q6_01 | AP2_Q6_01 | P53_01 |
| WT1_Q6 | WT1_Q6 | MZF1_02 |
| SP1_Q6 | ZF5_01 | PAX4_04 |
| SP1_Q4_01 | SP1_Q6 | GATA1_03 |
| SP1_Q2_01 | SP1_Q4_01 | CRX_Q4 |
Experimentally verified TF–tissue associations (indicated in bold) are found in liver and kidney when analyzing the top 500 genes for each tissue. While no verified associations are detected when performing the analysis on only HCP genes (all discovered motifs are GC rich) verified associations are found for all tissue sets when analyzing P-value matched LCP genes.
DISCUSSION
Traditionally, vertebrate genes are being divided into two distinct classes based on the CpG content of their promoters. While tissue-specific genes tend to possess CpG-depleted promoters, housekeeping genes (broadly expressed) usually have CpG-rich promoters. However, as shown here, this picture is less clear-cut than generally assumed with many tissue-specific genes falling into the HCP rather than the LCP class. We find that the amount of tissue-specific regulatory TF-binding signals around the TSS is thereby vastly different for LCP and HCP promoters. Consequently, any promoter content analysis assessing the overrepresentation of TF motifs should start by separate the two promoter classes.
In accordance with this paradigm, for set of tissue-specific genes with CpG-depleted promoters we find many well characterized TF-tissue associations such as hepatocyte nuclear factor (HNF1) with liver, and pancreas specific TF (PTF1) with pancreas. Successful predictions thereby stem from cis-regulatory elements located usually within only 200-bp upstream of the TSS. Analyzing HCP promoters proved to be much less successful. A notable exception is the association of neuron-restrictive silencing factor, NRSF, with brain specific genes of the HCP class. Interestingly, this association is not detected in the corresponding LCP class and also appears less significant when combining CpG-rich and CpG-depleted promoters indicating that NRSF acts preferentially on the transcription of CpG-rich promoters. In general, while the overall enrichment scores across all HCP categories are weak, motifs overrepresentation analysis of the HCP genes revealed an accumulation of core promoter elements in tissue-specific genes with CpG-rich promoters. For instance, within 200-bp upstream of the TSS we found NFY as the most enriched motif in liver and muscle, TATA in intestine and stomach and the CAAT box in lung. While these motifs represent the very opposite to tissue-specific signals, they demonstrate a general enrichment of such core promoter elements in CpG-rich promoters of tissue-specific genes. This suggests that such promoters might tend to be activated differently from CpG-rich promoters of broadly expressed genes.
A plausible explanation for the weak enrichment scores across HCP genes is that regulatory elements driving expression in these contexts are more likely to be outside of ‘conventional’ promoter regions, and a typical analysis in which a fixed sequence range around the TSS is analyzed either misses them or drowns them in a too large sequence space (42). An increasing amount of evidence indicates that many genes have key regulatory elements at large distances in both directions from the core promoter (31,43)—too large, in fact, for any approach that takes a fixed amount of upstream and/or downstream sequence to work. For these, the only hope for finding regulatory elements might come in the form of exhaustive genome-wide experimental TF-binding data from ChIP-seq and related technologies combined with e.g. chromatin capture assays (44).
Another problem with enrichment testing in proximal promoters might be caused by the presence of multiple alternative promoters as expression data often does not reveal which of them is used in a given context (45). Similarly, in a large subset of individual vertebrate core promoters, typically those overlapping a CpG island, TSS positions are not unique but rather broadly distributed (17). Therefore, taking a fixed amount of sequence around any given TSS is likely to result in a functionally heterogeneous set, on which the interpretation of TF content and their position relative to TSS becomes ambiguous. However, since the typical CpG-rich promoters have TSS positions spread over a span of only 50–200 bp, this imprecision cannot by itself account for the lack of tissue-specific signals reported here. In the worst case, it would result in a slightly weaker association due to the ambiguous determination of TSS position, and not the almost complete absence of it that is observed.
CpG islands are relatively easy to find in genomes of tetrapod vertebrates; in many fish genomes, however, they are much smaller and more difficult to detect, although the main distinction between CpG-depleted promoters with well defined TSSs and CpG-rich promoters with ambiguous start positions still holds (A.C. Previti and B. Lenhard, unpublished data). Of invertebrates, Drosophila species were shown to have multiple types of core promoters (46) that are associated with different responsiveness to long-range enhancers and different level of tissue-specificity (47). It remains to be seen if genome compaction has led to more of the promoters having the majority of their regulatory elements close to the TSS. Other model invertebrates were also shown to have a distinct subset of genes responsive to long range enhancers (48). It is tempting to conclude that the distinction between promoters responding to proximal and distal signals could be found in most metazoan genomes.
The specific enrichment of regulatory sequence elements in only CpG-depleted promoters points to the potential involvement of alternative mechanisms in the regulation of tissue-specific expression of HCP genes. These mechanisms likely include DNA methylation and distinct histone modifications. With the recent advent of technologies such as ChIP-seq new large-scale data will become available soon that will allow to associate specific histone modifications with specific expression patterns across a variety of different tissues.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biosapiens project (contract LHSG-CT-2003-503265); German National Genome Research Network (NGFN) and by the SFB project 618. Funding for open access charge: Max Planck Institute for Molecular Genetics.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang LW, Nagarajan R, Magee JA, Milbrandt J, Stormo GD. A systematic model to predict transcriptional regulatory mechanisms based on overrepresentation of transcription factor binding profiles. Genome Res. 2006;16:405–413. doi: 10.1101/gr.4303406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halperin Y, Linhart C, Ulitsky I, Shamir R. Allegro: analyzing expression and sequence in concert to discover regulatory programs. Nucleic Acids Res. 2009;37:1566. doi: 10.1093/nar/gkn1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conlon E. Integrating regulatory motif discovery and genome-wide expression analysis. Proc. Natl Acad. Sci. USA. 2003;100:3339–3344. doi: 10.1073/pnas.0630591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guhathakurta D. Computational identification of transcriptional regulatory elements in DNA sequence. Nucleic Acids Res. 2006;34:3585. doi: 10.1093/nar/gkl372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roider HG, Manke T, O'Keeffe S, Vingron M, Haas SA. PASTAA: identifying transcription factors associated with sets of co-regulated genes. Bioinformatics. 2009;25:435–442. doi: 10.1093/bioinformatics/btn627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roider HG, Kanhere A, Manke T, Vingron M. Predicting transcription factor affinities to DNA from a biophysical model. Bioinformatics. 2007;23:134–141. doi: 10.1093/bioinformatics/btl565. [DOI] [PubMed] [Google Scholar]
- 12.Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter – the gateway to transcription. Curr. Opin. Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 14.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita R, Suzuki Y, Sugano S, Nakai K. Genome-wide analysis reveals strong correlation between CpG islands with nearby transcription start sites of genes and their tissue specificity. Gene. 2005;350:129–136. doi: 10.1016/j.gene.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 18.Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ., Jr Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valen E, Pascarella G, Chalk A, Maeda N, Kojima M, Kawazu C, Murata M, Nishiyori H, Lazarevic D, Motti D, et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19:255–265. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann O, Caballero OL, Stevenson BJ, Chen YT, Cohen T, Chua R, Maher CA, Panji S, Schaefer U, Kruger A, et al. Genome-wide analysis of cancer/testis gene expression. Proc. Natl Acad. Sci. USA. 2008;105:20422–20427. doi: 10.1073/pnas.0810777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta S, Vingron M, Haas SA. T-STAG: resource and web-interface for tissue-specific transcripts and genes. Nucleic Acids Res. 2005;33:W654–W658. doi: 10.1093/nar/gki350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl Acad. Sci. USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahmann S, Muller T, Vingron M. On the power of profiles for transcription factor binding site detection. Stat. Appl. Genet. Mol. Biol. 2003;2 doi: 10.2202/1544-6115.1032. Article 7. [DOI] [PubMed] [Google Scholar]
- 25.Lodish HF, Berk A, Kaiser CA, Krieger M, Scott MP, Bretscher A, Ploegh H, Matsudaira P. Molecular Cell Biology. 6. N.Y., USA: W. H. Freeman and Company; 2008. [Google Scholar]
- 26.Qian J, Esumi N, Chen Y, Wang Q, Chowers I, Zack DJ. Identification of regulatory targets of tissue-specific transcription factors: application to retina-specific gene regulation. Nucleic Acids Res. 2005;33:3479–3491. doi: 10.1093/nar/gki658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber BR, Bulyk ML. Meta-analysis discovery of tissue-specific DNA sequence motifs from mammalian gene expression data. BMC Bioinformatics. 2006;7:229. doi: 10.1186/1471-2105-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X, Lin J, Zack DJ, Qian J. Computational analysis of tissue-specific combinatorial gene regulation: predicting interaction between transcription factors in human tissues. Nucleic Acids Res. 2006;34:4925–4936. doi: 10.1093/nar/gkl595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith AD, Sumazin P, Xuan Z, Zhang MQ. DNA motifs in human and mouse proximal promoters predict tissue-specific expression. Proc. Natl Acad. Sci. USA. 2006;103:6275–6280. doi: 10.1073/pnas.0508169103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith AD, Sumazin P, Zhang MQ. Tissue-specific regulatory elements in mammalian promoters. Mol. Syst. Biol. 2007;3:73. doi: 10.1038/msb4100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pennacchio LA, Loots GG, Nobrega MA, Ovcharenko I. Predicting tissue-specific enhancers in the human genome. Genome Res. 2007;17:201–211. doi: 10.1101/gr.5972507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wasserman WW, Fickett JW. Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol. 1998;278:167–181. doi: 10.1006/jmbi.1998.1700. [DOI] [PubMed] [Google Scholar]
- 33.Maeda T, Gupta MP, Stewart AF. TEF-1 and MEF2 transcription factors interact to regulate muscle-specific promoters. Biochem. Biophys. Res. Commun. 2002;294:791–797. doi: 10.1016/S0006-291X(02)00556-9. [DOI] [PubMed] [Google Scholar]
- 34.Petrucco S, Wellauer PK, Hagenbuchle O. The DNA-binding activity of transcription factor PTF1 parallels the synthesis of pancreas-specific mRNAs during mouse development. Mol. Cell Biol. 1990;10:254–264. doi: 10.1128/mcb.10.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 36.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Don J, Stelzer G. The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis. Mol. Cell Endocrinol. 2002;187:115–124. doi: 10.1016/s0303-7207(01)00696-7. [DOI] [PubMed] [Google Scholar]
- 38.Latham KE, Litvin J, Orth JM, Patel B, Mettus R, Reddy EP. Temporal patterns of A-myb and B-myb gene expression during testis development. Oncogene. 1996;13:1161–1168. [PubMed] [Google Scholar]
- 39.Mattei F, Schiavoni G, Borghi P, Venditti M, Canini I, Sestili P, Pietraforte I, Morse HC, 3rd, Ramoni C, Belardelli F, et al. ICSBP/IRF-8 differentially regulates antigen uptake during dendritic-cell development and affects antigen presentation to CD4+ T cells. Blood. 2006;108:609–617. doi: 10.1182/blood-2005-11-4490. [DOI] [PubMed] [Google Scholar]
- 40.Bassuk AG, Barton KP, Anandappa RT, Lu MM, Leiden JM. Expression pattern of the Ets-related transcription factor Elf-1. Mol. Med. 1998;4:392–401. [PMC free article] [PubMed] [Google Scholar]
- 41.Defrance M, Touzet H. Predicting transcription factor binding sites using local over-representation and comparative genomics. BMC Bioinformatics. 2006;7:396. doi: 10.1186/1471-2105-7-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akalin A, Fredman D, Arner E, Dong X, Bryne JC, Suzuki H, Daub CO, Hayashizaki Y, Lenhard B. Transcriptional features of genomic regulatory blocks. Genome Biol. 2009;10:R38. doi: 10.1186/gb-2009-10-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engstrom PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 46.Ohler U. Identification of core promoter modules in Drosophila and their application in accurate transcription start site prediction. Nucleic Acids Res. 2006;34:5943. doi: 10.1093/nar/gkl608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engstrom PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 2007;17:1898–1908. doi: 10.1101/gr.6669607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vavouri T, Walter K, Gilks WR, Lehner B, Elgar G. Parallel evolution of conserved non-coding elements that target a common set of developmental regulatory genes from worms to humans. Genome Biol. 2007;8:R15. doi: 10.1186/gb-2007-8-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.