Abstract
DEAD-box proteins play specific roles in remodeling RNA or ribonucleoprotein complexes. Yet, in vitro, they generally behave as nonspecific RNA-dependent ATPases, raising the question of what determines their specificity in vivo. SrmB, one of the five Escherichia coli DEAD-box proteins, participates in the assembly of the large ribosomal subunit. Moreover, when overexpressed, it compensates for a mutation in L24, the ribosomal protein (r-protein) thought to initiate assembly. Here, using the tandem affinity purification (TAP) procedure, we show that SrmB forms a complex with r-proteins L4, L24 and a region near the 5′-end of 23S rRNA that binds these proteins. In vitro reconstitution experiments show that the stability of this complex reflects cooperative interactions of SrmB with L4, L24 and rRNA. These observations are consistent with an early role of SrmB in assembly and explain the genetic link between SrmB and L24. Besides its catalytic core, SrmB possesses a nonconserved C-terminal extension that, we show, is not essential for SrmB function and specificity. In this regard, SrmB differs from DbpA, another DEAD-box protein involved in ribosome assembly.
INTRODUCTION
DEAD-box proteins are widely distributed in nature and they play important roles in nearly all processes involving RNA (1). They are characterized by a highly conserved structure, the ‘helicase core’, containing at least nine conserved amino acid motifs, most of which are involved in the binding of the substrates RNA and ATP. In vitro, they possess a RNA-dependent ATPase activity and many of them can dissociate short RNA duplexes in an ATP-dependent manner [‘RNA helicase’ activity (1)]; in addition, some DEAD-box proteins have been shown to displace proteins from RNA, or assist RNA folding or RNA–RNA annealing (2). In vivo, these proteins are believed to rearrange RNA or ribonucleoprotein (RNP) structures. Yet, in no case is this function precisely understood at the molecular level. An intriguing question is how they are targeted to their correct substrates. Indeed, despite the high sequence and structure conservation of the helicase core, individual helicases perform very specific functions in vivo (2). A well-understood case is the Escherichia coli RNA helicase DbpA, which is involved in the assembly of the large (50S) ribosomal subunit (Sharpe Elles,L. and Uhlenbeck,O.C., personal communication). This protein only shows ATPase and RNA helicase activities with RNA substrates carrying a specific motif, hairpin 92 of 23S rRNA (3,4). This specificity reflects the fact that DbpA carries a C-terminal extension that tightly binds this hairpin (5,6). However, to date, DbpA is the only DEAD-box protein exhibiting such RNA specificity. As for others, it has been postulated and sometimes shown that substrate specificity is conferred by accessory proteins or cofactors that bind the helicase and tether it to its site of action (7).
Besides DbpA, two other E. coli DEAD-box proteins play well-documented roles in the assembly of the large ribosomal subunit (8). Among them, SrmB was originally characterized by the fact that its overexpression suppresses a temperature-sensitive mutation in ribosomal-protein (r-protein) L24 (9). Later, we showed that SrmB co-sediments with 50S precursors; moreover, its absence leads to a severe deficit of free 50S subunits and to the accumulation of 40S particles corresponding to incompletely assembled 50S subunits (10). Among those r-proteins that are missing from the 40S particle stands L13, which together with L4, L20, L22 and L24 is essential and sufficient for the formation of the first intermediate during 50S assembly in vitro (11). On this basis, we suggested that SrmB is required at an early stage of the 50S assembly (10).
In spite of this in vivo specificity, SrmB shows no stringent preference for a particular RNA substrate in vitro, unlike for DbpA. Indeed, a variety of RNAs including the homopolymers poly(A) and poly(C) stimulate its ATPase activity (9,12), and 23S and 16S rRNAs are equally efficient in this respect [(6) and Iost,I., unpublished data]. Here, we show that in vivo and in vitro SrmB forms a specific RNP complex with the 5′ region of 23S rRNA and two r-proteins, including L24. These data shed light on the way this ‘nonspecific’ RNA helicase is targeted to its precise physiological function.
MATERIALS AND METHODS
Bacterial strains and plasmids
The srmB-TAP fusion gene was constructed by fusing two PCR fragments carrying the srmB ORF plus 212 nt upstream sequences, with the TAP tag (Supplementary Data). The product was cloned into the BamHI site of either pCL1920 (13) or pACYC184. Reverse PCR from the pCL1920-srmB-TAP plasmid was used to construct an in-frame deletion of the SrmB C-terminal extension (Supplementary Data). For TAP experiments, the BL21(DE3) derivative ENS133ΔsrmB (10) carrying the above plasmids were grown in LB containing 50 µg/ml spectinomycin (pCL1920 derivatives) or 30 µg/ml chloramphenicol (pACYC184 derivatives).
For overexpression purposes, the rplX gene encoding L24 was amplified by PCR and cloned into the pPROEX-HTa expression vector (Invitrogen). Similarly, the srmB-CBP fusion gene was cloned into NdeI–XhoI sites of pHL9-pET28a-HisTev (Supplementary Data), a pET28 derivative kindly provided by H. Le Hir. Plasmid pET11c-TthL4 encoding Thermus thermophilus L4 was a gift of M. Garber, Pushchino, Russia. The SrmB overexpressing plasmid is described elsewhere (12).
TAP purification
Extracts were routinely prepared from 1 l of culture grown to an OD600 of ∼1 at 30°C. Cells were pelleted, resuspended in disruption buffer (10 mM Tris–HCl pH 7.5, 100 mM NaCl, 0.2 mM EDTA, 10% glycerol, 0.5 mM DTT) and broken with a French press. After clarification, the extract was subjected to the two affinity purification steps (14).
For controlled digestion of the SrmB complex, the purified complex was first diluted 30 times with calmodulin binding buffer (14) and CaCl2 to restore binding conditions, then incubated on ice with 0.05–0.5 µg/ml of RNase A for 10 min, and finally purified on calmodulin–Sepharose beads.
Protein analysis
Proteins from TAP eluates were concentrated by TCA precipitation and separated by denaturing polyacrylamide gel electrophoresis (PAGE). For mass spectrometry, Coomassie-stained bands were cut out and proteins were digested overnight by trypsin. Digests were analyzed by MALDI-TOF and the resulting peptide spectra identified using MASCOT software (Plate-Forme Spectrométrie de Masse et Protéomique, Université Paris VI).
RNA analysis
The position of guanines was determined by partial nuclease T1 digestion. The 0.5- or 0.2-kb RNA fragment was first purified by urea-PAGE, dephosphorylated with calf intestinal alkaline phosphatase and 5′ labeled with γ-32P ATP. The labeled RNA was further gel purified and then digested with RNase T1 as in (15). The same RNA fragment was partially hydrolyzed by alkali to produce a 1-nt ladder. Reaction mixtures were analyzed on a 10% polyacrylamide-8 M urea gel.
Primer extension was carried out as in (10), using SuperscriptII RNaseH-Reverse Transcriptase (Invitrogen) and a primer complementary to residues 35–55 of 23S rRNA. Extension products were separated on 8% acrylamide-7 M urea gels. Northern blots were prepared and probed with 5′ labeled oligonucleotides as in (10), except that RNA was separated on 6% acrylamide-7M urea gels. Radioactive signals were visualized with a FLA-3000 PhosphorImager (Fuji).
In vitro transcription
Templates for in vitro transcription were amplified by PCR from plasmid pNO2680, which carries the rrnB operon (Supplementary Data). After transcription (RiboMAX System T7 from Promega) and DNase treatment, unincorporated nucleotides were removed by G50 filtration. RNA was then extracted with phenol:chloroform and precipitated. Purity was tested on agarose gels.
Purification of SrmB, L24 and L4
Proteins SrmB, L4, L24 and SrmB-CBP were overexpressed in the E. coli B strain BL21(DE3) after 3 h IPTG induction. SrmB was purified as in (12) except that the storage buffer was 0.3 M NaCl, 20 mM HEPES pH 7.5, 10% glycerol, 0.1 mM EDTA, 1 mM DTT. The His6-L24 protein was purified as SrmB except that buffers were supplemented with 6 M urea and the His6-tag was not removed. The storage buffer was either 100 mM KCl, 20 mM HEPES pH 7.5, 10% glycerol, 0.1 mM EDTA, 1 mM DTT or 100 mM NH4Cl, 20 mM HEPES pH 7.5, 4 mM MgCl2.
L4 from T. thermophilus was preferred to the E. coli protein because of its higher solubility. The two proteins bind E. coli 23S rRNA similarly (16). Cells overexpressing TthL4 were sonicated in 900 mM LiCl, 90 mM MgCl2, 50 mM Tris–HCl, pH 7.5, 2 mM beta-mercaptoethanol, 2 mM PMSF and the extract was centrifuged at 15 000 g for 15 min. The supernatant was heated at 65°C for 15 min. After centrifugation, the L4 protein was purified by heparin chromatography using a 0.2–2 M NaCl gradient in 50 mM Tris–HCl pH 7.5, 20 mM MgCl2. Peak fractions were >95% pure as judged by denaturing PAGE and used as such.
Protein SrmB-CBP was first purified on nickel column. After removing the His6 tag by TEV digestion, the protein was further purified on calmodulin affinity resin as described in (14). SrmB-CBP was stored in 1.5× PBS containing 10% glycerol, 1 mM Mg acetate and 2 mM DTT.
In vitro reconstitution of the SrmB complex
Experiments were performed as in (17) with slight modifications. Thirty-five picomoles of SrmB-CBP were mixed with 40–60 pmol of L4, 240 pmol of L24 and 40 pmol of spe-RNA or ctrl-RNA in binding buffer (BB-125) containing 20 mM HEPES pH 7.5, 125 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 1 mM DTT, 0.1% NP-40 and 5% glycerol, complemented with 2 mM ATP in a final volume of 60 µl. RNA was heated for 5 min at 70°C in 10 mM HEPES pH 7.5, 100 mM NaCl and 4 mM MgCl2 and immediately cooled to 4°C before addition to the mixture. An aliquot of one-sixth of the mixture was withdrawn before adding RNA, and used as the ‘Input’ control in the protein gels. After 30 min at 30°C, 12 µl of calmodulin resin (Stratagene) and 200 µl of BB-150 (150 mM NaCl) were added. After 2 h at 4°C under mild agitation, the resin was washed three times with 400 µl BB-150 and then eluted with 20 mM EGTA in 30 µl BB-150 for 5 min at 30°C. On denaturing PAGE 50% of the eluate was loaded and proteins were visualized by Coomassie or silver staining. The latter also allows the visualization of RNA. When stated, BB-125 was complemented with heparin (0.01 µg/µl). Moreover, in some experiments, ATP has been replaced by ADP in both BB-125 and BB-150.
Fluorescence anisotropy titrations
Binding of r-proteins to rRNA fragments was performed as in (16), with a 10% excess of r-proteins over RNA. SrmB was labeled with Alexa 488 as in (18). Anisotropy measurements were carried out at 30°C in dilution mode. Briefly, SrmB-Alexa488 (20 nM final) was mixed with either free rRNA or rRNA/r-proteins complex (4 µM final) in 20 mM Tris–HCl, pH 7.5, 150 mM KCl, 2.5 mM MgCl2, 1 mg/ml bovine serum albumin and 2 mM AMP-PNP, a nonhydrolyzable analog of ATP. The mixture was then serially diluted with the same buffer containing only 20 nM SrmB-Alexa488. Measurements were made at each dilution using a Varian Cary/Eclipse spectrofluorimeter in polarization mode, and anisotropy was calculated as in (18). Experiments were repeated twice with different batches of proteins and RNAs and yielded indistinguishable results.
Ribosome profile analysis
Extracts of ENS133ΔsrmB cells carrying pCL1920 or pCL1920 containing the wild-type srmB-TAP or srmB ΔC-TAP gene, were prepared and analyzed as in (10).
RESULTS
Identification of an SrmB RNP complex
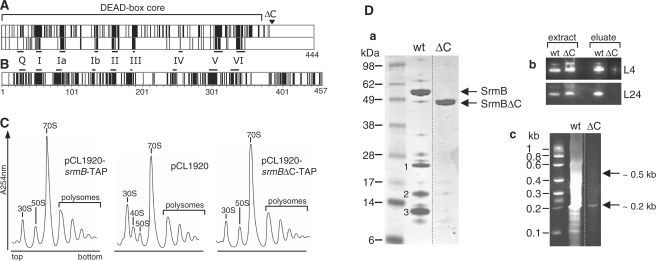
To isolate proteins that potentially interact with SrmB, we employed the TAP method, a technique of choice for purifying complexes under native conditions (14,19). The calmodulin binding peptide (CBP) and protein A affinity tags were fused in-phase to the carboxy-terminus of SrmB (Figure 1A) and the resulting construct was cloned under the srmB promoter in either pCL1920 or pACYC184 plasmids (around 5 and 10–15 copies/cell, respectively). The resulting plasmids were introduced in an E. coli B strain deleted for srmB. The SrmB-TAP fusion protein appears functional in these strains (‘TAP strains’) since it fully corrected the growth and ribosome assembly defects associated with the srmB deletion (see Figure 4C below). In contrast, no correction was observed with the empty plasmids (‘Control strains’). Since the role of SrmB in ribosome assembly is most prominent at low temperature (10), both TAP and control strains were grown at 30°C for TAP purification.
Figure 1.
Identification of SrmB partners by TAP purification. (A) Schematic representation of the SrmB-TAP fusion protein. The TAP tag consists of a calmodulin-binding peptide (CBP), a TEV cleavage site (TEV) and the IgG-binding domain of protein A (ProtA). (B) Visualization of the proteins interacting with SrmB. (Left) Proteins from crude extracts (‘Extract’, 1/1000 of the sample), after the first IgG column (‘IgG’, 1/50 of the sample) and from three TAP eluate fractions (whole sample) were resolved by denaturing PAGE and stained with Coomassie. C and T stand for Control (empty vector) and TAP (vector expressing SrmB-TAP protein) strains, respectively. Left and right panels correspond to two experiments yielding different protein patterns (see text). (C) Analysis of SrmB-interacting RNA species. RNA from the same elution fractions as in (B) was separated on a 1% agarose gel and stained with ethidium bromide. The left and right panels correspond to the same two experiments as in (B). (D) The ∼0.5-kb RNA species co-eluting with SrmB [(C), left panel] corresponds to domain I of 23S rRNA. Left, Primer extension (PE) analysis of the ∼0.5-kb RNA. A sequencing ladder (GCAT) obtained with the same primer from plasmid pNO2680, which carries the rrnB operon, was run in parallel. The 5′-end of mature 23S rRNA (23S 5′-end) is indicated. Right, Partial digestion of the ∼0.5-kb RNA with RNase T1, which cleaves 3′ to guanine residues. The digest (T1), together with a sample of the same RNA after limited alkaline hydrolysis (OH), was analyzed by urea–PAGE. The sequence of 23S rRNA from nucleotide 15 (bottom) to 50 (top) is shown on the right, with guanine residues (G) facing the corresponding RNase T1 fragments. In this and other figures, dotted, vertical lines mean that lanes from the same gel have been brought together at this position.
Figure 4.
The C-terminal extension of SrmB is dispensable for ribosome assembly. (A) SrmB-specific residues are essentially located within the helicase core. Upper frame: alignment of the E. coli SrmB (444 amino acids) with orthologs from representatives of four distant orders of gamma-proteobacteria. Vertical bars indicate identical residues. Besides E. coli K12 (Enterobacteriales), selected representatives were Vibrio vulnificus CMCP6 (Vibrionales), Pseudoalteromonas haloplanktis TAC125 (Alteromonadales), Aeromonas hydrophila ATCC7966 (Aeromonadales) and Haemophilus influenzae Rd KW20 (Pasteurellales) (the pattern remains largely invariant upon changing representatives within the same orders). Orthologs were identified by their high BLAST scores and aligned with CLUSTALW to the E. coli protein. Shown above the panel is the helicase DEAD-box core, assumed to extend down to amino acid 368, after a predicted alpha-helix that is conserved in known structures of DEAD-box proteins. The start of the ΔC truncation is indicated by an open triangle. Lower frame: same as upper frame, except that SrmB was aligned with the four other E. coli DEAD-box proteins: identity in this case is largely restricted to the DEAD-box motifs [horizontal bars; see (1)]. (B) Same as [(A), upper frame], except that orthologs of DbpA were aligned. The bacteria used for alignment are the same as in (A), except that Haemophilus influenzae, which lacks a DbpA ortholog (8) was replaced by Pseudomonas aeruginosa PAO1 (Pseudomonadales). The DbpA orthlogs used here (457–462 amino acids) are much closer in length than the SrmB orthologs (407–444 residues), due to the homogeneous size of their C-terminal extensions. (C) ΔsrmB cells transformed with either pCL1920 or its derivatives carrying the wild-type or srmB ΔC-TAP gene, were grown at 30°C. Ribosome profiles show the 30S and 50S subunits, 70S ribosomes (free couples and monosomes), polysomes, as well as the aberrant 40S particles. The ΔsrmB ribosome assembly defect (deficit of 50S subunits and accumulation of 40S particles) is fully corrected by the wild-type and ΔC proteins. (D) Deletion of the SrmB C-terminal extension weakens the SrmB complex. (a) Proteins co-eluting with wild-type and truncated SrmB during TAP purification were analysed as in Figure 1B. Previously identified bands 1–3 (see Figure 1B) are indicated. (b) Crude extracts and eluates from cells expressing wild-type or truncated SrmB were analyzed by western blotting using L4 and L24 antibodies. (c) RNA co-eluting with wild-type and truncated SrmB was visualized as in Figure 1C, except that a 6% polyacrylamide-urea gel stained was used.
After the two consecutive affinity chromatography steps (14), proteins in elution fractions were analyzed by denaturing PAGE. No obvious difference was noted whether pCL1920 or pACYC184 derivatives were used. In many experiments, only three major bands, numbered 1–3, were detected along with the band corresponding to SrmB (lanes T in Figure 1B, left). These bands were particularly visible in elution fraction 2, which is most enriched in proteins, and were absent in fractions from the control strain (lanes C in Figure 1B). The bands were excised from the gel, and tryptic digests were analyzed by MALDI-TOF mass spectrometry. Bands 1 and 3 correspond to 50S r-proteins L4 and L24, respectively. Occasionally, r-proteins L21 and L22 were also detected in band 3, but western analysis (data not shown) confirmed that L24 was predominant. The protein(s) corresponding to band 2 yielded only a few tryptic peptides, and they could not be identified. Beyond bands 1–3, minor bands were also analyzed and found to correspond to different r-proteins (data not shown).
SrmB, L4 and L24 are all RNA-binding proteins. To test whether the above complex (the ‘SrmB complex’) contains RNA, elution fractions shown in Figure 1B (left) were analyzed on an agarose gel after phenol extraction. As shown in Figure 1C (left; lane T), a smear with a prominent band migrating like the 0.5-kb RNA marker was detected. Extensive treatment of the extract with RNase A prior to TAP purification eliminated this pattern, suggesting that it corresponds to RNA. Moreover, L4 and L24 no longer co-purified with SrmB under these conditions, indicating that this RNA participates in the formation or stability of the complex (Figure S1). Since in vivo SrmB associates with a 50S precursor (10), the eluted RNA may correspond to fragments of the 23S rRNA. Consistently, northern blot analysis of the eluted RNA with a probe corresponding to the full-length (2.9 kb) 23S gene revealed the same pattern as ethidium bromide (EtBr) staining (data not shown). After purification, the major ∼0.5-kb RNA fragment was further characterized by primer extension and RNase T1 footprinting. As shown in Figure 1D (left), the 5′-end of the fragment corresponded to the 5′-end of 23S rRNA. Moreover, the T1 pattern unambiguously matched that of the 5′ proximal part of 23S rRNA, with the first readable G corresponding to nucleotide +15 (Figure 1D, right). Thus, the major RNA species co-purifying with SrmB-TAP extends from nucleotide (nt) 1 to ca 500 of 23S rRNA, i.e. it corresponds to domain I of this rRNA (for the boundaries of E. coli rRNA domains, see http://www.rna.ccbb.utexas.edu). Incidentally, domain I contains the binding sites for r-proteins L4 and L24 [(16); see Figure 2D and F].
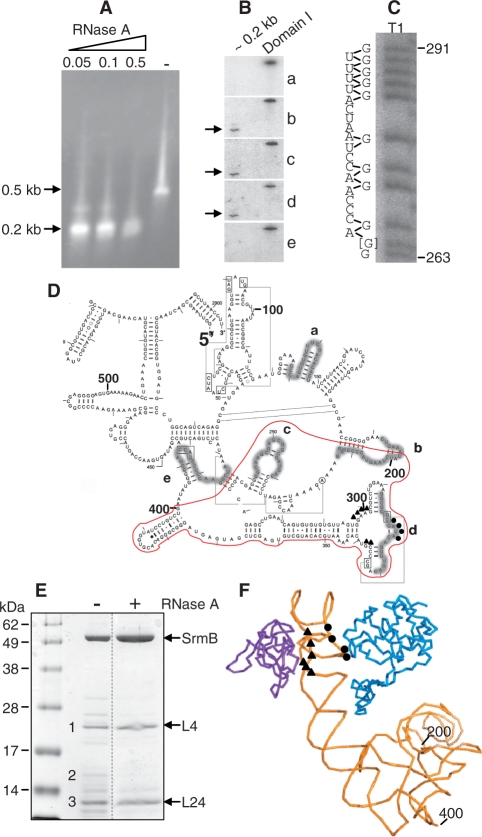
Figure 2.
Characterization of a minimal SrmB complex. (A) The SrmB complex was treated at 0°C with different RNase A concentrations [from 0 (−) to 0.5 µg/ml] and further purified on a calmodulin column. The RNA from elution fractions was analyzed as in Figure 1C. (B) The purified ∼0.2-kb fragment (0.05 µg/ml RNase A treatment) was run on a denaturing polyacrylamide gel, transferred to a nylon membrane and probed with the 5′-end-labeled oligonucleotides a-e (see below). Hybridization signals are indicated by arrows. An in vitro transcript corresponding to domain I of 23S rRNA was used as a positive control. (C) The ∼0.2-kb RNA species was labeled at its 5′ end and digested with RNase T1. The sequence of 23S rRNA from nt 263 (bottom) to nt 291 (top) is shown on the left of the gel, with guanine residues (G) facing the corresponding RNase T1 fragments. Note that the sequence at position 264 (marked [G]) is highly heterogeneous in E. coli. We assume that G predominates in the E. coli B strain used here, like in E. coli 0157:H7 but unlike in E. coli K12. (D) Secondary structure of E. coli 23S rRNA domain I. Regions complementary to the probes a–e used in the northern blot analysis (B) are highlighted. The fragment resulting from mild RNase A treatment of the SrmB complex is surrounded in red. Nucleotides that, based on biochemical evidence (16), interact with L4 (319–322) and L24 (298–301 and 337–338) are shown as circles and triangles, respectively. (E) Protein content of the SrmB complex after mild RNase treatment. (+) 0.05 µg/ml RNase A. (−) control without RNase. The previously identified bands 1–3 (Figure 1B) are indicated. (F) Part of the structure of the E. coli ribosome (PDB2aw4) showing nt 200–400 of 23S rRNA (yellow) together with L4 (blue) and L24 (purple).
Whereas many TAP experiments yielded the protein and RNA profiles shown in the left panels of Figure 1B and C, other experiments yielded larger RNA species (up to 2.9 kb in length), together with many more proteins (Figure 1B and C, right panels). Presumably, this variability can be rationalized as follows. Since in vivo SrmB associates with a 50S precursor, one would expect the whole 23S rRNA, together with all proteins present in the precursor, to be pulled down in these experiments. However, as no particular precautions were taken for controlling nucleases, the 23S rRNA is degraded to variable extent during the TAP procedure, explaining the different elution profiles: the more extensive the degradation, the smaller the rRNA fragments and the fewer the r-proteins that are pulled down with SrmB. Extensive degradation, as exemplified by the left panels of Figure 1B and C, is particularly useful here since it narrows the rRNA region and the number of proteins that may interact directly with SrmB.
Isolation of a minimal SrmB complex
To further narrow the rRNA region and the number of proteins implicated in SrmB binding, the purified SrmB–TAP complex was mildly digested with RNase A. This treatment should remove all rRNA regions that are not protected by SrmB, and all proteins that bind to these regions but do not interact directly with SrmB. The digested product was then re-purified on the calmodulin column (SrmB retains the CBP after TAP purification), and analyzed for its RNA and protein content. Whatever the RNase A concentration in the range 0.05–0.5 µg/ml, the ∼0.5-kb RNA fragment was converted into a ∼0.2-kb fragment (Figure 2A). After purification by urea-PAGE, this fragment was probed on a northern blot with oligonucleotide probes complementary to different regions of domain I (Figure 2D). An in vitro transcript encompassing domain I was run in parallel as a control. Whereas the ∼0.2-kb species hybridized with probes b (192–214 nt), c (240–259 nt) and d (312–331 nt), no signal was detected with probes a (128–147 nt) and e (409–428 nt) (Figure 2B). Thus, the ∼0.2- kb product corresponds to the central region of domain I. Using oligonucleotide c for primer extension, its 5′ extremity was mapped to nucleotide 198 (data not shown). Therefore, the fragment spans nt 198 to ∼400 of 23S rRNA. To further confirm this localization with a different technique, the ∼0.2-kb product was labeled at its 5′-end and partially digested with RNase T1, as above. The profile of the digest unambiguously matched the succession of Gs in the central region of domain I. This is illustrated on Figure 2C, where a part of the gel is aligned with the 23S sequence from G263 to G291.
The protein composition of the partially digested SrmB complex showed that, compared with the undigested control, the number and amount of minor proteins were greatly reduced, resulting in a complex containing essentially SrmB, L4 and L24 (compare lanes ‘−’and ‘+’ on Figure 2E). In particular, band 2 was no longer detected. Mass spectrometry confirmed the identity of the r-proteins. Of note, the binding sites for L4 and L24 both lie within the 0.2-kb RNA fragment [(16); Figure 2D], and they are in close proximity in the 3D-structure of the ribosome (Figure 2F).
Thus, the minimal SrmB complex comprises r-proteins L4 and L24 and a small rRNA fragment encompassing their binding sites. It must be relatively stable as it withstood TAP purification, incubation with RNase A, and finally an additional TAP purification step.
In vitro reconstitution of the SrmB complex
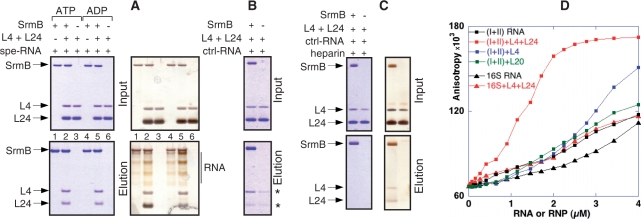
Next, we tested whether the minimal SrmB complex can be reconstituted in vitro from purified components. To this end, the method of Ballut et al. (17) was used. Briefly, the SrmB-CBP fusion protein was incubated with its potential partners and then affinity purified on calmodulin beads. Eventual complexes were visualized by electrophoresis, as in the last step of the TAP protocol. Proteins SrmB-CBP, L4 and L24 were purified as described in ‘Materials and methods’ section. A fragment of 23S rRNA (‘spe-RNA’; 185–399 nt), very close to the fragment contained in the minimal SrmB complex but slightly extended at its 5′-end to facilitate transcription by T7 RNA polymerase, was synthesized in vitro; another 23S fragment of similar size (‘ctrl-RNA’; 1–214 nt) was also prepared as a control. In the presence of spe-RNA, both L4 and L24 coeluted with SrmB (Figure 3A, lanes 2 or 5). Silver staining revealed also yellowish, multidisperse bands between SrmB and L4 that correspond to eluted spe-RNA (Figure 3A, right), as confirmed by EtBr staining (data not shown). Although L4 and (particularly) L24 were present in molar excess over SrmB–CBP in the input, the three proteins eluted in nearly stoichiometric amounts, as judged from densitometry of the Coomassie-stained gels. Elution patterns were identical whether ATP or ADP was present in the incubation and washing buffers (compare lanes 1–3 and 4–6 in Figure 3A; given the low ATPase activity of SrmB, only a small fraction of the input ATP is hydrolyzed during the assay). Importantly, no r-proteins or RNA were eluted in the absence of SrmB–CBP, demonstrating the specificity of the procedure (Figure 3A, lanes 3 and 6).
Figure 3.
Reconstitution of the SrmB complex in vitro. (A–C) Proteins SrmB-CBP, L4 and L24 were mixed together with a 215 nt rRNA fragment encompassing the L4 and L24 binding sites (spe-RNA) or a control fragment of similar size (ctrl-RNA). SrmB-CBP was then affinity purified, and the eluate was analysed by PAGE. The ‘Input’ samples (upper panels) correspond to aliquots (20%) that were withdrawn prior to RNA addition, whereas ‘elution’ samples (lower) correspond to 50% of the eluates. The composition of each mix is shown above the corresponding lane. Positions of SrmB-CBP (SrmB), L4 and L24 are shown by arrows. Blue and brownish backgrounds in (A) and (C) correspond to Coomassie or silver staining of the same gels. The stars shown in (B) pinpoint traces of L4 and L24 that are recovered in the control lacking SrmB-CBP. (D) Fluorescence anisotropy assays. The fluorescent anisotropy of Alexa-SrmB is plotted versus the concentrations of (I+II) or 16S RNA, with or without added r-proteins L4, L24 or L20 (‘RNP’ or ‘RNA’, respectively).
Thus, in vitro, SrmB forms a quaternary, apparently stoichiometric complex with L4, L24 and spe-RNA. Within this complex, L4 and L24 presumably interact specifically with their binding sites on spe-RNA (16); as for SrmB, it might either interact directly with its partners or simply binds nonspecifically to spe-RNA. To test for direct interactions between SrmB and the two r-proteins, these experiments were repeated with ctrl-RNA, which should bind L4 and L24 loosely if at all. Under these conditions, some L4 and trace amounts of L24 still co-purified with SrmB; oddly, however, small amounts of r-proteins were also detected in the control lacking SrmB (stars in Figure 3B). Presumably, in the absence of their specific RNA target, the highly basic r-proteins stick nonspecifically to the calmodulin beads, from which they can subsequently be eluted. To obviate this problem, the experiment was repeated in the presence of heparin, a highly charged anionic polymer that can challenge RNA-binding proteins. Heparin (0.01 µg/ml) largely diminished (Figure 3C, Coomassie staining) but did not abrogate (Figure 3C, silver staining) the co-elution of L4 and L24 with SrmB. In contrast, L4 or L24 were no longer detectable in the control. Of note, no ctrl-RNA was detected in the eluate, suggesting that any interaction of ctrl-RNA with SrmB is eliminated by heparin. Consistently, the same elution pattern was observed in the absence of ctrl-RNA (data not shown).
These latter results support the existence of direct, RNA-independent interactions between SrmB on one hand, L4 and L24 on the other. Yet, the yield of eluted r-proteins was higher in the presence of spe-RNA (compare Figure 3A with 3B and C). This fact may explain why, in crude extracts, extensive RNase treatment apparently abrogates the coelution of SrmB and r-proteins (Figure S1). Reciprocally, the yield of spe-RNA co-eluting with SrmB was consistently higher in the presence of r-proteins than in their absence (compare lanes 1 and 4 to lanes 2 and 5, respectively, in Figure 3A, silver stain). Thus, spe-RNA and L4+L24 reinforce the binding of each other to SrmB.
To further document these interactions, we turned to fluorescence anisotropy. This technique allows monitoring the progressive titration of a fluorescent protein with a ligand (18), provided the binding of the latter hinders the rotation of the fluorophore. For this experiment, spe-RNA was replaced by a 1.3-kb fragment covering domains I and II of 23S rRNA (subsequently called ‘I+II RNA’). In addition to L4 and L24, this longer fragment is known to bind r-protein L20 (18), thus permitting additional controls. A 1.5-kb transcript corresponding to 16S rRNA (‘16S RNA’) was used as a nonspecific control. SrmB, labeled at its N-terminus with the Alexa488 fluorophore, was incubated with various concentrations of these two RNAs, and anisotropy was measured. Under the conditions used, a plateau was not reached even at concentrations as high as 4 µM (I+II) or 16S RNA (Figure 3D). The addition of L4 and L24 to 16S RNA did not change this situation; in contrast, with I+II RNA, anisotropy increased markedly and reached a plateau well below 4 µM RNA. This effect was specific to L4+L24, since the addition of L20 or L24 alone had no effect, whereas L4 alone had but a modest effect (Figure 3D and data not shown). Altogether, these results confirm that the simultaneous presence of L4, L24 and an RNA fragment carrying their binding sites, creates a new interaction with SrmB that does not occur when any of these three molecules is missing.
The carboxy-terminal extension of SrmB is not essential to its function
As noted in the ‘Introduction’ section, the unique C-terminal extension of DbpA is essential to its specificity. Since SrmB also possesses a C-terminal extension, we tested whether it plays a similar role. An alignment of the E. coli SrmB sequence with orthologs from four distant orders of γ-proteobacteria revealed many identical residues (Figure 4A, upper frame) that are not conserved among unrelated DEAD-box proteins (e.g. the five E. coli DEAD-box proteins; Figure 4A, lower frame). However, most of these SrmB-specific residues lie within the helicase core: except for the presence of lysine stretches, the C-terminal extension shows very little conservation (Figure 4A, upper frame), and it is quite variable in length. In contrast, the C-terminal extension of DbpA is well conserved in length and sequence (Figure 4B). To assess the importance of the C-terminal extension of SrmB, an in-phase deletion (named ΔC) was constructed in the srmB-TAP gene, retaining the TAP sequence but removing the last 60 amino acids of SrmB (Figure 4A). This truncated protein was tested for its functionality in ribosome assembly. The ribosome profiles of the ΔsrmB strain expressing either the wild-type or truncated proteins were identical, i.e. the ΔC protein fully complements the ΔsrmB defect (Figure 4C).
The ΔC and wild-type proteins were then purified in parallel with the TAP protocol. In contrast with the wild-type SrmB, no r-proteins co-eluting with the ΔC protein could be detected by Coomassie staining (Figure 4Da) or western analysis (Figure 4Db). Consistently, only a small amount of RNA co-eluted with the ΔC mutant (Figure 4Dc). However, northern analysis showed that this RNA still corresponds to the central region of domain I (data not shown). A plausible interpretation is that the SrmB complex can still form but with a reduced stability, so that most of it does not survive the TAP procedure (‘Discussion’ section).
DISCUSSION
Although the E. coli ribosome can be assembled in vitro from purified components, assembly is far more efficient in vivo (20). Among the factors responsible for this difference stand nonribosomal proteins that transiently assist assembly (21). The DEAD-box RNA helicase SrmB was among the first such protein identified (10), but the basis for its specificity, as well as its exact role in assembly, remains obscure. A major finding from this work is that SrmB interacts specifically with a RNP complex located near the 5′-end of 23S rRNA.
The SrmB complex
Using the TAP purification procedure coupled with a mild RNase A treatment, we have shown that in cell extracts, SrmB associates with r-proteins L4 and L24 and a 0.2-kb fragment from domain I of 23S rRNA that encompasses the binding sites of these two proteins (Figure 2D and F). We propose that SrmB binds specifically to this region of 23S rRNA in vivo. The quaternary complex, referred to as the ‘minimal SrmB complex’, can be reconstituted in vitro from isolated components, indicating that its formation does not require additional factors (Figure 3A). Of note, L4, L24 and domain I are presumably among the first components of the 50S subunit to assemble together. The 5′ proximal domain I is the first 23S domain to be synthesized, and biochemical and electron microscopy work indicates that in vitro L24 binds to this domain without the help of other proteins, thereby initiating the cooperative assembly process (20,22). Thus, L24 may be the first protein to bind 23S rRNA in vivo. As for L4, it belongs (like L24) to a group of five proteins that are essential and sufficient for the formation of the first in vitro assembly intermediate (20). Altogether, these data suggest that the binding of SrmB to its specific target occurs soon after the onset of 23S synthesis, consistent with the proposed early role of SrmB in 50S subunit assembly (10).
The specificity of SrmB for one particular region of 23S rRNA in vivo contrasts with its absence of specificity in vitro in the absence of other proteins (‘Introduction’ section). The reconstitution assay has pinpointed one cause for this specificity: the two proteins L4 and L24 interact directly with SrmB, presumably via protein–protein contacts (Figure 3B and C). The capacity of the two r-proteins to specifically bind both SrmB and a RNA fragment encompassing their binding sites (spe-RNA), explains why SrmB binds L4+L24 and spe-RNA synergistically, as shown by reconstitution experiments and anisotropy assays (Figure 3). Indeed, the two r-proteins can bridge SrmB and spe-RNA, thus tightening the binding of SrmB to this particular RNA; conversely, spe-RNA can bridge SrmB and the two r-proteins, thus reinforcing their interaction. Altogether, the existence of these cooperative interactions seems sufficient to explain the stability of the SrmB complex. As judged by anisotropy measurements, the simultaneous presence of both r-proteins is needed for this stability, though L4 may have some effect on its own (Figure 3D).
Interestingly, the in vitro interaction of SrmB with its partners is the same whether ADP or ATP is present (Figure 3A), suggesting that SrmB remains tethered to its binding site upon ATP hydrolysis. That SrmB can bind RNA in the presence of either ATP or ADP is not inconsistent with its helicase activity: simply, in the two situations, RNA must interact differently with the RNA-binding track within the active site. RhlE, another E. coli DEAD-box helicase, also binds RNA with similar affinities in the ATP- and ADP-bound states (12).
When overexpressed, SrmB can suppress a temperature-sensitive L24 mutation (rplX19) that decreases L24 affinity for 23S rRNA (9,23). Conversely, the deletion of srmB considerably aggravates the rplX19 phenotype (10). Our results suggest a plausible explanation for these genetic links. As expected for an initiator protein, the wild-type L24 protein can bind its rRNA target without any assistance (16); however, this may not hold true for the mutant. We speculate that SrmB, by bridging L4 and L24, helps bringing the mutated L24 to its rRNA site at the time L4 binds to its own site. By eliminating the bridge, the deletion of SrmB would then reinforce the rplX19 phenotype; conversely, its overexpression would alleviate this phenotype by stabilizing the L4-SrmB-L24-rRNA interaction.
Role of SrmB C-terminal extension
Our results show that the C-terminal extension of SrmB is not essential for ribosome assembly in vivo (Figure 4C), but yet that it is important for the formation or for the stability of the SrmB complex (Figure 4D). It seems very unlikely that these two functions of SrmB can be dissociated, i.e. that SrmB can act on the nascent ribosome without being tethered to it by the SrmB complex. We therefore propose that, in vivo, SrmB lacking the C-terminal extension (ΔC) still forms the SrmB complex, but that the complex is not stable enough to survive the TAP purification. Consistent with this view, northern analysis shows that the small amount of RNA co-eluting with ΔC (Figure 4Dc) still corresponds to the central region of domain I, as expected if complex formation had occurred. As for the C-terminal region, it has been shown that the deletion of the last 51 amino acids reduces markedly the affinity of SrmB for RNA, as judged by a higher Km in ATPase assays (Bizebard,T., personal communication). Thus, this highly basic, lysine-rich region might stabilize the complex by interacting nonspecifically with rRNA. Consistent with a nonspecific role, it carries no phylogenetically conserved residues that might be implicated in the specific recognition of partners (Figure 4A).
According to this view, the residues important for specific interactions of SrmB with L4 and L24 would be located within the DEAD-box core, not within the C-terminal extension. This situation differs from that of DbpA, in which the C-terminal extension, by binding a particular RNA motif, is responsible for specificity (‘Introduction’ section). This difference is consistent with phylogenetic comparisons, which show many conserved residues in the C-terminal extension of DbpA, whereas for SrmB conservation is limited to the core (Figure 4A and B). Beyond SrmB and DbpA, other cases are known where either the helicase core or its extensions are responsible for specificity. Similar to DbpA, the yeast DEAD-box related proteins Prp16 and Prp2 use nonconserved extensions to recognize their site of action, the spliceosome (24,25). In contrast, the interaction of eIF4AIII with its protein partners within the exon-junction complex involves residues that are mainly located within the core (26). Interestingly, several of these residues are conserved amongst eIF4IIIA orthologs (26). It will be interesting to determine whether, similarly, those residues that are conserved amongst SrmB orthologs (Figure 4A) participate in the interaction with L4 and L24.
CONCLUSION
Although this work shows that SrmB binds a ribonucleoparticle near the 5′ extremity of 23S rRNA, it gives no clue on what it does once there. A search for 23S rRNA mutations that suppress the ΔsrmB phenotype—these mutations are expected to affect SrmB targets—pinpoint several sites downstream of domain I (Proux,F., Dreyfus,M. and Iost,I., unpublished data). The tethering of SrmB near the 5′-end of 23S rRNA would allow it to interact with these targets as soon as they form. Conceivably, SrmB may use ATP hydrolysis to actively rearrange the structure of these targets, as often assumed for DEAD-box proteins. However, because SrmB is an inefficient ATPase in vitro (12), we consider another scenario: like for eIF4AIII in the exon-junction complex, SrmB may simply clamp its RNA-binding track on the target RNA in an ATP-dependent manner, transiently stabilizing RNA structure. Its low ATPase activity would then reflect the need for stabilizing these structures long enough to match the kinetics of ribosome assembly.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ecole Normale Supérieure, Centre National de la Recherche Scientifique, Agence Nationale de la Recherche (grant 08-BLAN-0086-02 to M.D.); and Région Ile de France to (D.T.). Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs P. Romby for the gift of RNase T1, R. Brimacombe for antibodies against r-proteins, and M. Garber and B. Séraphin for plasmids pET11c-TthL4 and pBS1479, respectively. We thank Dr H. Le Hir for advices for the reconstitution experiment and Dr F. Bonnet for his help with HPLC purification. We are much indebted to Dr. T. Bizebard for the communication of unpublished results, and to Drs O.C. Uhlenbeck, M. Springer, and K. Nierhaus for their constant interest. I. I. thanks L. Minvielle-Sébastia and X. Darzacq for support.
REFERENCES
- 1.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Jankowsky E, Fairman ME. RNA helicases–one fold for many functions. Curr. Opin. Struct. Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicol SM, Fuller-Pace FV. The “DEAD box” protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc. Natl Acad. Sci. USA. 1995;92:11681–11685. doi: 10.1073/pnas.92.25.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsu CA, Kossen K, Uhlenbeck OC. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol. 2002;324:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 7.Silverman E, Edwalds-Gilbert G, Lin RJ. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 8.Iost I, Dreyfus M. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 2006;34:4189–4197. doi: 10.1093/nar/gkl500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishi K, Morel-Deville F, Hershey JWB, Leighton T, Schnier J. An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly. Nature. 1988;336:496–498. doi: 10.1038/336496a0. [DOI] [PubMed] [Google Scholar]
- 10.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2003;48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 11.Spillmann S, Dohme F, Nierhaus KH. Assembly in vitro of the 50 S subunit from Escherichia coli ribosomes: proteins essential for the first heat-dependent conformational change. J. Mol. Biol. 1977;115:513–523. doi: 10.1016/0022-2836(77)90168-1. [DOI] [PubMed] [Google Scholar]
- 12.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 13.Lerner CG, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 15.Donis-Keller H, Maxam AM, Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977;4:2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stelzl U, Nierhaus KH. A short fragment of 23S rRNA containing the binding sites for two ribosomal proteins, L24 and L4, is a key element for rRNA folding during early assembly. RNA. 2001;7:598–609. doi: 10.1017/s1355838201002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 18.Guillier M, Allemand F, Dardel F, Royer CA, Springer M, Chiaruttini C. Double molecular mimicry in Escherichia coli: binding of ribosomal protein L20 to its two sites in mRNA is similar to its binding to 23S rRNA. Mol. Microbiol. 2005;56:1441–1456. doi: 10.1111/j.1365-2958.2005.04644.x. [DOI] [PubMed] [Google Scholar]
- 19.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 20.Nierhaus KH. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 21.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 22.Tumminia SJ, Hellmann W, Wall JS, Boublik M. Visualization of protein-nucleic acid interactions involved in the in vitro assembly of the Escherichia coli 50 S ribosomal subunit. J. Mol. Biol. 1994;235:1239–1250. doi: 10.1006/jmbi.1994.1077. [DOI] [PubMed] [Google Scholar]
- 23.Cabezon T, Herzog A, Petre J, Yaguchi M, Bollen A. Ribosomal assembly deficiency in an Escherichia coli thermosensitive mutant having an altered L24 ribosomal protein. J. Mol. Biol. 1977;116:361–374. doi: 10.1016/0022-2836(77)90075-4. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Guthrie C. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA. 1998;4:1216–1229. [PMC free article] [PubMed] [Google Scholar]
- 25.Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol. Cell. Biol. 2004;24:10101–10110. doi: 10.1128/MCB.24.23.10101-10110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.