Abstract
The arrangement of the human telomeric quadruplex in physiologically relevant conditions has not yet been unambiguously determined. Our spectroscopic results suggest that the core quadruplex sequence G3(TTAG3)3 forms an antiparallel quadruplex of the same basket type in solution containing either K+ or Na+ ions. Analogous sequences extended by flanking nucleotides form a mixture of the antiparallel and hybrid (3 + 1) quadruplexes in K+-containing solutions. We, however, show that long telomeric DNA behaves in the same way as the basic G3(TTAG3)3 motif. Both G3(TTAG3)3 and long telomeric DNA are also able to adopt the (3 + 1) quadruplex structure: Molecular crowding conditions, simulated here by ethanol, induced a slow transition of the K+-stabilized quadruplex into the hybrid quadruplex structure and then into a parallel quadruplex arrangement at increased temperatures. Most importantly, we demonstrate that the same transitions can be induced even in aqueous, K+-containing solution by increasing the DNA concentration. This is why distinct quadruplex structures were detected for AG3(TTAG3)3 by X-ray, nuclear magnetic resonance and circular dichrosim spectroscopy: Depending on DNA concentration, the human telomeric DNA can adopt the antiparallel quadruplex, the (3 + 1) structure, or the parallel quadruplex in physiologically relevant concentrations of K+ ions.
INTRODUCTION
All healthy somatic cells have a defined number of possible cell divisions, the so-called Hayflick limit (1). More than thirty years ago it was proposed that this phenomenon (i.e. cellular senescence) is regulated by telomere shortening and the presence of a cellular enzyme responsible for telomere elongation, especially in cancer cells, was predicted (2,3). Both these predictions were confirmed (4). The enzyme, called telomerase, is usually active in cancer cells (5). Thus, telomere length predicts replication capacity of cells (6).
Telomeres contain a 3′ overhang of the G-rich DNA strand (7,8). The G-rich strand is able to form guanine quadruplexes, as was shown for the Tetrahymena telomeric sequence (TTGGG)n (9) and the vertebrate telomeric sequence (TTAGGG)n (10,11). Granotier et al. showed that a guanine quadruplex structure is present at the very ends of chromosomes (i.e. in the telomeric region) in vivo (12). Quadruplex formation in telomeric regions inhibits telomerase function (13). Thus, quadruplex stabilizing agents suppress telomere elongation (14) and proliferation of tumor cells (15). A number of ligands that specifically bind quadruplex DNA have been described (16) and the structure of a quadruplex–ligand complex has been determined (17). These agents may be of great importance for cancer treatment. To enhance the efficacy of these compounds, we must understand the structure and function of their target, the telomeric quadruplex.
In 1993, based on nuclear magnetic resonance (NMR) studies and molecular dynamics simulations, Wang and Patel proposed that the structure of quadruplex formed by human telomeric sequence AG3(TTAG3)3 in sodium solution was an antiparallel guanine quadruplex (18). A year later, Balagurumoorthy and Brahmachari detected, using circular dichrosim (CD) spectroscopy and chemical probing, intramolecular antiparallel quadruplexes formed by G3(TTAG3)3 and (TTAG3)4 sequences both in sodium and potassium solutions (19). In 2002, Parkinson et al. (20) observed an intramolecular parallel quadruplex in crystals of AG3(TTAG3)3 formed in the presence of potassium ions. However, platinum cross-linking studies in Na+ and K+ solutions demonstrated that AG3(TTAG3)3 forms basket-type antiparallel quadruplex (21), similar to the structure observed by NMR in Na+ (18). 125I-radioprobing confirmed the antiparallel basket arrangement in Na+, but a chair type quadruplex was present in K+ (22).
Several studies also demonstrated a mixture of parallel and antiparallel quadruplex in potassium solution (23,24), but it was later reported that AG3(TTAG3)3 did not form a parallel quadruplex in solution (25). In 2006, a new quadruplex type was revealed by NMR: AG3(TTAG3)3 and sequences with the same core but different flanking nucleotides form a hybrid (3 + 1) quadruplex with three parallel chains and one antiparallel quadruplex chain (26,27). Depending on the method and precise primary sequence, various arrangements of the human telomeric sequence in potassium solution have been reported (28–35). Recently, a new type of an antiparallel quadruplex of the natural human telomeric sequence G3(T2AG3)3T in K+ containing solution was proposed based on NMR data (36). It contains only two guanine tetrads capped with several layers of stacked guanines and adenines on both ends of the tetrad core. The parallel quadruplex was only observed in solution in the presence of polyethylene glycol (PEG), which simulates the overcrowded solvent conditions present inside a cell (37) or in the presence of Sr2+ ions (38). Considering these sometimes conflicting results, the relevant structure of the human telomere quadruplex in potassium solution is unclear. Also, the structure of long telomeric sequences remains undetermined. This paper describes data that resolves these apparently conflicting results.
MATERIALS AND METHODS
Synthetic oligonucleotides were purchased from VBC Biotech (Vienna, Austria). Lyophilized oligonucleotides were dissolved in 1 mM Na phosphate buffer (pH 7) with 0.3 mM EDTA. Before any measurements were made, the DNA samples were thermally denatured in this buffer. The oligonucleotides were heated in the relevant solution for 3 min at 90°C and then slowly annealed over the course of 4 h to room temperature. CD measurements were done in a Jobin-Yvon CD6 dichrograph (Longjumeau, France) in 1 cm to 0.001 cm path-length quartz Hellma cells placed in a thermostated holder. The scan rate was 0.5 nm/s. The measurement conditions and the procedure used with foldaway 0.01 and 0.001 cm cells are specified in figure legends. Precise DNA concentrations were determined on the basis of UV absorption at 260 nm of the sample measured in 1 mM Na phosphate buffer (pH 7), 0.3 mM EDTA at 90°C, using molar extinction coefficients calculated according to Gray (39). UV absorption spectra were measured on a UNICAM 5625 UV/VIS spectrometer (Cambridge, UK). CD signals are expressed as the difference in the molar absorption Δε of the right- and left-handed circularly polarized light. The molarity was related to nucleosides.
Experimental conditions were changed directly in the cells by adding concentrated solutions of sodium or potassium chloride or 96% ethanol and the final nucleoside concentration was corrected for the volume increase. The pH of the solution was kept constant at neutral value by addition of 10 mM sodium or potassium phosphate buffer (pH 7), depending on whether sodium or potassium chloride was added to the sample.
Native polyacrylamide gel electrophoresis (PAGE) was performed in a temperature-controlled electrophoretic apparatus (SE-600; Hoefer Scientific, San Francisco, CA, USA). Gel concentration was 16% (29:1 monomer to bis ratio, Applichem, Darmstadt). About 2 µg of DNA was loaded into each well of a 14 × 16 × 0.1 cm gel. Samples were electrophoresed at 23°C for 19 h at 30 V (∼2 Vcm−1). Gels were stained with Stains All (Sigma, St. Louis, MO, USA) after electrophoresis and scanned using a Personal Densitometer SI, model 375-A (Molecular Dynamics, Sunnyvale, CA, USA).
RESULTS AND DISCUSSION
Spectroscopic results reveal the same antiparallel folding of the basic G3(T2AG3)3 human telomeric quadruplex in Na+ and K+ solutions
The human telomeric sequence G3(T2AG3)3 yields a CD spectrum similar to that of AG3(T2AG3)3 in sodium solution; both are characteristic of antiparallel quadruplex as described by Wang and Patel (18). The spectrum contains positive peaks at 290 and 210 nm and a negative one at 265 nm (Figure 1). Addition of potassium ions to the sodium-stabilized quadruplex results in disappearance of the negative peak at 265 nm. Similar experiments with AG3(T2AG3)3 were previously explained by a structural transition of the antiparallel quadruplex to a hybrid (3 + 1) quadruplex structure (26,30,40). However, we argue that structures stabilized by Na+ and K+ ions have essentially the same antiparallel topology. Though CD spectroscopy does not provide direct evidence for the structural arrangement, the course of spectral changes enables one to clearly distinguish non-cooperative processes (structural changes within a single conformational state) from cooperative processes (conformational transitions between discrete structures separated by an energy barrier) (41,42).
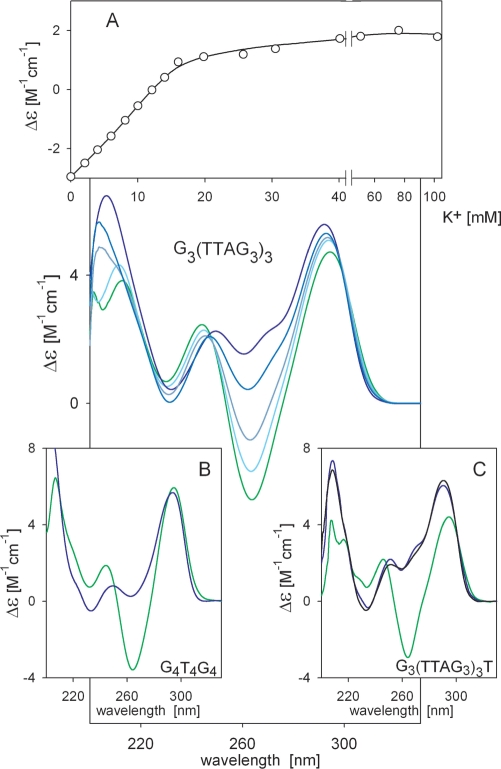
Figure 1.
CD spectra reflecting KCl-induced changes in the sodium-stabilized quadruplex of G3(TTAG3)3. The molecule was annealed in 10 mM sodium phosphate (pH 7) with 135 mM NaCl as described in ‘Materials and Methods’ section (green line). Blue lines (from light to dark) correspond to KCl concentrations of 4, 8, 16 and 102 mM. CD spectra were measured in 0.1 cm cells at 0°C immediately after salt addition (no changes took place over time). (A) KCl-induced spectral changes monitored at 265 nm. (B) CD spectra of G4T4G4 in 100 mM NaCl (green line) or 100 mM KCl (blue line) measured in 0.1 cm cells at 0°C. (C) CD spectra of 6 × 10−5 M G3(TTAG3)3T in 70 mM NaCl (green line) or 70 mM KCl (blue line) and at 0.055 M DNA [concentration used for NMR measurements (36)] in 70 mM KCl (black line) at 23°C.
The CD changes observed during addition of K+ to sodium-stabilized G3(T2AG3)3 were fast (no changes were observed with time) and nearly linear before saturation, thus non-cooperative (Figure 1A). In contrast, the transition from antiparallel to (3 + 1) arrangement should be a complicated process. The change in topology coupled with breaking and reunion of at least 12 hydrogen bonds and changes in glycosidic torsion angles should represent much higher energetic demands than the recently reported 1.4–2.4 kcal mol−1 energy barrier that separates the Na+- and K+-stabilized quadruplexes (43). Any transition between the discrete conformational states that must overcome an energetic barrier should be cooperative. Similar non-cooperative CD spectral changes caused by Na+ for K+ exchange were also observed for the bimolecular quadruplex of G4T4G4 (Figure 1B), whereas both NMR (10) and X-ray crystallography (44) revealed the same topology for G4T4G4 in both salts. This clearly shows that the extensive changes in the CD spectrum are not a consequence of a change in quadruplex topology. The same conclusion follows from Figure 1C, in which we show CD spectra of G3(T2AG3)3T in the presence of Na+ and K+ ions. The spectral change is similar to that observed for G3(T2AG3)3. This 22-mer was shown to adopt an antiparallel quadruplex even in the presence of K+ ions (36). The spectrum did not substantially change at DNA concentrations used for NMR (discussed below). Thus, the spectrum observed in the presence of K+ ions does not correspond to the (3 + 1) quadruplex. The fast non-cooperative changes in CD spectrum after addition of K+ ions to the Na+-stabilized quadruplexes probably reflect changes in stacking of quadruplex tetrads due to specific K+ ions coordination (45). The changes in stacking interactions are sensitively detected by CD (41).
A number of spectroscopic papers ascribe, in agreement with NMR results, the CD spectral changes caused by the exchange of Na+ for K+ ions to the transition from the basket-type antiparallel form to a hybrid (3 + 1) quadruplex (26,40,46). However, direct evidence of a hybrid (3 + 1) structure for the four-repeat human telomeric sequence in K+ solution has been provided only by NMR (26,27). In contrast, many experiments show that addition of potassium ions does not cause large structural changes: these analyses include structural and dynamics data based on single-molecule resonance energy transfer (23), studies incorporating 125I-radioprobing (22), NMR studies assisted by the incorporation of 8-bromoguanines (32), and studies of a comparably fast kinetics of conformational changes of short and long telomeric DNA (31). Also, platinum crosslinking of the same adenines and guanines in Na+- and K+-stabilized human telomere quadruplexes provided evidence that the same guanines occupy identical tetrads in the both salts (21). The same conclusion follows from our previous paper (47): We showed that substitutions of adenine for particular guanines in Na+- or K+-stabilized G3(T2AG3)3 quadruplexes exert a similar effect on guanines forming the upper tetrad and those forming the bottom tetrad as expected for the antiparallel quadruplex model of Wang and Patel (18).
In spite of the above arguments, NMR measurements have unambiguously shown that the four-repeat human telomeric sequence forms the (3 + 1) quadruplex in the presence of K+ ions. We explain this apparent discrepancy between optical and NMR observations in the following chapters.
Flanking nucleotides stabilize the hybrid (3 + 1) quadruplex but long telomere DNA molecules form antiparallel structure like G3(TTAG3)3
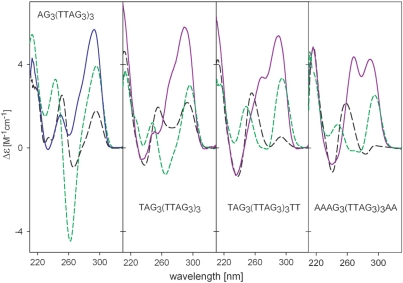
The presence of multiple G-quadruplex conformations in K+ solution under conditions of NMR measurements makes structural elucidation difficult (26). To stabilize a single arrangement, several nucleotides were appended to either 5′ or both ends. Figure 2 shows the CD spectra of AG3(T2AG3)3, TAG3(T2AG3)3, TAG3(T2AG3)3T2 and A3G3(T2AG3)3A2, studied earlier by NMR (26,27,48). The CD spectrum of the 22-mer AG3(T2AG3)3 was similar to that of G3(T2AG3)3 in K+ solution. The addition of the flanking nucleotides resulted in a shoulder on the short wavelength side of the 295 nm CD band. The height of this shoulder increased with the number of flanking nucleotides; two separated bands of comparable magnitude were observed at 260 and 295 nm for the 26-mer A3G3(T2AG3)3A2 (Figure 2). We previously observed a similar CD spectrum for G3(T2AG3)4, which contains one redundant T2AG3 repeat, and suggested that it corresponds to a quadruplex with three parallel and one antiparallel strands (28). Later, the hybrid (3 + 1) structure was reported to be adopted by all the sequences mentioned in Figure 2 in K+ solution (26,27,48). Our data indicate that increasing the number of flanking nucleotides shifts the equilibrium towards the (3 + 1) arrangement. The same was shown for addition of nucleotides to the 5′ end of telomeric oligonucleotides (49). However, this is not true for G3(T2AG3)3T (36), implying that the identity of the appended nucleotides is also important.
Figure 2.
Dependence of CD spectra on DNA sequence in 1 mM sodium phosphate (pH 7) (black dashes) in the presence of 10 mM sodium phosphate plus 135 mM NaCl (green dashes) and after addition of 100 mM KCl (blue or violet lines).
The changes in equilibrium between the antiparallel and hybrid (3 + 1) topology were also observed after addition of quadruplex stabilizing ligands (49,50). Ligands may not simply distinguish and stabilize the preferred topology, but might directly support the formation of a desired topology and shift the equilibrium towards it (51). Stabilization of the parallel quadruplex by a ligand was also recently described (52).
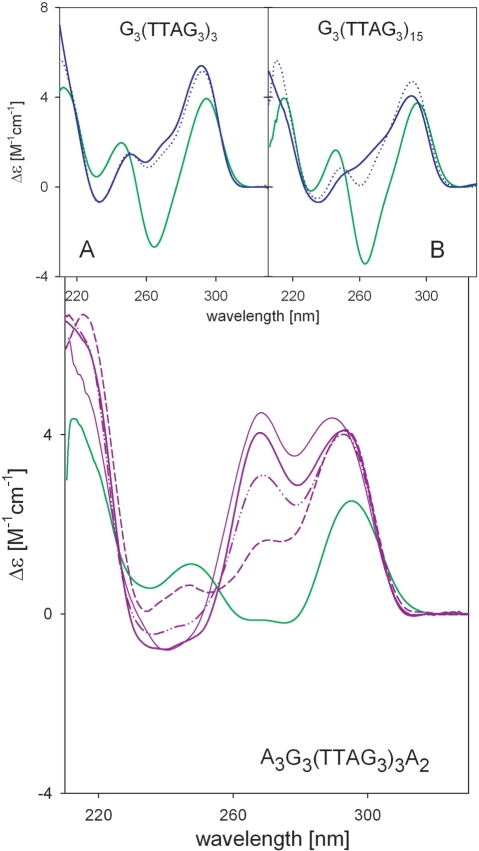
The 26-mer A3G3(T2AG3)3A2 adopted the hybrid (3 + 1) quadruplex structure even under conditions of our spectroscopic measurements. This allowed us to show that there is a complicated K+-induced transition to the (3 + 1) arrangement (Figures 3 and S1). The CD changes observed upon addition of K+ ions to the Na+-stabilized quadruplex of A3G3(T2AG3)3A2 consisted of two processes: The first was fast and was also observed with G3(T2AG3)3; the second corresponded to the slow transition into the hybrid (3 + 1) structure (Supplementary Figure S1). The second transition is cooperative and slow, consistent with a change in topology. This is contrast to G3(T2AG3)3. No changes were observed over time with G3(T2AG3)3 as K+ was gradually added (Figure 1) and changes upon one-shot addition of 100 mM K+ were fast (Figures 3A and Supplementary S1A).
Figure 3.
CD spectra of A3G3(TTAG3)3A2 in 10 mM sodium phosphate buffer (pH 7) with 135 mM NaCl (green line) and upon addition of 30 mM K+ (violet lines) measured immediately (dashes), after 3 h (dash-dot-dot), and 1 day (thick solid line), and with 100 mM K+ after 3 days (thin solid line). Upper CD spectra are of G3(TTAG3)3 and G3(TTAG3)15 in 10 mM sodium phosphate buffer (pH 7) with 150 mM NaCl (green line) and after addition of 100 mM K+ measured immediately (blue dots) and after 3 days (blue line). CD spectra were measured in 0.1 cm cells at 0°C.
It is to be noted that CD spectra of the oligonucleotides shown in Figure 2 differ even in Na+ solutions. The deep negative 260 nm band of AG3(T2AG3)3 diminished as flanking nucleotides were added. This may indicate destabilization of the antiparallel quadruplex arrangement.
The telomeric sequence in vivo is much longer then the four G tracts motifs usually studied. We previously suggested (28) that long telomeric sequences have a bead-like arrangement, similar to nucleosomes. Each bead is formed by a G3(T2AG3)3 quadruplex and the beads are linked by TTA triplets. The beads-on-a-string arrangement was later supported by other authors (53). Inserts in Figure 3 show that the long fragment G3(T2AG3)15 responded to Na+ for K+ ion exchange in the same way as the core G3(T2AG3)3 oligonucleotide: the negative 260 nm CD band of the Na+-stabilized quadruplex immediately diminished upon addition of 100 mM K+. Only slight changes occurred with time, but in contrast to the spectrum of A3G3(T2AG3)3A2, no positive band at 260 nm was observed in the spectrum of G3(T2AG3)15 even after a week (Figure 3B). Thus, G3(T2AG3)3 is the basic quadruplex motif; any flanking nucleotides are irrelevant for the arrangement of the long molecule. The same fast kinetics upon K+ addition was also reported for short and long (T2AG3)4 and (T2AG3)13 fragments (31). G3(T2AG3)16, containing one redundant repeat, yielded a spectrum very similar to that of G3(T2AG3)15 in both Na+ and K+ ions (data not shown). Thus, whereas flanking sequences dramatically influence the spectrum of the basic four-G-block telomeric sequence, redundant repeats exert nearly no effect on long telomeric DNA. We conclude that long human telomeric sequences adopt the antiparallel arrangement in K+ solutions under conditions of spectroscopic experiments.
Ethanol induces the transition from antiparallel to hybrid (3 + 1) and to parallel quadruplex in the presence of K+
Ethanol is a very potent inducer of quadruplex formation (54). Ethanol, like polyethylene glycol, simulates the crowded environment inside cells. These compounds effectively change the DNA concentration by a mechanism of excluded volume and by changing the activity of water (55). Addition of ethanol to G3(T2AG3)3 or AG3(T2AG3)3 in 150 mM KCl led to a slow formation of a CD spectrum corresponding to (3 + 1) quadruplex and to a parallel quadruplex upon denaturation (Supplementary Figure S2). A similar result was observed in PEG solutions (37).
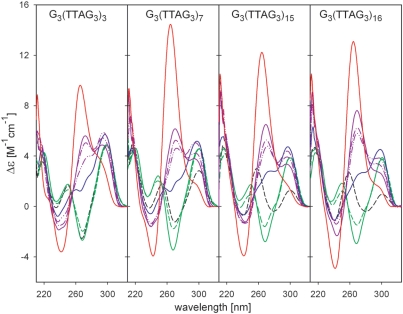
At low temperature and in the absence of K+ ions, low (20%) ethanol concentrations stabilized the antiparallel arrangement of G3(T2AG3)n telomeric sequences (Figure 4). This was especially obvious with the longer sequences, which do not form quadruplex at low ionic strength. Further increase in ethanol concentration diminished the negative 260 nm peak but no structural transition occurred, even after thermal denaturation (not shown). The negative 260 nm CD band was disappeared immediately after the first K+ addition and then, at 2 mM K+ concentration, the CD spectrum corresponding to the (3 + 1) quadruplex appeared with slow kinetics. The structural rearrangement was facilitated by sequence length (Figure 4), as expected for a cooperative transition. Further increases in K+ concentration did not significantly change the CD spectra. However, the spectrum of the longest fragment analyzed, G3(T2AG3)16, indicated a substantial amount of the parallel conformation after a 5-day incubation in the presence of 2 mM K+. Thermal denaturation (Figure 4) gave rise to the CD spectrum with a high-amplitude positive peak at 265 nm, characteristic of parallel quadruplex (Figure 4). The need for denaturation or an extremely long incubation time indicates a further energetic barrier between the hybrid (3 + 1) and the parallel quadruplex forms. The transition to a parallel quadruplex was greatly facilitated by increased temperature: at 37°C in 50% ethanol and 2 mM K+ the parallel quadruplex was formed within 24 h without denaturation (Supplementary Figure S3).
Figure 4.
CD spectra of G3(TTAG3)n in water/ethanol solution in the absence or presence of potassium ions. Spectra in 1 mM sodium phosphate buffer, pH 7 (black dashes), with 20% ethanol (green line), and 57% ethanol (green dashes); KCl was added to the oligonucleotides in 57% ethanol to 1 mM (blue line) and 2 mM concentration (violet lines) and measured immediately (dash-dot-dot), after 1 (dash-dot), and after 5 days (full line). CD spectra were measured in 1 cm cells at 0°C. Red lines correspond to the oligonucleotides in 50% ethanol and 2 mM KCl measured at 23°C after thermal denaturation.
The conformational isomerization from the antiparallel to (3 + 1) and parallel quadruplex with the increasing molecule length may be facilitated by decreasing stability of the antiparallel quadruplex as length increases (28). Also, quadruplex units within the long molecule may interact to support formation of parallel arrangements. An experimental proof has been recently provided for the existence of intramolecular quadruplex–quadruplex interactions (56) between two contiguous quadruplex motifs, which influence their overall structure.
Human telomeric quadruplex folding is determined by DNA concentration
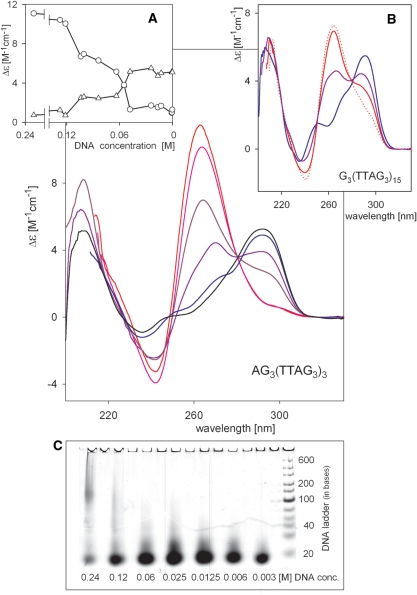
X-ray crystallography, NMR and CD spectroscopy evaluate DNA structure at different DNA concentrations. Using X-ray crystallography, AG3(T2AG3)3 was shown to form a parallel quadruplex in K+ solution, whereas NMR revealed a hybrid (3 + 1) quadruplex. Our CD spectroscopic results showed that both G3(T2AG3)3 and AG3(T2AG3)3 adopt an antiparallel basket arrangement in K+-containing solution at DNA concentrations usual for CD spectroscopy (0.05–1 mM; to be consistent with Δε expression, we state DNA concentration related to nucleosides).
Due to these discrepancies, we have studied the effect of human telomere DNA concentration on its structure. Using extremely short path-length cells we were able to obtain spectra at DNA concentrations up to 200 mM. Up to 40 mM, AG3(T2AG3)3 provided a CD spectrum similar to those measured at low concentrations in 1 cm cells (Figure 5A). However, at higher concentrations the amplitude of the 295 nm positive peak decreased and a 260 nm positive peak was observed. These spectral characteristics are similar to those of the hybrid (3 + 1) arrangement observed in ethanol solutions (Figure 4). NMR studies of the hybrid (3 + 1) structure employed DNA strand concentrations of 5 × 10−3 to 5 × 10−4 M (27) or 3.5 × 10−3 to 1.0 × 10−4 M (26). In this concentration range, CD spectra have two positive peaks. At very high DNA concentrations, around 100 mM in nucleosides, the parallel quadruplex arrangement of AG3(T2AG3)3 was stabilized as indicated by the high amplitude positive CD peak at 260 nm (Figure 5). It is to be noted that formation of intermolecular associates was observed at these extremely high concentrations (Figure 5, bottom insert), which might contribute to the positive 260 nm CD signal. However, the majority of oligonucleotides formed intramolecular structures at this concentration and the CD spectra did not reveal the presence of antiparallel quadruplex. We have tested various ways of preparing the concentrated samples, including preparation without annealing and omission of heating [to mimic NMR sample preparation (26,27)]. The sample preparation protocol used for the spectra shown in Figure 5 (i.e. denaturation in low salt and annealing in the presence of 0.15 M KCl) led to the lowest population of intermolecular associates in samples with DNA nucleoside concentrations close to 0.1 M.
Figure 5.
Dependence of CD spectra of AG3(TTAG3)3 on DNA concentration. AG3(TTAG3)3 in 10 mM potassium phosphate (pH 7.2) and 0.15 M KCl measured at room temperature in 0.001 cm to 1 cm cells. Every sample was prepared separately. Samples were first denatured at 90°C for 3 min in 1 mM sodium phosphate plus 0.3 mM EDTA, then cooled to room temperature. After addition of KCl, samples were heated for 3 min at 90°C and then cooled to room temperature over the course of 3 h. Sample concentrations were determined on the basis of defined dilution from the stock solution and refined by measuring absorption taking in account the difference in extinction coefficients in low salt and 90°C and in 150 mM KCl and room temperature. DNA concentrations in nucleosides: 0.82 mM (black), 16.4 mM (blue), 55 mM (violet), 100 mM (dark red), 121 mM (pink) and 232 mM (red). (A) Dependence of AG3(TTAG3)3 CD spectrum on DNA concentration monitored at 265 (circles) and 295 nm (triangles). (B) CD spectra of G3(TTAG3)15 prepared and measured in the same way as described for AG3(TTAG3)3. Concentration in nucleosides: 0.44 mM (blue), 88 mM (violet), 156 mM (red) and 204 mM (red dots indicate that the sample formed a gel). (C) PAGE of AG3(TTAG3)3 samples at indicated concentrations. The samples were prepared as for CD experiments. Concentrations were estimated based on dilution a stock solution. About 2 µg of DNA was loaded in each lane on the gel.
The spectra shown in Figure 5 indicate that human telomere DNA fragment AG3(T2AG3)3 can, depending on its concentration, adopt each of the three types of intramolecular quadruplex arrangements previously characterized at the atomic level (18,20,26,27). The plateau in the dependencies of Δε at 260 and 295 nm on DNA concentration (Figure 5A) indicates that the (3 + 1) structure is probably an inevitable intermediate between parallel and antiparallel quadruplex arrangements. The spectra of G3(T2AG3)3 21-mer (Supplementary Figure S4) and the long telomeric sequence G3(T2AG3)15 (Figure 5B) were similarly dependent on concentration. The positive 260 nm CD band of G3(T2AG3)15 had lower ellipticity values than 22- or 21-mers at comparable nucleoside concentrations (Figure 5B); strand concentrations are, however, four times lower. At nucleoside concentrations higher than 150 mM, G3(T2AG3)15 formed a gel.
Interestingly, the CD spectrum of G3(T2AG3)3T, which was shown by NMR to form antiparallel quadruplex, did not change with DNA concentration (Figure 1C). We recorded the spectra of the oligonucleotide at 11 mM and 55 mM concentrations to bracket the concentration range of the NMR analysis (36). The CD spectral shape was the same in K+ solution as with this oligonucleotide or with G3(T2AG3)3 at low DNA concentration. Thus, CD does not distinguish that the G3(T2AG3)3T quadruplex, formed at high DNA concentration, contains only two guanine tetrads as determined by NMR (36). It may be supposed that guanines in the deficient tetrad stack in a very similar way as in the standard three-tetrad quadruplex. This is supported by very high stability of the G3(T2AG3)3T quadruplex (36). As CD does not distinguish the three-tetrad from the two-tetrad quadruplex, it is possible that the K+-stabilized quadruplex of G3(T2AG3)3 transforms, with slow kinetics or at high DNA concentration, to the same quadruplex form as observed with G3(T2AG3)3T. Experiments are in progress to explore this possibility. In every case, however, the K+-stabilized quadruplex of G3(T2AG3)3 is antiparallel.
The concentration-dependent conformational transitions of the human telomere sequence are the same as those observed in the presence of ethanol. However, the DNA concentration used for these analyses, which leads to the structural changes, is too low to cause the crowding effect attributed to ethanol. The concentration-dependent structural changes are probably a consequence of weak interactions among the intramolecular quadruplexes. An analogical situation was observed with poly(dG)·poly(dC). Raman spectroscopy showed that the polynucleotide transformed in aqueous solution from B to A conformation depending on its concentration (57). This structural change took place in the same concentration region as observed in our work: The polymer adopted B-form at 10 mM and A-form at 200 mM DNA concentration. The authors note that a 0.2 M concentration of DNA in aqueous solution should not cause the dehydration needed to cause a B–A transition and suggested that the observed transition was a result of interduplex associations.
Figure 6 illustrates the observed structures of long telomere DNA molecules in physiologically relevant concentrations of K+. In the antiparallel arrangement, the tetrads of the beads are oriented parallel to the longitudinal axis of the molecule. This may be why elongation of the sequence does not, as it does for duplexes, contribute to stabilization of the quadruplex structure: Quadruplex stability actually decreases with the molecule length (28). For the (3 + 1) quadruplex, the bead tetrads are oriented across the longitudinal axis and are mutually parallel. In the parallel arrangement, the loops are on the sides of the beads and the tetrads may stack on each other as suggested by Haider et al. (58). Regrettably, neither the high DNA concentrations, evaluated in narrow foldaway cells, nor ethanol conditions allow us to verify the dependence of the parallel quadruplex thermal stability on the molecule length.
Figure 6.
Schematics of quadruplex structures of short and long human telomere molecules in physiologically relevant K+-containing solutions: (A) antiparallel quadruplex, (B) hybrid (3 + 1) quadruplex and (C) parallel quadruplex.
The described polymorphism of the human telomere quadruplex is restricted to K+-containing solutions. No analogous CD changes took place within a comparable concentration range in Na+-solutions (Supplementary Figure S4).
CONCLUSIONS
We show that the arrangement of the human telomeric quadruplex strongly depends on DNA concentration. This explains why distinct structures were reported by various methods. CD spectroscopy revealed very similar antiparallel arrangements of G3(T2AG3)3 both in K+ and Na+ solution at low DNA concentrations. In contrast, NMR studies revealed a hybrid (3 + 1) arrangement in K+ for extended G3(T2AG3)3 analogs. The flanking nucleotides stabilized the hybrid (3 + 1) arrangement. However, the long telomeric sequences behave in the same way as the basic quadruplex unit G3(T2AG3)3. The data reported here show that human telomeric DNA can adopt, depending on concentration, antiparallel, (3 + 1) and parallel quadruplex arrangements in physiologically relevant solvent conditions. There is no doubt that nature can take advantage of this conformational variability. Determining the function of particular arrangements will be a challenge for future research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant Agency of the Czech Republic (grant 204/07/0057, 204/00/D012) and the Grant Agency of the Academy of Sciences of the Czech Republic (grants IAA 100040701, AVOZ50040507, AVOZ50040702). Funding for open access charge: Grant Agency of the Academy of Sciences of the Czech Republic (grants IAA 100040701).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are indebted to Prof. Janos Sagi and Dr Jaroslav Kypr for careful reading the article and valuable comments.
REFERENCES
- 1.Hayflick L, Moorhead PS. Serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Olovnikov AM. Principle of marginotomy in template synthesis of polynucleotides. Dokl. Akad. Nauk SSSR. 1971;201:1496–1499. [PubMed] [Google Scholar]
- 3.Watson JD. Origin of concatemeric T7 DNA. Nat. New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 4.Harley CB, Futcher AB, Greider CW. Telomeres shorten during aging of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 5.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 6.Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl Acad. Sci. USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henderson ER, Blackburn EH. An overhanging 3′ terminus is a conserved feature of telomeres. Mol. Cell Biol. 1989;9:345–348. doi: 10.1128/mcb.9.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 10.Smith FW, Feigon J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- 11.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA-sequence, (TTAGGG)n, present at the telomeres of human-chromosomes. Proc. Natl Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granotier C, Pennarun G, Riou L, Hoffschir F, Gauthier LR, De Cian A, Gomez D, Mandine E, Riou JF, Mergny JL, et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005;33:4182–4190. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 14.Izbicka E, Nishioka D, Marcell V, Raymond E, Davidson KK, Lawrence RA, Wheelhouse RT, Hurley LH, Wu RS, Von Hoff DD. Telomere-interactive agents affect proliferation rates and induce chromosomal destabilization in sea urchin embryos. Anticancer Drug Des. 1999;14:355–365. [PubMed] [Google Scholar]
- 15.Zimmermann S, Martens UM. Telomeres and telomerase as targets for cancer therapy. Cell Mol. Life Sci. 2007;64:906–921. doi: 10.1007/s00018-007-6481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Cian A, Lacroix L, Douarre C, Temime-Smaali N, Trentesaux C, Riou JF, Mergny JL. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 19.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 20.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 21.Redon S, Bombard S, Elizondo-Riojas MA, Chottard JC. Platinum cross-linking of adenines and guanines on the quadruplex structures of the AG3(T2AG3)3 and (T2AG3)4 human telomere sequences in Na+ and K+ solutions. Nucleic Acids Res. 2003;31:1605–1613. doi: 10.1093/nar/gkg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He YJ, Neumann RD, Panyutin IG. Intramolecular quadruplex conformation of human telomeric DNA assessed with 125I-radioprobing. Nucleic Acids Res. 2004;32:5359–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ying LM, Green JJ, Li HT, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ourliac-Garnier I, Elizondo-Riojas MA, Redon S, Farrell NP, Bombard S. Cross-links of quadruplex structures from human telomeric DNA by dinuclear platinum complexes show the flexibility of both structures. Biochemistry. 2005;44:10620–10634. doi: 10.1021/bi050144w. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambrus A, Chen D, Dai JX, Bialis T, Jones RA, Yang DZ. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: An intramolecular (3+1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vorlickova M, Chladkova J, Kejnovska I, Fialova M, Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi JY, Shafer RH. Covalent ligation studies on the human telomere quadruplex. Nucleic Acids Res. 2005;33:3185–3192. doi: 10.1093/nar/gki632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorgan. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 31.Chang CC, Chien CW, Lin YH, Kang CC, Chang TC. Investigation of spectral conversion of d(TTAGGG)4 and d(TTAGGG)13 upon potassium titration by a G-quadruplex recognizer BMVC molecule. Nucleic Acids Res. 2007;35:2846–2860. doi: 10.1093/nar/gkm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsugami A, Xu Y, Noguchi Y, Sugiyama H, Katahira M. Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution. FEBS J. 2007;274:3545–3556. doi: 10.1111/j.1742-4658.2007.05881.x. [DOI] [PubMed] [Google Scholar]
- 33.Antonacci C, Chaires JB, Sheardy RD. Biophysical characterization of the human telomeric (TTAGGG)4 repeat in a potassium solution. Biochemistry. 2007;46:4654–4660. doi: 10.1021/bi602511p. [DOI] [PubMed] [Google Scholar]
- 34.Dai JX, Carver M, Punchihewa C, Jones RA, Yang DZ. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rujan IN, Meleney JC, Bolton PH. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim KW, Amrane S, Bouaziz S, Xu WX, Mu YG, Patel DJ, Luu KN, Phan AT. Structure of the Human Telomere in K+ Solution: A stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue Y, Kan ZY, Wang Q, Yao Y, Liu J, Hao YH, Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 38.Pedroso IA, Duarte LF, Yanez G, Baker AM, Fletcher TM. Induction of parallel human telomeric G-quadruplex structures by Sr2+ Biochem. Biophys. Res. Co. 2007;358:298–303. doi: 10.1016/j.bbrc.2007.04.126. [DOI] [PubMed] [Google Scholar]
- 39.Gray DM, Hung SH, Johnson KH. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995;246:19–34. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]
- 40.Gray RD, Chaires JB. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008;36:4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucl. Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vorlickova M. Conformational transitions of alternating purine-pyrimidine DNAs in perchlorate ethanol solutions. Biophys. J. 1995;69:2033–2043. doi: 10.1016/S0006-3495(95)80073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gray RD, Li J, Chaires JB. Energetics and kinetics of a conformational switch in G-quadruplex DNA. J. Phys. Chem. B. 2009;113:2676–2683. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haider S, Parkinson GN, Neidle S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 45.Hud NV, Plavec J. The role of cations in determining quadruplex structure and stability. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. 2006. pp. 100–130. [Google Scholar]
- 46.Dai JX, Carver M, Yang DZ. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomasko M, Vorlickova M, Sagi J. Substitution of adenine for guanine in the quadruplex-forming human telomere DNA sequence G3(T2AG3)3. Biochimie. 2009;91:171–179. doi: 10.1016/j.biochi.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 48.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaynutdinov TI, Neumann RD, Panyutin IG. Structural polymorphism of intramolecular quadruplex of human telomeric DNA: effect of cations, quadruplex-binding drugs and flanking sequences. Nucleic Acids Res. 2008;36:4079–4087. doi: 10.1093/nar/gkn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rezler EM, Seenisamy J, Bashyam S, Kim MY, White E, Wilson WD, Hurley LH. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 51.De Cian A, Mergny JL. Quadruplex ligands may act as molecular chaperones for tetramolecular quadruplex formation. Nucleic Acids Res. 2007;35:2483–2493. doi: 10.1093/nar/gkm098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Q, Xiang J, Li X, Chen L, Xu X, Tang Y, Zhou Q, Li L, Zhang H, Sun H, et al. Stabilizing parallel G-quadruplex DNA by a new class of ligands: two non-planar alkaloids through interaction in lateral grooves. Biochimie. 2009;91:811–819. doi: 10.1016/j.biochi.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 53.Yu HQ, Miyoshi D, Sugimoto N. Characterization of structure and stability of long telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2006;128:15461–15468. doi: 10.1021/ja064536h. [DOI] [PubMed] [Google Scholar]
- 54.Vorlickova M, Bednarova K, Kypr J. Ethanol is a better inducer of DNA guanine tetraplexes than potassium cations. Biopolymers. 2006;82:253–260. doi: 10.1002/bip.20488. [DOI] [PubMed] [Google Scholar]
- 55.Miyoshi D, Sugimoto N. Molecular crowding effects on structure and stability of DNA. Biochimie. 2008;90:1040–1051. doi: 10.1016/j.biochi.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 56.Schonhoft JD, Bajracharya R, Dhakal S, Yu ZB, Mao HB, Basu S. Direct experimental evidence for quadruplex-quadruplex interaction within the human ILPR. Nucleic Acids Res. 2009;37:3310–3320. doi: 10.1093/nar/gkp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishimura Y, Torigoe C, Tsuboi M. An A-form poly(dG).poly(dC) in H2O solution. Biopolymers. 1985;24:1841–1844. doi: 10.1002/bip.360240913. [DOI] [PubMed] [Google Scholar]
- 58.Haider S, Parkinson GN, Neidle S. Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophys. J. 2008;95:296–311. doi: 10.1529/biophysj.107.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.