Abstract
Both the ERCC1-XPF complex and the proteins involved in homoIogous recombination (HR) have critical roles in inter-strand cross-link (ICL) repair. Here, we report that mitomycin C-induced lesions inhibit replication fork elongation. Furthermore, mitomycin C-induced DNA double-strand breaks (DSBs) are the result of the collapse of ICL-stalled replication forks. These are not formed through replication run off, as we show that mitomycin C or cisplatin-induced DNA lesions are not incised by global genome nucleotide excision repair (GGR). We also suggest that ICL-lesion repair is initiated either by replication or transcription, as the GGR does not incise ICL-lesions. Furthermore, we report that RAD51 foci are induced by cisplatin or mitomycin C independently of ERCC1, but that mitomycin C-induced HR measured in a reporter construct is impaired in ERCC1-defective cells. These data suggest that ERCC1–XPF plays a role in completion of HR in ICL repair. We also find no additional sensitivity to cisplatin by siRNA co-depletion of XRCC3 and ERCC1, showing that the two proteins act on the same pathway to promote survival.
INTRODUCTION
Inter-strand cross-link (ICL)-inducing agents cause a unique class of DNA lesions that are extremely cytotoxic. ICLs covalently modify both strands of the DNA, thus preventing their separation and consequently blocking transcription, segregation and replication (1). ICL agents have been effectively used in anticancer chemotherapy, because their toxicity targets proliferating cells. Even though a variety of DNA adducts can be produced by cross-linking agents, their effective cytotoxicity is related to their capability of forming ICLs (2). Due to the complexity of the ICL-induced DNA damage, repair machinery that simply excises the damaged DNA and then uses replication from a template for nucleotide replacement is inadequate. This has been well demonstrated in Escherichia coli and Saccharomyces cerevisiae, where components of nucleotide excision repair (NER), homologous recombination (HR) and translesion synthesis (TLS) are all required for adequate ICL repair (3,4). However, it is shown that a replication and recombination independent ICL repair machinery is active on transcribed DNA (5,6).
HR and TLS are both needed for ICL repair in yeast and higher eukaryotes, but not all NER proteins are involved in ICL repair in mammals. Studies have proposed that among all the NER proteins only ERCC1 and XPF play a key role in ICL repair (7). The ERCC1 protein forms a highly conserved endonuclease heterodimeric complex with XPF (8) that stabilizes both proteins for their role in DNA repair (9,10). In mammalian cells, this structure-specific heterodimeric endonuclease complex, ERCC1-XPF, is recruited by XPA to the damaged DNA site (11) to create a nick at the 5′ side of the helix-distorting lesion. A structure specific endonuclease, XPG, compliments ERCC1-XPF by nicking the DNA on the other side of the lesion, thus excising the damaged DNA (12). These endonucleases also cleave many DNA structures, including bubbles, stem–loops, splayed arms and flaps (13–15). Importantly, mammalian XPG mutants were shown to be less sensitive to ICL damage when compared to ERCC1 and XPF mutants (7,16) implying that ICL repair in mammals is different from ICL repair in lower eukaryotes, where both endonuclease activities are necessary for efficient ICL repair (17).
Moreover, XPF patients’ cells were shown to proficiently process ICLs but failed to deal with mono-adducts via NER (18), suggesting that XPF functions in ICL repair in an NER independent manner. The ERCC1-XPF endonuclease complex has been reported to cleave an artificial substrate on both sides of a crosslink lesion, flanking a single-stranded 3′ flap, induced by psoralen (19). Therefore the ERCC1-XPF endonuclease function was designated as the initiation of the strand breaks adjacent to, and on each side of, the lesion in one strand of the DNA during ICL repair, known as the unhooking step. However, this contradicts cellular findings that ICL-inducing agents cause the same increased level of DNA double-strand break (DSB) formation in ERCC1 and XPF mutant cells as in wild-type cells (16,20). These findings suggest that ERCC1-XPF functions after formation of the DSB. In addition to their role in NER and ICL repair, the ERCC1–XPF complex has been found to function in some sub-pathways of homology-directed DSB repair, through single-strand annealing, gene conversion and homologous gene targeting (21–24).
The exact role of the ERCC1–XPF complex in ICL repair is still under debate (25), although most data suggest a link with HR, as discussed above. Here, we use ERCC1-defective or siRNA-depleted cells to study the role of the ERCC1–XPF complex and report that it is involved in the late stage of HR during ICL repair, but we also point to a role for ERCC1 in ICL repair that is distinct from HR. Furthermore, we provide more insight into the formation of replication-associated ICL-induced DSBs.
MATERIALS AND METHODS
Cell lines
All cell lines used were cultured in Dulbecco's; Modified Eagle's; Medium (DMEM) supplemented with 10% fetal calf serum, penicillin (60–100 µg/ml), streptomycin (100 µg/ml) at 37°C containing 5% CO2, in a humidified incubator. The medium was supplemented with hygromycin (0.05 mM) in order to maintain the DRneo vector and SCneo vector, and Blasticidin (3 µg/ml) to maintain the PEf6-V5-His- ERCC1 and PEF6-V5-His-Topo vector-containing cell lines. The 51D1SC.2, 51D1SC.4, AA8SN.6, AA8SN.10 and AA8SN.12 cell lines were created following the electroporation of the SCneo vector into the parental AA8 and Rad51D1–/– 51D1 cells and isolation of individual clones as described earlier (Table 1) (26). The SQ20B human head and neck cancer cell line was obtained from ATCC.
Table 1.
Genotype and origin of Chinese hamster cell lines used in this study and IC50 values for mitomycin C (MMC), cisplatin and melphalan
| Cell line | Genotype | Defect / modification | Origin | IC50 [MMC] (nM) | IC50 [cisplatin] (nM) | IC50 [melphalan] (nM) | Reference |
|---|---|---|---|---|---|---|---|
| 51D1 | RAD51D−/− | RAD51D knockout | AA8 | ND | ND | ND | (26) |
| 51D1SC.2, | RAD51D−/− | Integrated SCneo | 51D1 | 3.07 | 97.86 | 0.23 | This study |
| 51D1SC.4 | recombination reporter (hygR) | 3.44 | 116.10 | ND | |||
| AA8 | wt | wt | Ovary | 238.98 | ND | 6.94 | (35) |
| AA8SN.10, | wt | Integrated SCneo | AA8 | 275.05 | >1000 | ND | This study |
| AA8SN.12 | recombination reporter (hygR) | 290.46 | >1000 | ND | |||
| ERCC1.17, | wt | Integrated DRneo | UV4DR7 | 164.41 | >1000 | 6.97 | (22) |
| ERCC1.21 | recombination reporter (hygR) + ERCC1 expressing vector | 164.70 | >1000 | ND | |||
| irs1SF | XRCC3− | XRCC3−, deficient in HR | AA8 | 1.36 | ND | ND | (35) |
| PEF7 | ERCC1− | integrated DRneo recombination reporter (hygR) + empty vector | UV4DR7 | 1.49 | 61.24 | 0.33 | (22) |
| S8DN4 | wt | Integrated DRneo recombination reporter (hygR) | SPD8 | ND | 919.64 | ND | (22) |
| UV4DR7 | ERCC1− | Integrated DRneo recombination reporter (hygR) | AA8 | 1.45 | 77.61 | 0.40 | (22) |
| V79Z | wt | wt | Lung | ND | ND | 8.71 | (58) |
| V-C8 | BRCA2− | BRCA2−, deficient in HR | V79Z | ND | ND | 0.08 | (59) |
| V-C8+B2 | wt | Expressing BRCA2 from vector | V-C8 | ND | ND | 27.80 | (59) |
ND, not determined.
Survival assays for hamster cells
Five hundred cells from each cell line (V-C8, V-C8+B2, V79Z, AA8SN.10, AA8SN.12, 51D1SC.2, 51D1SC.4, UV4DR7, ERCC1.17, ERCC1.21 and PEF7) were seeded over night to be treated with different doses of MMC, cisplatin or melphalan on the following day. After that they were incubated for 7–14 days in the humidified 5% CO2 /37°C incubator, the colonies were fixed and stained with 0.4% methylene blue in methanol to be counted.
Pulsed-field gel electrophoresis
Flasks with either UV4DR7, ERCC1.17, or irs1SF cell lines, were inoculated with 4 × 106 cells for 4 h prior to a 24 h treatment with hydroxyurea (2 mM), 24-h treatment with Aphidocolin (3 µM), 4-h treatment with MMC (0.5, 5, 10 µM), or 24-h treatment with Aphidocolin (3 µM) in addition to a 4-h treatment with MMC (10 µM). After treatment, the cells were released from the flask by trypsinization and 1 × 106 cells were set into each agarose plug (75 ml, 1% InCert Agarose, BMA). Inserts were incubated in 0.5 M EDTA, 1% N-laurylsarcosyl and proteinase K (1 mg/ml) at room temperature for 48 h and thereafter washed four times in TE-buffer prior to loading onto an agarose separation gel (1% chromosomal grade agarose, Bio-Rad). Separation was performed on a CHEF DR III equipment (BioRad; 120_ field angle, 240 s switch time, 4 V/cm, 14°C) for 18 h. The gel was stained with ethidium bromide overnight and subsequently analysed by scanning fluorescence reader (FLA-3000, Fujifilm) using Image Gauge software.
Measurement of NER as incision mediated strand breaks
The incisions during the NER process generate SSBs that will give rise to single-stranded DNA (ssDNA) when treated with alkaline. Not incised labeled DNA is expected to be double stranded (dsDNA). The levels of NER in the samples were estimated by the ratio of ssDNA to dsDNA at a given UV- or MMC-dose.
Chinese hamster AA8 and UV4 cells with homogenously radiolabeled (3H-TdR, 7.4 kBq/ml) DNA were incubated with hydroxyurea (HU, 2 mM) and cytosine arabinocide (AraC, 20 µM) in DMEM for 30 min prior to UV-exposure in order to inhibit the polymerization step in NER. The amount of ssDNA was measured after 1-h post-treatment with HU/AraC by the alkaline DNA unwinding technique as described elsewhere (27,28).
Replication fork elongation assay
In each well of a 24-well plate, 1 × 105 cells were seeded and incubated over night. Medium were exchanged to 1 ml of DMEM containing, 37 kBq 3H-TdR, to each well and incubated in 37°C and 5% CO2 for 30 min. The labelled cells were washed with 500 µl Hanks balances salt solution with calcium, magnesium and 5 ml HEPES (HBSS++), the cells were then treated with MMC in 500 µl HBSS++ 15 min in 37°C. Thereafter each well was washed with 500 µl of HBSS++ prior to incubation at 37°C in fresh DMEM for 0, 0.5, 1, 2, 4 or 6 h. At the selected time points selected cells were washed twice with ice-cold 0.15M NaCl prior to 30 min unwinding in 500 µl 0.03M NaOH at 0°C in darkness. The unwinding were ended by neutralization by adding 1 ml NaH2PO4 to each well and the samples were sonicated for 15 se before the addition of SDS to a final concentration of 0.25% and storage in −20°C until elution as described elsewhere (27).
Recombination assay
To avoid contamination with spontaneous G418-resistant clones in the DRneo an SCneo recombination assays, 103 cells from each cell lines were separately expanded to confluency on ∼10 different Petri dishes for each cell line (Ø 100 mm). The spontaneous recombination frequency was determined for each cell population by selection of 2 × 105 cells/plate (Ø 100 mm) seeded in G418 (100 µg/ml). Only those cell populations that did not retrieve viable G418R colonies were used for further recombination assays, as they have low background frequency of spontaneous G418R cells.
In the SCneo recombination assay, 1.5 × 106 cells of each cell line (51D1SC.2, 51D1SC.4, AA8SN.6, AA8SN.10 and AA8SN.12) were seeded over night, and transiently transfected with or without pCMV3xnls-I-SceI expression vector (1 μg) according to manufacturer’s protocol (Lipofectamine2000TM, Invitrogen) to create DSBs. After 5 h of incubation in the humidified 5% CO2/37°C incubator, cells were changed to complete medium. Twenty-four hours after transfection, the cells were trypsinized and counted.
In the ICL recombination assay, 1.5 × 106 cells of each cell line (UV4DR7, S8DN.4, ERCC1.17, ERCC1.21 and PEF7) were seeded over night. Then S8DN.4, ERCC1.17 and ERCC1.21 were treated with 0 nM, 200 nM and 400 nM of MMC, while the UV4DR7 and PEF7 were treated with 0 nM, 2 nM, 5 nM and 10 nM MMC. After 24 h, the cells were trypsinized and counted.
For recombination frequency, 2 × 105 cells/plate (Ø 100 mm) were seeded and G418 (100 µg/ml) was added for selection to both treated and untreated plates. To determine the cloning efficiency, 500 cells/plate were seeded. Plates were incubated for 7 or 10 days for cloning and selection, respectively, after which they were fixed and stained with 0.4% methylene blue in methanol.
Immunofluorescence
0.3 × 106 of ERCC1.17 and PEF7 cells were plated onto coverslips and grown for 4 h before treatment, with 100 nM cisplatin or 10 μM MMC, or overnight if to be untreated. The medium was removed, the coverslips rinsed once in PBS (37°C) and fixed in 3% paraformaldehyde in PBS-T (PBS containing 0.1% Triton X-100) for 20 min. The coverslips were rinsed once in PBS-T prior to incubation with primary antibody, rabbit polyclonal anti- RAD51 (H-250, Santa Cruz) at a dilution of 1:1000, for 16 h at 4°C. The coverslips were then rinsed 4 × 15 min in PBS-T after which they were incubated for 1 h at room temperature with the secondary antibody, Cy-3-conjugated goat anti-rabbit IgG antibody (Zymed) at a concentration of 1:500, and then they were rinsed 4 × 15 min in PBS-T. The coverslips were then covered with SlowFade Antifade Kit (Molecular Probes). The DNA was stained with 1 mg/ml To Pro (Molecular Probes). The coverslips were then mounted on slides and sealed with nail varnish (slides were stored in the dark). Images were obtained using a Zeiss LSM 510 inverted confocal microscope using a planapochromat 633/NA 1.4 oil immersion objective, and excitation wavelengths 546 and 630 nm.
siRNA transfection and clonogenic survival assay
SQ20B cells were used at ∼70–80% confluency. siRNA targeting ERCC1 (siGENOME 006311) and XRCC3 (siGENOME 012067) were obtained from Dharmacon. siCONTROL Non-Targeting siRNA #2 (D-001210-02-20) from Dharmacon was used as the nontargeting control (NT). siRNAs were resuspended following manufacturer’s protocol. Transfection was carried out in 6-well plates (3 ml per well) using optimized transfection conditions and Dharmacon’s reverse transfection procedure. Seventy-five microliters of 2 µM siRNAs were used per transfection. Dharmafect1 was used at 2.25 µl per transfection. Cells were counted and resuspended in antibiotic free complete medium, and added to the siRNA/Dharmafect mix at 96 000 cells per well. Cells were changed to normal medium 24 h after transfection.
Forty-eight hours after transfection, cells were trypsinized, diluted to the appropriate cell density, plated in 60-mm dishes and allowed to adhere to culture dishes before exposure to drugs. Western blot was performed to check for knockdown. Cisplatin was prepared fresh and dissolved in 0.9% NaCl solution. Cisplatin and Mitomycin C at predetermined concentrations were added to the dishes for 2 h before changing to drug-free medium. Colonies were stained and counted ∼14 days after treatment.
Western blot analysis
Cells were washed twice with ice-cold PBS and lysed by adding RIPA buffer with protease inhibitors (Complete Mini, Roche). Samples were kept on ice for 5 min and lysate were gathered using a cell scraper. Lysate were centrifuged at 14 000 g for 15 min at 4°C, and stored at −80°C until analyzed. The protein concentration in the samples was determined by bicinchoninic acid protein assay (Pierce). Samples containing equal amounts of protein were run on Invitrogen Novex 10% Bis–Tris gels with MOPS running buffer and blotted onto nitrocellulose membranes. Membranes were blocked in PBS containing 0.1% Tween-20 and 5% milk before the addition of primary antibody. Membranes were probed with antibody directed against ERCC1 (Clone 3H11, Thermo Scientific) at 1:133 dilution or with antibody directed against XRCC3 (NB100-165SS, Novus Biologicals) at 1:5000 dilution or β-actin (clone AC-15, Sigma) at 1:4000 dilution. Detection of antibody binding was done using the enhanced chemiluminescence detection kit (Amersham) using the appropriate secondary antibody.
(V-C8, V-C8+B2, V79Z, AA8SN.10, AA8SN.12, 51D1SC.2, 51D1SC.4, UV4DR7, ERCC1.17, ERCC1.21 and PEF7).
RESULTS
RAD51D mutated cells have a defect in homology-directed repair of DNA DSBs
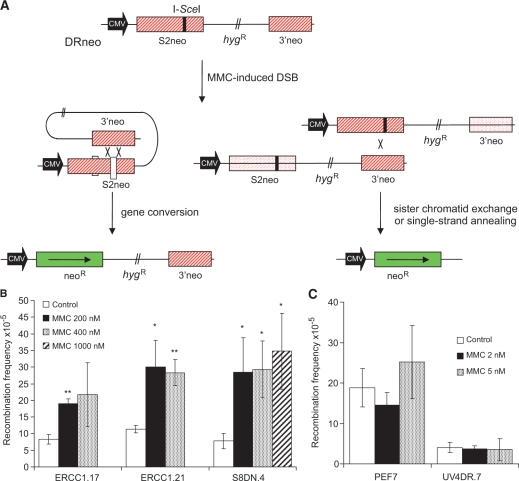
Use of the RAD51D−/− cells generated through a knockout approach in wild type CHO cells (29) provides a strong model system for DNA repair studies, as these isogenic mutants do not have the phenotypic complications associated with the random mutagenesis used to create other CHO DNA repair mutants. However, although these cells show similar phenotypes to other RAD51 paralogue mutant hamster cells, their HR proficiency has not been tested directly in a chromosomally integrated recombination reporter construct. Here, we transfected the SCneo recombination reporter (Figure 1A) (30) through electroporation into the RAD51D-defecient CHO cells, 51D1, and parental AA8 cell lines and isolated independent hygromycin-resistant clones stably carrying a single copy of the SCneo reporter; 51D1SC.2 and 51D1SC.4 from 51D1 cells and AA8SN.6, AA8SN.10 and AA8SN.12 from AA8 cells. To trigger HR in the SCneo substrate, we transiently transfected cells with the pCMV3xnlsISceI vector expressing the I-SceI restriction endonuclease to induce a specific DSB in the SCneo substrate. A functional neoR gene can be gained by HR, which is then selected for using G418. We found that RAD51D–/– clones have an 11–17-fold decrease in the recombination frequency when compared to wild-type AA8 clones (Figure 1B), showing that RAD51D is required for HR in mammalian cells, and verifying that these isogenic cells are useful as a model for HR deficiency in CHO cells.
Figure 1.
RAD51D cells are deficient in homologous recombination. (A) Schematic illustration of the SCneo recombination substrate. The SCneo substrate contains two non-functional copies of the neoR gene. After an I-SceI induced DSB, a functional neoR gene arises by HR via intrachromatid pairing or sister chromatid pairing. Single-strand annealing (SSA), a non-conservative HR sub-pathway, does not produce a functional neoR gene (60). (B) Recombination frequency to G418 resistance after transient transfection of the pCMV3xnlsI-SceI vector. Resistance to G418 reflects a recombination event of either intrachromatid pairing or sister chromatid pairing but not SSA. Columns depict the average and bars represent the standard deviation of at least three experiments.
Differential sensitivity to cross-linkers in ERCC1-defective and HR-defective cells
It has long been understood that ICL agents trigger HR in mammalian cells (31) and that recombination-defective cells show an extreme hypersensitivity to such cross-linkers (32,33). Similarly, it has been shown that ERCC1-defective cells are hypersensitive to cross-linkers (34) and that ERCC1 may play a role in ICL repair separate to that of its function in NER (16). Here, we confirm that both ERCC1- and HR-defective RAD51D–/– cells are hypersensitive to the cross-linkers mitomycin C, cisplatin and melphalan (Figure 2). We find that ERCC1 mutated cells are more sensitive to mitomycin C and cisplatin as compared to RAD51D–/– cells (Figure 2A and B), suggesting that HR and ERCC1 may have distinct roles in repair of mitomycin C-induced DNA lesions. This is likely depending on a separate role for the ERCC1 in repair of the intra-cross-links produced by mitomycin C and cisplatin that are repaired by NER (5,6). On the contrary to this, we find that RAD51D–/– and BRCA2-defective V-C8 cells are more sensitive to melphalan than the ERCC1-defective cells (Figure 1C), which confirms an earlier finding that XRCC3-defective cells are more sensitive than ERCC1-defective cells to this drug (35). These data support the differential use of DNA repair pathways when repairing different cross-link lesions. Furthermore, the data suggest that HR and ERCC1 have distinct roles in ICL repair, which may or may not be overlapping.
Figure 2.
ERCC1- and RAD51D-defective cells are hypersensitive to cross-linking agents mitomycin C and cisplatin. Survival was determined in colony outgrowth experiments in UV4DR7 (ERCC1 mutated) and ERCC1 complemented cells (ERCC1.17, ERCC1.21) or transfection with empty vector (PEF7), along with wild-type AA8 cells (AA8SN.10, AA8SN.12) or RAD51D targeted cells (51D1SC.2, 51D1SC.4) (26) as well as BRCA2-defective V-C8 cells and wild-type (V79Z) or BRCA2 complemented control (V-C8+B2) (59) following treatment with (A) mitomycin C (MMC) (B), cisplatin or (C) melphalan. The average and standard deviation of at least three experiments is depicted.
Mitomycin C-induced DSBs are independent of ERCC1-XPF and HR, but require active replication elongation
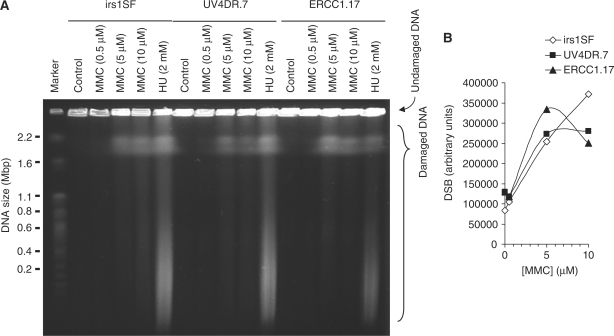
It is established that ICLs are converted into DSBs during repair in mammalian cells, although the extent of DSB formation varies widely between different crosslinkers (36). It has previously been demonstrated that the Mus81–Eme1 complex is involved in converting ICLs into DSBs (37) independently to the ERCC1–XPF endonuclease activity (16,20). Here, we examined the DSBs induced by mitomycin C in HR-defective irs1SF [XRCC3 mutated (38)] and ERCC1 mutated UV4DR7 cells and compared with the UV4DR7 cells complemented to a vector expressing wild-type ERCC1, ERCC1.17 (22). We found a dose-dependent increase in DSBs following a 4-h treatment with mitomycin C (Figure 3) and that DSBs were induced in ERCC1 mutated cells with equal efficiency, as compared to ERCC1 complemented cells, which was in agreement with previous observations (16,20). We also found that DSBs were produced equally efficiently in HR-defective irs1SF cells.
Figure 3.
DNA double-strand breaks caused by mitomycin C are independent of XRCC3 or ERCC1. (A) irs1SF and UV4DR7 cells, defective in XRCC3 and ERCC1, respectively, were treated with mitomycin C (MMC) for 4 h and DSBs then analysed by pulsed-field gel electrophoresis. ERCC1.17 cells were used as repair-proficient control; 2 mM hydroxyurea treatments (24 h) were used as positive control. (B) Quantification of MMC-induced DSBs.
It has previously been shown that DSBs are only formed by cross-linkers in S-phase cells (16,20), suggesting that they occur at crosslink-stalled replication forks. Here, we wanted to test if DSB formation also requires an active polymerase elongating the replication fork. To test this we inhibited replication elongation with the replication inhibitor aphidicolin, which specifically inhibits DNA polymerase α (39).
We found fewer DSBs when co-incubating mitomycin C and aphidicolin as compared with treatment with mitomycin C alone, showing that replication elongation is a requirement for converting most ICLs into a DSB (Figure 4). Furthermore, we found that replication elongation was required for mitomycin C-induced DSBs regardless of ERCC1 or HR status, confirming that these repair pathways act downstream of DSB formation.
Figure 4.
DNA double-strand breaks caused by MMC are dependent on replication elongation. (A) irs1SF, UV4DR7 and ERCC1.17 cells were treated with 10 μM MMC+/− 3 μM Aphidocolin for 4 h prior to analysis by pulsed-field gel electrophoresis; 2 mM hydroxyurea (HU) treated cells (24 h) were used as positive control. (B) Fragment analysis of released DNA. (C) Quantification of MMC-induced DSBs in presence or absence of Aphidocolin.
Mitomycin C inhibits replication fork elongation
Our data suggest that the elongating replication fork hits a MMC-induced DNA lesion that is then converted to DSBs. Such a model suggests that replication elongation is inhibited by MMC. Here, we wanted to test if replication elongation is slowed down with MMC. To test this, we used a method that uses the single-stranded DNA ends at a replication fork as starting points for DNA unwinding in alkaline solution (40). The cells were pulse-labelled for 30 min with 3H-thymidine and the speed of replication fork elongation is monitored as the time required for the labelled DNA to be progressed into the double-stranded DNA fraction following alkaline unwinding (Figure 5A). We treated cells with MMC for 15 min and followed replication elongation and found that replication elongation is inhibited by MMC (Figure 5B). We then tested if the decrease of replication elongation is depending on MMC dose and found that replication elongation is increasingly inhibited with an increased dose (Figure 5C).
Figure 5.
Mitomycin C inhibits replication elongation. (A) Replication elongation can be measured as the time it takes to prevent release of 3H-thymidine-labelled DNA onto the ssDNA fraction (37). (B) Time-course of replication fork progression in AA8 cells after a 15-min MMC treatment (100 µM). (C) Dose-dependent replication elongation inhibition in AA8 hamster cells 2 h following MMC treatments. The means and standard deviation (bars) of three independent experiments are shown. Statistical significance determined in t-test is indicated with one star (P < 0.05), two stars (P < 0.01) and three stars (P < 0.001).
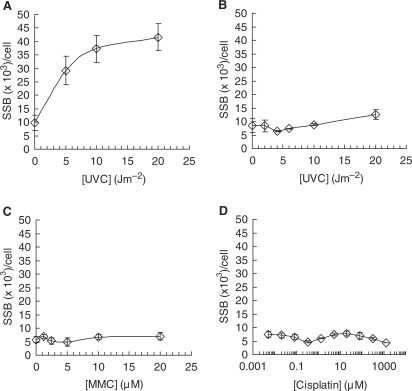
NER does not incise Mitomycin C or cisplatin-induced DNA lesions
It is possible to envision two nonexclusive models for DSB formation of an ICL lesion at a replication fork. The first model would be that NER incises the ICL in an attempt to repair the lesion (19), but fails as a consequence of the opposing strands being covalently cross-linked. The incised ICL lesion, which includes a DNA SSB (19), could then be encountered by an oncoming replication fork and convert the SSB into a one-sided DSB, similarly to the conversion of a camptothecin stabilized topoisomerase I complex into a one-ended DSB (41,42). This model is supported by the results showing replication elongation is required for DSB formation (Figure 4). An alternative nonexclusive model is that the replication fork is stalled at the cross-link lesion and awaits endonucleases to convert the stalled fork into a DSB, as suggested by the requirement for Mus81 for mitomycin C-induced DSBs (37), which is mediated by Snm1B (43). In an attempt to distinguish between these two distinct models of ICL repair, we quantified global genome NER-associated incisions following mitomycin C treatment. We added hydroxyurea and cytosine arabinocide to cells to prevent NER polymerization and ligation of any incised SSBs after mitomycin C exposures, and then used the alkaline DNA unwinding assay, which detects SSBs (27). As expected using this method we found robust accumulation of SSBs in wild-type AA8 cells following exposure to UVC treatments (Figure 6A), as these cells are proficient in global genome repair of UV lesions. In contrast, ERCC1 defective UV4 cells were unable to incise the UVC lesion (Figure 6B) as a result of their endonuclease defect. Interestingly, we found no SSB accumulation in wild-type AA8 cells following treatment with mitomycin C or cisplatin, showing no detectible level of NER incision by mitomycin C or cisplatin-induced DNA lesions (Figure 6C and D). These results are in contrast to data showing that ICL are excised by NER, in transcribed sequences on extrachromosomal plasmids (6). It is important to point out that we are only measuring incisions globally and are unable to detect few incisions in transcribed DNA. The ADU technique used here utilises the separation of the strands in order to detect the incised SSB. Thus, an ICL would prevent the unwinding in the direction across the ICL, but proceed normally in the opposite direction. To obtain an accurate estimation of the NER incisions of ICL as compared with UV lesions the mitomycin C incisions need to be multiplied by two. However, we cannot detect any ssDNA induced following mitomycin C treatments, which would have been expected if NER would incise at the mitomycin C-induced DNA lesions. Altogether, these data suggest that mitomycin C-induced DSBs occur enzymatically after the replication fork stalls rather than following replication run off at NER incised DNA lesions.
Figure 6.
Nucleotide excision repair does not incise mitomycin C-induced DNA lesions. The efficiency of nucleotide excision repair (NER) incisions can be measured by the amount of SSBs incised following DNA damage, as measured by alkaline DNA unwinding technique (27). The NER polymerization step is inhibited using hydroxyurea (2 mM) and cytosine arabinocide (20 µM). Incision of SSB by NER in (A) wild-type AA8 or (B) ERCC1-defective UV4 cells following exposure to increasing doses of UVC. Incision of SSB by NER in wild-type AA8 following exposure to increasing doses of (C) mitomycin C (MMC) or (D) cisplatin. Error bars designate standard deviation of at least three experiments.
ERCC1-XPF endonuclease is not required for RAD51 foci formation, but essential for completion of mitomycin C-induced HR
Since ERCC1-defective cells are more sensitive to ICLs than cells mutated in other NER genes, it is suggested that the role of ERCC1–XPF in ICL is separate from its role in NER (16,25). Our data, showing that mitomycin C-induced DNA lesions are poor substrates for NER, support this notion. ERCC1–XPF does have a role in single-strand annealing and gene conversion during HR (22), as well as a role in micro-homology-mediated end joining of DSBs (44), two functions for this protein complex that may be particularly important in the process of ICL repair. The RAD51 protein catalyzes the strand invasion process during HR and is relocated into nuclear foci following DNA damage (45). It is well established that proteins involved in HR (e.g. RAD51 paralogs and BRCA2) are required for RAD51 foci formation (46,47). Here, we find that RAD51 foci form in response to both cisplatin and mitomycin C (Figure 7), in agreement with that HR is important in repair of cross-link lesions (48,49). Furthermore, we find that ERCC1-defective UV4DR.7 cells form RAD51 foci equally well as the same cells complemented with wild-type ERCC1 (Figure 7B), suggesting that ERCC1 is not critical for RAD51 loading at DSBs and initiation of HR.
Figure 7.
ERCC1-defective cells form RAD51 foci in response to mitomycin C and cisplatin. (A) Representative image of cisplatin-induced RAD51 foci. (B) The percentage of cells containing more than 10 RAD51 foci in ERCC1.17 and UV4DR7 cells following a 24-h treatment of 100 nM cisplatin or 10 μM MMC. Error bars designate standard error of at least three experiments.
Although RAD51 foci form in ERCC1-defective cells, it is possible that the completion of ICL-induced HR is impaired. To test this, we investigated HR induced by mitomycin C in the ERCC1-defective UV4DR.7 cell line that also carry the DRneo recombination reporter (22). ERCC1-defective UV4DR.7 cells are only partially defective in an I-SceI-induced DSB, which is explained by ERCC1 being involved in completion of a subset of recombination events when 3′ ssDNA flaps are formed (22). Here, we investigated the recombination frequency induced by mitomycin C and found that HR is not induced in ERCC1-defective UV4DR.7 or in the plasmid control transfected PEF7 clone (Figure 8). In contrast, equally toxic doses of mitomycin C triggered HR in the ERCC1.17 and ERCC1.21 clones (derived from UVDR.7 cells complemented with an ERCC1WT expression vector) to a similar extent as in the wild-type S8DN.4 cells carrying the DRneo substrate. These results are in agreement with Zhang and co-workers (50), who reported that ICL-induced recombination is impaired in ERCC1 defective cells, using an extra-chromosomal plasmid-based assay.
Figure 8.
ERCC1-deficient cells are defective in mitomycin C-induced homologous recombination. (A) Structure of the DRneo recombination substrate containing two non-functional copies of the neoR gene. A functional neoR gene can be produced by SSA, sister chromatid exchange (SCE) or gene conversion upon induction of a DSB following expression of the I-SceI restriction endonuclease. Recombination induction of G418 resistance following DSB formation in the DRneo HR reporter vector following mitomycin C treatment in (B) wild-type cells or (C) ERCC1-defective cells. The average and standard errors of at least three experiments is depicted. Statistical significance determined in t-test is indicated with one star (P < 0.05) and two stars (P < 0.01).
The ERCC1-XPF endonuclease functions in the same pathway as HR in ICL repair
Since cells defective in ERCC1 appeared to initiate HR in a normal manner after ICL exposure, we wished to determine if ERCC1 might play a role during late stages of HR, after the formation of RAD51 nucleoprotein filaments. To test the possibility that the ICL repair function of ERCC1–XPF functions in concert with HR repair, we used siRNA to knock down the expression of ERCC1, XRCC3 and ERCC1 together with XRCC3 in SQ20B cancer cells of the human head and neck (Figure 9A) and investigated the clonogenic survival following treatments with cisplatin (Figure 9B). We found that both XRCC3 and ERCC1 siRNA depletion individually decreased clonogenic survival to cisplatin, in agreement with the sensitivity of the hamster cells defective in HR and ERCC1 (Figure 1). Furthermore, we found no additive effect of depleting XRCC3 in already ERCC1 siRNA-depleted cells, strongly suggesting that the two proteins act through the same pathway to promote survival in these cells. We found a slight increased survival to cisplatin in the XRCC3- and ERCC1-double-depleted cells. However, this is entirely explained by loss of viability in XRCC3 and ERCC1 siRNA-double-depleted cells and not a result of decreased sensitivity to cisplatin.
Figure 9.
XRCC3 acts in the same pathway as ERCC1 in ICL repair. (A) Western blot showing knockdown of ERCC1, XRCC3 or ERCC1+XRCC3 in SQ20B cells. NT, non-targeting. β-Actin was used to control for loading. (B) The effects on clonogenic survival of knocking down ERCC1, XRCC3 or ERCC1+XRCC3 in SQ20B to cisplatin treatment. NT siRNA was used as negative control. Cisplatin was added to the dishes for 2 h before changing to drug-free medium. Graph indicates average and standard deviation of three to eight experiments.
Interestingly, siRNA depletion of ERCC1 showed a more severe effect on clonogenic survival after exposure to cisplatin as compared to siRNA depletion of XRCC3, in agreement with survival studies comparing ERCC1-defective hamster cells with the HR-defective RAD51D-deficient cells (Figure 1B). These data highlight an additional role for ERCC1 in repair of ICL-induced lesions that is not totally explained by its role in XRCC3- or RAD51D-mediated HR.
DISCUSSION
There are several ICL agents used in anti-cancer treatment with a wide variety of specificities towards different types of tumours. For instance, cisplatin is highly efficient in treatment of testicular cancer, while other ICL agents are less effective (51). The reason for the difference in anti-cancer activity most likely depends on the particular types of DNA lesions formed. For instance, cisplatin does not form detectible DSBs, in contrast to mitomycin C (7,16). In this study we show that ERCC1-defective cells are more sensitive to mitomycin C and cisplatin than HR defective cells and that the opposite is true following treatments with melphalan (Figure 1). This observation underscores the difference in DNA lesions formed by ICL agents and highlights the complex interplay between DNA repair pathways in ICL repair that is most certainly unique in recognition and repair of each type of ICL lesion. It also suggests that, at least for lesions induced by mitomycin C, the role of ERCC1–XPF in ICL repair is not simply within a sub-pathway of HR repair. Indeed, ERCC1 has been shown to have a role in recombination-independent ICL repair, supporting this notion (6).
Previous studies have established that the formation of DSBs by ICL agents is linked with replication during the S-phase of the cell cycle (3,16,20,35,52). Here, we confirm these results and show that mitomycin C-induced DSBs are prevented by arrest of replication elongation. It has been shown that NER is able to incise at cross-links in vitro (19) and it has then been proposed that the associated SSBs may collapse replication forks into a one-sided DSB. Here, we show that global genome NER is unable to incise mitomycin C-induced DNA lesions in wild-type cells (Figure 7C), which argues against such a replication run-off model. Instead, our data support a model where the replication fork is first stalled by the ICL lesion, and then processed into a DSB, which is supported by Mus81 being required for mitomycin C-induced DSBs (37).
Interestingly, we find that the ICL-inhibition of replication elongation is not as efficient as equitoxic doses of UV damage (27). Thus, decreased BrdU incorporation in ICL-treated cells is explained primarily by loss of replication initiation rather than efficient inhibition of replication elongation.
Although we do not see any global genome wide NER incisions, it has previously been shown that ICL repair occurs on transcribed DNA independently of recombination, which obviously would require NER incision. This recombination-independent ICL repair requires CSA and CSB proteins that are exclusively involved in transcription-coupled repair (TCR) (6). Thus, we conclude that ICL lesions can be detected and incised in a transcription-dependent manner, which would be confined to transcribed DNA. This is also supported by the insensitivity of GGR defective XPC cells to the toxic effects of cisplatin (53).
The ERCC1–XPF complex is implicated in HR (21–24) and it has been suggested that this is the role for ERCC1–XPF in ICL repair (25,35,50). Here, we report that ERCC1-defective cells are proficient in forming RAD51 foci after cisplatin or mitomycin C treatments. This suggests that HR is initiated normally in ERCC1-defective cells, the ERCC1–XPF being likely to have a role in HR in a step subsequent to the activities of the HR proteins RAD51D, XRCC3 or BRCA2, as those mutant cells are deficient in RAD51 foci formation (46,47). Using a recombination reporter, we also found that ERCC1 mutant cells are impaired in mitomycin C-induced HR, and that the defect can be reverted by introduction of a functional ERCC1 gene expressed from a vector. These results are in agreement with previous data showing that ERCC1 is required for ICL-induced recombination on an extra-chromosomal plasmid (50). The complete deficiency of ERCC1 mutant cells to induce HR after mitomycin C treatment is not a simple reflection of ERCC1–XPF being required for all types of HR events, as a two-ended I-SceI-induced DSB still triggers recombination 100-fold in the UV4DR.7 cells (22). Thus, it is likely that the ERCC1–XPF complex is specifically involved in ICL-induced HR, likely involved in resolving late-stage recombination products. The double siRNA knockout experiment with ERCC1 and XRCC3 (Figure 9) provides strong evidence that ERCC1 and HR function in the same ICL repair pathway, and that additional knockdown of XRCC3 in already ERCC1-depleted cells shows no additional decrease in clonogenic survival.
The differential survival of ERCC1- and HR-defective cells to mitomycin C/cisplatin and melphalan (Figure 2) highlights the evidence that the ERCC1–XPF complex has additional roles in ICL repair beyond resolution of HR products, as discussed above. This is also supported by increased sensitivity to cisplatin in ERCC1 siRNA-depleted cells as compared with XRCC3-depleted cells (Figure 9). We speculate that a possible role for ERCC1 could be to unhook the ICL to allow TLS bypass (Figure 10). Theoretically, this could also be done by Mus81, but if the enzyme were to cut the stalled fork with the same polarity as the first stalled fork, the ICL would not be released to allow TLS.
Figure 10.
Model for ICL repair in mammalian cells. ICL repair can be initiated either at a replication fork (A) or at a stalled RNA polymerase (B). (A) ICLs in DNA initially stall replication forks that collapse into a one-sided DSB by Mus81 activity, mediated by Snm1B. The release one-sided DSB is likely resected by the Mre11-RAD50-Nbs1 complex and subsequently coated with the RAD51 protein to promote HR at a later stage. An opposing second stalled replication fork is possibly processed by the ERCC1–XPF complex to unhook the cross-link and to allow TLS, possibly with Rev1 or Polζ. The DNA molecule would be invaded by RAD51 during HR to initiate synthesis-dependent strand annealing repair and the final lesion removed by NER. (B) ICLs in DNA will stall RNA polymerase during transcription that will initiate TCR. The RNA polymerase will either backtrack or be degraded during subsequent repair, which proceed through TLS, using Polη, Rev1 or Rev3 (61). NER factors will be attracted for second incision that will remove the ICL to allow resumption of transcription.
In conclusion, we would like to present a model for ICL repair in mammalian cells (Figure 10), pointing out that ICL repair is initiated when either a replication forks or RNA polymerase runs into a ICL lesion. We also want to point out that different ICL agents cause different distortion to DNA and that this model is likely not true for all ICL agents. In replication-initiated ICL repair, replication forks will stall at the ICL lesion and the initial DSB is catalysed by Mus81 (37) and not by replication run off. The released one-sided DSB is likely resected by the Mre11-RAD50-Nbs1 complex (54) and subsequently coated with the RAD51 protein. An opposing second replication fork likely arrives at the crosslink (55), and the ERCC1–XPF complex could unhook this replication fork to allow TLS, possibly with Rev1 or Polζ (56). The ligated DNA molecule would be invaded by RAD51 to initiate synthesis-dependent strand annealing (57) and the final lesion removed by NER. In transcription-initiated ICL repair, ICL lesions are recognised by the RNA polymerase, as TCR factors are required for recombination-independent ICL repair in absence of global genome wide NER incision. This would recruit NER factors for both first and second incision, but would not generate a DSB intermediate.
FUNDING
The Swedish Cancer Society; the Swedish Children's; Cancer Foundation; the Swedish Research Council; the Swedish Pain Relief Foundation; and the Medical Research Council. Funding for open access charge: The Swedish Pain Relief Society.
Conflict of interest statement. None declared.
ACKNOWLEDEGEMENTS
We thank Drs Larry Thompson, Maria Jasin and Margret Zdzienicka for materials.
REFERENCES
- 1.Lawley PD, Phillips DH. DNA adducts from chemotherapeutic agents. Mutat. Res. 1996;355:13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- 2.Palom Y, Suresh Kumar G, Tang LQ, Paz MM, Musser SM, Rockwell S, Tomasz M. Relative toxicities of DNA cross-links and monoadducts: new insights from studies of decarbamoyl mitomycin C and mitomycin C. Chem. Res. Toxicol. 2002;15:1398–1406. doi: 10.1021/tx020044g. [DOI] [PubMed] [Google Scholar]
- 3.Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat. Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 4.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol. Cell Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol. Cell Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defects in interstrand cross-link uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic Acids Res. 2002;30:3848–3856. doi: 10.1093/nar/gkf479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park CH, Sancar A. Formation of a ternary complex by human XPA, ERCC1, and ERCC4(XPF) excision repair proteins. Proc. Natl Acad. Sci. USA. 1994;91:5017–5021. doi: 10.1073/pnas.91.11.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biggerstaff M, Szymkowski DE, Wood RD. Co-correction of the ERCC1, ERCC4 and xeroderma pigmentosum group F DNA repair defects in vitro. EMBO J. 1993;12:3685–3692. doi: 10.1002/j.1460-2075.1993.tb06043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Vuuren AJ, Appeldoorn E, Odijk H, Yasui A, Jaspers NG, Bootsma D, Hoeijmakers JH. Evidence for a repair enzyme complex involving ERCC1 and complementing activities of ERCC4, ERCC11 and xeroderma pigmentosum group F. EMBO J. 1993;12:3693–3701. doi: 10.1002/j.1460-2075.1993.tb06044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 12.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 13.Bessho T, Sancar A, Thompson LH, Thelen MP. Reconstitution of human excision nuclease with recombinant XPF-ERCC1 complex. J. Biol. Chem. 1997;272:3833–3837. doi: 10.1074/jbc.272.6.3833. [DOI] [PubMed] [Google Scholar]
- 14.de Laat WL, Appeldoorn E, Jaspers NG, Hoeijmakers JH. DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 1998;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- 15.Houtsmuller AB, Rademakers S, Nigg AL, Hoogstraten D, Hoeijmakers JH, Vermeulen W. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science. 1999;284:958–961. doi: 10.1126/science.284.5416.958. [DOI] [PubMed] [Google Scholar]
- 16.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat. Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 18.Zhang N, Zhang X, Peterson C, Li L, Legerski R. Differential processing of UV mimetic and interstrand crosslink damage by XPF cell extracts. Nucleic Acids Res. 2000;28:4800–4804. doi: 10.1093/nar/28.23.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 20.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adair GM, Rolig RL, Moore-Faver D, Zabelshansky M, Wilson JH, Nairn RS. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 2000;19:5552–5561. doi: 10.1093/emboj/19.20.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 2001;20:6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sargent RG, Meservy JL, Perkins BD, Kilburn AE, Intody Z, Adair GM, Nairn RS, Wilson JH. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 2000;28:3771–3778. doi: 10.1093/nar/28.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergstralh DT, Sekelsky J. Interstrand crosslink repair: can XPF-ERCC1 be let off the hook? Trends Genet. 2008;24:70–76. doi: 10.1016/j.tig.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson F, Lagerqvist A, Erixon K, Jenssen D. A method to monitor replication fork progression in mammalian cells: nucleotide excision repair enhances and homologous recombination delays elongation along damaged DNA. Nucleic Acids Res. 2004;32:e157. doi: 10.1093/nar/gnh154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erixon K, Ahnstrom G. Single-strand breaks in DNA during repair of UV-induced damage in normal human and xeroderma pigmentosum cells as determined by alkaline DNA unwinding and hydroxylapatite chromatography: effects of hydroxyurea, 5-fluorodeoxyuridine and 1-beta-D-arabinofuranosylcytosine on the kinetics of repair. Mutat. Res. 1979;59:257–271. doi: 10.1016/0027-5107(79)90164-7. [DOI] [PubMed] [Google Scholar]
- 29.Hinz JM, Tebbs RS, Wilson PF, Nham PB, Salazar EP, Nagasawa H, Urbin SS, Bedford JS, Thompson LH. Repression of mutagenesis by Rad51D-mediated homologous recombination. Nucleic Acids Res. 2006;34:1358–1368. doi: 10.1093/nar/gkl020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson RD, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 31.Moore PD, Holliday R. Evidence for the formation of hybrid DNA during mitotic recombination in Chinese hamster cells. Cell. 1976;8:573–579. doi: 10.1016/0092-8674(76)90225-7. [DOI] [PubMed] [Google Scholar]
- 32.Jones NJ, Cox R, Thacker J. Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res. 1987;183:279–286. doi: 10.1016/0167-8817(87)90011-3. [DOI] [PubMed] [Google Scholar]
- 33.Caldecott K, Jeggo P. Cross-sensitivity of gamma-ray-sensitive hamster mutants to cross-linking agents. Mutat. Res. 1991;255:111–121. doi: 10.1016/0921-8777(91)90046-r. [DOI] [PubMed] [Google Scholar]
- 34.Thompson LH, Mooney CL, Burkhart-Schultz K, Carrano AV, Siciliano MJ. Correction of a nucleotide-excision-repair mutation by human chromosome 19 in hamster-human hybrid cells. Somat. Cell Mol. Genet. 1985;11:87–92. doi: 10.1007/BF01534738. [DOI] [PubMed] [Google Scholar]
- 35.Clingen PH, De Silva IU, McHugh PJ, Ghadessy FJ, Tilby MJ, Thurston DE, Hartley JA. The XPF-ERCC1 endonuclease and homologous recombination contribute to the repair of minor groove DNA interstrand crosslinks in mammalian cells produced by the pyrrolo[2,1-c][1,4]benzodiazepine dimer SJG-136. Nucleic Acids Res. 2005;33:3283–3291. doi: 10.1093/nar/gki639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defects in interstrand cross-link uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic Acids Res. 2002;30:3848–3856. doi: 10.1093/nar/gkf479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tebbs RS, Zhao Y, Tucker JD, Scheerer JB, Siciliano MJ, Hwang M, Liu N, Legerski RJ, Thompson LH. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc. Natl Acad. Sci. USA. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikegami S, Taguchi T, Ohashi M, Oguro M, Nagano H, Mano Y. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature. 1978;275:458–460. doi: 10.1038/275458a0. [DOI] [PubMed] [Google Scholar]
- 40.Johansson F, Allkvist A, Erixon K, Malmvarn A, Nilsson R, Bergman A, Helleday T, Jenssen D. Screening for genotoxicity using the DRAG assay: investigation of halogenated environmental contaminants. Mutat. Res. 2004;563:35–47. doi: 10.1016/j.mrgentox.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 41.Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 42.Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell Biol. 2000;20:3977–3987. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bae JB, Mukhopadhyay SS, Liu L, Zhang N, Tan J, Akhter S, Liu X, Shen X, Li L, Legerski RJ. Snm1B/Apollo mediates replication fork collapse and S Phase checkpoint activation in response to DNA interstrand cross-links. Oncogene. 2008;27:5045–5056. doi: 10.1038/onc.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol. Cell Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl Acad. Sci. USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 48.Hussain S, Witt E, Huber PA, Medhurst AL, Ashworth A, Mathew CG. Direct interaction of the Fanconi anaemia protein FANCG with BRCA2/FANCD1. Hum. Mol. Genet. 2003;12:2503–2510. doi: 10.1093/hmg/ddg266. [DOI] [PubMed] [Google Scholar]
- 49.Cummings M, Higginbottom K, McGurk CJ, Wong OG, Koberle B, Oliver RT, Masters JR. XPA versus ERCC1 as chemosensitising agents to cisplatin and mitomycin C in prostate cancer cells: role of ERCC1 in homologous recombination repair. Biochem. Pharmacol. 2006;72:166–175. doi: 10.1016/j.bcp.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 50.Zhang N, Liu X, Li L, Legerski R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair (Amst) 2007;6:1670–1678. doi: 10.1016/j.dnarep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams SD, Loehrer PJ, Sr, Einhorn LH. Chemotherapy of advanced testicular cancer. Semin. Urol. 1984;2:230–237. [PubMed] [Google Scholar]
- 52.Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell Biol. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furuta T, Ueda T, Aune G, Sarasin A, Kraemer KH, Pommier Y. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res. 2002;62:4899–4902. [PubMed] [Google Scholar]
- 54.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raschle M, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Ford DK, Yerganian G. Observation of the chromosomes of Chinese hamster cells in tissue culture. J. Natl Cancer Inst. 1958;21:393–425. [PubMed] [Google Scholar]
- 59.Kraakman-van der Zwet M, Overkamp WJ, van Lange RE, Essers J, van Duijn-Goedhart A, Wiggers I, Swaminathan S, van Buul PP, Errami A, Tan RT, et al. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol. Cell Biol. 2002;22:669–679. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen X, Jun S, O'N;eal LE, Sonoda E, Bemark M, Sale JE, Li L. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA) J. Biol. Chem. 2006;281:13869–13872. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]