Abstract
Topoisomerase I is essential for DNA metabolism in nuclei and mitochondria. In yeast, a single topoisomerase I gene provides for both organelles. In vertebrates, topoisomerase I is divided into nuclear and mitochondrial paralogs (Top1 and Top1mt). To assess the meaning of this gene duplication, we targeted Top1 to mitochondria or Top1mt to nuclei. Overexpression in the fitting organelle served as control. Targeting of Top1 to mitochondria blocked transcription and depleted mitochondrial DNA. This was also seen with catalytically inactive Top1 mutants, but not with Top1mt overexpressed in mitochondria. Targeting of Top1mt to the nucleus revealed that it was much less able to interact with mitotic chromosomes than Top1 overexpressed in the nucleus. Similar experiments with Top1/Top1mt hybrids assigned these functional differences to structural divergences in the DNA-binding core domains. We propose that adaptation of this domain to different chromatin environments in nuclei and mitochondria has driven evolutional development and conservation of organelle-restricted topoisomerase I paralogs in vertebrates.
INTRODUCTION
Replication and transcription of nuclear and mitochondrial genomes create DNA-topological stress that needs to be released in order to allow these DNA metabolic processes to proceed. Therefore, the ability of DNA topoisomerases (Top) to release topological stress from DNA is essential in the nuclear as well as the mitochondrial compartment (1). Topoisomerases employ transient breakage and religation of either one (Top1 and Top3) or both DNA strands (Top2) to allow swiveling of or strand passage through the double helix, thereby changing DNA topology (2).
Five different enzymes (Top1, Top2α, Top2β, Top3α and Top3β) provide topoisomerase activity for the nucleus of vertebrate cells. Two of these are also targeted to the mitochondria via posttranscriptional mechanisms: mitochondrial Top3α is created by alternative translation initiation of the same mRNA that encodes nuclear Top3α (3); mitochondrial Top2 seems derived directly from the nuclear enzyme Top2β by proteolysis (4). In contrast, the nuclear and mitochondrial versions of topoisomerase I (Top1 and Top1mt) are encoded by separate genes (5).
Duplication and diversification of the TOP1 gene into nuclear and mitochondrial paralogs is conserved in vertebrates, whereas invertebrate eukaryotes do not possess a genetically distinct, mitochondria-targeted topoisomerase I (6,7). It is believed that a single form of Top1 is functional in nuclei and mitochondria of invertebrates, because genetic silencing of the single TOP1 gene in Saccharomyces cerevisiae suppresses mitochondrial Top1 activity (8,9). In silico analysis predicts that the single Top1 form of Schizosaccharomyces pombe is also targeted to nuclei and mitochondria (3). It is unclear why vertebrates do not employ the same topoisomerase I in nuclei and mitochondria and instead maintain genetically distinct paralogs of the enzyme dedicated to either organelle. Transcription and replication machineries acting on mitochondrial DNA (mtDNA) differ significantly between yeast and mammals (10). Therefore, evolution of multicellular organisms could have required the development of a specialized Top1mt with properties distinct from its nuclear counterpart. On the other hand, differences in nuclear chromatin organization known to exist between yeast and mammals could have incited the development of a topoisomerase I that is adapted to nuclear DNA metabolism but incompatible with mtDNA maintenance.
Here, we investigated the biological meaning of the duplication of the topoisomerase I gene into TOP1 and TOP1mt by studying mitochondria-targeted Top1 and nuclear-targeted Top1mt in human cells. We demonstrate that Top1 and Top1mt differ significantly in their interaction with nuclear and mtDNA. This difference is encoded in the core domain of the two enzymes. As a consequence, Top1 is incompatible with stable mtDNA propagation, while Top1mt is incapable of interacting with nuclear metaphase chromosomes.
MATERIALS AND METHODS
DNA constructs
For mitochondrial targeting of fluorescent fusion proteins, YFP in the vector pMC-EYFP-N (11) was extended in frame at the 5′-end with the sequence encoding the mitochondrial targeting sequence (MTS) from subunit VIII of cytochrome C oxidase (COX) (12) using linker PCR, thus generating pMC-MTS-EYFP-N. Likewise, the coding sequence for Top1mt (5) was inserted in frame at the C-terminal end of MTS-YFP. For mitochondrial targeting of Top1, its open reading frame was cloned into the MluI and ApaI sites of the pMC-MTS-EYFP-N vector. Variants of this vector, containing truncated versions of Top1 (Top1191–765 and the corresponding active site mutant (Y723F) Top1191–765*) were constructed in the same way. Exchange of regions between Top1 and Top1mt was accomplished by overlap-extension PCR (13). For nuclear localization of Top1mt and chimeric Top1mt/Top1, their open reading frames were supplemented with the nuclear localization signal (NLS) of SV40. Construction and characterization of vectors for stable expression of YN-Top1191–765 have been described previously (14,15). For lentiviral expression of MY or MY-Top1 in HT-1080 cells, the sequences were cloned into the lentiviral pCL1P vector. The pCL1P is a derivative of the pCL1EG vector (16), in which the EGFP open reading frame was replaced by the selection marker puromycin-N-acetyl transferase and a multicloning site (BamHI-ClaI-EcoRI-AgeI-NheI-XhoI-PstI) introduced between the lentiviral cPPT and the internal SFFV U3 promotor.
Cell culture
Transfections and selection of HT-1080 cells (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) followed published procedures (15). Cells were selected using 0.6 µg/ml puromycin and grown in DMEM supplemented with 50 µg/ml uridine, if not specified otherwise. For each construct, at least 12 cell clones supporting stable expression of the desired fusion proteins were picked and expanded. For the preparation of virus particles, vector plasmids pCL1 (5 µg), helper plasmid pCD/NL-BH (5 µg) and the envelope plasmid pczVSV-Gwt (5 µg) (17) were cotransfected into HEK-293T cells using polyethylene (Sigma, Taufkirchen, Germany). The virus stocks were harvested 48 h after transfection, filtered (0.45 µm) and stored at –80°C. HT-1080 cells were transduced and medium was replaced 12 h after transduction with fresh medium containing 50 µg/ml uridine and 0.6 µg/ml puromycin.
Cell growth and cell-cycle analysis
For determination of cell growth, cells were repeatedly split 1:2 upon confluency. Cell cycle progression was monitored using a flow cytometer (FACSCalibur, BD Bioscience, Heidelberg, Germany). Briefly, cells were harvested and fixed in 70% ethanol, treated with 50 µg/ml RNAse and DNA was stained with 50 µg/ml propidium iodide. A total of 10 000 cells per sample were analyzed.
Life cell imaging and senescence-associated β-galactosidase staining
Live cell imaging was done with a Zeiss LSM 510 META inverted confocal laser-scanning microscope equipped with a Zeiss incubator XL and a 40x/1.3 NA Plan-NeoFluar® oil immersion objective. Cells were cultured under the microscope in CO2-independent medium (18045-045, Invitrogen, Karlsruhe, Germany). For visualization of mitochondria, cells were incubated for 5 min with 10 nM MitoTracker®Red CMXRos (Invitrogen), washed with PBS and then supplied with fresh medium prior to inspection. Staining for β-galactosidase was done as described (18) and pictures were taken with a Zeiss Axiovert135 microscope with a 20x/1.3 Apochromat objective.
Preparation of whole cell or mitoplast lysates and immunoblotting
Whole cell lysates were prepared from 3 × 106 cells as described previously (19). To isolate the inner-membrane mitochondrial fraction (mitoplast), a protocol for mitochondria isolation (20) was modified by addition of digitonin, a detergent disrupting all cellular membranes except the inner mitochondrial membrane. Briefly, cells from two confluent 175 cm2 tissue culture flasks were washed with D-PBS (Invitrogen), scraped off and resuspended in 5 ml mitochondria isolation buffer (0.3 M sucrose, 1 mM EGTA, 5 mM MOPS, 5 mM KH2PO4, 1 mg/ml BSA, pH 7.4) supplemented with 0.1 mg/ml digitonin, incubated for 10 min, homogenized with 25 strokes in a Dounce homogenizer and centrifuged twice at 2600g for 7 min. The supernatant was centrifuged at 15 000g for 10 min and pelleted mitoplasts were resuspended in 0.1 ml mitochondria isolation buffer without digitonin. All steps were performed on ice. Mitoplast lysates were prepared by addition of an equal volume of 2-fold lysis buffer (250 mM Tris–HCl, pH 6.8, 2% glycerol, 4% SDS, 20 mM DTT, 1.4 M urea, 20 mM EDTA, 5 mM AEBSF, 0.04% bromophenol blue). Cell or mitoplast lysates equivalent to 105 cells were subjected to SDS–PAGE, and transferred to PVDF membranes (Immobilon P, Millipore, Bedford, MD). Blots were probed with antibodies against YFP (clone JL8, Clontech, Heidelberg, Germany).
Banddepletion assay
For immunoband depletion, cells were first cultured with 100 µm topotecan (TPT) for 30 min prior to harvesting. The same concentration of drug was also added to the lysis buffer.
Mitoplast extraction and plasmid relaxation
Mitoplasts were extracted for 10 min at 4°C (0.3 M sucrose, 500 mM NaCl, 1 mM EGTA, 5 mM Mops, 5 mM KH2PO4, 0.25 mM AEBSF, 0.5 mM DTT, 0.1% Triton, 1 mg/ml BSA, pH 7.4). Extracts were cleared (15 000g, 10 min), and total protein concentrations were adjusted to 1.7 mg/ml. Extracts were diluted 10-fold with relaxation buffer (10 mM Tris–HCl, pH 8, 70 mM KCl, 3 mM MgCl2, 0.3 mM EDTA, 0.3 mg/ml BSA and 0.5 mM DTT) and 170 ng total protein were incubated with 0.4 µg supercoiled pUC18 DNA at 37°C. Reactions were stopped by adding 0.5% SDS. Samples were further analyzed as described (21).
MtDNA copy number
Total DNA was isolated using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). MtDNA copy number was determined according to the quantitative TaqMan-PCR method (22). Briefly, 100 ng total DNA was subjected to amplification reactions performed as 25 µl duplicates in a 96-well microplate with a forward primer (5′-GATTTGGGTACCACCCAAGTATTG-3′) and a reverse primer (5′-TTGACTCACCCATCAACAACC-3′), each recognizing a unique mtDNA sequence. To determine the abundance of mtDNA in the sample, amplification was performed in parallel with different concentrations of standard plasmids.
2D neutral agarose gel electrophoresis and Southern blotting
For analysis of replication intermediates (RIs), mitochondria were isolated and mtDNA prepared as described (23). For 2D neutral agarose-gel electrophoresis, 1.5 µg of mtDNA was digested with HincII. The first dimension was run in a 0.4% agarose gel in TBE buffer, and the second dimension was run with 0.1% EtBr in a 0.95% agarose gel in TBE. Southern blotting was carried out using standard procedures (23). The fragment 13 636-1006 bp containing the control region was detected by hybridization with a 32P-labeled cytochrome b probe (nucleotides 14 846–15 358) (23).
RNA extraction and quantification of mitochondrial transcripts by RT-PCR
Total RNA was isolated using RNeasy Mini Kit (Qiagen) according to manufacturer’s instructions. For quality assurance, the RNA integrity was determined with the Agilent 2100 Bioanalyzer (RNA 6000 LabChip, Agilent Technologies, Waldbronn, Germany). Quantitative RT-PCR reactions were performed in duplicates in a reaction volume of 25 µl using the QuantiTect SYBR Green one-step RT-PCR Kit (Qiagen). COXI and 12S rRNA, were amplified from 5 ng total RNA, whereas 5 pg samples were used for quantification of the internal standard 18S rRNA. Used primer pairs: COXI (5′-TACCTATTATTCGGCGCATGAGCTGGA-3′, 5′-TGCATGGGCTGTGACGATAACGTTGTA-3′), 12S rRNA (5′-GGTTGGTCAATTTCGTGCC-3′, 5′-GAGTTTTTTACAACTCAGGTG-3′), SDHB (5′-AGTTGACTCTACTTTGACCTTCCGAAG-3′, 5′-GACCTTATTGAGGTTGGTGTCAATCCT-3′) and 18s rRNA (5′-ATTAGAGTGTTCAAAGCAGGCCCGAGC-3′, 5′-CGTCCCTCTTAATCATGGCCTCAGTTC-3′; or 5′-GTCCAAAGCAGGCCCGAG-3′, 5′-CCCTCTTAATCATGGCCTC-3′). For quantification, standard plasmids were used, which contained the amplified gene sequences in a pCR2.1-TOPO vector (Invitrogen).
Modeling of the Top1 structure
Cartoon ribbon drawings of the X-ray crystal structure (PDB entry 1K4T) by Staker et al. (24) were created using the MACPyMOL software (Delano Scientific LLC).
Statistics
Quantitative results are given as mean values ± SEM of multiple measurements carried out with independent cell clones expressing the same construct. The actual number of clones investigated is stated in each figure. Data sets were compared by the t-test for unpaired samples. P < 0.05 was considered significant.
RESULTS
Constitutive expression of mitochondria-targeted constructs of Top1 in HT-1080 cells
In order to simultaneously target and track Top1 and variants thereof, we generated constructs for the expression of N-terminal YFP-tagged proteins with a MTS from COX subunit VIII (Figure 1A). Mitochondrial targeting was performed for full-length Top1 (MY-Top1) and a N-terminally truncated version, in which residues 1–190 were deleted (MY-Top1191–765). The truncated version is devoid of all NLSs (25,26). MY-Top1191–765 closely resembles Top1mt, since it lacks most amino acids encoded by the first eight exons of the TOP1 gene (residues 1–205), which are unique to Top1 (7). The active site tyrosine (Y723F) in MY-Top1191–765 was replaced with phenylalanin to generate a catalytically inactive enzyme (MY-Top1191–765*). The MY tag was also appended at the N-terminus of Top1mt (MY-Top1mt). The basic vector encoding for mitochondria-targeted YFP alone (MY) served as control for an inert protein not binding DNA. When transfected into human HT-1080 cells, all constructs gave rise to viable cell clones. Clones expressing MY or MY-Top1mt exhibited growth rates and gross morphologies similar to untransfected cells. Cells expressing any of the mitochondria-targeted versions of Top1 (including the inactive mutant MY-Top1191–765*) were growth-arrested, unless the culture media was supplemented with uridine (see below).
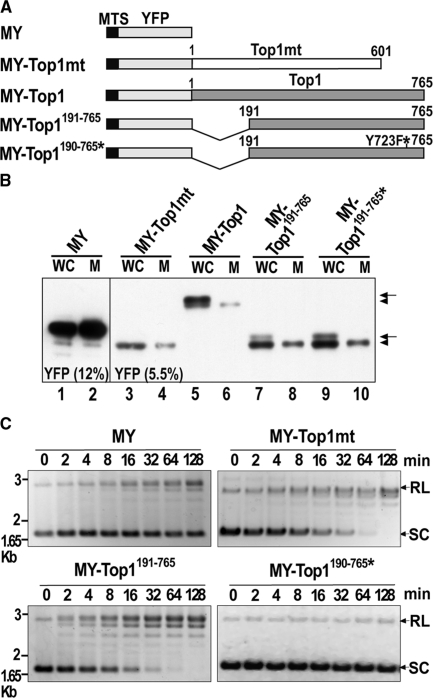
Figure 1.
Expression and activity of mitochondria-targeted YFP fusion proteins. (A) Schematic outline of constructs: MTS-YFP (MY); MTS-YFP-Top1mt (MY-Top1mt); MTS-YFP-Top1 (MY-Top1); MTS-YFP-Top1191–765 (MY-Top1191–765); and MTS-YFP-Top1191–765, Y723F (MY-Top1191–765*). All constructs contained a MTS. Top1 and Top1mt indicate the nuclear and mitochondrial paralogs, respectively; MY-Top1191–765* is a catalytically inactive Top1 mutant (Y723F). (B) Western blot analysis of lysates from whole cells (WC) or isolated mitoplasts (M) from representative HT-1080 cell clones expressing the constructs indicated above. Samples were subjected to SDS–PAGE (left, 12% gel; right, 5.5% gel) and western blotting using YFP antibodies. Proteins with (arrow) or without presequence (arrowhead) are indicated. (C) Relaxation over time of supercoiled pUC18 plasmid DNA by mitoplast extracts (170 ng total proteins) prepared from cell clones expressing the indicated constructs. Positions of relaxed (RL) and supercoiled (SC) plasmid forms are indicated on the right. Time points 0 represent DNA not exposed to mitoplast extracts.
To assess the integrity and mitochondrial targeting of the fusion proteins, we subjected whole cells and isolated mitoplasts (mitochondrial matrix surrounded by the inner membrane) to immunoblotting with YFP antibodies. MY and MY-Top1mt were detected as single protein bands of expected size in lysates of whole cells and mitoplasts (Figure 1B). Mitochondria-targeted versions of Top1 were detected in lysates from whole cells as a protein doublet, of which only the faster migrating band was detected in corresponding mitoplast lysates (Figure 1B). The slower migrating band probably represents a protein fraction not imported into mitochondria and still possessing the MTS presequence. Consistent with this, YFP-fluorescence of these fusion proteins was not only exclusively localized in mitochondria, but also found in the nucleus to various extents (see Figure 2A and Supplementary Figure 1). In the case of MY-Top1 bearing multiple endogenous NLSs in the N-terminal domain (25,26) and showing a prominent nuclear localization, the slower migrating band is the prevalent one. In contrast, the faster migrating band was predominant in all mitochondria-targeted constructs of Top1 lacking major portions of the N-terminal domain and exhibiting only a weak nuclear localization (MY-Top1191–765 and MY-Top1191–765*). It should finally be noted that all fusion proteins were expressed at similar levels which allowed their comparative analysis.
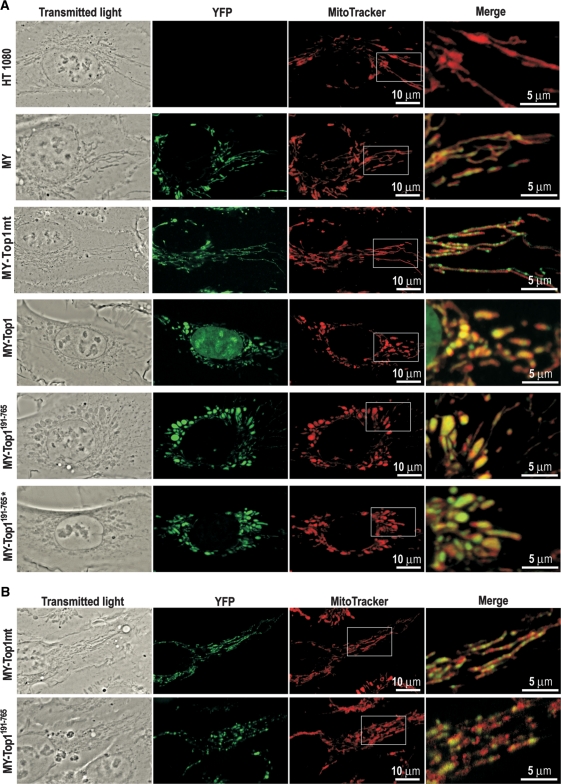
Figure 2.
In vivo localization of mitochondria-targeted YFP fusions proteins. Each row shows representative confocal images of living cells stably (A) or transiently (B) expressing the indicated constructs. Shown are images of transmitted light, YFP- and MitoTrackerRed fluorescence. Merged signals of YFP- and MitoTrackerRed fluorescence in the indicated areas are shown at 3-fold magnification (right).
To ascertain that topoisomerase I constructs were active after mitochondrial import, we measured their DNA relaxation activity in salt extracts of isolated mitoplasts. Mitoplast extracts from cells expressing MY or the catalytically inactive mutant MY-Top1191–765* exhibited only a very weak DNA relaxation activity, which was similar to that found in untransfected cells (data not shown) and probably due to endogenous Top1mt (Figure 1C). Extracts from cells expressing MY-Top1mt or MY-Top1191–765 exhibited enhanced DNA relaxation activity. A comparable increase in DNA relaxation activity was observed in mitoplast extracts from cells expressing MY-Top1 (data not shown). Thus, mitochondrial targeting did not compromise catalytic activity of the respective topoisomerase I constructs.
Targeting of Top1 to mitochondria induces mtDNA depletion and a ρ− phenotype
Subcellular localization of the various mitochondria-targeted topoisomerase I constructs and their impact on mitochondrial morphology was studied by live cell confocal microscopy. Staining of untransfected HT-1080 cells with the mitochondria-specific dye MitoTracker revealed mitochondria forming a largely connected reticulum typical of mammalian cells [Figure 2A; (27)]. A similar pattern was seen in cells expressing MY or MY-Top1mt. YFP fluorescence corresponding to MY and MY-Top1mt largely coincided with MitoTracker staining, which confirmed complete mitochondrial import of these proteins. Merged and enlarged pictures revealed that MY had a diffuse homogenous mitochondrial distribution, which was similar to MitoTracker and consistent for a protein not interacting with mitochondrial structures. In contrast, MY-Top1mt accumulated at speckles within the mitochondrial compartment (Figure 2A). Counterstaining of DNA confirmed that these speckles were identical or associated with mitochondrial nucleoids (data not shown), indicating proper incorporation of MY-Top1mt into its native microenvironment.
MY-Top1 as well as active and inactive truncated variants thereof (MY-Top1191–765 and MY-Top1191–765*) were also localized in mitochondria (Figure 2A). MY-Top1 showed a dual localization in mitochondria and cell nucleus due to multiple NLSs in the N-terminal domain (25,26). Truncation of this domain in the constructs MY-Top1191–765 and MY-Top1191–765* induced a predominant mitochondrial localization with only traces of nuclear staining remaining detectable (Supplementary Figure 1). Most notably, all three constructs derived from Top1 induced rounded and ballooned mitochondria with enhanced fragmentation (Figure 2A). A similar morphology has previously been observed in ρ− cells depleted of mtDNA (28). This coincidence suggests that targeting of Top1 to mitochondria could induce a loss of mtDNA. This notion is also supported by the observation that all constructs derived from Top1 were diffusely distributed in mitochondria and not accumulated in speckled substructures, possibly indicating an absence of nucleoids in these mitochondria (Figure 2A). Additional phenotypic similarities between ρ− cells and cells expressing mitochondria-targeted Top1 are summarized in the data supplement: both cell types require uridine supplementation for growth due to a deficiency in de novo pyrimidine synthesis [(29); Supplementary Figure 2A]. In the absence of uridine, they are arrested in G2 phase, have elevated levels of polyploid and hypodiploid cells (30) (Supplementary Figure 2C) and exhibit features of cellular senescence such as enhanced activity of senescence-associated β-galactosidase and increased cell size [(30); Supplementary Figure 2B].
To analyze whether mtDNA content was indeed affected by the continuous expression of mitochondria-targeted Top1 constructs, we performed quantitative PCR experiments. As summarized in Table 1, mtDNA content was not altered in cells overexpressing MY-Top1mt as compared with control cells expressing MY alone. Mitochondrial targeting of full-length Top1 (MY-Top1) led to a strong depletion of mtDNA. This effect was also seen with the N-terminally truncated Top1 (MY-Top1191–765) or the active site mutant (MY-Top1191–765*). These data demonstrate that a continued presence of Top1 in the mitochondrial matrix results in a complete loss of mtDNA and that this effect does not rely on catalytic activity or the N-terminal domain, which is the most distinctive structural feature of the nuclear paralog.
Table 1.
MtDNA depletion in cells expressing topoisomerase I constructs
| Constructs | MtDNA copies (±SEM) | Number of clones investigated | P-value |
|---|---|---|---|
| MY | 1.0 ± 0.33 | 13 | – |
| MY-Top1mt | 0.94 ± 0.25 | 27 | – |
| MY-Top1 | 7.23 × 10−5 ± 5.56 × 10−5a | 2 | <0.0001 |
| MY-Top1191–765 | 1.97 × 10−4 ± 3.0 × 10−4a | 3 | <0.0001 |
| MY-Top1191–765* | 1.19 × 10−4 ± 2.59 × 10−5a | 3 | <0.0001 |
MtDNA copy numbers were determined by quantitative real time PCR.
aValues differed significantly from the control (MY) according to an unpaired t-test. Values of control cells (MY) were arbitrarily set to 1.0.
Association of MY-Top1191–765 with mitochondrial nucleoids precedes mtDNA depletion
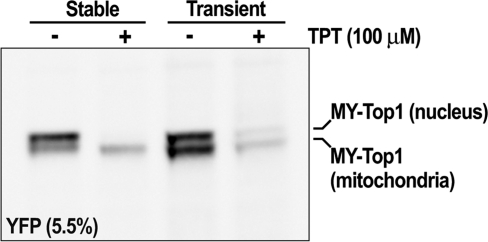
Because the results up to here stem from stably transfected cell clones, they describe an endpoint where mtDNA was terminally depleted. Therefore, they do not allow conclusions regarding the process leading to this endpoint. It remains unclear whether mtDNA depletion occurs instantaneously or gradually, whether it is a direct effect due to active degradation of mtDNA by Top1 constructs and whether the nuclear enzymes associated with mtDNA and nucleoids in the same way as MY-Top1mt (Figure 2A). To address these questions, we studied localization of MY-Top1mt and MY-Top1191–765 by transient transfection. Thirty-six hours after transfection, MY-Top1mt accumulated at mitochondrial speckles in a pattern similar to constitutively expressing cells clones (compare Figure 2B with A). Within the same time frame of transient expression, MY-Top1191–765 concentrated at nucleoid-like speckled structures in a fashion similar to MY-Top1mt. This is clearly different from the homogeneous distribution of MY-Top1191–765 seen upon stable expression (compare Figure 2B with A), suggesting that the enzyme interacts with mtDNA prior to inducing its depletion. It should also be noted that 36 h after transient transfection with MY-Top1191–765, the morphology of mitochondria was already disordered to some extent (Figure 2A and B). To ascertain that the mitochondrial portion of MY-Top1 indeed interacts with mtDNA during transient expression, we treated the cells with TPT, a specific topoisomerase I poison known to access both nuclear and mitochondrial compartments (31) and to stabilize the transient covalent complexes between Top1 and DNA. Since such complexes are retained in the gel slots, Top1-specific signals get depleted from immunoblotting analysis upon effective interaction of the enzyme with DNA and TPT. Therefore, interactions of MY-Top1 with mtDNA should be detectable as TPT-inducible depletion of the lower protein band presumably corresponding to the mitochondrial portion of the enzyme. As shown in Figure 3, this was indeed the case in cells transiently expressing MY-Top1. Both bands were depleted upon TPT treatment suggesting that the enzyme was actively engaged in DNA catalysis in the nucleus and the mitochondria. In contrast, TPT treatment of cells constitutively expressing MY-Top1 (and devoid of mtDNA) only led to a depletion of the upper protein band corresponding to the nuclear portion of enzyme, whereas the lower band representing mitochondrial located enzyme was not diminished. Taken together, these findings indicate that upon targeting to mitochondria Top1 indeed interacts with mtDNA, which possibly triggers mtDNA depletion occurring subsequently.
Figure 3.
MY-Top1 interacts with mtDNA prior to inducing its depletion. Immunoband depletion assay of cells stably or transiently expressing MY-Top1. Lysates of whole cells were subjected to SDS–PAGE (5.5% gel) and western blotting. Blots were probed with YFP antibodies. Cells were cultured in absence (−) or presence of 100 µM TPT (+) for 30 min before lysis.
Nuclear topoisomerase I depletes mtDNA via blockade of RNA transcription
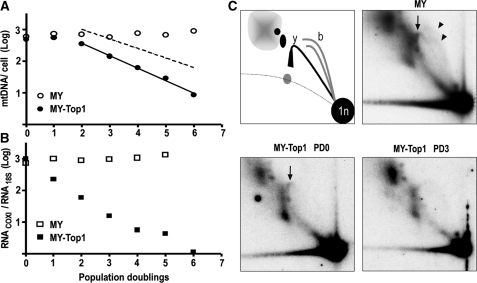
For a more precise analysis of the time course of mtDNA depletion, we transduced HT-1080 cells with lentiviral-based vectors facilitating expression of MY alone or MY-Top1, and monitored cellular mtDNA content during subsequent cell population doublings (PDs). To avoid growth arrest, cells were supplemented with uridine at all times. During the first two PDs, mtDNA copy number was not significantly diminished by expression of either construct (Figure 4A). Subsequently, it remained unchanged in MY-expressing control cells, whereas it declined exponentially in cells expressing MY-Top1 (Figure 4A). The delay in onset of mtDNA depletion excluded that MY-Top1 directly blocked mtDNA replication or actively degraded mtDNA. The exponential manner of subsequent mtDNA depletion suggested rather a mechanism related to cell doubling. Overall, the time course of mtDNA depletion in cells expressing MY-Top1 was best approximated by a model (Figure 4A), assuming a lag phase of ∼2 PD followed by a exponential decay phase, in which mtDNA replication was abolished and mtDNA was reduced 2-fold after each cell division.
Figure 4.
MY-Top1-mediated inhibition of mtDNA transcription precedes mtDNA loss. (A) Kinetics of mtDNA loss. Mitochondria-targeted YFP alone (open circles) or MY-Top1 (closed circles) were expressed in HT-1080 cells through a lentiviral vector system. Transduced cells were grown in the presence of uridine, and mtDNA copy numbers were determined after each PD. Time zero (∼24 h after infection) was defined as the moment at which YFP fluorescence became detectable in mitochondria by fluorescence microscopy and western blotting (data not shown) and in all cultures used, at least 99% of the cells were YFP positive. The full line represents the regression of experimentally determined mtDNA copy numbers from PD 2–6. The hatched line represents a one phase exponential decay (N = N0·e−kt) resulting from a theoretical 2-fold dilution of mtDNA after each cell division. (B) Kinetics of mtRNA loss. In parallel to the mtDNA quantification in (A), COXI mRNA was determined by quantitative RT-PCR and normalized to the level of 18S rRNA in cells expressing MY (open squares) or MY-Top1 (closed squares). (C) Analysis of mtDNA RIs. 2D neutral agarose gel electrophoretic analysis of mtDNA isolated from cells expressing MY at PD 3 (top right) or MY-Top1 at PD 0 (bottom left) and PD 3 (bottom right). The mtDNA was digested with HincII and the fragment containing the control region was detected with a radioactively labeled cytochrome b probe. Positions of bubble arcs (initiation of replication) and the y-arc (ongoing replication) are indicated by arrowheads and arrows, respectively. Schematic illustration of detected RIs (top left): spot (1n), fragments not being replicated; the bubble arcs (b), indicative of fragments with replication initiation; the y-arc (y), fragments on which the replication machinery passes through; positions of RNA-containing RIs are depicted in gray.
These findings suggested an indirect effect of Top1 on mtDNA replication. One mechanism compatible with this would be blockade of mtDNA transcription, which is connected to mtDNA replication, because the transcription machinery synthesizes RNA primers required for replication initiation (10,32). Thus, a strong inhibition of mitochondrial transcription could eventually abolish mtDNA replication. To investigate this possibility, we determined the levels of mtRNA transcripts during MY-Top1-induced mtDNA depletion. Expression of MY-Top1 induced a reduction of COXI mRNA levels by ∼77, ∼94, ∼98 and >99% after 1, 2, 3 and 4 PDs, respectively (Figure 4B). Similar depletion kinetics were observed for mitochondrial 12S rRNA (data not shown). In contrast, MY alone had no such effect (Figure 4B). Comparison of the depletion kinetics of mtDNA and mtRNA transcripts revealed that expression of MY-Top1 inhibited mtDNA transcription instantaneously, whereas reduction in mtDNA copy numbers followed one or two PDs later. Treatment with the specific mtDNA replication inhibitor dideoxycytidine (33) showed a reverse order of mtDNA and mtRNA depletion, and treatment with ethidium bromide that inhibits transcription and replication (34) simultaneously induced depletion of mtDNA and mtRNA (Supplementary Figure 3A and B). These experiments show that the kinetics of inhibition of mtDNA transcription and replication were clearly distinct and that mitochondrial targeting of Top1 first induced a strong inhibition of mtDNA transcription followed by a gradual decline in mtDNA content.
Because mitochondrial targeting of Top1 induced instant depletion of mtRNA and a delayed depletion of mtDNA, we hypothesized that these effects could be caused by an inhibition of RNA-primer synthesis required for mtDNA replication. An implicit prediction of this hypothesis is that, upon mitochondrial targeting of Top1, initiation of mtDNA replication should become suppressed, whereas the already initiated replication processes continue until completion. We, therefore, analyzed replication intermediates (RIs) of mtDNA after expression of MY-Top1. To this end, the major noncoding region of mtDNA, containing the principal initiation and termination sites for replication, was released by digestion with HincII and after separation by 2D neutral agarose gel electrophoresis detected in Southern blots with a cytochrome b probe. During the first PD, when cells expressing MY-Top1 had not yet entered the mtDNA depletion phase, the bubble arcs, indicative of replication initiation, became significantly reduced as opposed to cells expressing MY (Figure 4C). At this stage, the y-arc, indicative of ongoing replication, was only marginally reduced. These differences became more pronounced during the phase of exponential mtDNA depletion, when the bubble arcs as well as the y-arc were completely lacking (Figure 4C). RIs indicative of stalled replication forks were absent at all stages, confirming that Top1 inhibited the initiation of mtDNA replication but not the continuation of started replication forks.
MtDNA depletion is linked to the N-terminal portion of the core domain of Top1
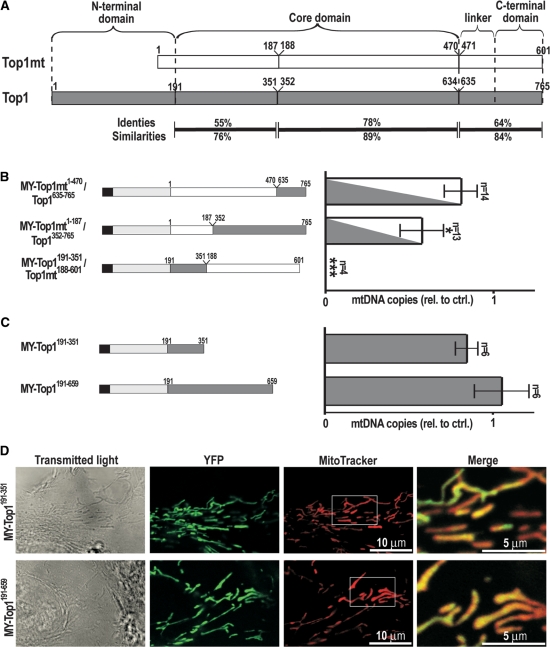
Our observation that Top1 is incompatible with stable mtDNA propagation suggests that the two genetically distinct paralogs of topoisomerase I have undergone evolutionary specialization to the divergent requirements of nuclear and mitochondrial genomes. To determine which evolutionary modifications of the primary structure were crucial in this, we expressed hybrid proteins in which corresponding regions were exchanged between Top1 and Top1mt. In order to simplify, we here designated residues 191–634 of Top1 as the core domain. By this definition, the core includes a part (residues 191–210) of the N-terminal domain. Sequence alignment identified the N-terminal half of the core domain and a C-terminal region encompassing linker and C-terminal domains as regions with the greatest sequence differences between Top1 and Top1mt [55 and 64% identical residues, respectively, Figure 5A; (6,7)]. These regions were selected for exchange. The construct MY-Top1mt1–470/Top1635–765 is identical to Top1mt up to residue 470 and then continues with residues 635–765 of Top1 (Figure 5B, left), whereas MY-Top1mt1–187/Top1352–765 contained residues 1–187 of Top1mt followed by residues 352–765 of Top1. We also exchanged the divergent N-terminal half of the core domain by replacing residues 1–187 in Top1mt with 191–351 of Top1 creating MY-Top1191–351/Top1mt188–601. These constructs were transfected into HT-1080 cells, and several cell clones with stable expression were selected. Immunoblotting and measurement of DNA relaxation activity confirmed that the expressed proteins were of correct size and catalytically active (Supplementary Figure 4A and B). Furthermore, confocal fluorescence microscopy verified their mitochondrial localization (Supplementary Figure 4C). In conclusion, the hybrid proteins were imported into mitochondria, correctly folded and active.
Figure 5.
MtDNA depletion is inflicted by residues 191–351 in Top1. (A) Comparison of human Top1mt (white) and Top1 (gray). Topoisomerase I is composed of N-terminal, core, linker and C-terminal domains as indicated above. Numbers indicate the residues flanking the sites chosen for exchanging regions between Top1 and Top1mt. Amino acid identities and similarities for each region are indicated below. For simplicity, residues 191–634 are designated in the present study as the core domain. (B) MtDNA copy numbers in cell clones expressing Top1/Top1mt hybrids. Left: schematic synopsis of hybrid constructs. All constructs were fused to MY at their N-terminal end: MY-Top1mt1–470/Top1635–765; MY-Top1mt1–187/Top1352–765; and MY-Top1191–351/Top1mt188–601. Right: mtDNA copy numbers in cell clones expressing the constructs indicated on the left were determined using quantitative PCR. Values were normalized to the mean value of clones expressing MY alone, which was arbitrarily set to 1.0. Number of clones investigated is indicated (n), and error bars indicate the SEM. Significant differences between each bar and the control cells MY are marked with asterisks (*P < 0.05, ***P < 0.0001) according to an unpaired t-test. (C) MtDNA copy numbers in cell clones expressing C-terminally truncated versions of Top1. Left: schematic synopsis of MY-Top1191–351 and MY-Top1191–659. Right: mtDNA copy numbers determined as in B. (D) Homogenous mitochondrial distribution of MY-Top1191–351 and MY-Top1191–659. Shown are confocal images of transmitted light (left panel), YFP fluorescence (middle left panel), MitoTrackerRed fluorescence (middle right panel) and merged signals of latter two restricted to the area indicated in the MitoTrackerRed column at 3-fold magnification (right panel).
Analysis of the effects of these constructs on mtDNA content revealed that cells expressing the hybrid MY-Top1mt1–470/Top1635–765, where the C-terminal end of Top1mt was replaced with a corresponding Top1 sequence, displayed no significant reduction of mtDNA levels (Figure 5B, right). Even extending the exchange position into the middle of the DNA-binding core domain (MY-Top1mt1–187/Top1352–765) resulted only in a minor reduction of mtDNA copy numbers. However, replacement of the N-terminal half of the Top1mt core domain with the corresponding portion of the nuclear enzyme (MY-Top1191–351/Top1mt188–601) resulted in complete depletion of mtDNA (Figure 5B). Thus, mtDNA-repressive properties of Top1 are fully retained in hybrids bearing the N-terminal half of the core domain of Top1 (residues 191–351), whereas replacement of all other residues of Top1 with corresponding Top1mt regions failed to induce mtDNA depletion.
Nevertheless, it remained unclear whether the mtDNA-repressive effect is a property of the N-terminal half of the Top1 core domain itself or exerted only when this region is placed in the context of a complete topoisomerase I enzyme. To investigate this, we targeted C-terminal truncations of Top1 to mitochondria. One construct (MY-Top1191–351) was truncated behind residue 351 and thus only contained the minimal region of Top1 conferring the complete mtDNA-repressive effect (Figure 5C). Since this fragment constitutes the entire core subdomain II, but only part of the core subdomain I and none of core subdomain III of Top1 (2), it was uncertain if it would fold into a proper Top1-substructure. Therefore, we created a second construct containing the entire core and part of the linker domain (MY-Top1191–659). This second Top1 fragment is catalytically inactive but its activity can be reconstituted in vitro by addition of a recombinant fragment encompassing residues 658–765 (35), indicating that residues 191–659 are able to form a functional substructure of the Top1 molecule. However, neither MY-Top1191–351 nor MY-Top1191–659 depleted mtDNA (Figure 5C, right) indicating that the mtDNA-repressive potential residing in residues 191–351 of Top1 is only exerted, when this portion is embedded in the entire context of core-, linker- and C-terminal domains of the enzyme.
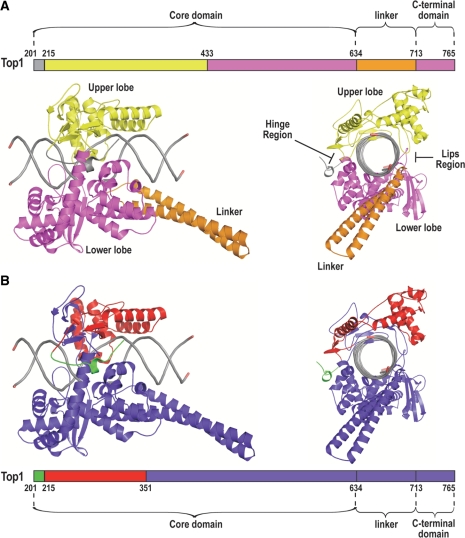
Together these three domains form a bi-lobed protein that clamps around DNA [See illustration in Figure 7A; (2,24)]. Top1 reconstituted by mixing two recombinant fragments encompassing the three domains (Top1175–659 + Top1658–765) is active but has a ∼20-fold lower DNA affinity than the holoenzyme (35). This is thought to indicate the importance of the C-terminal domain for enabling Top1 to clamp around the DNA. MY-Top1191–659 lacking the C-terminal domain is probably unable to interact with mtDNA and therefore unable to repress mtDNA. If this reasoning was correct, C-terminally truncated Top1 constructs should not notably accumulate at mitochondrial nucleoids. As shown in Figure 5D, this was indeed the case for MY-Top1191–351 and MY-Top1191–659. Both were homogenously distributed within the mitochondria and did not exhibit any signs for a speckled accumulation within the mitochondrial matrix. This is in clear contrast with constructs having the entire C-terminal domain such as MY-Top1mt and MY-Top1191–765 (under conditions of transient expression), which have a speckled distribution reflecting association with nucleoids (Figure 2). Therefore, in the case of MY-Top1191–351 and MY-Top1191–659, the absence of speckles is most likely due to the fact that these constructs do not properly bind to DNA.
Figure 7.
Ribbon diagram of the bi-loped structure of Top1 in covalent complex with DNA viewed with the DNA axis horizontally orientated (left) or looking down the axis of the DNA (right). Diagrams are based on the X-ray crystal structure by Staker et al. (24) of a ternary complex of Top1, DNA and TPT, which provides resolution of residues 201–765. TPT is omitted for clarity. (A) Representation of upper lobe in yellow, lower lobe in magenta, the linker region in orange and the DNA in gray. The hinge region that opens and closes the enzyme around the DNA and the lips region where the upper and lower lobes interact upon DNA binding are indicated. (B) Representation of the mtDNA repressive regions (residues 201–214 and 215–351) in green and red, respectively.
On chromosomes Top1mt is less capable of DNA-interaction and DNA-binding than Top1
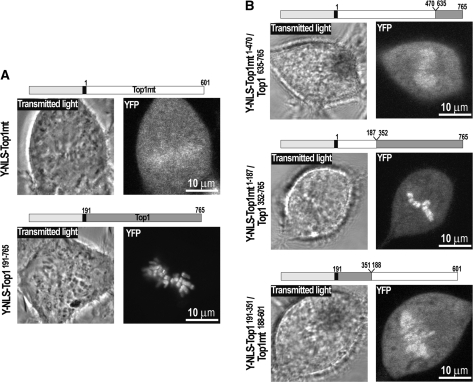
We conclude from these experiments that the N-terminal half of the DNA-binding core domain of Top1 (residues 191–351) suffices for conferring the complete mtDNA-repressive effect. However, downstream regions also play a role, since C-terminal truncations, bearing the mtDNA-repressive N-terminal sequence (i.e. MY-Top1191–351 and MY-Top1191–659), did not cause mtDNA depletion (Figure 5C). The most likely reason for this is their lack of a proper DNA clamping activity that prevents efficient mtDNA interaction. Moreover, hybrid enzymes where the C-terminal half of the DNA-binding core domain of Top1mt was replaced with a corresponding sequence of Top1 (MY-Top1mt1–187/Top1352–765) induced mtDNA depletion, although to a reduced extent (Figure 5B). In summary, mtDNA-repressive properties of Top1 appear to arise from the concerted action of the N- and C-terminal halves of the DNA-binding core domain, which together seem to form a tighter DNA clamp. Consequently, one would expect that Top1mt has a weaker DNA-binding activity than Top1. Consistently, Top1mt can be extracted from mtDNA at lower salt concentration than Top1 from nuclear DNA (H.Zhang and Y.Pommier, unpublished data). We therefore compared binding of the enzymes with native chromatin, e.g. chromosomes in living cells. To this end, we targeted Top1mt to the cell nucleus by addition of an N-terminal YFP-NLS tag (Y-NLS-Top1mt) and compared its localization pattern with a corresponding Top1 construct (Y-NLS-Top1191–765). Y-NLS-Top1191–765 was tightly bound to chromosome during mitosis, as opposed to Y-NLS-Top1mt (Figure 6A). Thus, Top1mt has a much weaker capability to bind DNA than Top1 in a native chromosomal context.
Figure 6.
Interaction of Top1mt, Top1 and Top1mt/Top1 hybrids with metaphase chromosomes. (A) Confocal images of transmitted light (left) and YFP fluorescence (right) of living cells stably expressing Y-NLS-Top1mt and Y-NLS-Top1191–765. Design of expressed construct is shown above. (B) Confocal images of living cells stably expressing Y-NLS-Top1mt1–470/Top1635–765, Y-NLS-Top1mt1–187/Top1352–765 and Y-NLS-Top1191–351/Top1188–601. Design of expressed construct is shown above.
Chromosome interactions of Top1 and Top1mt are determined by the core domains
Based on the differential capabilities of Top1 and Top1mt to bind to chromosomal and mtDNA (Figures 5B and 6A), we speculated that the different domain sequences that confer the mtDNA-repressive potential also enable Top1 to interact with and accumulate on mitotic chromosomes. To test this, we targeted the Top1/Top1mt hybrids used to study the contribution of the individual enzyme domains on mtDNA depletion (Figure 5B) to the cell nucleus and studied their accumulation on mitotic chromosomes. Again, exchange of the linker and C-terminal regions (Y-NLS-Top1mt1–470/Top1635–765) that had not affected mtDNA depletion in MY-Top1 also did not improve chromosome accumulation (Figure 6A and B). In contrast, an exchange in the middle of the core domain combining the N-terminal half of the Top1mt core with the C-terminal half of the Top1 core (Y-NLS-Top1mt1–187/Top1352–765) or vice versa (Y-NLS-Top1191–351/Top1mt188–601) resulted in hybrid enzymes that were clearly more concentrated on chromosomes than Y-NLS-Top1mt but less concentrated than Y-NLS-Top1191–765 (Figure 6B). Because each enzyme equipped with either half of the Top1 core domain has a capability to interact with chromosomes that is half way between Top1 and Top1mt, both halves of the core domain seem to contribute equally to the interaction with metaphase chromosomes. In summary, these data unambiguously demonstrate that the capability of Top1 to interact with chromosomes and the capability of Top1mt to securely maintain mtDNA transcription and replication have coevolved by structural modifications of those enzyme domains that are crucial for DNA affinity and clamping.
DISCUSSION
In this study, we investigated the meaning of the evolutionary duplication of the TOP1 gene into nuclear (Top1) and mitochondrial paralogs (Top1mt). This duplication is conserved in all vertebrates, while invertebrate eukaryotes do not possess a genetically distinct, mitochondria-targeted version of the enzyme (7). This unique feature suggests that Top1mt has a particular significance for mtDNA metabolism in vertebrates. However, Top1mt is not essential for mouse development, even though its loss may have still unknown negative effects in the adult mouse [H.Zhang and Y.Pommier, unpublished data; (36)]. Thus, the existence of two topoisomerase I paralogs could serve to improve the regulation of Top1mt activity. Alternatively, specialized topoisomerase I enzymes could be needed to meet divergent requirements of the two genomes.
The findings presented here support the latter assumption. The two enzyme paralogs of vertebrates have undergone functional specialization to such an extent that they cannot substitute for each other, as opposed to the single enzyme form in lower eukaryotes that provides topoisomerase I activity for both genomes. In yeast, mitochondrial overexpression of the single yeast topoisomerase I is fully compatible with the maintenance of yeast mtDNA (31), whereas we show here that in human cells, mitochondrial targeting of Top1 is incompatible with mtDNA maintenance. Conversely, nuclear targeting of human Top1mt revealed that the mitochondrial enzyme has lost the ability to interact properly with human chromosomes.
Nonphysiological Top1mt protein levels and the resulting increase in topoisomerase I activity in the mitochondrial matrix had no effect on mtDNA copy numbers, suggesting that significantly increased levels of the mitochondria dedicated enzyme paralog are well tolerated. Equivalent levels of the nuclear enzyme paralog in the mitochondria induce complete loss of the mitochondrial genome, although we cannot exclude that much lower levels of Top1 in the mitochondria could be compatible with mtDNA stability. This difference in tolerance shows clearly that the functional properties of the two enzyme paralogs differ significantly. Although the most prominent structural difference between the two enzyme forms is located in the N-terminal domain, this region is mainly responsible for organelle-specific targeting (5,26), but not involved in the functional differentiation of Top1 and Top1mt. We mapped the divergent functional properties of Top1 or Top1mt rather to the enzyme’s core domains and found that the relevant structural deviations are discrete and spread across the entire core region. The most relevant alterations are located in the proximal half of the core domain, i.e. residues 191–351 in Top1 (Figure 7B). Within this region, residues 215–351 (red colored segment in Figure 7B) constitute a part of the upper lobe of the DNA-clamp (a substructure formed by residues 215–434 of Top1, yellow colored segment in Figure 7A). Furthermore, residues 201–214 (green colored segment in Figure 7B) regulate opening and closing of the DNA clamp (2), where three clustered tryptophan residues (Trp203, 205, 206) seem to play a key role (37). Hence, the N-terminal half of Top1’s core domain crucially influences DNA-binding. Additionally, C-terminal deletion mutants, which are inactive and unable to properly clamp around the DNA double helix, do also not repress mtDNA replication, further substantiating our interpretation. In addition, mtDNA-repressive properties are retained in Y723F mutants of Top1. This mutant is also inactive, but has a fully operational DNA clamp as well as normal DNA affinity in vitro (38), and hence possesses an undiminished propensity to interact with mitotic chromosomes (39). Moreover, Top1mt is bound more loosely to mtDNA than Top1 to nuclear DNA (H.Zhang and Y.Pommier, unpublished data) and two out of the three tryptophan residues found to play a key role in Top1 function (37) are not conserved in Top1mt (5). Thus, modifications of the enzyme’s DNA clamp are involved in making Top1mt compatible with mtDNA. The same alterations also diminish the ability of Top1mt to bind mitotic chromosomes. In summary, this suggests that different abilities of Top1 and Top1mt to bind and clamp around DNA cause their divergent functionality on nuclear and mtDNA. The enhanced ability of Top1 to clamp around DNA appears necessary for proper interaction with mitotic chromosomes but, on the other hand, mediates repressive effects on mtDNA. Conversely, in Top1mt DNA binding seems reduced to ascertain compatibility with mtDNA transcription and replication at the cost of a proper function in the chromosome context. Ultimately, in vitro studies using recombinant Top1 and Top1mt will be required to discern in detail how and to what extent, DNA interaction properties differ between the two enzymes.
The dominant-negative effect of Top1 constructs on mtDNA replication is not without precedent. For example, heterologous expression of mutated DNA polymerase-γ or Twinkle helicase strongly decreases mtDNA copy number in HEK-293 cells (23,40,41). However, the molecular events leading to mtDNA depletion in this case appear to be quite different. While mutated Twinkle and DNA polymerase-γ reduce mtDNA copy number by severe replication stalling (23), we demonstrate here by comparison with mtDNA and mtRNA depletion kinetics that the primary effect of Top1 is a repression of mtDNA transcription. Moreover, time resolved analysis of mtDNA RIs indicates that already initiated mtDNA replication proceeds in the presence of Top1, whereas initiation of replication is blocked. This suggests that Top1 depletes mtDNA by inhibiting the RNA transcription necessary for mtDNA replication and that mtDNA depletion is a secondary effect. Such a linkage between blockade of mtDNA transcription and replication has also been observed in Drosophila melanogaster Schneider cells upon knock down of mitochondrial transcription factor B2, which resulted in reduced mtDNA transcription and concomitant decreases in mtDNA copy number (42).
While mtDNA depletion by mitochondria-targeted Top1 is clearly linked to a repression of mitochondrial RNA synthesis, the precise molecular mechanism of this effect can only be deduced by exclusion of unlikely candidates. Clearly, mtDNA depletion is not a direct response to altered mtDNA topology, as the Y723F mutant incapable of removing supercoils from mtDNA also inflicts mtDNA depletion. Given the multiple indications of a stronger DNA binding, it is likely that Top1 coats mtDNA in an obstructive manner, presenting a physical barrier to the progression of the transcription machinery. However, such obstructive coating should also lead to replication stalling of which we found no signs in cells expressing mitochondria-targeted Top1. Moreover, this event should result in a faster decline of RNA transcripts distal (e.g. COXI mRNA) rather than proximal (e.g. 12S rRNA) to the initiation site, whereas we observed identical time courses of decline for COXI mRNA and 12S rRNA in cells expressing MY-Top1. Because our results are inconsistent with a blockade of transcription progression, repression of transcription initiation is the most likely underlying mechanism.
This notion is corroborated by comparison of the known effects of Top1 on nuclear RNA transcription. In yeast, Top1 facilitates nuclear RNA transcription by removing transcription-generated supercoils during proliferation (43), whereas it represses transcription during the stationary growth phase of yeast (44). In reconstituted transcription assays employing human RNA polymerase II, human Top1 has been shown to act both as a cofactor of activator-dependent transcription and a repressor of basal transcription (45,46). The ability to repress transcription is fully conserved in topoisomerase-inactive Y723F mutants and hence independent of Top1 DNA cleavage and relaxation activity. This transcriptional repression is promoter-specific, which strongly indicates a blockade of initiation as the underlying mechanism (46). The similarities between the repressive effects of Top1 on RNA polymerase II transcription and mtDNA transcription suggest a similar underlying mechanism, namely repression of transcription initiation at the promoter. The promoters for mtDNA transcription are located in the major noncoding region of mtDNA that also contains OH, the major origin of replication for leading strand synthesis. This region also contains the regulatory displacement loop (D-loop) that is thought to be involved in transcription and replication initiation as well as nucleoid organization (10,47). Several proteins have been described to bind specifically to the D-loop. In fact, this structure attract and direct Top1mt, since Top1mt cleavage sites are clustered downstream of the D-loop (36). Thus, it is feasible that Top1 inhibits mtDNA transcription by a dominant-negative interaction with the D-loop region.
Whatever the precise mechanism of this dominant-negative interaction may be, we clearly show that it is associated with features that enhance the association of Top1 with condensed chromatin DNA. Attenuation of this property renders Top1mt compatible with mtDNA metabolism. Interestingly, the reduced interaction with condensed chromatin is a feature of all topoisomerases found in mitochondria of vertebrates so far. In human, alternative translation of the mRNA encoding topoisomerase Top3α creates mitochondrial Top3α or nuclear Top3α (3). Incidentally, Top3α hardly interacts with mitotic chromosomes (48) as opposed to Top3β that is exclusively nuclear (49). Along the same lines, the only Top2 found to be associated with mammalian mtDNA appears to be a C-terminally truncated derivative of nuclear Top2β (4), which again is the isoform that hardly interacts with mitotic chromosomes as opposed to Top2α (19,50).
These findings suggest that a divergence in chromatin association is a common event during adaptation of topoisomerases to divergent requirements in nuclear and mitochondrial genomes of vertebrates. Our study clearly shows that such a divergence of requirements must exist, since it has incited the development of specialized topoisomerase I paralogs. However, the evolutionary driving force directing the adaptation process remains unclear. On the one hand, changes in the mtDNA transcription process could have prompted the attenuation of DNA association in Top1mt. On the other hand, an increased complexity of the vertebrate nuclear DNA could have necessitated the development of a more robust Top1, which then became incompatible with mtDNA transcription.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (grants CH 713/1-1, HA2322/2-1, SFB 728 and GRK 1033); Intramural Program of the Center for Cancer Research, National Cancer Institute, National Institute of Health. Funding for open access charge: Deutsche Forschungsgemeinschaft (SFB 728 and GRK 1033).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We are indebted to Stephanie Borkens and Yvonne Linka (Department of Pediatric Oncology, Hematology and Immunology, Heinrich-Heine-University) for providing the pCL1P vector.
REFERENCES
- 1.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Champoux JJ. DNA Topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Lyu YL, Wang JC. Dual localization of human DNA topoisomerase IIIalpha to mitochondria and nucleus. Proc. Natl Acad. Sci. USA. 2002;99:12114–12119. doi: 10.1073/pnas.192449499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Low RL, Orton S, Friedman DB. A truncated form of DNA topoisomerase IIbeta associates with the mtDNA genome in mammalian mitochondria. Eur. J. Biochem. 2003;270:4173–186. doi: 10.1046/j.1432-1033.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, Barcelo JM, Lee B, Kohlhagen G, Zimonjic DB, Popescu NC, Pommier Y. Human mitochondrial topoisomerase I. Proc. Natl Acad. Sci. USA. 2001;98:10608–10613. doi: 10.1073/pnas.191321998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Meng LH, Pommier Y. Mitochondrial topoisomerases and alternative splicing of the human TOP1mt gene. Biochimie. 2007;89:474–481. doi: 10.1016/j.biochi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Meng LH, Zimonjic DB, Popescu NC, Pommier Y. Thirteen-exon-motif signature for vertebrate nuclear and mitochondrial type IB topoisomerases. Nucleic Acids Res. 2004;32:2087–2092. doi: 10.1093/nar/gkh525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Kearney K, Derby M, Wernette CM. On the relationship of the ATP-independent, mitochondrial associated DNA topoisomerase of Saccharomyces cerevisiae to the nuclear topoisomerase I. Biochem. Biophys Res. Commun. 1995;214:723–729. doi: 10.1006/bbrc.1995.2345. [DOI] [PubMed] [Google Scholar]
- 9.Tua A, Wang J, Kulpa V, Wernette CM. Mitochondrial DNA topoisomerase I of Saccharomyces cerevisiae. Biochimie. 1997;79:341–350. doi: 10.1016/s0300-9084(97)80028-4. [DOI] [PubMed] [Google Scholar]
- 10.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in Mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 11.Christensen MO, Barthelmes HU, Feineis S, Knudsen BR, Andersen AH, Boege F, Mielke C. Changes in Mobility Account for Camptothecin-induced Subnuclear Relocation of Topoisomerase I. J. Biol. Chem. 2002;277:15661–15665. doi: 10.1074/jbc.C200066200. [DOI] [PubMed] [Google Scholar]
- 12.Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr. Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- 13.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 14.Christensen MO, Barthelmes HU, Boege F, Mielke C. The N-terminal domain anchors human topoisomerase I at fibrillar centers of nucleoli and nucleolar organizer regions of mitotic chromosomes. J. Biol. Chem. 2002;277:35932–35938. doi: 10.1074/jbc.M204738200. [DOI] [PubMed] [Google Scholar]
- 15.Christensen MO, Barthelmes HU, Boege F, Mielke C. Residues 190-210 of human topoisomerase I are required for enzyme activity in vivo but not in vitro. Nucleic Acids Res. 2003;31:7255–7263. doi: 10.1093/nar/gkg927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rio P, Meza NW, Gonzalez-Murillo A, Navarro S, Alvarez L, Surralles J, Castella M, Guenechea G, Segovia JC, Hanenberg H, et al. In vivo proliferation advantage of genetically corrected hematopoietic stem cells in a mouse model of Fanconi anemia FA-D1. Blood. 2008;112:4853–4861. doi: 10.1182/blood-2008-05-156356. [DOI] [PubMed] [Google Scholar]
- 17.Mochizuki H, Schwartz JP, Tanaka K, Brady RO, Reiser J. High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J. virol. 1998;72:8873–8883. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linka RM, Porter AC, Volkov A, Mielke C, Boege F, Christensen MO. C-terminal regions of topoisomerase IIalpha and IIbeta determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 2007;35:3810–3822. doi: 10.1093/nar/gkm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coffey G, Campbell C. An alternate form of Ku80 is required for DNA end-binding activity in mammalian mitochondria. Nucleic Acids Res. 2000;28:3793–3800. doi: 10.1093/nar/28.19.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barthelmes HU, Habermeyer M, Christensen MO, Mielke C, Interthal H, Pouliot JJ, Boege F, Marko D. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J. Biol. Chem. 2004;279:55618–55625. doi: 10.1074/jbc.M405042200. [DOI] [PubMed] [Google Scholar]
- 22.Koch H, Wittern KP, Bergemann J. In human keratinocytes the common deletion reflects donor variabilities rather than chronologic aging and can be induced by ultraviolet A irradiation. J. Invest. Dermatol. 2001;117:892–897. doi: 10.1046/j.0022-202x.2001.01513.x. [DOI] [PubMed] [Google Scholar]
- 23.Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl Acad. Sci. USA. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mo YY, Wang C, Beck WT. A novel nuclear localization signal in human DNA topoisomerase I. J. Biol. Chem. 2000;275:41107–41113. doi: 10.1074/jbc.M003135200. [DOI] [PubMed] [Google Scholar]
- 26.Alsner J, Svejstrup JQ, Kjeldsen E, Sorensen BS, Westergaard O. Identification of an N-terminal domain of eukaryotic DNA topoisomerase I dispensable for catalytic activity but essential for in vivo function. J. Biol. Chem. 1992;267:12408–12411. [PubMed] [Google Scholar]
- 27.Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 28.Gilkerson RW, Margineantu DH, Capaldi RA, Selker JM. Mitochondrial DNA depletion causes morphological changes in the mitochondrial reticulum of cultured human cells. FEBS letters. 2000;474:1–4. doi: 10.1016/s0014-5793(00)01527-1. [DOI] [PubMed] [Google Scholar]
- 29.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 30.Park SY, Choi B, Cheon H, Pak YK, Kulawiec M, Singh KK, Lee MS. Cellular aging of mitochondrial DNA-depleted cells. Biochem. Biophys. Res. Commun. 2004;325:1399–1405. doi: 10.1016/j.bbrc.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 31.Diaz de la Loza MC, Wellinger RE. A novel approach for organelle-specific DNA damage targeting reveals different susceptibility of mitochondrial DNA to the anticancer drugs camptothecin and topotecan. Nucleic Acids Res. 2009;37:e26. doi: 10.1093/nar/gkn1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wanrooij S, Fuste JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl Acad. Sci. USA. 2008;105:11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson MV, Chin CD, Keilbaugh SA, Lin TS, Prusoff WH. Studies on the inhibition of mitochondrial DNA replication by 3′-azido-3′-deoxythymidine and other dideoxynucleoside analogs which inhibit HIV-1 replication. Biochem. Pharmacol. 1989;38:1033–1036. doi: 10.1016/0006-2952(89)90245-1. [DOI] [PubMed] [Google Scholar]
- 34.Seidel-Rogol BL, Shadel GS. Modulation of mitochondrial transcription in response to mtDNA depletion and repletion in HeLa cells. Nucleic Acids Res. 2002;30:1929–1934. doi: 10.1093/nar/30.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart L, Ireton GC, Champoux JJ. Reconstitution of human topoisomerase I by fragment complementation. J. Mol. Biol. 1997;269:355–372. doi: 10.1006/jmbi.1997.1056. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Pommier Y. Mitochondrial Topoisomerase I Sites in the Regulatory D-Loop Region of Mitochondrial DNA. Biochemistry. 2008;47:11196–11203. doi: 10.1021/bi800774b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laco GS, Pommier Y. Role of a tryptophan anchor in human topoisomerase I structure, function and inhibition. Biochem. J. 2008;411:523–530. doi: 10.1042/BJ20071436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madden KR, Stewart L, Champoux JJ. Preferential binding of human topoisomerase I to superhelical DNA. Embo. J. 1995;14:5399–5409. doi: 10.1002/j.1460-2075.1995.tb00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mo YY, Wang P, Beck WT. Functional expression of human DNA topoisomerase I and its subcellular localization in HeLa cells. Exp. Cell Res. 2000;256:480–490. doi: 10.1006/excr.2000.4864. [DOI] [PubMed] [Google Scholar]
- 40.Jazayeri M, Andreyev A, Will Y, Ward M, Anderson CM, Clevenger W. Inducible expression of a dominant negative DNA polymerase-gamma depletes mitochondrial DNA and produces a rho0 phenotype. J. Biol. Chem. 2003;278:9823–9830. doi: 10.1074/jbc.m211730200. [DOI] [PubMed] [Google Scholar]
- 41.Spelbrink JN, Toivonen JM, Hakkaart GA, Kurkela JM, Cooper HM, Lehtinen SK, Lecrenier N, Back JW, Speijer D, Foury F, et al. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J. Biol. Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- 42.Matsushima Y, Garesse R, Kaguni LS. Drosophila mitochondrial transcription factor B2 regulates mitochondrial DNA copy number and transcription in schneider cells. J. Biol. Chem. 2004;279:26900–26905. doi: 10.1074/jbc.M401643200. [DOI] [PubMed] [Google Scholar]
- 43.Brill SJ, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54:403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 44.Choder M. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 1991;5:2315–2326. doi: 10.1101/gad.5.12a.2315. [DOI] [PubMed] [Google Scholar]
- 45.Kretzschmar M, Meisterernst M, Roeder RG. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc. Natl Acad. Sci. USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 47.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Temime-Smaali N, Guittat L, Wenner T, Bayart E, Douarre C, Gomez D, Giraud-Panis MJ, Londono-Vallejo A, Gilson E, Amor-Gueret M, et al. Topoisomerase IIIalpha is required for normal proliferation and telomere stability in alternative lengthening of telomeres. EMBO J. 2008;27:1513–1524. doi: 10.1038/emboj.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi M, Hanai R. M phase-specific association of human topoisomerase IIIbeta with chromosomes. Biochem. Biophys. Res. Commun. 2001;287:282–287. doi: 10.1006/bbrc.2001.5580. [DOI] [PubMed] [Google Scholar]
- 50.Christensen MO, Larsen MK, Barthelmes HU, Hock R, Andersen CL, Kjeldsen E, Knudsen BR, Westergaard O, Boege F, Mielke C. Dynamics of human DNA topoisomerases IIα and IIβ in living cells. J. Cell Biol. 2002;157:31–44. doi: 10.1083/jcb.200112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.