Abstract
Escherichia coli DbpA is an ATP-dependent RNA helicase with specificity for hairpin 92 of 23S ribosomal RNA, an important part of the peptidyl transferase center. The R331A active site mutant of DbpA confers a dominant slow growth and cold sensitive phenotype when overexpressed in E. coli containing endogenous DbpA. Ribosome profiles from cells overexpressing DbpA R331A display increased levels of 50S and 30S subunits and decreased levels 70S ribosomes. Profiles run at low Mg2+ exhibit fewer 50S subunits and accumulate a 45S particle that contains incompletely processed and undermodified 23S rRNA in addition to reduced levels of several ribosomal proteins that bind late in the assembly pathway. Unlike mature 50S subunits, these 45S particles can stimulate the ATPase activity of DbpA, indicating that hairpin 92 has not yet been sequestered within the 50S subunit. Overexpression of the inactive DbpA R331A mutant appears to block assembly at a late stage when the peptidyl transferase center is formed, indicating a possible role for DbpA promoting this conformational change.
INTRODUCTION
Biogenesis of the two subunits of the Escherichia coli 70S ribosome involves coordinated transcription, processing and folding of three ribosomal RNA (rRNA) molecules and sequential binding of the 54 ribosomal proteins (r-proteins) (1,2). Ribosome assembly proceeds in vivo through at least two known small subunit (30S) precursors (3,4) and three known large subunit (50S) precursors (3–6) that are short-lived at normal growth conditions (7). Reconstitution of subunits from rRNA and total r-proteins (TP30 and TP50) in vitro is characterized by intermediate particles (8–10) that are similar to the precursors observed in vivo (2,11,12). The resulting assembly maps that define the sequence of r-protein binding events during in vitro reconstitution of both subunits (13,14) are generally thought to be good models for the biogenesis of ribosomes in vivo. However, successful reconstitution of subunits in vitro requires non-physiological conditions such as long incubation times and elevated temperatures to overcome kinetic barriers due to misfolding of the rRNA (15).
Several proteins have been identified as assembly cofactors that interact transiently with the maturing ribosome and may serve to overcome similar kinetic barriers in vivo (16,17). More than 20 E. coli ribosomal assembly factors identified to date can be classified in three groups: maturation factors, GTPases and DEAD-box proteins (16). Although the precise roles in assembly for most of these factors are not well defined, a majority of these ribosomal assembly factors have been implicated in 50S subunit assembly, possibly due to its more complex tertiary structure (18,19) compared to the 30S subunit.
One 50S assembly cofactor that is particularly well characterized is DbpA, an E. coli DEAD-box protein with ATPase activity that is strongly stimulated by 23S rRNA (20,21). The C-terminal domain of DbpA resembles an RNA Recognition Motif (RRM) and binds tightly and specifically to hairpin 92, which is part of the peptidyl transferase center (PTC) of 23S rRNA (20–24). Two N-terminal RecA-like domains bind RNA weakly and non-specifically and form the ATP binding pocket between them (22). In the presence of 23S rRNA, intact DbpA binds tightly to hairpin 92 and its ATPase activity is activated by nearby rRNA residues that contact the RecA-like domains. When in the presence of other RNAs, DbpA is difficult to saturate but can show much lower rates of ATPase activity (21,25). DbpA has been demonstrated to be a non-essential gene at various growth conditions (26,27). However, an active site mutant, dbpA R331A, confers a dominant negative slow growth phenotype when overexpressed in a wild-type background (28). The DbpA R331A protein binds rRNA normally but is severely impaired in ATPase and helicase activities compared to wild-type DbpA protein. The ATPase activity of DbpA is not stimulated by mature 50S subunits or 70S ribosomes, leading to the hypothesis that DbpA is involved in ribosome assembly rather than translation (21).
Here, we examine the role of DbpA in 50S assembly by characterizing 45S particles that accumulate in the DbpA R331A mutant strain. Formation of the 45S particle is accompanied by defects in rRNA processing and modification. Analysis of the protein composition of the 45S particle reveals depleted levels of proteins, including the late-binding protein L16, which binds near the PTC and the 23S rRNA binding site of DbpA. These particles stimulate DbpA ATPase activity but this activity alone is not sufficient to reconstitute mature 50S subunits. Taken together these results suggest a possible mechanism of DbpA action during the timeline of 50S subunit maturation.
MATERIALS AND METHODS
Protein, RNA and DNA preparation
Site-directed mutagenesis and purification of DbpA, DbpA R331A, DbpA K53A and DbpA E154A proteins were performed as described previously (28,29).
BL21(DE3) ΔdbpA was a generous gift from I. Iost. The deletion was originally constructed in the strain WJW45 by replacing the dbpA ORF with a kanamycin resistance gene (30). The dbpA deletion was tagged with Tn10 using the CAG12081 strain (zda-3061::tn10) and then P1-transduced into the BL21(DE3) strain. The resulting strain is kanamycin and tetracycline resistant.
Native E. coli ribosomal RNA (23S + 16S rRNA), RNA from ribosomal subunits collected from sucrose gradients (described below), and the 23S rRNA transcript transcribed from pCW1 (21) were prepared as described previously (30). The 32 nucleotide minimal substrate for DbpA (31,32) and 2′-O-methyl U2552 derivative were purchased from IDT DNA.
DNA primers were purchased from IDT DNA and 5′-32P labeled with polynucleotide kinase and γ-32P ATP. Labeled DNA was purified by 20% denaturing PAGE, extracted from the gel by passive elution overnight at 4°C and then precipitated with ethanol. Primers 1, 2 and 3 were complementary to residues 35–55, 2567–2592 and 2519–2545 of 23S rRNA, respectively.
Ribosome profiles
Plasmids containing wild-type dbpA or mutants dbpA R331A, dbpA K53A, or dbpA E154A were transformed into E. coli Tuner™ (DE3) pLacI (Novagen) modified to be RecA− (a gift from M.E. Saks), grown to OD600 ∼ 0.3–0.4, quick-cooled by pouring over ice, and harvested by centrifugation. Cells were lysed as described previously (33), except the lysis buffer contained 20 mM Hepes pH 7.5, 30 mM NH4Cl, 10 mM MgCl2 and 4 mM β-Mercaptoethanol. Clarified lysate was layered over 40% sucrose in buffer A (10 mM MgCl2, 20 mM Hepes KOH pH 7.5, 150 mM NH4Cl, 4 mM β-mercaptoethanol) and centrifuged twice in a Type 70.1 Ti rotor in a Beckman Optima LE-80K ultracentrifuge at 42 000 rpm and 4°C for 15 h to purify the ribosomes. Pelleted ribosomes were resuspended in buffer B (10 mM MgCl2, 50 mM Hepes–KOH pH 7.5, 50 mM KCl), aliquoted, and used immediately or flash frozen and at −80°C. To separate polysomes, 70S ribosomes, and the individual subunits, clarified lysate or purified ribosomes were loaded onto linear gradients containing 10–50% sucrose in buffer A and centrifuged in a Beckman SW-41 rotor at 24 000 r.p.m. and 4°C for 13.4 h. To completely dissociate all polysomes and 70S ribosomes into the individual subunits, clarified lysate or purified ribosomes were loaded onto linear gradients containing 20–40% sucrose in buffer C (1 mM MgCl2, 20 mM Hepes–KOH pH 7.5, 150 mM NH4Cl, 4 mM β-mercaptoethanol) and centrifuged in a Beckman SW-41 rotor at 31 500 r.p.m. and 4°C for 14 h. Gradients were analyzed and fractionated using a Teledyne Isco density gradient system with a UA-6 detector and Foxy Jr. fraction collector. Fractions were collected in a 96-well flat bottom UV–Vis plate and analyzed with a Molecular Devices plate reader at 254 nm. Fractions corresponding to 70S, 50S, 45S, or 30S were pooled and pelleted by centrifuging in a Beckman Type 70.1 Ti rotor at 40 000 r.p.m. and 4°C for 15 h. Pelleted ribosomes and subunits were resuspended in buffer B and loaded on subsequent gradients at the same conditions to further purify the subunits or flash frozen and stored at −80°C. Analytical ribosome profiles were analyzed by pumping the gradient through a flow cell cuvette and measuring the absorbance at 254-nm every 2 s over a total of 18 min with a SpectraMax Plus 384 spectrophotometer (Molecular Devices). Purified ribosome subunits and particles from wild-type rrmJ (HB24) and ΔrrmJ (HB23) strains (33) were collected by similar methods.
Recombination of 70S ribosomes from subunits was performed by incubating equal concentrations of 30S subunits from MRE600 cells with DbpA 50S subunits or 45S particles in buffer B at 37°C for 30 min. This was then loaded onto sucrose gradients containing 10 mM Mg2+ and analyzed with a flow cell cuvette as explained above. Additional recombination experiments were performed in the presence of 1.8 µM DbpA and 2 mM ATP or 10 pmol tRNAfmet and 10 pmol of a 27 nucleotide mRNA (34) and analyzed similarly.
Partial reconstitution of 50S subunits was performed by adding 1 µM DbpA and 15 mM ATP to fresh DbpA R331A lysates, incubating at 24°C for 60 min and analyzed by running on 20–40% sucrose gradients containing 1 mM Mg2+ followed by measuring the absorbance of the gradient in a flow cell cuvette as described earlier.
Primer extension
Detection of single nucleotide modifications was performed by primer extension in the presence of a low concentration of dNTPs based on experiments described previously (35). 5′-32P-end labeled 23S DNA primers 2 or 3 (300 nM) was annealed to 23S + 16S rRNA or rRNA purified from 50S subunits or 45S particles (150 nM) in annealing buffer (500 mM Tris–HCl pH 7.5, 500 mM KCl and 100 mM DTT) by heating at 95°C for 2 min then 50°C for 5 min and cooling on ice for 5 min. Reverse transcription was performed in the presence of high (1 mM) or low (4 µM) dNTPs and 1 U AMV RT enzyme (Promega) and 1× reaction buffer (Promega) at 37°C for 1 h. To each reaction tube, 1 µg of RNaseA was added and then incubated for 1 h at 37°C. Formamide load dye was added and the entire reaction loaded on a 20% acrylamide sequencing gel and run at 100 W for ∼8 h. Gels were exposed to phosphor screens overnight.
Primer extension to determine the length of the 5′-end of 23S rRNA from purified 50S subunits and 45S particles was performed using 5′-32P end labeled DNA primer 1 annealed to rRNA purified from 50S subunits or 45S particles as above. Reverse transcription was performed as above with 1 U of AMV RT in the presence of 1 mM dNTPs.
LiCl washed 50S subunits
MRE600 cells grown to midlog at 37°C were lysed and run on 20–40% sucrose gradients containing 1 mM Mg2+ as explained earlier. Fractions containing 50S subunits were collected and purified on 40% sucrose cushions as explained earlier. Purified 50S subunits were washed with 0.4, 0.6, 0.8, or 1.0 M LiCl in wash buffer (1 M Tris–HCl pH 7.5, 1 M MgOAc) for 5 h on ice and pelleted as described previously (11,36). Particles were resuspended in buffer B, flash frozen and stored at −80°C.
Quantitative analysis of protein levels using mass spectrometry
Complete ribosomal subunits and sub-ribosomal particles collected from sucrose gradients were analyzed for ribosomal protein content by quantitative electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS). Total proteins from wild-type 50S, DbpA R331A 50S, DbpA R331A 45S and LiCl washed particles were first purified by TCA precipitation. Aliquots containing 50–100 pmol of total protein were then mixed with an equal amount of a standard mixture consisting of stoichiometric amounts of total proteins isolated from 15N-labeled 50S subunits. The mixture of unlabeled sample proteins and fully 15N-labeled standard proteins was digested overnight with trypsin, and then analyzed using liquid chromatography coupled ESI-TOF MS, essentially as described previously (37).
The ESI-TOF MS data consists of pairs of peaks corresponding to unlabeled and fully labeled copies of peptides. Individual peaks are identified by their charge and mass/charge ratio and peak pairs are determined by differences in mass/charge ratios, which correspond to the number of nitrogens in the peptide. Typically, data is obtained for multiple peptides for each protein, although this number varies between samples and with the size of the protein.
The relative amounts of unlabeled sample and labeled standard for each pair of peaks are determined by fitting theoretical isotope distributions to the experimental data as previously described (37). The unlabeled : labeled ratio for each protein is determined by averaging the ratios obtained for all peptides from that protein, including multiple charge states for a single peptide.
The magnitude of these ratios is dependent upon the actual amounts of sample and standard mixed together, so they are normalized such that the value obtained for L3 is one and all other values are relative to the L3 content of the particle. L3 is chosen as the basis for normalization as it is both a primary binder expected to be consistently present at near wild-type levels and is a large protein for which data from multiple peptides are consistently obtained. Proteins that are relatively less abundant in the sample than in the standard will have normalized unlabeled fractions less than one. In some cases proteins have normalized ratios slightly larger than one, and this is attributed to experimental errors and the imperfect use of L3 as a benchmark as opposed to these proteins actually being present in super-stoichiometric amounts.
To compensate for differences between strains and laboratories (the samples and standard mixture are prepared in two different laboratories), the normalized ratios from each sample are divided by the normalized ratios obtained for wild-type 50S particles on a protein-by-protein basis. The resulting values give the relative amount of each protein present in the sample as a fraction of the amount present in wild-type subunits.
As a control to demonstrate the accuracy of the ESI-TOF MS method, both unlabeled and labeled versions of the standard mixture were mixed together and the resulting ratios normalized to the value obtained for L3. Data were obtained for 30 proteins, with an average ratio of 0.97 ± 0.06, which is a good estimate for the error inherent to the method itself (Supplementary Figure S1).
ATPase assay
The rate of ATP hydrolysis by DbpA in the presence of 32-mer RNA with and without the 2′-O-methyl at U2552 was measured using the previously described high-throughput coupled spectroscopic assay (22).
Endpoint or kinetic rates of ATP hydrolysis by DbpA in the presence of ribosomal subunits or particles were measured using a TLC plate assay as described previously (38) with the following modifications. In a total volume of 25 µl, 100 nM of RNA (transcribed 23S rRNA, purified ribosome subunits or particles, or LiCl cores) was incubated with 40 nM DbpA and 20 µM ATP plus 0.1 µCi α-32P ATP in buffer (50 mM Hepes pH 7.5, 20 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.1% Tween 20) for 30 min at 24°C. At various time points, 2 µl aliquots (or the entire reaction) were quenched with 500 mM EDTA pH 8, and 2 µl aliquots were spotted on a PEI cellulose TLC plate, run in a LiCl : Formic acid buffer (750 mM : 1 M), dried, and exposed to a phosphor screen. The fraction of ATP hydrolyzed was quantitated and converted to the concentration of ATP hydrolyzed per DbpA. This value was plotted versus time, and the resulting linear slopes were determined as the rates of ATP hydrolysis.
RESULTS
DbpA mutant strains accumulate incomplete large subunits
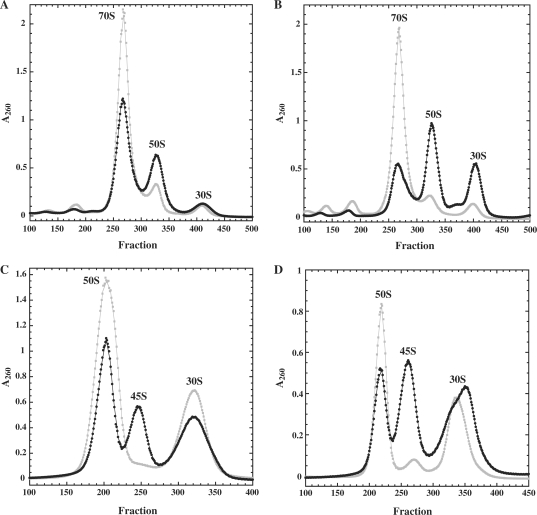
Ribosomes and polysomes in strains overexpressing either wild-type or mutant DbpA R331A were analyzed and compared using sucrose gradient ultracentrifugation. At 37°C, cells overexpressing the mutant exhibited increased levels of individual subunits and decreased levels of 70S ribosomes, compared to cells overexpressing wild-type DbpA (Figure 1A). At 22°C, where the slow growth phenotype is more severe (28), the proportion of 70S ribosomes is further reduced in the mutant overexpressing strain (Figure 1B). Therefore, the slow growth of cells overexpressing DbpA R331A may reflect the reduced rate of protein synthesis resulting from fewer actively translating 70S ribosomes.
Figure 1.
Ribosomes from cells overexpressing wild-type DbpA (grey squares) or the DbpA R331A mutant (black circles) grown at 37°C (A and C) or 22°C (B and D) and analyzed on gradients containing 10 mM Mg2+, 10–50% sucrose (A and B) or 1 mM Mg2+, 20–40% sucrose (C and D). Approximately, 1A260 unit of ribosomes was loaded on each gradient.
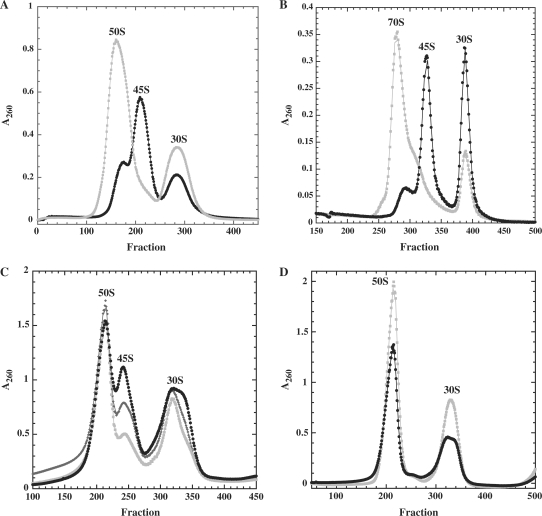
Sucrose gradient profiles in low magnesium conditions, where polysomes and ribosomes dissociate into subunits, reveal a 45S particle in the strain overexpressing DbpA R331A mutant at both 37°C (Figure 1C) and 22°C (Figure 1D) that is virtually absent in the strain expressing wild-type DbpA. The presence of this 45S particle correlates with the higher proportion of free subunits observed at high magnesium conditions, which suggests that it is unable to be incorporated into 70S subunits. To test this, the 50S and 70S peaks from the DbpA R331A strain were collected at high magnesium and reanalyzed under conditions where the subunits dissociate (low magnesium). As shown in Figure 2A, the 70S ribosomes dissociate into clearly defined 30S and 50S peaks, while the 50S subunit is primarily a 45S particle. Both 45S and 50S particles from the DbpA R331A overexpressing strain were also tested for their ability to form 70S ribosomes by incubating them in vitro with wild-type 30S subunits at high magnesium concentration. While the 50S particles could assemble into 70S ribosomes, the 45S particle could not (Figure 2B). Incubation of the DbpA R331A 45S particle with either DbpA and ATP or DbpA, ATP, mRNA, and tRNAfmet prior to the addition of 30S subunits did not promote 70S ribosome formation (data not shown). Taken together, these results indicate that the large subunits isolated from the cells expressing DbpA R331A are a mixture of 50S subunits that can assemble into 70S ribosomes and of partially-assembled 45S particles that cannot.
Figure 2.
(A) DbpA R331A 50S subunits (black circles) and 70S ribosomes (grey squares) collected from gradients containing 10 mM Mg2+ analyzed on a second gradient containing 1 mM Mg2+. (B) DbpA R331A 50S (grey circles) and R331A 45S (black circles) incubated with native 30S subunits and analyzed on gradients containing 10 mM Mg2+. (C) Ribosomes from cells overexpressing DbpA mutants, DbpA R331A (black circles), DbpAK53A (dark grey diamonds) or DbpA E154A (grey squares) were analyzed on sucrose gradients containing 1 mM Mg2+. (D) BL21(DE3) (grey squares) and BL21(DE3) ΔdbpA (black circles) were grown to midlog at 22°C and analyzed on sucrose gradients containing 1 mM Mg2+.
Ribosomes from cells that overexpress two different active site mutants of DbpA, DbpA K53A and DbpA E154A also showed accumulation of 45S particles. These mutants were chosen because they have less severe growth defects and slightly greater RNA-dependent ATPase activity than the DbpA R331A mutant (27). At 37°C, cells overexpressing DbpA K53A and DbpA E154A grow similarly to cells expressing wild-type DbpA. However, at 22°C, the doubling times of DbpA K53A and DbpA E154A (130 min) are slightly slower than wild-type (110 min), but faster than cells expressing the DbpA R331A mutant (250 min) (27). Ribosomes isolated from cells overexpressing DbpA K53A and DbpA E154A grown at 37°C resemble wild-type ribosomes (data not shown), but when they are grown at 22°, they clearly show enhanced amounts of 45S particles, although less than in the cells overexpressing DbpA R331A (Figure 2C). Therefore, the severity of the slow growth phenotype and the accompanying accumulation of 45S particles is roughly correlated with the degree to which the biochemical activity of the overexpressed protein is impaired.
As several groups have observed with other dbpA disruption strains (26,27,39), E. coli BL21(DE3) cells with a deletion of the chromosomal copy of dbpA (ΔdbpA) grow normally at both 22 and 37°C. Ribosomes isolated from ΔdbpA dissociate completely into 50S and 30S subunits similar to wild-type BL21(DE3) cells (Figure 2D). Therefore, it is not the absence of DbpA activity, but rather the presence of large amounts of inactive DbpA that causes ribosome assembly defects and specific accumulation of the 45S particle. However, ribosome assembly is not completely blocked in these ΔdbpA strains or in any of the strains overexpressing inactive DbpA because some mature 50S and 70S ribosomes are produced.
Characterization of the rRNA component in 45S particles
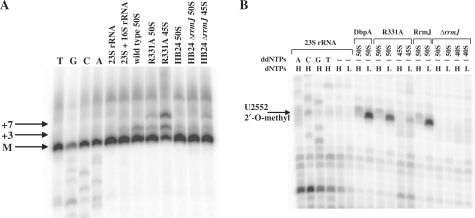
The rRNA present in the 50S subunits and 45S particles from cells overexpressing wild-type DbpA or DbpA R331A mutant was analyzed for possible alterations in rRNA processing or modification. Primer extension analysis of rRNA in 50S subunits from cells overexpressing wild-type DbpA or DbpA R331A showed the mature 5′-end of 23S rRNA and small amounts of a +3 nucleotide extension from incomplete processing (Figure 3A), as had been observed previously (33,40,41). However, a similar analysis of the 5′-ends of the rRNA from DbpA R331A 45S particles revealed reduced amounts of mature 23S rRNA and substantial amounts of incompletely processed +3 and +7 nucleotide products (Figure 3A). Corresponding results have been observed in other particles isolated from deletion strains of different ribosome assembly factors (33,40,42,43), consistent with observations that the final steps of 23S rRNA processing do not occur until after a 70S translation initiation complex is formed (41,44). Primer extension experiments on the rRNA from the broader 30S peak observed for DbpA R331A grown at 22°C (Figure 1D) revealed that a small amount of pre-23S rRNA was present (data not shown), suggesting the presence of a second much smaller precursor particle that could not be resolved from the 30S subunits. It will be interesting to purify this particle and determine its protein and RNA composition.
Figure 3.
RNA from wild-type DbpA 50S subunits, DbpA R331A 50S subunits, DbpA R331A 45S particles, RrmJ 50S subunits, ΔrrmJ 50S subunits and ΔrrmJ 40S particles was analyzed by primer extension using 5′-end labeled primers 1 and 2. (A) The positions of the mature 5′-end (M) of 23S rRNA and the longer products (+3 and +7) of pre-23S rRNA are as indicated. (B) Primer 2 is complementary to nucleotides 3′ of hairpin 92 of 23S rRNA and permits detection of the 2′-O-methyl at U2552 by reverse transcription in the presence of low dNTP concentration. H and L indicate high (1 mM) and low (4 µM) dNTP concentrations. Primer extension stops at modification sites are seen as bands in the low dNTP lanes. On both gels, the first four lanes show a sequencing ladder obtained using the same primers and 23S rRNA transcript. Control experiments were performed with 23S rRNA transcript and 23S + 16S rRNA purified from MRE600 cells.
Similar primer extension experiments were performed with a primer specific for the 5′-end of 5S rRNA in 45S and 50S particles, revealing mature 5′-ends in both the wild-type DbpA and DbpA R331A overexpressing strains (data not shown). Primer extension analysis of the 5′-end of 16S rRNA from 30S subunits of cells overexpressing wild-type DbpA was fully processed, but the 16S rRNA from cells overexpressing DbpA R331A contained a small amount of a +115 nucleotide product. This minor product is likely to be the 17S precursor of 16S rRNA that is matured late in assembly, possibly after subunit association (45). The presence of immature 5′-end of 16S rRNA in cells overexpressing DbpA R331A may be an indirect consequence of the defect in 50S subunit assembly, which leads to an accumulation of 30S subunits that do not form 70S ribosomes, which is necessary for complete 16S rRNA processing. A similar indirect effect on 30S biogenesis has been observed in deletion strains of other E. coli large ribosomal subunit assembly factors (33,40,42,43).
Numerous covalent modifications of rRNA occur during various stages of ribosome maturation (46). In the region of 23S rRNA where DbpA footprints (29), three methylation sites, Um2552, m2A2503 and Cm2498, can all be detected as primer extension stops in the presence of low dNTP concentrations (35,47). The 23S rRNA isolated from 50S subunits from E. coli HB24 cells (48) or cells overexpressing wild-type DbpA or DbpA R331A showed clear primer extension stops at all three expected positions (Figure 3B). However, pre-23S rRNA from the 45S particles showed normal primer extension stops at A2503 and C2498 but only a very weak primer extension stop corresponding to U2552 (Figure 3B), suggesting that this modification is absent in the 45S particles. The 2′-O-methylation at position 2552 is generated by the methyltransferase RrmJ late in assembly (48). As expected, 23S rRNA from either 50S subunits or 40S particles isolated from an rrmJ deletion strain (33) is completely missing the U2252 2′-O-methyl modification (Figure 3B).
Given that U2552 is in hairpin 92, the primary binding site of DbpA, the undermethylation in 45S particles raised the possibility of a functional relationship between DbpA and RrmJ methyltransferase. To determine whether the 2′-O-methyl modification at U2552 had any effect on the binding or activity of DbpA, synthetic 32-nucleotide minimal rRNA substrates (31,32) with and without the U2552 modification were tested for activity in the DbpA ATPase assay. The kmax and Kapp,RNA values for these two substrates are identical, as shown in Table 1, confirming that the 2′-O-methyl group has no effect on DbpA function in vitro. Earlier experiments comparing DbpA activity in the presence of modified and unmodified 23S rRNA also suggested that there was no preference (21).
Table 1.
ATPase activities of DbpA with 32-mer RNA with and without the 2′-O-methyl at U2552
| kmax (min−1) | Kapp,RNA(nM) | |
|---|---|---|
| 32-mer | 45 ± 3.6 | 220 ± 32 |
| 2′-O-methyl 32-mer | 43 ± 2.6 | 230 ± 31 |
The Kapp,RNA and kmax values were measured in the presence of saturating (5 mM) ATP, 60 nM DbpA and 0–2 μM RNA in buffer (50 mM Hepes pH 7.5, 10 mM MgCl2, 50 mM KC1, 100 μM DTT, 200 μM NADH, 1 mM phospho(enol)pyruvate and 10 mM phosphate kinase/lactate dehydrogenase) at 24°C. Lines were fit to the Michaelis–Menten equation.
Characterization of the r-protein composition of 45S particles
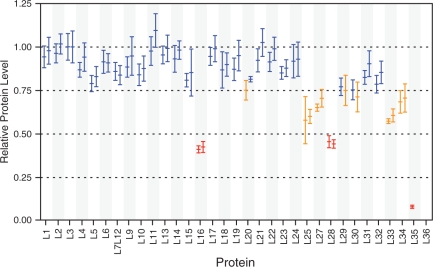
It is unclear from the sedimentation coefficient alone whether the 45S particles from DbpA R331A cells are missing certain r-proteins or sediment slower due to an extended or more open conformation. Slower sedimenting precursor particles that are present during assembly (4) or that accumulate in mutant strains (33) often contain incomplete complements of r-proteins. However, other slower sedimenting particles present in 50S biogenesis and in vitro reconstitution experiments contain a full complement of L-proteins, and as a result must have an altered conformation (8). The 45S particles and 50S subunits from cells overexpressing the DbpA R331A mutant were purified from two successive sucrose gradients and the L-protein composition was quantitatively analyzed using ESI-TOF MS. In order to observe any differences in L-protein composition, the relative protein levels for the 14N-45S particles were compared to the wild-type 15N-50S subunits (Figure 4). Seven proteins (L16, L25, L27, L28, L33, L34 and L35) were significantly reduced (relative abundance <0.75) in the DbpA R331A 45S particles. Three additional proteins (L20, L29 and L30) were reduced in only one of two replicates (levels were slightly above 75% in the other replicate). L26 is present at a reduced level, but this may be due to differential segregation of L26 between subunits in the 45S particle compared to the wild-type 50S subunits, as L26 is also protein S20 from the small subunit (49). No data was obtained for L36 because it is a small protein for which peptides are not routinely found in the new LC/MS data. The relative abundance of these proteins varies significantly, but only L35 is nearly completely absent, indicating the 45S particles are heterogeneous in r-protein composition, each missing only a subset of the proteins present in reduced quantities. With the exception of L20, none of the proteins present at reduced levels assemble early in assembly experiments in vitro, and none of them are required for subsequent protein binding, aside from the dependency of L33 on L28 (14,49,50).
Figure 4.
ESI-TOF MS analysis of the L-protein content of 45S particles from cells overexpressing DbpA R331A. Shown are the normalized relative protein levels compared to wild-type 50S subunits for two sample replicates. Proteins present at near-native levels are shown in blue, proteins present at moderately decreased levels are shown in orange, and proteins present at significantly decreased levels are shown in red. Error bars indicate differences in measurements obtained for multiple peptides from the same protein. No data was obtained for L36.
In order to detect the presence of DbpA in isolated ribosomal subunits, Western blots were performed using a polyclonal antibody against DbpA (28). While no endogenous DbpA was observed in 50S or 30S subunits isolated from wild-type E. coli cells (28), significant levels of DbpA were detected in 30S and 50S subunits from cells overexpressing wild-type DbpA and in 30S, 45S and 50S subunits from cells overexpressing DbpA R331A. After purifying particles through a second sucrose gradient, only very low levels of wild-type or mutant DbpA was observed in any of the particles. This suggests that the observed association of the over-expressed DbpA with ribosomes is the result of non-specific interaction of the basic protein with the polyanionic ribosomes. The absence of stable association of DbpA with any specific ribosomal subunit is in contrast to results demonstrating that two other E. coli DEAD-box proteins, SrmB and CsdA, selectively associate with 40S precursor particles on sucrose gradients when added to cell lysates from ΔsrmB and ΔcsdA mutant strains (33,40).
DbpA R331A 45S particles closely resemble other precursor particles
Accumulation of 50S precursors has been previously observed in cells containing mutations of other proteins proposed to participate in ribosome assembly. Deletions of two other E. coli DEAD-box proteins, SrmB and CsdA, result in slow growth and accumulation of 40S precursor particles which contain incompletely processed 23S rRNA (33,40). However, these particles are somewhat smaller, containing reduced levels of at least 14 different L-proteins, and they presumably correspond to intermediates from earlier stages in ribosome assembly. Deletion or mutation of two different E. coli GTPases, CgtAE and EngA, also results in the accumulation of 50S and 30S subunits and precursor particles that contain incompletely processed 23S rRNA (42,43,51,52). A strain containing a mutant form of CgtAE (G80E D85N) clearly resembles cells overexpressing DbpA R331A, where a 40S particle accumulates, containing reduced levels of L33, L34 and to a lesser extent L16 (43). The large subunit precursor particle that most closely resembles the DbpA R331A 45S particle accumulates in the ΔrrmJ strain (48,53). Since RrmJ targets the same hairpin that binds and activates DbpA, the protein composition of ribosomes from a ΔrrmJ strain was directly compared with ribosomes from cells overexpressing DbpA R331A. Although the ΔrrmJ precursor particle was termed 40S in previous experiments (48), it runs at a similar position on sucrose gradients as the DbpA R331A 45S particle. Quantitation of the protein levels in the ΔrrmJ 40S particle shows that proteins L6, L7/L12, L10, L16, L25, L27, L28, L31, L33 and L35 are observed at reduced levels (Supplementary Figure S2), which is similar to those reduced in the DbpA R331A 45S particle (Figure 4).
Because the DbpA R331A 45S particles have reduced levels of L-proteins that are primarily late-stage assembly proteins, they can also be compared with 50S subunits treated with increasing concentrations of LiCl, which successively removes L-proteins in an order that is inversely correlated to 50S assembly (11,14). Treatment of 50S subunits with 0.4, 0.6, 0.8 and 1.0 M LiCl produced three different core particles which were isolated, and their L-protein content was determined using quantitative mass spectrometry as explained earlier. The 0.4 M LiCl cores showed only very slightly reduced levels of a few proteins, the 0.6 M LiCl cores contained reduced amounts of L28 and the 0.8 M LiCl cores contained clearly reduced amounts of L16 and L28 and slightly reduced amounts of L7/L12 and L10 (Supplementary Figure S3). The 1.0 M LiCl core particles contained severely reduced levels of L16 and L28 and slightly reduced levels of L25, and in addition, proteins L9, L10, L11, L7/L12 that are not reduced in the DbpA R331A 45S particle. The 0.8 M LiCl particle is the closest representation of DbpA R331A 45S particles because it has a similar sedimentation coefficient and protein compositions and also does not bind 30S subunits to form 70S ribosomes (data not shown).
DbpA R331A 45S particles and LiCl core particles are substrates for DbpA
Mature 70S ribosomes and 50S subunits do not stimulate the ATPase activity of DbpA because the binding site of DbpA is likely buried (21). Therefore, the incomplete 45S particles from DbpA R331A and ΔrrmJ strains along with the LiCl core particles were tested for their ability to stimulate the ATPase activity of DbpA using a TLC assay (38). Both the DbpA R331A 45S and ΔrrmJ 40S particles stimulate ATP hydrolysis by DbpA to an extent that is intermediate (∼10 min−1) to the rate observed in the presence of 50S subunits (0.4 min−1) and the rate observed with 23S rRNA (25 min−1) (Table 2). Consequently, the DbpA binding site in hairpin 92 must be much more accessible in 45S and 40S particles than it is in mature 50S subunits. However, since the rate of ATP hydrolysis by DbpA is still not as high as that observed with 23S rRNA, hairpin 92 may still be slightly occluded by L-proteins or other rRNA helices. The 0.4, 0.6 and 0.8 M LiCl core particles stimulate ATP hydrolysis by DbpA to rates of 4.5, 15.3 and 19 min−1, respectively (Table 2). The 0.4 M LiCl particles are apparently somewhat accessible to DbpA, even though they have a nearly complete complement of proteins. The particles from higher LiCl concentrations give rates that approach the observed rates for 23S rRNA, suggesting the DbpA binding site is even less occluded than in 45S particles.
Table 2.
ATP hydrolysis rates of rRNA and particles
| Strain | Substrate | ATP hydrolyzed per DbpA (min−1) | +40 nM R331A | +100 nM R331A | +200 nM R331A |
|---|---|---|---|---|---|
| ATP only | 0.05 ± 0.01 | ||||
| 23S rRNA | 25.1 ± 0.9 | 18.6 ± 1.1 | 10.4 ± 1.2 | 4.6 ± 1.0 | |
| Tuner + dbpA | 50S | 0.4 ± 0.1 | |||
| Tuner + R331A | 45S | 10.6 ± 1.5 | 7.3 ± 1.3 | 5.0 ± 0.4 | 4.0 ± 0.8 |
| 50S | 2.1 ± 0.2 | ||||
| rrmJ | 50S | 0.5 ± 0.2 | |||
| ΔrrmJ | 40S | 10.3 ± 1.4 | |||
| 50S | 5.1 ± 1.5 | ||||
| LiCl cores | 0.4M | 4.5 ± 0.6 | |||
| 0.6M | 15.3 ± 2.0 | ||||
| 0.8M | 19.3 ± 1.4 |
ATP hydrolyzed per DbpA was monitored over time in the presence of 100 nM 23S rRNA or ribosome particles, 40 nM DbpA and 20 μM ATP in buffer (50 mM Hepes pH 7.5, 20 mM KC1, 5 mM MgCl2, 1 mM DTT, 0.1% Tween 20) at 24°C.
The inactive DbpA R331A mutant can bind to full 23S rRNA and 23S rRNA fragments equally as well as wild-type DbpA (28). This suggests that the accumulation of the 45S particle results from the large amounts of DbpA R331A protein binding to hairpin 92 and blocking access to active DbpA during ribosome assembly. To confirm that DbpA R331A can effectively compete with wild-type DbpA for substrates, competition assays were performed where the activity of DbpA stimulated by saturating amounts of 23S rRNA or 45S particles was measured in the presence of DbpA R331A. The addition of increasing concentrations of the mutant DbpA R331A protein significantly reduces the rate of ATP hydrolysis in the presence of 23S rRNA from 25 min−1 to 4.6 min−1 (Table 2). Similarly, the rate of ATP hydrolysis in the presence of 45S particles is reduced from 10.6 min−1 in the absence of DbpA R331A protein to 4.0 min−1 when DbpA R331A protein is added (Table 2). This reduction in ATP hydrolysis rate confirms that the inactive DbpA R331A mutant can compete with active DbpA and therefore can block its function in vivo where the levels of wild-type DbpA are much lower than the overexpressed mutant protein (28).
Since the DbpA R331A 45S particles are substrates for DbpA in vitro, it is possible that in the presence of wild-type DbpA a structural rearrangement could occur, which would allow the particle to continue on the assembly pathway and form 50S subunits in vitro. As mentioned above, this type of rearrangement of 45S particles does not occur in the absence of other components, such as L-proteins, because the addition of DbpA and ATP alone to 45S particles did not permit the formation of 70S ribosomes (Figure 2B). Therefore, DbpA R331A lysates were supplemented with high concentrations of DbpA and ATP, enough to compete with the DbpA R331A protein and relieve the block in assembly, in an environment where L-proteins are available. Fresh clarified lysates of DbpA R331A cells were incubated with up to 1 µM DbpA protein and 1.5 mM ATP for 60 min at 24°C and then run on sucrose gradients containing low magnesium. The resulting ribosome profiles showed no difference and were similar to the profiles of extracts not treated with DbpA (Figure 1C and D).
DISCUSSION
DEAD-box proteins catalyze structural rearrangements in many RNA–protein complexes (54–56), so it is reasonable to propose that DbpA carries out a similar role in the large subunit assembly pathway. However, cells lacking DbpA grow at wild-type rates and show no defect in ribosome assembly, and thus any conformational rearrangement stimulated by DbpA must not be limiting for assembly under normal growth conditions. Ribosome assembly in the absence of DbpA may proceed through a slightly different pathway, possibly involving other DEAD-box proteins (57), but still resulting in normal active ribosomes. If DbpA is not essential for 50S maturation, why do 45S particles accumulate in E. coli when inactive mutant DbpA is overproduced but not when active DbpA is overproduced? One attractive explanation is that the inactive enzyme is unable to catalyze an rRNA conformational change in the 50S assembly pathway and as a result prevents further addition of ribosomal proteins and leads to accumulation of an intermediate particle. Because this intermediate can presumably still bind DbpA, the presence of a high concentration of inactive protein can saturate the site and block any alternative assembly pathways used by the dbpA deletion strains. Presumably, the lower amounts of intact 50S subunits that are observed in the mutant strain are either the result of occasional successful competition by an alternative pathway or the activity of the low concentration of endogenous wild-type DbpA. Conversely, when wild-type DbpA is overproduced, the necessary conformational change does occur and additional ribosomal proteins bind rapidly and block further access to DbpA, allowing 50S subunits to form along the normal pathway. Therefore, although the mutant and wild-type proteins both bind and release rRNA rapidly and show similar binding constants (28), only the overproduction of the inactive mutant protein leads to an assembly defect because it cannot catalyze the conformation change.
It is possible that the 45S particle that accumulates in cells overexpressing the inactive R331A mutant of DbpA is an intermediate in the 50S assembly pathway upon which DbpA normally acts. As shown above, this 45S particle exhibits reduced levels of seven L-proteins (L16, L25, L27, L28, L33, L34 and L35). Since the molecular weight of the 45S particle is only slightly less than the 50S subunit, its slower sedimentation rate is primarily due to an extended, or more open, conformation that is also observed with natural 50S precursors and reconstitution intermediates that are in the final stages of assembly (8). Indeed, many of the proteins that are reduced in the 45S subunit bind late in the proposed ribosome assembly pathways (Supplementary Figure S4), and most of them bind near the peptidyl transferase center of the 50S subunit where DbpA also binds (14,29). Of particular interest is the reduced amount of L16 in the 45S particles. In in vitro ribosome assembly, the binding of L16 to a core particle induces a large conformational change, suggesting that it acts as ‘molecular glue’ to hold the 50S subunit in its compact structure (58). Additionally, it is striking that L16 makes contacts with 23S rRNA at helix 89, which has also been identified as a site of DbpA binding in the presence of ATP and thus a putative site of DbpA action (29,59). Thus, it seems possible that DbpA could promote structural changes in the 45S particle that permits binding of L16 and other late proteins that allows the final assembly of the compact 50S subunit.
Because these 45S particles contain little to no inactive DbpA after purification over two sucrose gradients, it was possible to assay them for stimulation of ATPase activity by wild-type DbpA. Although robust ATPase activity was observed, this only establishes that hairpin 92 is accessible in the 45S particle, not that the 45S particle is the natural substrate of DbpA. Because DbpA binds tightly to and reacts with 23S rRNA in the absence of L-proteins (21), it is possible that it acts early in the assembly pathway and, when inactive, prevents the further maturation of the 50S subunit. In this scenario, the 45S particle is a misfolded, dead-end intermediate that has accumulated many of the L-proteins but cannot progress further in assembly because of the absence of an early rRNA isomerization step. Additionally, the 45S particle, which appears to be a heterogeneous mixture of particles, may also be the result of the dissociation of proteins from an improperly assembled even larger particle that is not stable in the low MgCl2 conditions used for purification. An important future goal is to establish whether or not this 45S particle is able to form active ribosomes. Although our failure to convert 45S particles into 50S ribosomes by adding active DbpA to crude extracts suggests that they are dead end intermediates, pulse chase experiments using either labeled RNA or proteins will be more definitive.
Another possible explanation for the observed effects of overexpression of DbpA R331A is that the methylation of U2552 in hairpin 92 by RrmJ is blocked, which may be an essential event for the final maturation of the 50S subunit. This model is supported by the observation that the 2′-O-methylation at U2552 is absent in DbpA R331A 45S particles and by the fact that ΔrrmJ strains also accumulate 40S particles that are very similar to the 45S particles obtained by overproducing the DbpA R331A mutant (Supplementary Figure S2). However, since RrmJ methylates 50S particles and not 40S particles from ΔrrmJ strains, it is proposed to act late in assembly after the 50S subunit is nearly complete (48,60). Therefore, the unmethylated U2552 in DbpA R331A mutant 45S particles would presumably not be recognized as a substrate for RrmJ and would not be modified until later. Consequently, inhibition of U2552 methylation seems unlikely as the explanation for the phenotype of the DbpA R331A mutant.
In summary, although the absence of DbpA activity does not affect ribosome assembly under normal conditions, the overproduction of inactive DbpA mutants cause a block in assembly resulting in slow growth due to an accumulation of incomplete 50S subunits. These 45S particles contain reduced amounts of L-proteins that are found near the DbpA binding site, which is consistent with the absence of a conformational rearrangement necessary for their binding at this site. However, the point at which DbpA acts in assembly of the 50S subunit is not precisely defined and future studies will be important in determining the nature of the conformational change as well as the stage in assembly at which it occurs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of General Medical Sciences Award Number (F32GM083510 to M.T.S.); the National Institutes of Health (Grant R37-GM53757 to J.R.W.); and the National Institutes of Health (Grant R01-GM60268-08 to O.C.U.). Funding for the open access charge: Discretionary funds from Northwestern University (O.C.U.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Ivelitza Garcia and Dr Margaret E. Saks for helpful discussions and support. The DbpA deletion strain was generously supplied by Dr Isabelle Iost and the DbpA antibody by Dr Frances Fuller-Pace. RrmJ and ΔrrmJ strains were generously provided by Dr Ursula Jakob.
REFERENCES
- 1.Sieber G, Nierhaus KH. Kinetic and thermodynamic parameters of the assembly in vitro of the large subunit from Escherichia coli ribosomes. Biochemistry. 1978;17:3505–3511. doi: 10.1021/bi00610a013. [DOI] [PubMed] [Google Scholar]
- 2.Spillmann S, Dohme F, Nierhaus KH. Assembly in vitro of the 50S subunit from Escherichia coli ribosomes: proteins essential for the first heat-dependent conformational change. J. Mol. Biol. 1977;115:513–523. doi: 10.1016/0022-2836(77)90168-1. [DOI] [PubMed] [Google Scholar]
- 3.Mangiarotti G, Apirion D, Schlessinger D, Silengo L. Biosynthetic precursors of 30S and 50S ribosomal particles in Escherichia coli. Biochemistry. 1968;7:456–472. doi: 10.1021/bi00841a058. [DOI] [PubMed] [Google Scholar]
- 4.Nierhaus KH, Bordasch K, Homann HE. In vivo assembly of Escherichia coli ribosomal proteins. J. Mol. Biol. 1973;74:587–597. doi: 10.1016/0022-2836(73)90049-1. [DOI] [PubMed] [Google Scholar]
- 5.Hayes F, Hayes DH. Biosynthesis of ribosomes in E. coli. I. Properties of ribosomal precursor particles and their RNA components. Biochimie. 1971;53:369–382. doi: 10.1016/s0300-9084(71)80104-9. [DOI] [PubMed] [Google Scholar]
- 6.Osawa S, Otaka E, Itoh T, Fukui T. Biosynthesis of 50s ribosomal subunit in Escherichia coli. J. Mol. Biol. 1969;40:321–351. doi: 10.1016/0022-2836(69)90158-2. [DOI] [PubMed] [Google Scholar]
- 7.Lindahl L. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J. Mol. Biol. 1975;92:15–37. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- 8.Dohme F, Nierhaus KH. Total reconstitution and assembly of 50 S subunits from Escherichia coli Ribosomes in vitro. J. Mol. Biol. 1976;107:585–599. doi: 10.1016/s0022-2836(76)80085-x. [DOI] [PubMed] [Google Scholar]
- 9.Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl Acad. Sci. USA. 1974;71:4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traub P, Nomura M. Mechanism of assembly of 30 s ribosomes studied in vitro. J. Mol. Biol. 1969;40:391–404. doi: 10.1016/0022-2836(69)90161-2. [DOI] [PubMed] [Google Scholar]
- 11.Homann HE, Nierhaus KH. Ribosomal proteins. Protein compositions of biosynthetic precursors and artifical subparticles from ribosomal subunits in Escherichia coli K 12. Eur. J. Biochem. 1971;20:249–257. doi: 10.1111/j.1432-1033.1971.tb01388.x. [DOI] [PubMed] [Google Scholar]
- 12.Nierhaus KH. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 13.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli further studies. J. Biol. Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 14.Herold M, Nierhaus KH. Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J. Biol. Chem. 1987;262:8826–8833. [PubMed] [Google Scholar]
- 15.Williamson JR. After the ribosome structures: how are the subunits assembled? RNA. 2003;9:165–167. doi: 10.1261/rna.2164903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaczanowska M, Rydén-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 18.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 19.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 20.Nicol SM, Fuller-Pace FV. The “DEAD box” protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc. Natl Acad. Sci. USA. 1995;92:11681–11685. doi: 10.1073/pnas.92.25.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsu CA, Uhlenbeck OC. Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry. 1998;37:16989–16996. doi: 10.1021/bi981837y. [DOI] [PubMed] [Google Scholar]
- 22.Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J. Biol. Chem. 2005;280:35499–35505. doi: 10.1074/jbc.M506815200. [DOI] [PubMed] [Google Scholar]
- 23.Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol. 2002;324:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Hu Y, Overgaard MT, Karginov FV, Uhlenbeck OC, McKay DB. The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA. 2006;12:959–967. doi: 10.1261/rna.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boddeker N, Stade K, Franceschi F. Characterization of DbpA, an Escherichia coli DEAD box protein with ATP independent RNA unwinding activity. Nucleic Acids Res. 1997;25:537–545. doi: 10.1093/nar/25.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iost I, Dreyfus M. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 2006;34:4189–4197. doi: 10.1093/nar/gkl500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharpe Elles LM, Uhlenbeck OC. Mutation of the arginine finger in the active site of Escherichia coli DbpA abolishes ATPase and helicase activity and confers a dominant slow growth phenotype. Nucleic Acids Res. 2008;36:41–50. doi: 10.1093/nar/gkm926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karginov FV, Uhlenbeck OC. Interaction of Escherichia coli DbpA with 23S rRNA in different functional states of the enzyme. Nucleic Acids Res. 2004;32:3028–3032. doi: 10.1093/nar/gkh640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Buchholz F, Muyrers JPP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 31.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsu CA, Kossen K, Uhlenbeck OC. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2003;48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 34.Fahlman RP, Dale T, Uhlenbeck OC. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 35.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 36.Nierhaus KH, Montejo V. A protein involved in the peptidyltransferase activity of Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA. 1973;70:1931–1935. doi: 10.1073/pnas.70.7.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sperling E, Bunner AE, Sykes MT, Williamson JR. Quantitative analysis of isotope distributions in proteomic mass spectrometry using least-squares Fourier transform convolution. Anal. Chem. 2008;80:4906–4917. doi: 10.1021/ac800080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuller-Pace FV, Nicol SM, Reid AD, Lane DP. DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peil L, Virumae K, Remme J. Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J. 2008;275:3772–3782. doi: 10.1111/j.1742-4658.2008.06523.x. [DOI] [PubMed] [Google Scholar]
- 40.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srivastava AK, Schlessinger D. Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc. Natl Acad. Sci. USA. 1988;85:7144–7148. doi: 10.1073/pnas.85.19.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang J, Inouye M. The tandem GTPase, Der, is essential for the biogenesis of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2006;61:1660–1672. doi: 10.1111/j.1365-2958.2006.05348.x. [DOI] [PubMed] [Google Scholar]
- 43.Jiang M, Datta K, Walker A, Strahler J, Bagamasbad P, Andrews PC, Maddock JR. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 2006;188:6757–6770. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirdeshmukh R, Schlessinger D. Why is processing of 23 S ribosomal RNA in Escherichia coli not obligate for its function? J. Mol. Biol. 1985;186:669–672. doi: 10.1016/0022-2836(85)90139-1. [DOI] [PubMed] [Google Scholar]
- 45.Srivastava AK, Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Ann. Rev. Microbio. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- 46.Ofengand J, del Campo M. In: EcoSal—Escherichia coli and Salmonella: Cellular and Molecuclar Biology. Björk GR, editor. Washington, DC: ASM Press; 2004. [Google Scholar]
- 47.Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- 48.Bügl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JCA, Jakob U. RNA methylation under heat shock control. Mol. Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- 49.Rohl R, Nierhaus KH. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA. 1982;79:729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roth HE, Nierhaus KH. Assembly map of the 50-S subunit from Escherichia coli ribosomes, covering the proteins present in the first reconstitution intermediate particle. Eur. J. Biochem. 1980;103:95–98. doi: 10.1111/j.1432-1033.1980.tb04292.x. [DOI] [PubMed] [Google Scholar]
- 51.Bharat A, Jiang M, Sullivan SM, Maddock JR, Brown ED. Cooperative and critical roles for both G domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA. J. Bacteriol. 2006;188:7992–7996. doi: 10.1128/JB.00959-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato A, Kobayashi G, Hayashi H, Yoshida H, Wada A, Maeda M, Hiraga S, Takeyasu K, Wada C. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells. 2005;10:393–408. doi: 10.1111/j.1365-2443.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- 53.Caldas T, Binet E, Bouloc P, Richarme G. Translational defects of Escherichia coli mutants deficient in the Um2552 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem. Biophys. Res. Commun. 2000;271:714–718. doi: 10.1006/bbrc.2000.2702. [DOI] [PubMed] [Google Scholar]
- 54.Silverman E, Edwalds-Gilbert G, Lin RJ. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 55.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol. Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 56.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 57.Jain C. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA. 2008;14:381–389. doi: 10.1261/rna.800308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishimura M, Yoshida T, Shirouzu M, Terada T, Kuramitsu S, Yokoyama S, Ohkubo T, Kobayashi Y. Solution structure of ribosomal protein L16 from Thermus thermophilus HB8. J. Mol. Biol. 2004;344:1369–1383. doi: 10.1016/j.jmb.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 59.Polach KJ, Uhlenbeck OC. Cooperative binding of ATP and RNA substrates to the DEAD/H Protein DbpA. Biochemistry. 2002;41:3693–3702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- 60.Caldas T, Binet E, Bouloc P, Costa A, Desgres J, Richarme G. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23S ribosomal RNA methyltransferase. J. Biol. Chem. 2000;275:16414–16419. doi: 10.1074/jbc.M001854200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.