Abstract
With an increase in the importance of umbilical cord blood (CB) as an alternative source of haematopoietic progenitors for allogenic transplantation, donor lymphocyte infusion (DLI) with donor CB-derived activated CD4+ T cells in the unrelated CB transplantation setting is expected to be of increased usefulness as a direct approach for improving post-transplant immune function. To clarify the characteristics of activated CD4+ T cells derived from CB, we investigated their mRNA expression profiles and compared them with those of peripheral blood (PB)-derived activated CD4+ T cells. Based on the results of a DNA microarray analysis and quantitative real-time reverse transcriptase–polymerase chain reaction (RT-PCR), a relatively high level of forkhead box protein 3 (Foxp3) gene expression and a relatively low level of interleukin (IL)-17 gene expression were revealed to be significant features of the gene expression profile of CB-derived activated CD4+ T cells. Flow cytometric analysis further revealed protein expression of Foxp3 in a portion of CB-derived activated CD4+ T cells. The low level of retinoic acid receptor-related orphan receptor γ isoform t (RORγt) gene expression in CB-derived activated CD4+ T cells was speculated to be responsible for the low level of IL-17 gene expression. Our data indicate a difference in gene expression between CD4+ T cells from CB and those from PB. The findings of Foxp3 expression, a characteristic of regulatory T cells, and a low level of IL-17 gene expression suggest that CB-derived CD4+ T cells may be a more appropriate source for DLI.
Keywords: CD4, cord blood, donor lymphocyte infusion, forkhead box protein 3, interleukin 17, T cell
Introduction
Donor lymphocyte infusion (DLI) is a direct and useful approach for improving post-transplant immune function. DLI has been shown to exert a graft-versus-leukaemia (GVL) effect and has emerged as an effective strategy for the treatment of patients with leukaemia, especially chronic myelogenous leukaemia, who have relapsed after unrelated haematopoietic stem cell transplantation (HSCT).1 In addition, DLI has been successfully used for some life-threatening viral infections, including Epstein–Barr virus and cytomegalovirus infections after HSCT.2
Although DLI frequently results in significant acute and/or chronic graft-versus-host disease (GVHD), several groups have demonstrated that depletion of CD8 T cells from DLIs efficiently reduces the incidence and severity of GVHD while maintaining GVL activity.3,4 Therefore, selective CD4 DLI is expected to provide an effective and low-toxicity therapeutic strategy for improving post-transplant immune function. Actually, selective CD4 DLI based on a recently established method for ex vivo T-cell expansion using anti-CD3 monoclonal antibody and interleukin (IL)-2 is now becoming established as a routine therapeutic means of resolving post-transplant immunological problems in Japan.5
The importance of umbilical cord blood (CB) as an alternative source of haematopoietic progenitors for allogenic transplantation, mainly in patients lacking a human leucocyte antigen (HLA)-matched marrow donor, has increased in recent years. Because of the naïve nature of CB lymphocytes, the incidence and severity of GVHD are reduced in comparison with the allogenic transplant setting. In addition, CB is rich in primitive CD16− CD56+ natural killer (NK) cells, which possess significant proliferative and cytotoxic capacities, and so have a substantial GVL effect.6
In contrast, a major disadvantage of CB transplantation is the low yield of stem cells, resulting in higher rates of engraftment failure and slower engraftment compared with bone marrow transplantation. In addition, it was generally thought to be difficult to perform DLI after CB transplantation using donor peripheral blood (PB), with the exception of transplantations from siblings. However, the above-described method for the ex vivo expansion of activated T cells can produce a sufficient amount of cells for therapy using the CB cell residues in an infused bag, which has solved this problem and made it possible to perform DLI with donor CB-derived activated CD4+ T cells in the unrelated CB transplantation setting.5 It has also been reported that CB-derived T cells can be expanded ex vivo while retaining the naïve and/or central memory phenotype and polyclonal T-cell receptor (TCR) diversity,7 and thus potential utilization for adoptive cellular immunotherapy post-CB transplantation has been suggested.8
There are functional differences between CB and PB lymphocytes, although the details remain unclear. In an attempt to clarify the differences in characteristics between activated CD4+ T cells derived from CB and those derived from PB, we investigated gene expression profiles. In this paper we present evidence that CB-derived CD4+ T cells are distinct from PB-derived CD4+ T cells in terms of gene expression.
Materials and methods
Cell culture and preparation
CB was distributed by the Tokyo Cord Blood Bank (Tokyo, Japan). The CB was originally collected and stored for stem cell transplantation. Stocks that were inappropriate for transplantation because they contained too few cells were distributed for research use with informed consent, with the permission of the ethics committee of the bank. In addition, all of the experiments in this study using distributed CB were performed with the approval of the local ethics committee. The mononuclear cells were isolated by Ficoll-Paque centrifugation and cultured in the presence of an anti-CD3 monoclonal antibody and interleukin (IL)-2 using TLY Culture Kit 25 (Lymphotec Inc., Tokyo, Japan) as described previously.5 Although several different methods for T-cell stimulation have been reported, this method is currently being used clinically in Japan. Thus we selected this method in this study. After 14 days of culture, CD4+ cells were isolated using a magnetic-activated cell sorting (MACS) system (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. As a control, mononuclear cells isolated from the peripheral blood of healthy volunteers were similar examined.
Polymerase chain reaction (PCR)
Total RNA was extracted from cells using an RNeasy kit (Qiagen, Valencia, CA) and reverse-transcribed using a First-Strand cDNA synthesis kit (GE Healthcare Bio-Science Corp., Little Chalfont, Buckinghamshire, UK) according to the manufacturer’s instructions. Using cDNA synthesized from 150 ng of total RNA as a template for one amplification, real-time reverse transcriptase (RT)-PCR was performed using SYBR® Green PCR master mix, TaqMan® Universal PCR master mix and TaqMan® gene expression assays (Applied Biosystems, Foster City, CA), and an inventoried assay carried out on an ABI PRISM® 7900HT sequence detection system (Applied Biosystems) according to the instructions provided. Either the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene or the β-actin gene was used as an internal control for normalization. The sequences of gene-specific primers for real-time RT-PCR are listed in Table 1.
Table 1.
The sequences of gene-specific primers for reverse transcriptase–polymerase chain reaction (RT-PCR) and real-time RT-PCR used in this study
| Primer | Sequence |
|---|---|
| IL-4 forward | CACAGGCACAAGCAGCTGAT |
| IL-4 reverse | CCTTCACAGGACAGGAATTCAAG |
| IL-6 forward | GTAGCCGCCCCACACAGA |
| IL-6 reverse | CCGTCGAGGATGTACCGAAT |
| IL-10 forward | GCCAAGCCTTGTCTGAGATGA |
| IL-10 reverse | CTTGATGTCTGGGTCTTGGTTCT |
| IL-17 forward | GACTCCTGGGAAGACCTCATTG |
| IL-17 reverse | TGTGATTCCTGCCTTCACTATGG |
| IL-17F forward | GCTTGACATTGGCATCATCAA |
| IL-17F reverse | GGAGCGGCTCTCGATGTTAC |
| IL-23 forward | GAGCCTTCTCTGCTCCCTGATAG |
| IL-23 reverse | AGTTGGCTGAGGCCCAGTAG |
| IL-23R forward | AACAACAGCTCGGCTTTGGTATA |
| IL-23R reverse | GGGACATTCAGCAGTGCAGTAC |
| IFNG forward | CATCCAAGTGATGGCTGAACTG |
| IFNG reverse | TCGAAACAGCATCTGACTCCTTT |
| GM-CSF forward | CAGCCCTGGAGCATGTG |
| GM-CSF reverse | CATCTCAGCAGCAGTGTCTCTACr |
| RORγt forward | TGGGCATGTCCCGAGATG |
| RORγt reverse | GCAGGCTGTCCCTCTGCTT |
| STAT-3 forward | GGAGGAGGCATTCGGAAAGT |
| STAT-3 reverse | GCGCTACCTGGGTCAGCTT |
| FOXP3 forward | GAGAAGCTGAGTGCCATGCA |
| FOXP3 reverse | GCCACAGATGAAGCCTTGGT |
IL, interleukin; IFNG, interferon γ; FOXP3, forkhead box protein 3; GM-CSF, granulocyte–macrophage colony-stimulating factor; RORγt, retinoic acid receptor-related orphan receptor γ isoform t; STAT, signal transducer and activator of transcription.
DNA microarray analysis
The microarray analysis was performed as previously described.9 Total RNA isolated from cells was reverse-transcribed and labelled using One-Cycle Target Labeling and Control Reagents as instructed by the manufacturer (Affymetrix, Santa Clara, CA). The labelled probes were hybridized to a Human Genome U133 Plus 2·0 Array (Affymetrix). The arrays were used in a single experiment and analysed with genechip operating software 1.2 (Affymetrix). Background subtraction and normalization were performed using genespring gx 7.3 software (Agilent Technologies, Santa Clara, CA). The signal intensity was pre-normalized based on the positive control genes (GAPDH and β-actin) for all measurements on that chip. To account for differences in detection efficiency between spots, the pre-normalized signal intensity of each gene was normalized to the median of pre-normalized measurements for that gene. The data were filtered as follows. (i) Genes that were scored as absent in all samples were eliminated. (ii) Genes with a signal intensity of < 90 were eliminated. (iii) Genes that exhibited increased (fold-change > 2) or decreased (fold-change > 2) expression in CB-derived CD4+ T cells compared with PB-derived CD4+ T cells were selected by comparing the mean value of signal intensities in each condition.
Immunofluorescence study
After periods of cultivation, cells were collected and stained with fluorescence-labelled monoclonal antibodies and analysed by flow cytometry (FC500; Beckman/Coulter, Fullerton, CA). A four-colour immunofluorescence study was performed with a combination of fluorescein isothiocyanate (FITC)-conjugated anti-CD3, phycoerythrin (PE)-conjugated anti- forkhead box protein 3 (Foxp3), phycoerythrin-cyanine-5 (PC5)-conjugated anti-CD4 and PC7-conjugated anti-CD8 (Beckman/Coulter). After staining of cell surface antigens, cells were permeabilized with IntraPrep (Dako, Glostrup, Denmark) and intracellular antigen (Foxp3) was further stained.
Statistical analysis
The statistical analysis was performed using a Student’s t-test and a P-value < 0·05 was considered to be statistically significant.
Results
Expression profiles of activated CD4+ T cells derived from human CB and PB
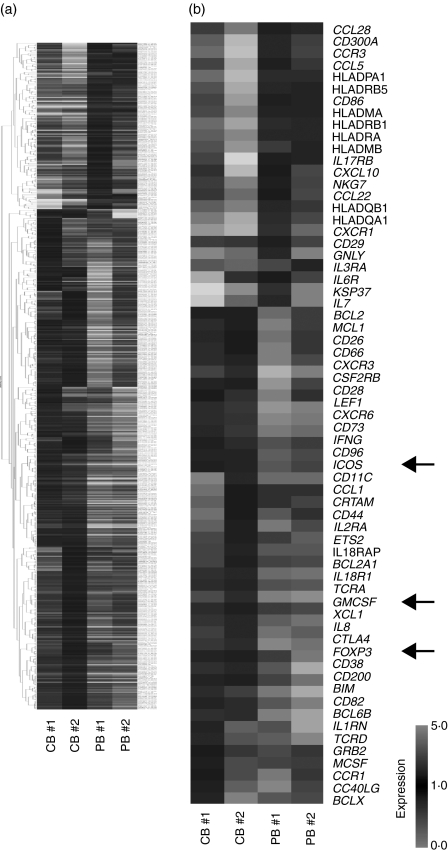
To compare the gene expression patterns of CB-derived CD4+ cells and PB-derived CD4+ cells, we performed DNA microarray analysis using the Affymetrix Human Genome U133 Plus 2·0 Array. After background subtraction, comparison of the gene expression profiles of two independent CB-derived CD4+ samples and PB-derived CD4+ samples was performed using a gene cluster analysis. The genes differentially expressed (fold-change > 2) between the activated CD4+ T cells derived from CB and those derived from PB were selected, and 396 probes were found to exhibit higher levels of expression in CB-derived CD4+ samples while 131 probes exhibited higher levels in PB-derived CD4+ samples. Parts of the data are summarized and presented in Fig. 1a and Tables 2–4.
Figure 1.
Comparison of the gene expression profiles of cord blood (CB)- and peripheral blood (PB)-derived CD4+ T cells. Hierarchical clustering of results from a microarray analysis for CB- and PB-derived CD4+ T cells is indicated. (a) A total of 529 genes characterizing CD4+ T cells (396 genes for CB-derived CD4+ T cells and 131 genes for PB-derived CD4+ T cells) were used to create the gene tree. The gene list is presented in Tables 3 and 4. (b) Genes related to T-cell development (40 genes for CB-derived CD4+ T cells and 26 genes for PB-derived CD4+ T cells) are presented. The arrows indicate the expression pattern of T-cell lineage-specific genes including inducible T-cell co-stimulator (ICOS), granulocyte-macrophage colony-stimulating factor (GM-CSF) and forkhead box protein 3 (FOXP3).
Table 2.
The microarray results for T-cell-related genes
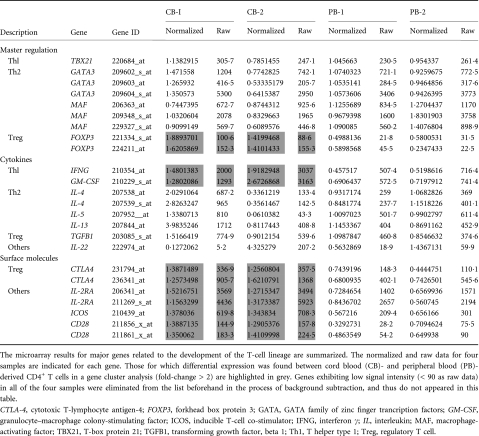 |
Table 4.
Genes up-regulated in CD4+ T cells from peripheral blood (PB)
| Fold change |
||||||
|---|---|---|---|---|---|---|
| Affi ID | Gene abbreviation | CB 1 | CB 2 | PB 1 | PB 2 | Gene name |
| Apoptosis | ||||||
| 1553681_a_at | PRF1 | 0·66 | 0·51 | 1·41 | 1·34 | Perforin 1 (pore-forming protein) |
| B- and T-cell development | ||||||
| 224499_s_at | AICDA | 0·06 | 0·44 | 1·56 | 3·47 | Activation-induced cytidine deaminase |
| 205495_s_at | GNLY | 0·40 | 0·51 | 1·49 | 6·34 | Granulysin |
| 217478_s_at | HLA-DMA | 0·67 | 0·39 | 1·33 | 1·35 | Major histocompatibility complex, class II, DM alpha |
| 203932_at | HLA-DMB | 0·64 | 0·31 | 2·02 | 1·36 | Major histocompatibility complex, class II, DM beta |
| 211991_s_at | HLA-DPA1 | 0·50 | 0·14 | 1·54 | 1·50 | Major histocompatibility complex, class II, DP alpha 1 |
| 212671_s_at | HLA-DQA1 | 0·44 | 0·23 | 1·56 | 2·56 | Major histocompatibility complex, class II, DQ alpha 1 |
| 211656_x_at | HLA-DQB1 | 0·63 | 0·48 | 1·37 | 7·07 | Major histocompatibility complex, class II, DQ beta 1 |
| 210982_s_at | HLA-DRA | 0·58 | 0·37 | 1·50 | 1·42 | Major histocompatibility complex, class II, DR alpha |
| 208306_x_at | HLA-DRB1 | 0·51 | 0·24 | 1·49 | 1·61 | Major histocompatibility complex, class II, DR beta 3 |
| 204670_x_at | HLA-DRB5 | 0·63 | 0·22 | 1·47 | 1·37 | Major histocompatibility complex, class II, DR beta 5 |
| 211634_x_at | IGHV1-69 | 0·69 | 0·77 | 1·23 | 1·99 | Immunoglobulin heavy variable 1–69 |
| 211645_x_at | IgK | 0·15 | 0·49 | 1·51 | 6·62 | Immunoglobulin kappa light chain (IGKV) |
| 221651_x_at | IGKC | 0·46 | 0·68 | 1·32 | 5·57 | Immunoglobulin kappa constant |
| 215379_x_at | IGLC2 | 0·62 | 0·41 | 1·38 | 4·26 | Immunoglobulin lambda joining 2 |
| 209031_at | IGSF4 | 0·50 | 0·03 | 2·33 | 1·50 | Immunoglobulin superfamily, member 4 |
| 205686_s_at | CD86 | 0·70 | 0·23 | 1·30 | 1·39 | CD86 antigen (CD28 antigen ligand 2, B7-2 antigen) |
| 204698_at | ISG20 | 0·68 | 0·49 | 1·32 | 1·64 | Interferon stimulated exonuclease gene, 20 kDa |
| 213915_at | NKG7 | 0·72 | 0·42 | 1·28 | 2·31 | Natural killer cell group 7 sequence |
| Cell growth and maintenance | ||||||
| 201334_s_at | ARHGEF12 | 0·74 | 0·50 | 1·26 | 1·96 | Rho guanine nucleotide exchange factor (GEF) 12 |
| 230292_at | CHC1L | 0·70 | 0·56 | 1·30 | 2·02 | Regulator of chromosome condensation (RCC1) |
| 205081_at | CRIP1 | 0·56 | 0·73 | 1·27 | 1·75 | Cysteine-rich protein 1 (intestinal) |
| 31874_at | GAS2L1 | 0·77 | 0·52 | 1·23 | 2·35 | Growth arrest-specific 2 like 1 |
| 202364_at | MXI1 | 0·43 | 0·73 | 1·27 | 1·44 | MAX interactor 1 |
| 219304_s_at | PDGFD | 0·65 | 0·71 | 1·29 | 3·68 | Platelet-derived growth factor D |
| 213397_x_at | RNASE4 | 0·64 | 0·46 | 1·36 | 2·21 | Ribonuclease, RNase A family, 4 |
| 213566_at | RNASE6 | 0·69 | 0·39 | 1·49 | 1·31 | Ribonuclease, RNase A family, k6 |
| 219077_s_at | WWOX | 0·40 | 0·78 | 1·25 | 1·22 | WW domain containing oxidoreductase |
| Cytokine and chemokine | ||||||
| 207861_at | CCL22 | 0·76 | 0·52 | 1·24 | 2·47 | Chemokine (C–C motif) ligand 22 |
| 238750_at | CCL28 | 0·74 | 0·45 | 1·26 | 1·41 | Chemokine (C–C motif) ligand 28 |
| 1555759_a_at | CCL5 | 0·71 | 0·23 | 1·29 | 1·92 | Chemokine (C–C motif) ligand 5 |
| 208304_at | CCR3 | 0·50 | 0·12 | 1·50 | 2·35 | Chemokine (C–C motif) receptor 3 |
| 205898_at | CX3CR1 | 0·30 | 0·20 | 1·70 | 4·16 | Chemokine (C–X3–C motif) receptor 1 |
| 204533_at | CXCL10 | 0·80 | 0·16 | 1·20 | 2·53 | Chemokine (C–X–C motif) ligand 10 |
| 219255_x_at | IL-17RB | 0·73 | 0·04 | 1·27 | 1·29 | Interleukin 17 receptor B |
| 206148_at | IL-3RA | 0·60 | 0·54 | 2·46 | 1·40 | Interleukin 3 receptor, alpha (low affnity) |
| 226333_at | IL-6R | 0·22 | 0·79 | 1·21 | 2·43 | Interleukin-6 receptor |
| 206693_at | IL-7 | 0·09 | 0·54 | 1·46 | 5·86 | Interleukin-7 |
| Signal transduction | ||||||
| 204497_at | ADCY9 | 0·76 | 0·40 | 1·24 | 2·40 | Adenylate cyclase 9 |
| 206170_at | ADRB2 | 0·58 | 0·35 | 1·42 | 3·97 | Adrenergic, beta-2-, receptor, surface |
| 202096_s_at | BZRP | 0·50 | 0·54 | 1·59 | 1·46 | Benzodiazapine receptor (peripheral) |
| 230464_at | EDG8 | 0·04 | 0·09 | 1·91 | 2·42 | Endothelial differentiation, sphingolipid G-protein-coupled receptor 8 |
| 223423_at | GPR160 | 0·54 | 0·68 | 1·40 | 1·32 | G protein-coupled receptor 160 |
| 227769_at | GPR27 | 0·07 | 0·08 | 1·92 | 244 | G protein in-coupled receptor 27 |
| 210095_s_at | IGFBP3 | 0·27 | 0·20 | 1·73 | 5·25 | Insulin-like growth factor binding protein 3 |
| 38671_at | PLXND1 | 0·08 | 0·65 | 1·35 | 2·57 | Plexin D1 |
| 226101_at | PRKCE | 0·56 | 0·43 | 1·72 | 1·44 | Protein kinase C. epsilon |
| 232629_at | PROK2 | 0·01 | 0·13 | 1·87 | 2·09 | Prokineticin 2 |
| 203329_at | PTPRM | 0·36 | 0·62 | 1·38 | 1·93 | Protein tyrosine phosphatase, receptor type, M |
| 204731_at | TGFBR3 | 0·78 | 0·55 | 1·22 | 2·04 | Transforming growth factor, beta receptor III (betaglycan, 300 kDa) |
| Transcription | ||||||
| 203129_s_at | KIF5C | 0·67 | 0·09 | 1·33 | 3·43 | Kinesin family member 5C |
| 213906_at | MYBL1 | 0·75 | 0·51 | 1·25 | 3·63 | V-myb myeloblastosis viral oncogene homologue (avian)-like 1 |
| 209815_at | PTCH | 0·59 | 0·27 | 1·41 | 4·17 | Patched homologue (Drosophila) |
| 213891_s_at | TCF4 | 0·74 | 0·65 | 2·06 | 1·26 | Transcription factor 4 |
| 238520_at | TRERFI | 0·70 | 0·77 | 1·23 | 2·30 | Transcriptional regulating factor 1 |
| 203603_s_at | ZFHX1B | 0·74 | 0·61 | 1·26 | 3·63 | Zinc finger homobox 1b |
| 213218_at | ZNF187 | 0·74 | 0·69 | 1·26 | 1·76 | Zinc finger protein 187 |
| 221123_x_at | ZNF395 | 0·38 | 0·71 | 1·63 | 1·29 | Zinc finger protein 395 |
Among these genes, those closely correlated to T-cell function and development were selected (Fig. 1b). The genes exhibiting higher levels of expression in CB-derived CD4+ samples included those encoding cell cycle regulators, including cyclin-dependent kinase (CDKN)2A and 2B, transcriptional regulators and signal transduction factors (Tables 2 and 3). The genes for cytokines, chemokines and their receptors such as Interferon γ (IFNG), granulocyte-macrophage colony-stimulating factor (GM-CSF) and for T-cell transcriptional regulators (FOXP3) as well as the genes related to T-cell development including CD28, cytotoxic T lymphocyte antigen-4 (CTLA4) and inducible T-cell co-stimulator (ICOS) were also found among the genes exhibiting higher levels of expression in CB-derived CD4+ samples (Fig. 1b). The factors reported to be essential for negative selection in CD4+ CD8+ thymocytes such as BCL2-like 11 (BIM)10 as well as other apoptotic regulators were also found among the genes exhibiting higher expression levels in CB-derived CD4+ samples.
Table 3.
Genes up-regulated in CD4+ T cells from cord blood samples 1 and 2 (CB 1 and CB 2, respectively)
| Fold change |
||||||
|---|---|---|---|---|---|---|
| Affi ID | Gene abbreviation | CB 1 | CB 2 | PB 1 | PB 2 | Gene name |
| Apoptosis | ||||||
| 1555372_at | BimL | 1·39 | 1·52 | 0·61 | 0·42 | BCL2-like 11 (apoptosis facilitator) |
| 237837_at | BCL2 | 1·27 | 1·32 | 0·49 | 0·73 | B-cell CLL/lymphoma 2 |
| 205681_at | BCL2A1 | 1·91 | 1·53 | 0·39 | 0·47 | BCL2-related protein A1 |
| 1558143_a_at | BCL2L11 | 1·68 | 1·74 | 0·32 | 0·32 | BGL2-like 11 (apoptosis facilitator) |
| 228311_at | BCL6B | 1·36 | 3·39 | 0·64 | 0·26 | B-cell CLL/lymphoma 6, member B (zinc finger protein) |
| 215037_s_at | BCLX | 2·56 | 1·27 | 0·73 | 0·56 | BCL2-like 1 |
| 224414_s_at | CARD6 | 2·65 | 1·34 | 0·56 | 0·66 | Caspase recruitment domain family, member 6 |
| 201631_s_at | IER3 | 1·62 | 2·95 | 0·38 | 0·31 | Immediate early response 3 |
| 218000_s_at | PHLDA1 | 2·34 | 1·21 | 0·53 | 0·79 | Pleckstrin homology-like domain, family A, member 1 |
| 209803_s_at | PHLDA2 | 2·87 | 1·32 | 0·31 | 0·68 | Pleckstrin homology-like domain, family A. member 2 |
| 203063_at | PPMIF | 1·26 | 1·53 | 0·74 | 0·64 | Protein phosphatase IF (PP2C domain containing) |
| 205214_at | STK17B | 1·78 | 1·26 | 0·74 | 0·71 | Serine/threonine kinase 17b (apoptosis-inducing) |
| 217853_at | TENS1 | 1·63 | 6·00 | 0·04 | 0·37 | Tensin 1 |
| B- and T-cell development | ||||||
| 211861_x_at | CD28 | 1·35 | 1·41 | 0·49 | 0·65 | CD28 antigen(Tp44) |
| 207892_at | CD40LG | 3·67 | 1·32 | 0·45 | 0·68 | C040 ligand (TNF superfamily, member 5, hyper-IgM syndrome) |
| 206914_at | CRTAM | 2·76 | 1·60 | 0·40 | 0·36 | Class I MHC-restricted T-cell-associated molecule |
| 210557_x_at | CSF1 | 3·79 | 1·22 | 0·78 | 0·70 | Colony-stimulating factor 1 (macrophage) |
| 210229_s_at | CSF2 | 1·28 | 2·67 | 0·69 | 0·72 | Colony-stimulating factor 2 (granulocyte–macrophage) |
| 205159_at | CSF2RB | 2·33 | 1·60 | 0·18 | 0·40 | Colony-stimulating factor 2 receptor |
| 231794_at | CTLA4 | 1·39 | 1·26 | 0·74 | 0·44 | Cytotoxic T-lymphocyte-associated protein 4 |
| 204232_at | FCER1G | 1·63 | 2·14 | 0·28 | 0·37 | Fc fragment of IgE, high affinity 1, receptor for; gamma polypeptide |
| 210439_at | ICOS | 1·38 | 1·34 | 0·57 | 0·66 | Inducible T-cell costimulator |
| 210354_at | IFNG | 1·48 | 1·92 | 0·46 | 0·52 | Human mRNA for HuIFN -gamma interferon |
| 230536_at | PBX4 | 1·48 | 1·26 | 0·50 | 0·74 | Pre-B-cell leukaemia transcription factor 4 |
| 215540_at | TCRA | 1·25 | 1·87 | 0·67 | 0·75 | T-cell antigen receptor alpha |
| 234440_al | TCRD | 7·51 | 1·48 | 0·50 | 0·52 | Human T-cell receptor delta-chain |
| Cell growth and maintenance | ||||||
| 213497_at | ABTB2 | 2·06 | 1·34 | 0·66 | 0·63 | Ankyrin repeat and BTB (POZ) domain containing 2 |
| 201236_s_at | BTG2 | 1·60 | 1·23 | 0·60 | 0·77 | BTG family, member 2 |
| 235287_at | CDK6 | 1·50 | 1·32 | 0·44 | 0·68 | Cyclin-dependent kinase 6 |
| 209644_x_at | CDKN2A | 2·90 | 1·21 | 0·67 | 0·79 | Cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) |
| 236313_at | CDKN2B | 3·24 | 1·28 | 0·58 | 0·72 | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| 241984_at | CHES1 | 1·38 | 1·34 | 0·66 | 0·63 | Checkpoint suppressor 1 |
| 202552_s_at | CRIM1 | 1·94 | 1·39 | 0·32 | 0·61 | Cysteine-rich transmembrane BMP regulator 1 (chordin-like) |
| 204844_at | ENPEP | 1·64 | 1·75 | 0·09 | 0·36 | Glutamyl aminopeptidase (aminopeptidase A) |
| 205418_at | FES | 1·39 | 1·80 | 0·61 | 0·25 | Feline sarcoma oncogene |
| 228572_at | GRB2 | 4·69 | 1·21 | 0·79 | 0·78 | Growth factor receptor-bound protein 2 |
| 207688_s_at | INHBC | 1·46 | 1·25 | 0·51 | 0·75 | Inhibin, beta C |
| 209744_x_at | ITCH | 1·30 | 1·47 | 0·63 | 0·70 | Itchy homolog E3 ubiquitin protein ligase (mouse) |
| 201548_s_at | JARID1B | 1·27 | 1·92 | 0·73 | 0·46 | Jumonji, AT-rich interactive domain IB (RBP2-like) |
| 203297_s_at | JARID2 | 1·42 | 1·28 | 0·54 | 0·72 | Jumonji, AT-rich interactive domain 2 |
| 41387_r_at | JMJD3 | 1·82 | 1·24 | 0·76 | 0·65 | Jumonji domain containing 3 |
| 205569_at | LAMP3 | 2·32 | 1·24 | 0·76 | 0·50 | Lysosomal-associated membrane protein 3 |
| 214039_s_at | LAPTM4B | 1·41 | 1·49 | 0·49 | 0·59 | Lysosomal-associated protein transmembrane 4 beta |
| 205857_x_at | MSH3 | 1·79 | 1·28 | 0·58 | 0·72 | MutS homolog 3 (E. coli) |
| 209550_at | NDN | 3·42 | 1·38 | 0·17 | 0·62 | Necdin homolog (mouse) |
| 207943_x_at | PLAGL1 | 1·37 | 1·43 | 0·57 | 0·63 | Pleiomorphic adenoma gene-like 1 |
| 204748_at | PTGS2 | 1·65 | 1·78 | 0·14 | 0·35 | Prostaglandin-endoperoxide synthase 2 |
| 201482_at | QSCN6 | 1·32 | 1·23 | 0·38 | 0·77 | Quiescin Q6 |
| 203743_s_at | TDG | 1·47 | 1·23 | 0·54 | 0·77 | Thymine-DNA glycosylase |
| 204227_s_at | TK2 | 2·12 | 1·26 | 0·56 | 0·74 | Thymidine kinase 2, mitochondrial |
| Cytokines and chemokines | ||||||
| 207533_at | CCL1 | 1·67 | 1·48 | 0·52 | 0·49 | Chemokine (C-C motif) ligand 1 |
| 205099_s_at | CCR1 | 4·70 | 1·21 | 0·61 | 0·79 | Chemokine (C-C motif) receptor 1 |
| 207681_at | CXCR3 | 1·51 | 1·33 | 0·41 | 0·67 | Chemokine (C-X-C motif) receptor 3 |
| 211469_s_at | CXCR6 | 1·58 | 1·95 | 0·32 | 0·42 | Chemokine (C-X-C motif) receptor 6 |
| 206613_at | IL-18R1 | 2·32 | 1·38 | 0·61 | 0·62 | Interleukin-18 receptor 1 |
| 207072_at | IL-18RAP | 2·16 | 1·44 | 0·46 | 0·56 | Interleukin-18 receptor accessory protein |
| 212657_s_at | IL-1RN | 1·44 | 3·12 | 0·56 | 0·37 | Interleukin 1 receptor |
| 206341_at | IL-2RA | 1·52 | 1·27 | 0·73 | 0·66 | Interleukin-2 receptor alpha |
| 202859_x_at | IL-8 | 1·31 | 3·75 | 0·38 | 0·69 | Interleukin-8 |
| 202643_s_at | TNFAIP3 | 1·61 | 1·25 | 0·67 | 0·75 | Tumour necrosis factor, alpha-induced protein 3 |
| 202687_s_at | TNFSF10 | 2·83 | 1·23 | 0·67 | 0·77 | Tumour necrosis factor (ligand) superfamily member 10 |
| 205599_at | TRAF1 | 2·25 | 1·32 | 0·68 | 0·61 | Tumour necrosis factor receptor-associated factor 1 |
| 202871_at | TRAF4 | 1·43 | 1·58 | 0·57 | 0·48 | Tumour necrosis factor receptor-associated factor 4 |
| 206366_x_at | XCL1 | 1·24 | 2·66 | 0·46 | 0·76 | Chemokine (C motif) ligand 1 |
| Signal transduction | ||||||
| 210538_s_at | AIP1 | 1·35 | 1·54 | 0·65 | 0·61 | Baculoviral IAP repeat-containing 3 |
| 209369_at | ANXA3 | 1·39 | 6·82 | 0·61 | 0·05 | Annexin A3 |
| 1554343_a_at | BRDG1 | 1·45 | 1·67 | 0·52 | 0·55 | BCR downstream signalling 1 |
| 225946_at | C12orf2 | 3·20 | 1·77 | 0·23 | 0·23 | Ras association (RaIGDS/AF-6) domain family 8 |
| 204392_at | CAMK1 | 1·26 | 1·62 | 0·74 | 0·54 | Calcium/calmodulin-dependent protein kinase I |
| 231042_s_at | CAMK2D | 1·31 | 1·63 | 0·25 | 0·69 | Calcium/calmodulin-dependent protein kinase (CaM kinase) II delta |
| 205692_s_at | CD38 | 1·37 | 1·29 | 0·71 | 0·48 | CD38 antigen (p45) |
| 231747_at | CYSLTR1 | 3·16 | 1·45 | 0·55 | 0·43 | Cysteinyl leukotriene receptor 1 |
| 211272_s_at | DGKA | 1·43 | 1·23 | 0·77 | 0·54 | Diacylglycerol kinase alpha 80 kDa |
| 200762_at | DPYSL2 | 1·35 | 1·40 | 0·37 | 0·65 | Dihydropyrimtdinase-like 2 |
| 208370_s_at | DSCR1 | 1·23 | 1·90 | 0·63 | 0·77 | Down syndrome critical region gene 1 |
| 204794_at | DUSP2 | 1·55 | 2·57 | 0·39 | 0·45 | Dual specificity phosphatase 2 |
| 204015_s_at | DUSP4 | 1·35 | 2·66 | 0·65 | 0·39 | Dual specificity phosphatase 4 |
| 211333_s_at | FASLG | 1·20 | 1·37 | 0·49 | 0·80 | Fas ligand (TNF superfamily, member 6) |
| 211535_s_at | FGFR1 | 1·23 | 2·79 | 0·70 | 0·77 | Fibroblast growth factor receptor 1 |
| 224148_at | FYB | 1·50 | 1·21 | 0·45 | 0·79 | FYN binding protein (FYB-120/130) |
| 209304_x_at | GADD45B | 1·55 | 1·29 | 0·65 | 0·71 | Growth arrest and DNA-damage-inducible beta |
| 234284_at | GNG8 | 1·50 | 3·16 | 0·50 | 0·35 | Guanine nucleotide binding protein (G protein), gamma 8 |
| 224285_at | GPR174 | 1·91 | 1·42 | 0·56 | 0·58 | G protein-coupled receptor 174 |
| 223767_at | GPR84 | 4·41 | 1·44 | 0·05 | 0·56 | G protein-coupled receptor 84 |
| 211555_s_at | GUCY1B3 | 1·66 | 1·73 | 0·34 | 0·03 | Guanylate cyclase 1, soluble, beta 3 |
| 38037_at | HBEGF | 1·54 | 1·36 | 0·55 | 0·64 | Heparin-binding EGF-like growth factor |
| 203820_s_at | IMP-3 | 1·83 | 2·18 | 0·17 | 0·17 | IGF-II-mRNA-binding protein 3 |
| 203006_at | INPP5A | 1·40 | 1·86 | 0·60 | 0·52 | Inositol polyphosphate-5-phosphatase, 40 kDa |
| 231779_at | IRAK2 | 1·93 | 1·46 | 0·46 | 0·54 | Interleukin-1 receptor associated kinase 2 |
| 32137_at | JAG2 | 1·58 | 1·29 | 0·71 | 0·64 | Jagged 2 |
| 203904_x_at | KAI1 | 1·65 | 1·59 | 0·41 | 0·25 | CD82 antigen |
| 235252_at | KSR | 1·72 | 1·56 | 0·43 | 0·44 | Kinase suppressor of ras 1 |
| 210948_s_at | LEF1 | 1·21 | 1·64 | 0·41 | 0·79 | Hypothetical protein LOC641518 |
| 203236_s_at | LGALS9 | 1·48 | 1·27 | 0·73 | 0·51 | Lectin, galactoside-binding, soluble, 9 (galectin 9) |
| 220253_s_at | LRP12 | 1·27 | 1·30 | 0·31 | 0·73 | Low-density lipoprotein-related protein 12 |
| 206637_at | P2RY14 | 1·32 | 1·48 | 0·39 | 0·68 | Purinergic receptor P2Y, G-protein coupled, 14 |
| 210837_s_at | PDE4D | 1·35 | 1·31 | 0·62 | 0·69 | Phosphodiesterase 4D, cAMP-specific |
| 206726_at | PGDS | 6·45 | 1·40 | 0·60 | 0·43 | Prostaglandin D2 synthase, haematopoietic |
| 210617_at | PHEX | 1·53 | 4·08 | 0·21 | 0·47 | Phosphate regulating endopeptidase homologue, X-linked |
| 206370_at | PIK3CG | 1·23 | 1·32 | 0·50 | 0·77 | Phosphoinositide-3-kinase, catalytic, gamma polypeptide |
| 205632_s_at | PIP5K1B | 1·32 | 1·42 | 0·64 | 0·68 | Phosphalidylinositol-4-phosphate 5-kinase, type 1 beta |
| 215195_at | PRKCA | 2·17 | 1·36 | 0·64 | 0·61 | Protein kinase C, alpha |
| 210832_x_at | PTGER3 | 4·44 | 1·47 | 0·07 | 0·53 | Prostaglandin E receptor 3 (subtype EP3) |
| 1553535_a_at | RANGAP1 | 1·58 | 1·39 | 0·58 | 0·61 | Ran GTPase activating protein 1 |
| 234344_at | RAP2C | 1·75 | 1·26 | 0·46 | 0·74 | RAP2C, member of RAS oncogene family |
| 223809_at | RGS18 | 2·12 | 1·67 | 0·15 | 0·33 | Regulator of G-protein signalling 18 |
| 209882_at | RIT1 | 1·74 | 1·32 | 0·63 | 0·68 | Ras-like without CAAX 1 |
| 209451_at | TANK | 1·34 | 1·20 | 0·42 | 0·80 | TRAF family member-associated NFKB activator |
| 204924_at | TLR2 | 1·60 | 2·52 | 0·36 | 0·40 | Toll-like receptor 2 |
| 217979_at | TM4SF13 | 1·21 | 2·47 | 0·30 | 0·79 | Tetraspanin 13 |
| 209263_x_at | TM4SF7 | 2·05 | 1·41 | 0·58 | 0·59 | Tetraspanin 4 |
| Transcription | ||||||
| 1566989_at | ARID1B | 1·42 | 1·27 | 0·09 | 0·73 | AT-rich interactive domain 1B (SWIl-like) |
| 203973_s_at | CEBPD | 3·06 | 1·51 | 0·33 | 0·49 | CCAAT/enhancer binding protein (C/EBP), delta |
| 221598_s_at | CRSP8 | 1·60 | 1·29 | 0·71 | 0·68 | Cofactor required for Spl transcriptional activation, subunit 8, 34 kDa |
| 205249_at | EGR2 | 1·33 | 4·27 | 0·67 | 0·60 | Early growth response 2 (Krox-20 homologue, Drosophila) |
| 206115_at | EGR3 | 1·31 | 6·15 | 0·69 | 0·48 | Early growth response 3 |
| 201328_at | ETS2 | 1·57 | 1·72 | 0·43 | 0·40 | V-ets erythroblastosis virus E26 oncogene homologue 2 (avian) |
| 218810_at | FLJ23231 | 2·13 | 1·37 | 0·63 | 0·63 | Zinc finger CCCH-type containing 12A |
| 209189_at | FOS | 21·56 | 1·31 | 0·13 | 0·69 | V-fos FBJ murine osteosarcoma viral oncogene homologue |
| 223408_s_at | FOXK2 | 2·26 | 1·22 | 0·48 | 0·78 | Forkhead box K2 |
| 202723_s_at | FOXO1A | 1·47 | 1·27 | 0·57 | 0·73 | Forkhead box O1A (rhabdomyosarcoma) |
| 224211_at | FOXP3 | 1·62 | 1·41 | 0·59 | 0·23 | Forkhead box P3 |
| 207156_at | HIST1H2AG | 1·73 | 1·30 | 0·41 | 0·70 | Histone 1, H2ag |
| 220042_x_at | HIVEP3 | 1·26 | 1·65 | 0·74 | 0·56 | Human immunodeficiency virus type I enhancer binding protein 3 |
| 207826_s_at | ID3 | 1·34 | 8·64 | 0·60 | 0·66 | Inhibitor of DNA binding 3, dominant negative helix-loop-hetix protein |
| 204549_at | IKBKE | 2·33 | 1·29 | 0·71 | 0·66 | Inhibitor of kappa light polypeptide gene enhancer in B cells |
| 219878_s_at | KLF13 | 1·89 | 1·26 | 0·34 | 0·74 | Kruppel-like factor 13 |
| 207667_s_at | MAP2K3 | 1·33 | 1·28 | 0·72 | 0·57 | Mitogen-activated protein kinase kinase 3 |
| 201502_s_at | NFKBIA | 2·31 | 1·29 | 0·71 | 0·57 | Nuclear factor of κ light polypeptide gene enhancer in B cells inhibitor |
| 222105_s_at | NKIRAS2 | 1·84 | 1·21 | 0·69 | 0·79 | NFKB inhibitor interacting Ras-like 2 |
| 204622_x_at | NR4A2 | 1·35 | 4·31 | 0·65 | 0·63 | Nuclear receptor subfamily 4, group A, member 2 |
| 207978_s_at | NR4A3 | 1·33 | 3·53 | 0·62 | 0·67 | Nuclear receptor subfamily 4, group A, member 3 |
| 202600_s_at | NRIPI | 1·86 | 1·39 | 0·26 | 0·61 | Nuclear receptor interacting protein 1 |
| 216841_s_at | SOD2 | 1·25 | 1·73 | 0·36 | 0·75 | Superoxide dismutase 2, mitochondrial |
| 201416_at | SOX4 | 1·53 | 2·21 | 0·47 | 0·38 | SRY (sex determining region Y)-box 4 |
| 223635_s_at | SSBP3 | 2·12 | 1·25 | 0·75 | 0·62 | Single-stranded DNA binding protein 3 |
| 206506_s_at | SUPT3H | 1·47 | 1·31 | 0·57 | 0·69 | Suppressor of Ty 3 homologue (S. cerevisiae) |
| 221618_s_at | TAF9L | 1·25 | 1·49 | 0·47 | 0·75 | TAF9-like RNA polymerase II |
| 203177_x_at | TFAM | 1·63 | 1·23 | 0·77 | 0·57 | Transcription factor A, mitochondrial |
| 213943_at | TWIST1 | 1·89 | 3·14 | 0·04 | 0·11 | Twist homologue 1 (acrocephalosyndactyly 3; Saethre-Chotzen syndrome) |
| 219836_at | ZBED2 | 1·33 | 4·76 | 0·67 | 0·21 | Zinc finger, BED-type containing 2 |
| 211965_at | ZFP36L1 | 2·02 | 1·47 | 0·29 | 0·53 | Zinc finger protein 36, C3H type-like 1 |
| 230760_at | ZFY | 1·41 | 1·25 | 0·75 | 0·02 | Zinc finger protein, Y-linked |
| 228854_at | ZNF145 | 3·26 | 1·21 | 0·40 | 0·79 | Transcribed locus |
| 235121_at | ZNF542 | 2·68 | 1·33 | 0·63 | 0·67 | Zinc finger protein 542 |
The genes with a higher level of expression in the PB-derived CD4+ T cells included those encoding transcriptional regulators, signal transduction factors, major histocompatibility complex (MHC) class II molecules (HLADMA, HLADMB, HLADPA1, HLADQB1, HLADRA, HLADRB1 and HLADRB5), and cytokines, chemokines and their receptors (IL-7, IL-17RB), as well as genes that characterize the T-cell lineage (CD29, CD86) (Fig. 1b, Tables 2, 4).
Notably, microarray studies showed that the expression of several regulatory T cell (Treg)-related genes was significantly higher in the CB-derived T cells. Foxp3 is an important T-cell transcription factor and is considered to be a marker of Tregs. Cytotoxic T-lymphocyte antigen-4 (CTLA-4) and ICOS, which belong to the CD28 family of receptors and play a crucial role in the activation of T cells, were reported to be highly expressed in activated Tregs.11,12 All of the above genes were expressed at higher levels in the CB-derived CD4 T cells (Fig. 1).
The microarray results for major genes related to the development of the T-cell lineage, including those not appeared in Fig. 1, are summarized in Table 2. As shown in Table 2, the expression of T-cell lineage master regulator genes, such as TBX21, GATA3 and MAF, and T cell-related cytokines, such as IL-4, IL-5, IL-13, IL-22 and TGFB1, revealed no significant difference between CB-derived CD4+ cells and PB-derived CD4+ cells. However, other T cell-related genes, including IL-2, IL-6, IL-9, IL-10 and IL-17, were eliminated from the list in the course of background subtraction because the signal intensity of each gene was low (< 90 as raw data) in all of the samples.
Differences in the expression patterns of T-cell lineage-specific genes between CB-derived and PB-derived CD4+ T cells
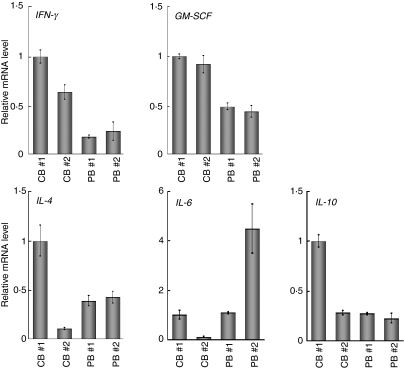
To further confirm the characteristic gene expression in CB- and PB-derived CD4+ T cells, we performed a real-time RT-PCR analysis. Consistent with the microarray data, when the mRNA levels of the genes related to the T helper type 1 (Th1) and Th2 phenotypes were examined, higher levels of GM-CSF and IFNG were observed in CB-derived T cells, while IL-4 revealed no significant tendency (Fig. 2). We also examined IL-6 and IL-10 and no significant tendency was observed either in the expression of these genes (Fig. 2).
Figure 2.
Quantitative polymerase chain reaction (PCR) analysis of the genes related to the T helper type 1 (Th1) and Th2 phenotypes. The expression of the genes indicated was examined by real-time reverse transcriptase (RT)-PCR using the same sample specimens as in Fig 1. Data are normalized to the mRNA level in PB 1 which is arbitrarily set to 1. The signal intensity was normalized using that of a control housekeeping gene [the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene]. Data are relative values with the standard deviation (SD) for triplicate wells.
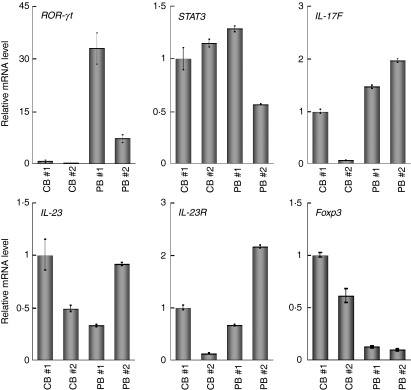
Next we examined the expression of the genes related to Tregs and observed a higher level of Foxp3, but lower levels of retinoic acid receptor-related orphan receptor γ isoform t (RORγt); and IL-17F, in CB-derived T cells (Fig. 3). In contrast, there was no significant tendency in the expression of genes encoding signal transducer and activator of transcription 3 (STAT-3), IL-23 and IL-23 receptors. In the case of the IL-17 gene, clear amplification was detected in PB-derived T cells whereas no amplification was observed in the samples of CB-derived T cells (data not shown).
Figure 3.
Quantitative polymerase chain reaction (PCR) analysis of the forkhead box protein 3 gene (FOXP3) and the genes related to the secretion of interleukin (IL)-17. The expression of the genes indicated was examined as in Fig 2. Data are normalized to the mRNA level in peripheral blood sample 1 (PB 1) as in Fig.2. The signal intensity was normalized using that of a control housekeeping gene [the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene]. Data are relative values with the standard deviation for triplicate wells.
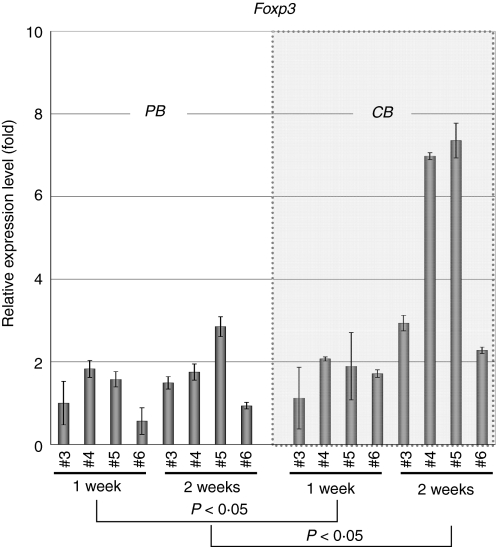
To further investigate whether increased expression of the FOXP3 gene is a general feature of CB-derived CD4+ T cells, we tested four samples of CB-derived CD4+ T cells by real-time RT-PCR analysis and compared the results with those for equivalent numbers of PB-derived samples. As shown in Fig. 4, two CB-derived samples (CB 4 and 5, at 2 weeks) revealed significantly increased gene expression of FOXP3 when compared with PB-derived samples, whereas the remaining two samples (CB 3 and 6; termed ‘additional’ samples below) did not. We also tested FOXP3 gene expression at an earlier time-point in the same samples and observed no significant increase of FOXP3 gene expression in CB-derived CD4+ T cells at 1 week (Fig. 4). When the data were analysed statistically, expression of the FOXP3 gene was found to be significantly higher in CB-derived CD4+ T cells in comparison with equivalent PB-derived CD4+ T cells at both 1 week (P<0·05) and 2 weeks (P<0·05) (Fig. 4).
Figure 4.
Quantitative polymerase chain reaction (PCR) analysis of the forkhead box protein 3 gene (FOXP3) in additional samples. Additional peripheral blood (PB) and cord blood (CB) samples were prepared and RNAs were extracted at 1 and 2 weeks. The expression of the FOXP3 gene was examined as in Fig. 2. Data are normalized to the mRNA level in the sample of PB 3 at 1 week, which is arbitrarily set to 1. The signal intensity was normalized using that of a control housekeeping gene (the human β-actin gene). Data are relative values with the standard deviation for triplicate wells. The data were analysed statistically and FOXP3 gene expression in CB-derived CD4+ T cells was found to be significantly higher in comparison with equivalent PB-derived CD4+ T cells at both 1 week (P<0·05) and 2 weeks (P<0·05).
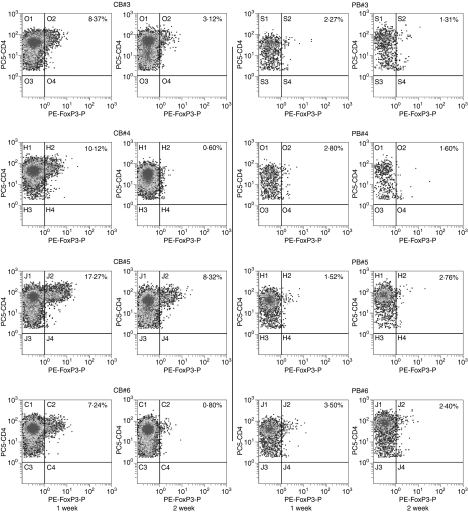
Next we assessed the expression of the Foxp3 protein in CB-derived CD4+ T cells. When the same samples as described above were examined by flow cytometry using a specific antibody, the Foxp3 protein was certainly detected in a portion of cells in all of four CB-derived samples while not detected in any of the PB-derived samples tested (Fig. 5). Inconsistent with the results of real-time RT-PCR, expression level of Foxp3 proteins was higher in CB-derived CD4+ T cells at 1 week than at 2 weeks.
Figure 5.
Protein expression of forkhead box protein 3 (Foxp3) in activated CD4+ T cells. The protein expression of Foxp3 in same sample specimens as in Fig. 4 was examined by flow cytometry. The CD4 versus Foxp3 cytogram of the population gated with CD3+ and CD4+ in each sample is presented.
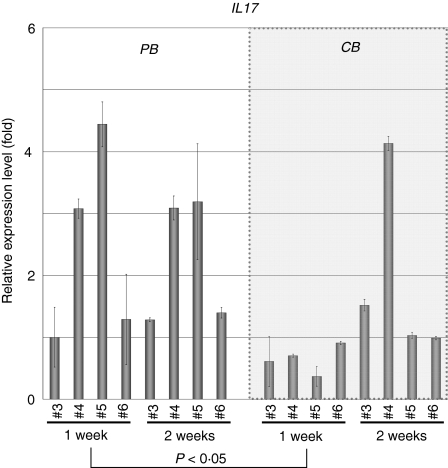
To investigate whether increased expression of the IL-17 gene is a general feature of PB-derived CD4+ T cells, we also tested IL-17 gene expression in the above-described additional samples by real-time RT-PCR analysis. As shown in Fig. 6, all of four PB-derived CD4+ T-cell samples revealed significantly increased gene expression of IL-17 when compared with the CB-derived samples at 1 week. At 2 weeks, however, IL-17 gene expression in PB-derived CD4+ T cells was diminished while some of the CB-derived CD4+ T cells (such as sample CB 4) exhibited increased IL-17 gene expression. When the data were analysed statistically, expression of the IL-17 gene was found to be significantly higher in PB-derived CD4+ T cells in comparison with equivalent CB-derived CD4+ T cells at 1 week (P < 0·05) but not at 2 weeks (Fig. 6).
Figure 6.
Quantitative polymerase chain reaction (PCR) analysis of interleukin (IL)-17 in additional samples. The expression of the IL-17 gene in the same sample specimens as in Fig. 4 was examined and presented as in Fig 2. The data were analysed statistically and IL-17 gene expression in peripheral blood (PB)-derived CD4+ T cells was found to be significantly higher in comparison with equivalent CB-derived CD4+ T cells at 1 week (P<0·05) but not at 2 weeks.
Discussion
Although it is generally believed that there are functional differences between CB and PB lymphocytes, the details are obscure. For instance, Azuma et al.13 reported that the phenotype and function of expanded CB lymphocytes were essentially equivalent to those of expanded PB lymphocytes when evaluated in in vitro experiments. In the present study, however, we have shown that CB-derived CD4+ T cells revealed a distinct expression profile of genes important for the function of particular T-cell subsets compared with PB-derived CD4+ T cells.
CD4+ T cells can be classified into distinct subsets, including effector CD4+ cells and Tregs, according to their functional characteristics as well as differentiation profiles.14–16 Typically, effector CD4+ T cells have been further divided into two distinct lineages on the basis of their cytokine production profiles, namely Th1 and Th2. Th1 cells producing cytokines such as IL-2, IFN-γ and GM-CSF have evolved to enhance the eradication of intracellular pathogens and are thought to be potent activators of cell-mediated immunity. In contrast, Th2 cells secreting cytokines such as IL-4, IL-5, IL-6, IL-9 and IL-13 have evolved to enhance the elimination of parasitic infections and are thought to be potent activators of B-cell immunoglobulin E production, eosinophil recruitment, and mucosal expulsion. Th1-type responses to self or commensal floral antigens can promote tissue destruction and chronic inflammation, whereas dysregulated Th2-type responses can cause allergy and asthma. The development of Th1 is specified by the transcription factor T-bet (also known as Tbx-21) and master regulators of Th2 differentiation are GATA-3 and c-maf.
As shown in Fig. 2 and Table 2, the gene expression profiles of CB- and PB-derived CD4+ T cells revealed no significant differences regarding cytokines related to the definition of Th1 and Th2, with the exceptions of IFN-γ and GM-CSF. The mRNA levels of IFN-γ and GM-CSF tended to be higher in CB-derived CD4+ T cells than in PB-derived CD4+ T cells. The mRNA expression of the transcription factors T-bet, GATA-3 and c-maf, which regulate Th1 and Th2 cell differentiation, did not differ significantly between CB- and PB-derived CD4+ T cells.
In addition to Th1 and Th2 cells, IL-17 (also known as IL-17A)-producing T lymphocytes have been recently shown to comprise a distinct third subset of T helper cells, termed Th17 cells, in the mouse immune system. Th17 cells exhibit pro-inflammatory characteristics and act as major contributors to autoimmune disease. A number of experiments using animal models support a significant role for IL-17 in the response to allografts.14,16,17 There is as yet no direct evidence for the existence of discrete Th17 cells in humans, although helper T cells secreting IL-17 have clearly been detected in the human immune system.18 Several studies have shown a correlation between allograft rejection and IL-17. For example, IL-17 levels are elevated in human renal allografts during subclinical rejection and there are detectable mRNA levels in the urinary mononuclear cell sediments of these patients.19,20 In human lung organ transplantation, IL-17 levels have also been reported to be elevated during acute rejection.21 Interestingly, in this study, most of the PB-derived CD4+ T-cell samples expressed higher levels of IL-17 mRNA than the CB-derived CD4+ T-cell samples, suggesting that PB-derived CD4+ T cells frequently include potent IL-17-secreting T cells.
Th17 cells expand independently of T-bet or STAT-1. Ivanov et al.22 have shown that the orphan nuclear receptor RORγt is the key transcription factor orchestrating the differentiation of the effector lineage. RORγt induces transcription of the gene encoding IL-17 in naïve CD4+ T helper cells and is required for its expression in response to IL-6 and transforming growth factor (TGF)-β, the cytokines known to induce IL-17 expression. IL-23 is also involved in Th17 cell differentiation, but naïve T cells do not have the IL-23 receptor and are relatively refractory to IL-23 stimulation.23,24 Although IL-23 seems to be an essential survival factor for Th17 cells, it is not required during their differentiation. It has been suggested that IL-23R expression is up-regulated on RORγt+ Th17 cells in an IL-6-dependent manner. IL-23 may therefore function subsequent to IL-6/TGF-β-induced commitment to the Th17 lineage to promote cell survival and expansion and, potentially, the continued expression of IL-17 and other cytokines that characterize the Th17 phenotype. As presented in Fig. 3, the expression of the RORγt gene was significantly weaker in CB-derived CD4+ T cells, whereas the expression of genes encoding IL-23 and the IL-23 receptor did not differ significantly between the CD4+ T cells. Based on the above findings of others, it is possible that the low-level expression of the RORγt gene in CB-derived CD4+ T cells is responsible for the absence of IL-17 mRNA expression in those cells.
Tregs are another functional subset of T cells having anti-inflammatory properties and can cause quiescence of autoimmune diseases and prolongation of transplant function. In vitro, Tregs have the ability to inhibit the proliferation and production of cytokines by responder (CD4+ CD25− and CD8+) T cells subjected to polyclonal stimuli, as well as to down-regulate the responses of CD8+ T cells, NK cells and CD4+ cells to specific antigens.25,26 These predicates translate in vivo to a great number of functions other than the maintenance of tolerance to self-components (prevention of autoimmune disease), such as the ability to prevent transplant rejection. Indeed, donor-specific Tregs can prevent allograft rejection in some models of murine transplant tolerance through a predominant effect on indirect alloresponses.
Foxp3 is thought to be responsible for the development of the Treg population and can act as a phenotypic marker of this fraction.27 Tregs constitutively express CTLA-4 and there are suggestions that signalling through this pathway may be important for their function, as antibodies to CTLA-4 can inhibit Treg-mediated suppression.28 As shown above, most of the CB-derived CD4+ T cells were found to express either the FOXP3 gene or the Foxp3 protein at higher levels compared with PB-derived CD4+ T cells, suggesting that CB-derived CD4+ T cells frequently include a potent Treg population.
As described above, IL-17 mRNA was more detectable in PB-derived CD4+ cells while FOXP3 mRNA expression was higher in CB-derived CD4+ cells. Post-transcriptional regulation, as well as differences in mRNA and protein turnover rates, can cause discrepancies between mRNA and protein expression and thus the differences observed in the mRNA expression do not necessary directly indicate those in protein expression.29 Indeed, we observed some discrepancy between the levels of mRNA and protein with regard to Foxp3 expression in CB-derived CD4+ T cells, as presented above. Nevertheless, changes in mRNA expression are mediated by the alteration of transcriptional regulation, and thus should indicate the differentiation ability of the cells. Therefore, our data indicate that CB-derived CD4+ T cells tend frequently to include potent Tregs, while PB-derived CD4+ T cells tend to include potent IL-17-secreting cells. As described above, DLI with donor CB-derived activated CD4+ T cells is currently becoming established as a routine therapeutic strategy in Japan. It has been proposed that the skewing of responses towards Th17 or Th1 cells and away from Tregs may be responsible for the development and/or progression of autoimmune diseases or acute transplant rejection, and it may thus also be speculated that CB-derived CD4+ T cells are more appropriate for DLI than PB-derived CD4+ T cells.
However, our data also indicate the presence of individual, donor-dependent variations in the characteristics of activated CD4+ T cells derived from CB and PB. Moreover, activated CD4+ T cells do not consist of a single population and should include several distinct functional subsets of CD4+ T cells. Therefore, it is important to clarify the characteristics of activated CD4+ T cells in each preparation to predict the therapeutic effect of DLI in each clinical case.
In summary, our findings demonstrate a difference in gene expression between activated CD4+ T cells derived from CB and those derived from PB. The higher level of FOXP3 gene expression and the lower level of IL-17 gene expression in CB-derived CD4+ T cells may indicate that these cells have potential as immunomodulators in DLI therapy. Further detailed analysis should reveal the advantages of activated CD4+ T cells from CB in DLI.
Acknowledgments
We thank the Tokyo Cord Blood Bank for the distribution of cord blood for research use. This work was supported by a grant from the Japan Health Sciences Foundation for Research on Publicly Essential Drugs and Medical Devices (KHC2032), Health and Labour Sciences Research Grants (the 3rd term comprehensive 10-year strategy for cancer control H19-010, Research on Children and Families H18-005, Research on Human Genome Tailor-made and Research on Publicly Essential Drugs and Medical Devices H18-005), and a Grant for Child Health and Development from the Ministry of Health, Labour and Welfare of Japan. It was also supported by CREST, JST.
Glossary
Abbreviations:
- BIM
BCL2-like 11
- CB
cord blood
- CTLA-4
cytotoxic T-lymphocyte antigen-4
- CDKN
cyclin-dependent kinase inhibitor
- DLI
donor lymphocyte infusion
- Foxp3
forkhead box protein 3
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GM-CSF
granulocyte–macrophage colony-stimulating factor
- GVHD
graft-versus-host disease
- GVL
graft-versus-leukaemia
- HSCT
haematopoietic stem cell transplantation
- ICOS
inducible T-cell co-stimulator
- IFNG
interferon γ
- IL
interleukin
- PB
peripheral blood
- RORγt
retinoic acid receptor-related orphan receptor γ isoform t
- RT
reverse transcriptase
- TCR
T-cell receptor
- Th
T helper cell
- Treg
regulatory T cell
Disclosures
No competing personal or financial interests exist for any of the authors in relation to this manuscript.
References
- 1.Loren AW, Porter DL. Donor leukocyte infusions after unrelated donor hematopoietic stem cell transplantation. Curr Opin Oncol. 2006;18:107–14. doi: 10.1097/01.cco.0000208781.61452.d3. [DOI] [PubMed] [Google Scholar]
- 2.Roush KS, Hillyer CD. Donor lymphocyte infusion therapy. Transfus Med Rev. 2002;16:161–76. doi: 10.1053/tmrv.2002.31464. [DOI] [PubMed] [Google Scholar]
- 3.Alyea EP, Soiffer RJ, Canning C, et al. Toxicity and efficacy of defined doses of CD4(+) donor lymphocytes for treatment of relapse after allogeneic bone marrow transplant. Blood. 1998;91:3671–80. [PubMed] [Google Scholar]
- 4.Giralt S, Hester J, Huh Y, et al. CD8-depleted donor lymphocyte infusion as treatment for relapsed chronic myelogenous leukemia after allogeneic bone marrow transplantation. Blood. 1995;86:4337–43. [PubMed] [Google Scholar]
- 5.Tomizawa D, Aoki Y, Nagasawa M, et al. Novel adopted immunotherapy for mixed chimerism after unrelated cord blood transplantation in Omenn syndrome. Eur J Haematol. 2005;75:441–4. doi: 10.1111/j.1600-0609.2005.00535.x. [DOI] [PubMed] [Google Scholar]
- 6.Cohena Y, Nagler A. Hematopoietic stem-cell transplantation using umbilical-cord blood. Leuk Lymphoma. 2003;44:1287–99. doi: 10.1080/1042819031000077016. [DOI] [PubMed] [Google Scholar]
- 7.Parmar S, Robinson SN, Komanduri K, et al. Ex vivo expanded umbilical cord blood T cells maintain naive phenotype and TCR diversity. Cytotherapy. 2006;8:149–57. doi: 10.1080/14653240600620812. [DOI] [PubMed] [Google Scholar]
- 8.Robinson KL, Ayello J, Hughes R, van de Ven C, Issitt L, Kurtzberg J, Cairo MS. Ex vivo expansion, maturation, and activation of umbilical cord blood-derived T lymphocytes with IL-2, IL-12, anti-CD3, and IL-7. Potential for adoptive cellular immunotherapy post-umbilical cord blood transplantation. Exp Hematol. 2002;30:245–51. doi: 10.1016/s0301-472x(01)00781-0. [DOI] [PubMed] [Google Scholar]
- 9.Miyagawa Y, Okita H, Nakaijima H, et al. Inducible expression of chimeric EWS/ETS proteins confers Ewing’s family tumor-like phenotypes to human mesenchymal progenitor cells. Mol Cell Biol. 2008;28:2125–37. doi: 10.1128/MCB.00740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werlen G, Hausmann B, Naeher D, Palmer E. Signaling life and death in the thymus: timing is everything. Science. 2003;299:1859–63. doi: 10.1126/science.1067833. [DOI] [PubMed] [Google Scholar]
- 11.Riley JL, June CH. The CD28 family: a T-cell rheostat for therapeutic control of T-cell activation. Blood. 2005;105:13–21. doi: 10.1182/blood-2004-04-1596. [DOI] [PubMed] [Google Scholar]
- 12.Woo EY, Yeh H, Chu CS, Schlienger K, Carroll RG, Riley JL, Kaiser LR, June CH. Cutting edge: regulatory T cells from lung cancer patients directly inhibit autologous T cell proliferation. J Immunol. 2002;168:4272–6. doi: 10.4049/jimmunol.168.9.4272. [DOI] [PubMed] [Google Scholar]
- 13.Azuma H, Yamada Y, Shibuya-Fujiwara N, et al. Functional evaluation of ex vivo expanded cord blood lymphocytes: possible use for adoptive cellular immunotherapy. Exp Hematol. 2002;30:346–51. doi: 10.1016/s0301-472x(02)00776-2. [DOI] [PubMed] [Google Scholar]
- 14.Afzali B, Lombardi G, Lechler RI, Lord GM. The role of T helper 17 (Th17) and regulatory T cells (Treg) in human organ transplantation and autoimmune disease. Clin Exp Immunol. 2007;148:32–46. doi: 10.1111/j.1365-2249.2007.03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castellino F, Germain RN. Cooperation between CD4+ and CD8+ T cells: when, where, and how. Annu Rev Immunol. 2006;24:519–40. doi: 10.1146/annurev.immunol.23.021704.115825. [DOI] [PubMed] [Google Scholar]
- 16.Reiner SL. Development in motion: helper T cells at work. Cell. 2007;129:33–6. doi: 10.1016/j.cell.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 17.Bi Y, Liu G, Yang R. Th17 cell induction and immune regulatory effects. J Cell Physiol. 2007;211:273–8. doi: 10.1002/jcp.20973. [DOI] [PubMed] [Google Scholar]
- 18.Fossiez F, Djossou O, Chomarat P, et al. T cell interleukin-17 induces stromal cells to produce proinflammatory and hematopoietic cytokines. J Exp Med. 1996;183:2593–603. doi: 10.1084/jem.183.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loong CC, Hsieh HG, Lui WY, Chen A, Lin CY. Evidence for the early involvement of interleukin 17 in human and experimental renal allograft rejection. J Pathol. 2002;197:322–32. doi: 10.1002/path.1117. [DOI] [PubMed] [Google Scholar]
- 20.Van Kooten C, Boonstra JG, Paape ME, et al. Interleukin-17 activates human renal epithelial cells in vitro and is expressed during renal allograft rejection. J Am Soc Nephrol. 1998;9:1526–34. doi: 10.1681/ASN.V981526. [DOI] [PubMed] [Google Scholar]
- 21.Vanaudenaerde BM, Dupont LJ, Wuyts WA, et al. The role of interleukin-17 during acute rejection after lung transplantation. Eur Respir J. 2006;27:779–87. doi: 10.1183/09031936.06.00019405. [DOI] [PubMed] [Google Scholar]
- 22.Ivanov, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 23.Langrish CL, Chen Y, Blumenschein WM, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–40. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oppmann B, Lesley R, Blom B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–25. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 25.Dieckmann D, Plottner H, Berchtold S, Berger T, Schuler G. Ex vivo isolation and characterization of CD4(+) CD25(+) T cells with regulatory properties from human blood. J Exp Med. 2001;193:1303–10. doi: 10.1084/jem.193.11.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wing K, Lindgren S, Kollberg G, Lundgren A, Harris RA, Rudin A, Lundin S, Suri-Payer E. CD4 T cell activation by myelin oligodendrocyte glycoprotein is suppressed by adult but not cord blood CD25+ T cells. Eur J Immunol. 2003;33:579–87. doi: 10.1002/eji.200323701. [DOI] [PubMed] [Google Scholar]
- 27.Wan YY, Flavell RA. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Pro Natl Acad Sci USA. 2005;102:5126–31. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Read S, Greenwald R, Izcue A, Robinson N, Mandelbrot D, Francisco L, Sharpe AH, Powrie F. Blockade of CTLA-4 on CD4+ CD25+ regulatory T cells abrogates their function in vivo. J Immunol. 2006;177:4376–83. doi: 10.4049/jimmunol.177.7.4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hack CJ. Integrated transcriptome and proteome data: the challenges ahead. Brief Funct Genomic Proteomic. 2004;3:212–9. doi: 10.1093/bfgp/3.3.212. [DOI] [PubMed] [Google Scholar]