Abstract
Mutations in Cu, Zn superoxide dismutase (SOD1) cause the neurodegenerative disease familial amyotrophic lateral sclerosis from an as-yet-unidentified toxic property(ies). Analysis in Saccharomyces cerevisiae of a broad range of human familial amyotrophic lateral sclerosis–linked SOD1 mutants (A4V, G37R, G41D, H46R, H48Q, G85R, G93C, and I113T) reveals one property common to these mutants (including two at residues that coordinate the catalytic copper): Each does indeed bind copper and scavenge oxygen-free radicals in vivo. Neither decreased copper binding nor decreased superoxide scavenging activity is a property shared by all mutants. The demonstration that shows that all mutants tested do bind copper under physiologic conditions supports a mechanism of SOD1 mutant-mediated disease arising from aberrant copper-mediated chemistry catalyzed by less tightly folded (and hence less constrained) mutant enzymes. The mutant enzymes also are shown to acquire the catalytic copper in vivo through the action of CCS, a specific copper chaperone for SOD1, which in turn suggests that a search for inhibitors of this SOD1 copper chaperone may represent a therapeutic avenue.
Mutations in Cu, Zn superoxide dismutase (SOD1) are responsible for ≈10% of the inherited cases of the fatal human motor neuron degenerative disease amyotrophic lateral sclerosis (1, 2). Overwhelming evidence has demonstrated that the disease arises from a gain of toxic property rather than a loss of superoxide scavenging activity. A series of transgenic mouse models expressing human familial amyotrophic lateral sclerosis (FALS)-linked SOD1 mutations exhibit late-onset motor neuron degeneration despite the presence of endogenous mouse SOD1 (for example, refs. 3 and 4) whereas mice homozygous for the deletion of the SOD1 gene do not develop motor neuron disease (5). Furthermore, despite over 43 different point mutations (depicted in Fig. 1A) known in human disease (6), there are no null mutations that preclude the synthesis of full length, or nearly full length, SOD1 polypeptides.
Figure 1.
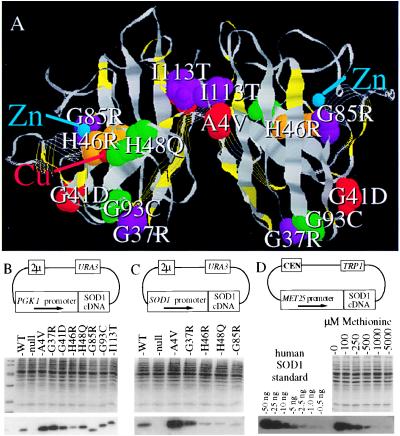
Representative human FALS–SOD1 mutants expressed in sod1Δ yeast. (A) Homodimeric Cu, Zn SOD1 enzyme with FALS-associated point mutations shown in color, carboxyl–terminal truncations illustrated as strands, and the FALS–SOD1 point mutations characterized in yeast shown as space-filling atoms. (B-D) cDNAs encoding human wild-type SOD1 and FALS–SOD1 mutants were subcloned into yeast expression plasmids under the control of the PGK1 promoter (B), the yeast SOD1 promoter (C), or the methionine repressible promoter MET25 promoter (D). After transformation into sod1Δ yeast, each culture was grown to mid log phase, and 10 μg of each protein extract was electrophoresed on an SDS/PAGE gel and was stained with Coomassie blue (B-D, Middle) or was transferred to nitrocellulose and was immunoblotted with an SOD1 antibody (B-D, Bottom). The immunoblot in D is of the MET25 human wild-type SOD1 titration.
The major function of SOD1 is believed to be the detoxification or dismutation of superoxide radicals (O2−). However, SOD1 also has been demonstrated to possess nonspecific peroxidase activity in reacting with H2O2 to oxidize substrates in vitro (7), to react with peroxynitrite (ONOO−) to catalyze the nitration of phenolic groups in vitro (8), and to buffer against copper toxicity (9). Although loss of superoxide scavenging activity does not appear to be the cause of FALS, alterations in these other (or additional, yet to be discovered) SOD1 activities may be involved.
Of the proposed hypotheses to explain how mutations scattered throughout the SOD1 gene can mediate the same neurodegenerative disease, the most prominent one involves the catalytic copper. The mutant enzymes may catalyze aberrant chemistry because of less restrictive active sites arising from less tightly folded enzymes that conceivably could allow greater access of substrates such as peroxynitrite (10), hydrogen peroxide (11), or other small substances to the reactive copper. The presence of copper in the active site is essential for SOD1-mediated peroxidation (7), nitration (8), and superoxide scavenging activity (12, 13). For aberrant copper-mediated chemistry to be a viable hypothesis, all FALS–SOD1 mutants must bind copper in vivo.
Although some FALS–SOD1 mutants have been demonstrated to confer significant SOD1 activity in vitro after extraction from a mammalian cell line (14) or in vivo when expressed in yeast (15), several findings have questioned whether a copper-mediated toxicity is a viable hypothesis for a common FALS-mutant property. In an in situ activity assay after native gel electrophoresis, SOD1 mutant G85R displays no activity (14), possibly reflecting little or no copper binding in vivo. Furthermore, two of the mutants (H46R and H48Q) modify residues that directly coordinate the catalytic copper; these mutations have been predicted to lower or eliminate efficient copper binding (16).
Although several groups (see, for example, refs. 16 and 17) have examined the characteristics of some FALS–SOD1 metalloproteins in vitro, very little is known about the in vivo properties of these mutants. We demonstrate in this study that a broad range of human FALS–SOD1 mutants do scavenge oxygen-free radicals and bind copper in at least one in vivo context (the yeast cell). We also show that, although the copper-binding site of a subset of these mutants may bind aberrantly to copper or may be occupied less fully than in wild-type SOD1, this is not a property shared by all mutants. Furthermore, for all mutants tested, the binding of copper is facilitated in vivo by CCS, a specific SOD1 copper chaperone (13), which provides support for the hypothesis that aberrant copper-mediated chemistry may be responsible for motor neuron disease and suggests that a search for inhibitors of this chaperone may represent a useful therapeutic approach.
MATERIALS AND METHODS
Yeast Strains, Media, and Growth Conditions.
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. The majority of strains (KS104, KS100, VC107, and VC277) contain multiple copies of the CUP1 copper metallothionein (as do most laboratory strains) whereas the isogenic strains JS003 and JS004 lack this metallothionein and consequently are exquisitely sensitive to copper. JS003 was generated by creating a Ura− derivation of the cup1Δ∷URA3 strain 51.2C (9) by selecting for resistance on fluoroorotic acid (20). All sod1Δ strains were generated by substituting the SOD1 coding sequence +18 to +314 with SOD1 replacement plasmids pKS1 (sod1Δ∷LEU2) (9), pKS3 (sod1Δ∷TRP1) (9), or pSΔ1403 (sod1Δ∷HIS3) (X. Lin and V.C.C., unpublished material) from the respective SOD1 parental strains listed in Table 1. Standard yeast media and methods were used (20). Anaerobic cultures were maintained in an O2-depleted culture jar as described (21).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference | Parental Strain (Ref.) |
|---|---|---|---|
| KS104 | MATa leu2 his4 trp1 ura3 sod1Δ∷LEU2 | This study | 1783 (18) |
| KS100 | MATa leu2 his3 trp1 ura3 sod1Δ∷LEU2 sod2Δ∷TRP1 | (9) | |
| JS003 | MATa trp1 ura3ade his leu2 met13 cup1Δ∷URA3 5FOAR | This study | 51.2C (9) |
| JS004 | MATa trp1 ura3ade his leu2 met13 cup1Δ∷URA3 5FOAR sod1Δ∷TRP1 | This study | JS003 |
| VC107 | MATa ura3 ade1 his3 leu2 sod1Δ∷LEU2 | This study | SY1699 (19) |
| VC277 | MATa ura3 ade1 his3 leu2 sod1Δ∷HIS3 lys7Δ∷LEU2 | This study | SY2950 (19) |
Analysis of Three-Dimensional Location of FALS-Causing SOD1 Mutations.
In an effort to compare the 43 of the point mutations and carboxyl–terminal deletions that cause FALS (6), the three-dimensional locations of these mutations and deletions within the crystalized human SOD1 (accession no. 134611) were highlighted by using the three-dimensional computer imaging software rasmol (R. Sayle, Glaxo).
FALS–SOD1 Expression Constructs.
Wild-type or FALS mutant human SOD1 cDNAs [WT, A4V, G37R, G41D, H46R, H48Q, G85R, G93C, and I113T (14)] were subcloned into yeast expression vectors containing a phosphoglycerol kinase (PGK1) promoter (pSM703 kindly provided by S. Michaelis, Johns Hopkins University Medical School), the yeast SOD1 promoter (sequences −700 to +16), or the methionine repressible promoter MET25 (22) (Fig. 1B). To integrate single copies, SOD1-promoted FALS expression cassettes were removed from the 2μ vector (Fig. 1C), were subcloned into the integrating vector pRS306 (23), were linearized, and were integrated at the URA3 locus by transformation.
Quantitation of SOD1 Protein Accumulation.
Lysates of FALS–SOD1 harboring yeast were prepared (13), and accumulated SOD1 was quantified by immunoblot by using a polyclonal SOD1 antibody either against residues 124–136 of human SOD1 or against the whole human SOD1 as described (14). A dilution series of either human SOD1 (Sigma) or yeast SOD1 (kindly provided by Dan Kosman, State University of New York at Buffalo) was used as a standard for quantitation.
SOD1 Titrations.
To quantify the level of SOD1 polypeptide necessary to rescue the lysine auxotrophy and paraquat sensitivities of the sod1Δ (KS104) strain, MET25–SOD–CEN-harboring KS104 was used. To test the SOD1 level needed to complement the lysine auxotrophy, SC–Met–Trp–Lys medium was diluted to an OD600 of 0.1 and aliquoted into six tubes with increasing levels of methionine (0, 100, 250, 500, 1000, and 5000 μM). To test the level needed to complement the paraquat sensitivity, SC–Met–Trp medium containing 10 mM paraquat was diluted to an OD600 of 0.1 and aliquoted into six tubes with increasing levels of methionine (0, 100, 250, 500, 1000, and 5000 μM). Growth was measured by optical density, cultures were harvested, and accumulated SOD1 levels were quantified. The lowest level of SOD1 polypeptide that was sufficient to give growth equal to wild-type yeast was taken as the minimal SOD1 requirement.
In Vitro Superoxide Scavenging Activity.
To measure superoxide scavenging activity in vitro, lysates of PGK–SOD1-harboring KS100 yeast were made and assayed by solution assay at pH 7.0 (24). Specified quantities of purified human SOD1 (Sigma) added to sod1Δsod2Δ extracts were used as standards for comparison. Activity also was determined by using an in situ assay after electrophoresis on a nondenaturing polyacrylamide activity gel (14).
RESULTS
Because of the difficulty of assaying FALS–SOD1 metalloproteins in mammalian cells already containing endogenous cytosolic Cu, Zn SOD1, extracellular Cu, Zn SOD3, and mitochondrial Mn SOD2, we exploited in vivo SOD1 assays in S. cerevisiae lacking Cu, Zn SOD1. Because human and yeast Cu, Zn SOD1 contain 54% amino acid identity and the wild-type human SOD1 can substitute functionally for the yeast SOD1 in complementing the various deficiencies of a sod1 null yeast strain [including slow growth in air (15), sensitivity to redox cycling drugs (15), a lysine biosynthetic defect (as shown below), and copper sensitivity (as shown below)], yeast is a reasonable in vivo system in which to characterize the properties of the human FALS–SOD1 mutants.
A broad range of FALS–SOD1 mutants were selected for study, including two at the dimer interface (A4V and I113T), three at the turns of the β barrel (G37R, G41D, and G93C), one at the base of the active site channel (G85R), and two at copper-coordinating residues in the active site (H46R and H48Q) (see Fig. 1A). After transformation into a sod1Δ strain, human SOD1 metalloproteins reproducibly accumulate to a level of ≈0.1–0.3% of the total protein when expression is driven from the PGK1 promoter (Fig. 1B), to a level of 0.03–0.10% when expression is driven from the endogenous SOD1 promoter (Fig. 1C), and to titratable levels ranging from ≈0.001 to 0.1% of total protein when expression is driven from the MET25 promoter (Fig. 1D). The higher levels of human SOD1 achieved with the PGK1 promoter are comparable to the levels of endogenous yeast SOD1 [which quantitative immunoblotting reveals to be ≈0.1–0.5% of total yeast protein (data not shown)].
All FALS–SOD1 Mutants Tested Confer Superoxide Scavenging Activity.
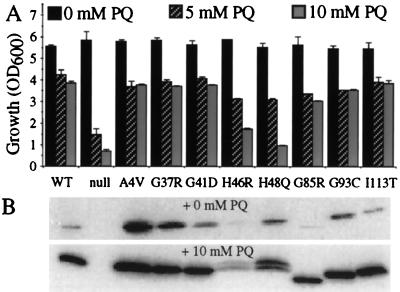
As an initial characterization of the in vivo superoxide scavenging activity of these FALS–SOD1 mutants, yeast harboring the different mutants expressed from multicopy PGK1-promoted constructs (Fig. 1B) were subjected to increasing concentrations of the free radical-generating drug paraquat. Whereas yeast lacking SOD1 failed to grow at 5-mM levels of paraquat, yeast harboring the human wild-type SOD1 as well as yeast expressing each of the eight different mutants were resistant to this 5-mM level (Fig. 2A), demonstrating that all do yield in vivo SOD1 activity. Relative to other mutants, however, the two mutants at the histidines that coordinate the catalytic copper (H46R and H48Q) conferred a decreased level of resistance, which was seen clearly by partial sensitivity to 10 mM paraquat. [Also noteworthy were the reproducible doublets that appeared in the H46R and H48Q lanes after growth in the presence of paraquat (Fig. 2B), possibly indicating covalent modification(s) to these enzymes through self-catalyzed oxidative damage.]
Figure 2.
All FALS–SOD1 mutants retain in vivo free radical scavenging activity in sod1Δ yeast. (A) sod1Δ yeast (JS004) harboring each of the nine PGK–SOD1 expression plasmids or vector alone (null) were diluted to a low density in rich media, the indicated level of paraquat (PQ) was added, and, after 36 hr, culture growth was measured spectrophotometrically. Values represent the average and range of two or three individual cultures in a single experiment. (B) Immunoblot of SOD1 levels in one representative set of cultures. [Note that added paraquat resulted in significant elevations in wild-type or mutant SOD1 accumulations (B, Bottom). Because the PGK1 promoter is not known to be affected by oxidative stress, the apparent induction of SOD1 by paraquat probably reflects selection for yeast cells with higher copy numbers of the 2μ expression plasmid, which are naturally quite variable in number.]
Because it was possible that the decreased paraquat resistance of the histidine mutants simply was caused by the lower levels of accumulation of these mutants, we next determined how much SOD1 is required to rescue either the paraquat sensitivity or the lysine auxotrophy of an sod1Δ strain. To do this, a 100-fold range of expression of human wild-type SOD1 was achieved by titrating transcription from a low copy MET25-promoted human wild-type SOD1 gene (Fig. 1D). Surprisingly, this revealed that as little as 0.01% of total protein (equivalent to 2% of normal yeast SOD1 levels) is sufficient to provide full protection against 10 mM paraquat and to rescue the lysine biosynthetic defect. A comparable level (2% of normal wild-type level) of mutants A4V and G37R was sufficient to provide such paraquat resistance, indicating in vivo-specific activities comparable to the wild-type SOD1. Tenfold higher levels (20% of normal levels) of G85R and I113T were necessary to provide an equivalent level of protection, so these two mutants appeared to possess specific activities of ≈10% that of the wild type. The histidine mutants H46R and H48Q were the least active, displaying apparent in vivo specific activities for protection from paraquat of ≈1% that of wild-type SOD1.
As a secondary measure of superoxide radical scavenging activity of these FALS–SOD1 mutants, extracts were assayed in vitro for their ability to prevent the reduction of cytochrome C by xanthine oxidase in vitro. Solution activity assays of extracts from sod1Δsod2Δ yeast harboring each mutant revealed that the six mutants outside the copper-coordinating residues routinely retained 80–100% specific activity of wild-type SOD1 whereas measurements of the two histidine mutants (H46R and H48Q) varied between 30 and 60% of wild-type specific activity.
The various FALS–SOD1 mutants also were assayed for their ability to rescue the aerobic slow growth defect and the aerobic lysine biosynthetic deficiency of the sod1Δ yeast. The six mutants outside the active site (A4V, G37R, G41D, G85R, G93C, and I113T) completely complemented the aerobic slow growth phenotype (lowering doubling time from the 2 hr of the null strain to the 1.5 hr characteristic of the wild-type strain) and restored lysine prototrophy whereas the active site mutants (H46R and H48Q) did not, suggesting (not unexpectedly) that the histidine mutants represent an SOD1 subclass of their own. However, in vitro activity assays coupled with in vivo paraquat resistance offer compelling evidence that the FALS–SOD1 mutants tested have some activity, and hence some degree of copper binding, under physiological conditions.
FALS–SOD1 Mutants Can Buffer Against High Levels of Intracellular Copper.
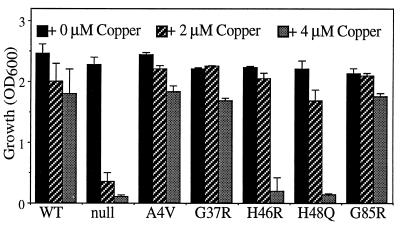
Independent of a role in detoxifying oxygen-free radicals, the abundant yeast Cu, Zn SOD1 also has been implicated in protecting or buffering against increased intracellular copper (9). In fact, Cu, Zn SOD1 was isolated in a genetic screen for copper metallothioneins in yeast (9), and there is evidence that Cu, Zn SOD1 may play a similar copper-buffering role in mammalian cells (25). To determine the ability of the FALS–SOD1 mutants to buffer copper independently of their antioxidant roles, yeast cultures were maintained anaerobically. All six nonactive site mutants were as efficient as the human wild-type SOD1 in protecting from high concentrations (450 μM) of CuSO4 whereas the two histidine mutants at best conferred only a partial resistance in sod1Δ yeast containing the CUP1 metallothionein (data not shown). Because copper metallothionein and SOD1 normally play overlapping roles in the cell (9, 26) and to some degree are coregulated (27), we next tested copper buffering in a yeast strain deleted not only in SOD1 but also in the major copper metallothionein (CUP1). When transformed with an expression series of the FALS–SOD1 mutants, the ability of this highly metal-sensitive (sod1Δ cup1Δ) strain to survive at increasing levels of extracellular copper was taken as an indication of in vivo copper binding and buffering of the corresponding mutant SOD1. As shown in Fig. 3, at 2 μM copper, the strains harboring each FALS–SOD1 mutant grew as well as the strain with wild-type SOD1 whereas growth of the sod1Δ cup1Δ strain was slowed severely. At 4 μM copper, the mutants outside the active site still provided protection comparable to wild-type SOD1 whereas the copper-coordinating mutants were not capable of buffering this increased copper concentration. The copper-coordinating mutants did accumulate to levels comparable to other mutants, suggesting that the 4 μM copper levels are simply beyond the threshhold of what the histidine mutants can buffer, and the copper overload in cells consequently prevents growth at 4 μM copper. Altogether, these approaches demonstrate that all mutants can provide some degree of copper buffering in vivo, albeit the histidine mutants retain a diminished capacity.
Figure 3.
FALS–SOD1 metalloproteins retain in vivo copper buffering. (A) sod1Δcup1Δ (JS004) yeast harboring yeast SOD1-promoted FALS–SOD1 expression plasmids were diluted to a low density in minimal media, the indicated level of copper sulfate was added, cultures were permitted to grow for 3 days, and culture growth was measured spectrophotometrically. All values represent the average and range of three independent cultures in a single experiment.
FALS–SOD1 Metalloenzymes Are Capable of Retaining Activity When Copper Is Limiting.
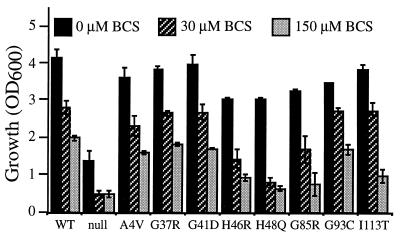
To test further the avidity of copper binding by the different FALS–SOD1 mutants, yeast harboring each of the FALS–SOD1 metalloproteins were grown under copper-limiting conditions. Previous studies demonstrated that yeast deficient in a high affinity copper transporter (CTR1) are unable to import copper into the cell, resulting in a phenotype identical to an sod1Δ strain (28) despite the presence of a normal level of the SOD1 polypeptide. A similar effect can be seen in wild-type yeast if trace copper is eliminated from media components and copper chelators such as bathocuproinedisulfonic acid (BCS) are added. To assay how much SOD1 activity is retained by the FALS mutants when intracellular copper levels are low, yeast harboring each of the FALS–SOD1 metalloproteins was first grown in the presence of the strong copper chelator BCS, paraquat was added, and the growth of the cultures was monitored. The ability of the culture to grow after paraquat addition was taken as an indication of remaining SOD1 activity, itself a reflection of copper binding by the FALS–SOD1 mutants. Under such conditions, the human wild type and all of the FALS–SOD1 mutants display diminishing resistance to paraquat because available copper is made more limiting (Fig. 4). Although not unexpected, the active site mutants (H46R, H48Q) as well as G85R [which is in close proximity to the active site (see Fig. 1A)] but not the other FALS–SOD1 mutants reproducibly lost their paraquat resistance more readily (Fig. 4). Loss of paraquat resistance could be prevented easily by preincubating cells in excess copper, demonstrating that the BCS effect on the cell is specific to copper availability (data not shown). Although such results do suggest that three of the FALS–SOD1 mutants have decreased copper loading in vivo even in the presence of the specific SOD1 copper chaperone (see below), decreased copper binding is not a common property of all FALS–SOD1 mutants, at least not in this in vivo setting.
Figure 4.
FALS–SOD1 metalloenzymes retain variable degrees of free radical scavenging activity when copper is limited by the addition of a copper chelator. sod1Δcup1Δ (JS004) yeast harboring the PGK–SOD1 expression plasmids or vector alone (null) were diluted to a low density in rich media, the indicated level of BCS (a copper chelator) was added, paraquat (a free radical-generating drug) was added to 5 mM, and culture growth was measured spectrophotometrically after 48 hr. Values represent the average and range of two independent cultures in one experiment.
FALS–SOD1 Mutants Require the SOD1 Copper Chaperone for Efficient Copper Loading In Vivo.
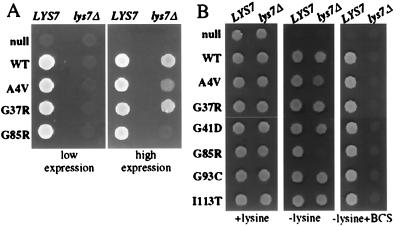
A striking difference between SOD1 behavior in vivo and in vitro is the recent discovery that the apoprotein acquires its active site copper through action of a copper chaperone in vivo (13) whereas the simple addition of copper to purified SOD1 is enough to charge the apoprotein in vitro (for example, ref. 12). In yeast lacking the yeast copper chaperone (LYS7), no yeast SOD1 activity or copper binding to SOD1 is detectable despite normal levels of full length, endogeneous polypeptide (13). To determine whether the copper loading of the wild-type human protein and the FALS–SOD1 mutants are also copper chaperone-dependent, the abilities of each to rescue the sod1Δ phenotype were monitored in a lys7Δsod1Δ strain. Although the human wild-type and FALS–SOD1 mutants examined (A4V, G37R, and G85R) were able to rescue the lysine biosynthetic defect of a sod1Δ strain, none could complement a lys7Δsod1Δ yeast when expressed at low levels (≈5–10% normal endogeneous SOD1 levels) from an integrated SOD1 promoter (Fig. 5A, Left). However, with the exception of G85R, when expressed at 10-fold higher levels (from an SOD1 promoter on a high copy plasmid), all mutants tested rescued the lysine auxotrophy (Fig. 5A, Right). A similar effect was seen with other FALS–SOD1 mutants (G41D, G93C, and I113T in addition to A4V and G37R) expressed at even higher levels (equal to endogenous SOD1) from the PGK1 promoter (Fig. 5B, Left and Center).
Figure 5.
FALS–SOD1 mutants depend on the SOD1 copper chaperone for efficient copper loading in vivo. (A) Cells (106) of sod1Δ (VC107) or sod1Δlys7Δ (VC277) yeast harboring integrated SOD1-promoted FALS–SOD1 constructs (Left) or high copy SOD1-promoted plasmids (Right) were spotted onto plates lacking lysine and permitted to grow for 2 days. (B) sod1Δ and sod1Δlys7Δ yeast harboring the high copy PGK–SOD1 constructs were spotted onto SC plates with lysine (Left), without lysine (Center), or without lysine and with 250 μM BCS (Right).
Despite the ability of high levels of wild-type or mutant SOD1 to complement lysine auxotrophy in the absence of CCS, the requirement for the CCS could be restored by limiting available copper. After the addition of the copper chelator BCS in the absence of the chaperone, all mutants (as well as human wild-type SOD1) are unable to grow on plates lacking lysine (Fig. 5B, Right). Because <2% of normal SOD1 levels are sufficient to rescue the lysine auxotrophy, we interpret these results to mean that, under these conditions, <2% of accumulated SOD1 is charged with copper in the absence of the copper chaperone. Immunoblots revealed that neither the addition of BCS nor the absence of the copper chaperone affect the accumulation of any mutants expressed from the PGK1 promoter. Moreover, expression of a human copper chaperone for SOD1 (CCS) (13) functionally substituted for the yeast LYS7 chaperone in loading human wild-type and mutant SOD1 with copper. Thus, both yeast and human copper chaperones facilitate copper acquisition by the human wild-type and FALS–SOD1 apoproteins and are essential for loading copper onto SOD1 in this in vivo context when either SOD1 polypeptide levels or available copper levels are below a certain threshold.
DISCUSSION
We demonstrate that neither loss of activity nor loss of (or decreased) copper binding is a common in vivo property shared by the FALS–SOD1 mutants examined, and, thus, neither loss of superoxide scavenging activity nor decreased copper buffering is likely to play a role in mediating disease. Rather, a property common to the several classes of mutants tested (including those at the copper-coordinating residues) is that all retain copper binding in vivo, as reflected in their ability to confer resistance to the free radical generator paraquat and superoxide scavenging activity scored by solution assay of extracts containing these mutants. Resistance to elevated copper levels under anaerobic conditions provides independent evidence for in vivo copper binding by the mutants.
Our evidence that G85R and H46R do have significant activity may seem discordant with previous reports that have failed to identify activity in gel activity assays for G85R (14) and H46R (16) or in solution assays of H46R performed at a nonphysiological pH of 8.2 (16). We believe that the discrepancy simply reflects the artificiality of the in vitro assays used. Indeed, we, too, have found that H46R-, H48Q-, and G85R-containing extracts score positive for superoxide scavenging activity in solution at a neutral pH but possess no detectable activity by such in situ gel activity assays. We suspect that the difference may be that these three mutants (H46R, H48Q, and G85R) have a lower affinity for copper. Not only do they appear to lose activity (presumably because of a loss of the catalytic copper and/or conformation) during electrophoresis on nondenaturing gels, but, when in vivo copper is limited (by the addition of a copper chelator), these mutants lose the ability to confer paraquat resistance more readily than wild-type SOD1. This result provides evidence for such weakened copper binding in vivo. On the other hand, modest (between 0.7- and 2.6-fold) differences in copper binding have been found in in vitro binding assays for four of the mutants (A4V, A4T, I113T, and L38V) (17). Because these assays were performed under partially denaturing conditions (2 M urea), it seems likely that more physiologic settings would yield even smaller differences. When coupled with our inability to find an increase in copper chelator sensitivity of some FALS–SOD1 mutants compared with the wild-type protein (when tested over a broad range of BCS levels), the collective evidence argues strongly against significantly decreased copper binding as a property shared among the FALS mutants and thus argues against decreased copper buffering as a mechanism of disease.
An additional argument for copper binding as a shared property is the emerging evidence for mutant SOD1-mediated nitration of tyrosines, presumably through more efficient use of peroxynitrite as a substrate (10). The evidence here includes the demonstration of elevated levels of nitrotyrosine contemporaneous with the earliest pathology and continuing throughout disease progression in affected regions of mice expressing mutation G37R but not in mice expressing a comparable level of wild-type human SOD1 (29). A similar finding was reported in end stage mice expressing mutation G93A (30). Increased levels of nitrotyrosine also have been reported using immunocytochemistry in FALS (31), which seems to reflect that toxicity arising in human disease correlates with increased nitrotyrosination.
We emphasize that our evidence does not support the claim that the histidine mutants are fully active and/or bind copper in a manner indistinguishable from wild-type SOD1 protein (or the other mutants). Although these mutants do protect against increased free radical production arising from paraquat, they do not complement either the aerobic slow growth defect or the aerobic lysine auxotrophy of the sod1Δ strain. With our determination that both paraquat resistance and complementation of the lysine auxotrophy each require about the same level of overall Cu, Zn SOD1 (≈2% of the normal amount), the differences in complementation raise two intriguing possibilities.
First, although the slow growth defect and amino acid auxotrophies are exhibited only when the sod1Δ yeast are grown aerobically and thus presumably are caused by oxidative damage, it is possible that these phenotypes arise not from superoxide radicals per se but rather, some other (currently unidentified) reactive species that SOD1 normally detoxifies. In this sense, the paraquat sensitivity and lysine complementation assays may be measurements of separate in vivo Cu, Zn SOD1 properties.
Second, the data provide evidence for the possibility that the mutants may protect differentially against different sources of intracellular superoxide radicals as a consequence of localized free radical generation and/or nonuniform SOD1 subcellular activity. Mitochondria often are sited as the major contributors to in vivo superoxide radical generation production (32). However, sod1Δ strains completely devoid of respiratory function (through deletion of mitochondrial DNA) still show the amino acid auxotrophies (J. Bode and V.C.C., unpublished work) and demonstrate exquisite sensitivity to oxygen toxicity (33). Thus, nonrespiratory sources of free radicals also may contribute to toxicity in sod1Δ cells, yet these sources remain elusive, as does the source of electrons relevant to paraquat toxicity. What is clear is that the damaging species responsible for the lysine biosynthetic defect in sod1Δ yeast is not the same as paraquat-generated superoxide because we are not able to induce a lysine auxotrophy in wild-type yeast by simply increasing paraquat levels. In addition, recent studies suggest that, in vivo, as much as 35% of SOD1 may exist as apoprotein (devoid of copper) and that, once charged with copper, at least the wild-type SOD1 does not exchange the copper readily (34). It is plausible that H46R and H48Q polypeptides release their copper more readily and have to be recharged repeatedly, which may be efficient only in certain cytosolic locations. This could produce nonuniform intracellular SOD1 activities and complementation of only a subset of phenotypes linked to sod1Δ.
Our evidence offers further proof that the mechanism through which the copper is added to SOD1 involves catalyzed addition, albeit the evidence thus far is limited to the context of a yeast cell. Adding to the earlier evidence that wild-type yeast SOD1 remains completely inactive despite normal levels of polypeptide in the absence of the copper-loading chaperone LYS7 (13), we have found that this also is true for the human wild-type and FALS mutants when expressed at low levels or when copper is limiting. Unlike yeast SOD1, for the human polypeptides, this may not be absolute, however, because increasing mutant (or human wild-type) SOD1 levels do yield an amount of activity sufficient to complement, with the unexpected exception of G85R, which remains completely dependent on the yeast or human chaperone for acquiring even low levels of activity. The fact that copper loading (and perhaps even zinc loading and SOD1 polypeptide folding) is chaperone-assisted in vivo makes studies such as this one critical for understanding the properties of ALS-causing SOD1 mutants. Altogether, these findings provide support for the hypothesis that FALS–SOD1-mediated toxicity arises, at least in part, from aberrant copper-mediated chemistry catalyzed by less tightly folded (and hence less constrained) mutant enzymes. Indeed, evidence from Bredesen and colleagues (11) suggests that an enhanced apoptosis induced by some mutants is alleviated partially by the addition of metal chelators. This evidence, in turn, leads to the suggestion that the search for inhibitors of the specific SOD1 copper chaperone may represent a therapeutic avenue.
Acknowledgments
We thank D. Borchelt, H. Slunt, and members of the Hieter and Culotta labs for assistance, advice, and various reagents. This work was supported by National Institutes of Health Grants NS 27036 (to D.W.C.) and GM 50016 (to V.C.C.) and by the Johns Hopkins University National Institute on Environmental Health Sciences Center. Salary support for D.W.C. is provided by the Ludwig Institute for Cancer Research.
ABBREVIATIONS
- FALS
familial amyotrophic lateral sclerosis
- SOD1
superoxide dismutase
- BCS
bathocuproinedisulfonic acid
- PGK
phosphoglycerol kinase
- CCS
copper chaperone for SOD1
References
- 1.Rosen D R, Siddique T, Patterson D, Figlewicz D A, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan J P, Deng H X, et al. Nature (London) 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 2.Deng H-X, Hentati A, Tainer J A, Iqbal Z, Cayabyab A, Hung W-Y, Getzoff E D, Hu P, Herzfeldt B, Roos R P, et al. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 3.Wong P C, Pardo C A, Borchelt D R, Lee M K, Copeland N G, Jenkins N A, Sisodia S S, Cleveland D W, Price D L. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 4.Bruijn L I, Becher M W, Lee M K, Anderson K L, Jenkins N A, Copeland N G, Sisodia S S, Rothstein J D, Borchelt D R, Price D L, et al. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 5.Reaume A B, Elliott J L, Hoffman E K, Kowall N W, Ferrante R J, Siwek D F, Wilcox H M, Flood D G, Beal M F, Brown R H, et al. Nat Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 6.Siddique, T., Nijhawan, D. & Hentati, A. (1996) Neurology 47, Suppl. 2, S27–S34. [DOI] [PubMed]
- 7.Hodgson E K, Fridovich I. Biochemistry. 1975;14:5294–5299. doi: 10.1021/bi00695a010. [DOI] [PubMed] [Google Scholar]
- 8.Ischiropoulos H, Zhu L, Chen J, Tsai M, Martin J C, Smith C D, Beckman J S. Arch Biochem Biophys. 1992;298:431–437. doi: 10.1016/0003-9861(92)90431-u. [DOI] [PubMed] [Google Scholar]
- 9.Culotta V C, Joh H D, Lin S J, Slekar K H, Strain J. J Biol Chem. 1995;270:29991–29997. doi: 10.1074/jbc.270.50.29991. [DOI] [PubMed] [Google Scholar]
- 10.Beckman J S, Carson M, Smith C D, Koppenol W H. Nature (London) 1993;364:584. doi: 10.1038/364584a0. [DOI] [PubMed] [Google Scholar]
- 11.Wiedau-Pazos M, Goto J J, Rabizadeh S, Gralla E D, Roe J A, Valentine J S, Bredesen D E. Science. 1996;271:515–518. doi: 10.1126/science.271.5248.515. [DOI] [PubMed] [Google Scholar]
- 12.McCord J, Fridovich I. J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 13.Culotta V C, Klomp L W J, Strain J, Casaren R L B, Krems B, Gitlin J D. J Biol Chem. 1997;272:23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 14.Borchelt D R, Lee M K, Slunt H S, Guarnieri M, Xu Z S, Wong P C, Brown R H, Jr, Price D L, Sisodia S S, Cleveland D W. Proc Natl Acad Sci USA. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabizadeh S, Gralla E B, Borchelt D R, Gwinn R, Valentine J S, Sisodia S, Wong P, Lee M, Hahn H, Bredesen D E. Proc Natl Acad Sci USA. 1995;92:3024–3028. doi: 10.1073/pnas.92.7.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carri M T, Battistoni A, Polizio F, Rotilio G. FEBS Lett. 1994;356:314–316. doi: 10.1016/0014-5793(94)01295-4. [DOI] [PubMed] [Google Scholar]
- 17.Crow J P, Sampson J B, Zhuang Y, Thompson J A, Beckman J S. J Neurochem. 1997;69:1936–1944. doi: 10.1046/j.1471-4159.1997.69051936.x. [DOI] [PubMed] [Google Scholar]
- 18.Slekar K H, Kosman D J, Culotta V C. J Biol Chem. 1996;271:28831–28836. doi: 10.1074/jbc.271.46.28831. [DOI] [PubMed] [Google Scholar]
- 19.Horecka J, Kinsey P T, Sprague G F. Gene. 1995;162:87–92. doi: 10.1016/0378-1119(95)00325-z. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 21.Liu X F, Elashvili I, Gralla E B, Valentine J S, Lapinskas P, Culotta V C. J Biol Chem. 1992;267:18298–18302. [PubMed] [Google Scholar]
- 22.Mumberg D, Muller R, Funk M. Nucleic Acids Res. 1995;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flohe L, Otting F. Methods Enzymol. 1984;105:93–104. doi: 10.1016/s0076-6879(84)05013-8. [DOI] [PubMed] [Google Scholar]
- 25.Petrovic N, Comi A, Ettinger M J. J Biol Chem. 1996;271:28335–28340. doi: 10.1074/jbc.271.45.28335. [DOI] [PubMed] [Google Scholar]
- 26.Tamai K T, Gralla E B, Ellerby L M, Valentine J S, Thiele D J. Proc Natl Acad Sci USA. 1993;90:8013–8017. doi: 10.1073/pnas.90.17.8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gralla E B, Thiele D J, Silar P, Valentine J S. Proc Natl Acad Sci USA. 1991;88:8558–8562. doi: 10.1073/pnas.88.19.8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dancis A, Yuan S, Haile D, Askwith C, Eide D, Moehle C, Kaplan J, Klausner R. Cell. 1994;76:393–402. doi: 10.1016/0092-8674(94)90345-x. [DOI] [PubMed] [Google Scholar]
- 29.Bruijn L I, Beal M F, Becher M W, Schulz J B, Wong P C, Price D L, Cleveland D W. Proc Natl Acad Sci USA. 1997;94:7606–7611. doi: 10.1073/pnas.94.14.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrante R J, Shinobu L A, Schulz J B, Matthews R T, Thomas C E, Kowall N W, Gurney M E, Beal M F. Ann Neurol. 1997;42:326–334. doi: 10.1002/ana.410420309. [DOI] [PubMed] [Google Scholar]
- 31.Beal M F, Ferrante R J, Browne S E, Matthews R T, Kowall N W, Brown R H. Ann Neurol. 1997;42:644–654. doi: 10.1002/ana.410420416. [DOI] [PubMed] [Google Scholar]
- 32.Longo V D, Gralla E B, Valentine J S. J Biol Chem. 1996;271:12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- 33.Guidot D M, Repine J E, Kitlowski A D, Flores S C, Nelson S K, Wright R M, McCord J M. J Clin Invest. 1995;96:1131–1136. doi: 10.1172/JCI118100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrovic N, Comi A, Ettinger J J. J Biol Chem. 1996;271:28331–28334. doi: 10.1074/jbc.271.45.28331. [DOI] [PubMed] [Google Scholar]