Abstract
Important optical properties of discrete pairs of DNA tethered gold nanoparticles, including their scattering cross-section and resonance wavelength, depend both on the dimer structure and the refractive index of their immediate environment. We show that far-field polarization microscopy aids the optical identification and interpretation of structural changes including hinge motions and nanoscale distance changes in individual assemblies. Grecco and Martinez have shown in their theoretical work that the interparticle separation dependent polarization anisotropy of discrete nanoparticle dimers enables nanoscale distance measurements (Optics Express 2006, 14, 8716 – 8721). Here we implement this approach experimentally and evaluate measured polarization anisotropies in the framework of a dipolar coupling model. We use polarization sensitive darkfield microscopy to resolve simultaneous distance and orientation changes during the compaction of discrete pairs of DNA tethered gold nanoparticles by fourth generation polyamidoamino (PAMAM) dendrimers. The relative contributions from interparticle separation and refractive index variations to changes in the light polarization and scattering intensity are quantified and compared.
I. Introduction
DNAs and RNAs are important building blocks for active nanostructures that undergo structural changes in response to external stimuli. In nature nucleic acids with defined structures have evolved that enable diverse biological activities ranging from catalysis to post-transcriptional regulation. Prominent examples of functional nucleic acids include RNA ribozymes,[1] RNA riboswitches,[2] and repressor controlled DNA operons[3]. Inspired by the functionality of these active biological molecules, a series of active engineered nucleic acid based nanostructures have been realized.[4–7] Nucleic acid based nano- and microstructures have additional applications in the material sciences where they are used as scaffolds to assemble inorganic nanoparticles into regular assemblies[8–10] in which the distance dependent optical[11, 12], magnetic[13] or electronic[14] properties can be utilized to achieve functionalities beyond that of the individual particles. One important hybrid material that Mirkin and coworkers have pioneered for colorimetric sensing are DNA linked gold nanoparticles.[15, 16] The DNA linked gold nanoparticles function in this case as selective chemical transducers which indicate changes in the average interparticle separation as spectral shifts in the gold nanoparticle plasmon resonance.
The sensitivity of the colorimetric detection can be further increased by monitoring interparticle separations in individual pairs of DNA linked noble metal nanoparticles.[17] Due to the distance dependence of interparticle coupling,[11, 18, 19] discrete dimers of biopolymer tethered gold nanoparticles represent dynamic molecular rulers[12, 17, 20, 21]. With decreasing interparticle separation, the plasmon resonance wavelength λR in these “plasmon rulers” red-shifts and the scattering cross-section σscat increases.[12] The direction of the polarization of the light scattered off individual gold nanoparticle dimers provides additional information about the orientation of the long dimer axis, enabling simultaneous distance and orientation measurements. Dimers of nucleic acid tethered gold nanoparticles are active nanostructures that can potentially detect and quantify conformational changes in biopolymers due to bending, twisting, and stretching and could thus provide important insights into the mechanical properties and fundamental functions of biopolymers in various biological processes.
Gold nanoparticles exhibit a higher photostability and larger light emission power than organic dyes,[22–24] and allow the monitoring of structural changes in individual biopolymers – in principle – without limitation in observation time. The detection of small distance changes using plasmon coupling is, however, complicated by the fact that the optical response of the particle dimers also depends on the refractive index of the surroundings.[25–28] Fluctuations in the micro-environment[25] of the nanoparticles lead to variations in λR and σscat, and control experiments are mandatory to assess the effect of refractive index changes under the respective experimental conditions.[20, 21] In order to be able to unambiguously discern changes in the optical properties due to structural changes in the biopolymer tether from refractive index fluctuations in their micro-environment, additional information besides λR or σscat are desirable. The polarization of the scattered light is a potential source of such information.
For interparticle separations shorter than ~1 particle diameter the optical response of two biopolymer tethered gold nanoparticles is dominated by the longitudinal plasmon mode, leading to a high degree of polarization along the long dimer axis.[29] Different cross-sections along the long and short dimer axis result in a polarization anisotropy (η) for the particle dimer which following the notation of Grecco and Martinez is introduced here as:[30]
| (1) |
where is the scattering cross-section along the long dimer axis and is the scattering cross-section along the short dimer axis.
Grecco and Martinez have performed discrete dipole approximation (DDA)[31] simulations that show that the polarization anisotropy of light scattered off individual pairs of gold nanoparticles depends on the interparticle separation and can be directly converted into absolute distances.[30] In addition, the direction of the polarization of the scattered light depends on the orientation of the long dimer axis and thus contains information about the dimer structure as well. Far-field polarization microscopy has been previously used to probe the orientation of individual fluorescent dyes or quantum dots[32–38] and surface confined anisotropic noble metal nanoparticles, such as gold nanorods.[39] We show herein that for pairs of polymer tethered gold nanoparticles, a polarization sensitive detection is not only helpful in discerning structural changes from refractive index fluctuations, but it also enables the detection and identification of structural changes that otherwise remain hidden. We apply the polarization sensitive detection to simultaneously monitor distance and orientation changes in individual dimers of DNA linked gold nanoparticles during compaction through fourth generation polyamidoamine (PAMAM) dendrimers.
II. Methods
Assembly of DNA tethered Gold Nanoparticle Dimers
Single stranded DNA (ssDNA) tethered gold nanoparticle dimers were synthesized using a sequential ligand exchange procedure.[20, 21] In short, bis(p-sulphonatophenyl)phenylphosphine (BSPP) functionalized gold nanoparticles (NP) with a mean diameter of D = 40 ± 8 nm were reacted with thiolated single stranded DNA handles (30 nucleotides) in a ratio 30:1 (DNA:NP) in buffer 1 (40 mM NaCl, 10 mM Tris-HCl pH8). Two batches of particles with different handles were prepared. The handle in batch 1 was 5’ thiolated, the handle of batch 2 was 3’ thiolated. Batch 1 was then passivated with polyethylene glycol (PEG) propionate. The particles of batch 2 were in addition functionalized with a low number of biotinylated PEGs (acid-PEG/biotin-PEG ratio: 20/1). After incubation overnight, both batches were washed by centrifugation and batch 1 was resuspended in buffer 2 (70 mM NaCl, 10 mM Tris-HCl pH8). Then ssDNA (80 nucleotides) with 3’ and 5’ termini complementary to the handles of batches 1 & 2, respectively, were added in excess to batch 1 and annealed overnight. Subsequently, batch 1 was cleaned by repeated centrifugation and resuspension. Batches 1 and 2 were finally resuspended in buffer 3 (120 mM NaCl, 10 mM Tris-HCl pH8) and combined to anneal. The formed dimers were purified by gel-electrophoresis in a 1 % agarose gel using 0.5x tris borate EDTA (TBE) as running buffer and isolated from the gel by electroelution. We confirmed by TEM that the isolated assemblies were dimers (see Figure S1, Supporting Information). The oligonucleotide sequences and a chart of the resulting DNA tether are shown in Figure S2.
Surface Immobilization of Plasmon Rulers
All experiments were performed in rectangular glass capillaries functionalized with a biotin/Neutravidin chemistry. First biotinylated bovine serum albumin (BSA-biotin) was attached to the glass surface, then – after one washing step with buffer 4 (50 mM NaCl, 10 mM Tris-HCl pH7) – Neutravidin was flushed into the chamber to bind to the BSA-biotin. To minimize any non-specific binding, the surface was incubated with a blocking buffer (Pierce, Superblock). After 5 min incubation with Superblock, buffer 4 was used to wash the chamber. Finally, a diluted solution of DNA tethered gold nanoparticles in buffer 4 was flushed into the chamber to bind to the Neutravidin coated glass surface. Control experiments with acid-PEG coated gold particles showed negligible binding on the functionalized glass surface, confirming that the biotin functionalized dimers bound specifically to the Neutravidin functionalized glass surface.
The low non-specific binding affinity of the nanoparticles and the high purity of the self-assembled DNA gold nanoparticle dimers after isolation facilitated an effective immobilization of active plasmon rulers on the surface of the flow chamber.
III. Results and Discussion
Polarization Resolved Plasmon Coupling Microscopy of Plasmon Rulers
The plasmon rulers used in this study were asymmetrically functionalized with biotin (see Figure 1a); only one of the two particles in each dimer was biotinylated and could attach to the Neutravidin functionalized glass surface with a dissociation constant of Kd ≈ 10−14 - 10−15 M for each individual biotin – Neutravidin contact.[40] The surface immobilized particle serves as an anchor for the second DNA tethered particle which is not biotinylated and that performs a hindered Brownian motion in a confined space. The accessible volume is determined by the dimer structure, in particular the ratio of nanoparticle diameter to spacer length, and the orientation of the long dimer axis in space. The maximum displacement during this motion is defined by the length and flexibility of the DNA tether. As we will discuss in more detail below, the end-to-end distance of the DNA tether (Re) used in this study corresponds to approximately half a particle diameter (D) in length. If the long dimer axis is oriented perpendicular to the chamber surface, the diffusing particle can explore a cone around the center of the immobilized particle with a solid angle (Ω) of ~ 110°. However, since only one hemisphere above the chamber surface is accessible to the particle, the space explored by the particle decreases if the long dimer axis is tilted into the surface plane.
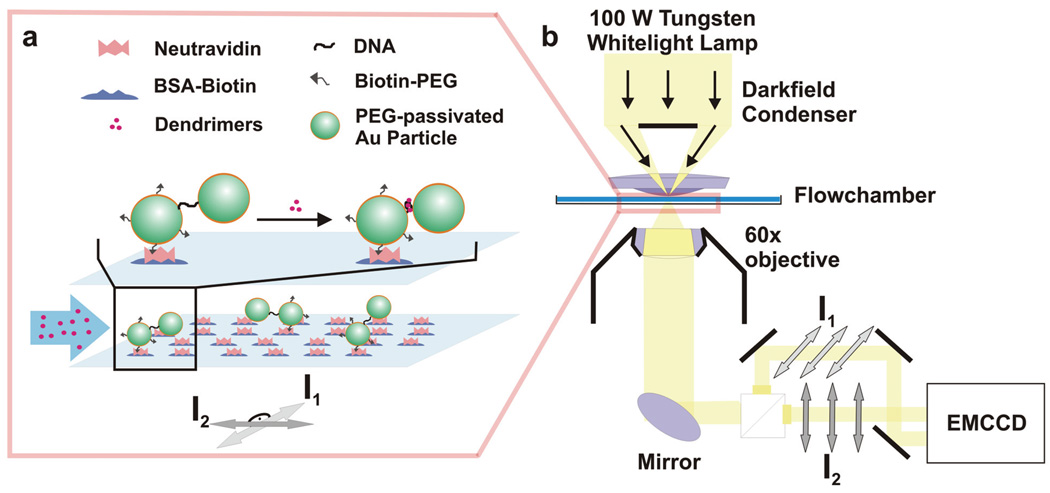
Figure 1. Experimental Set-Up.
a) Discrete pairs of DNA tethered gold nanoparticles (“plasmon rulers”) are anchored to the surface of a flowchamber. Fourth generation PAMAM dendrimers are added to trigger structural responses in the dimers. b) Far-field polarization darkfield microscopy is used to detect and identify structural changes in the gold nanoparticle dimers. The light scattered off individual plasmon rulers is collected with a 60x objective (NA = 0.75) and channeled through a polarizing beam splitter. The spatially separated polarization channels are reimaged on an enhanced CCD (EMCCD) detector.
With Re ≈ ½ D, the distance between the particles is sufficiently small to allow for significant interparticle coupling.[11, 41] Consequently, the light scattered off the dimers is polarized along the long axis. The direction of polarization varies from dimer to dimer and depends on the orientation of the dimer. Strongly coupled pairs of nanoparticles lying in the plane of the chamber surface can be considered to show similar light polarization propensities observed for gold nanorods[39]. In a dimer whose long axis has an out-of-plane component, the polarization direction and magnitude are determined by the projection of the long dimer axis into the chamber plane. The measured polarization orientation is defined by the azimuthal orientation of the long dimer axis.
Unlike rods, dimers of DNA tethered gold nanoparticles are active nanostructures that can respond to external stimuli with a change in interparticle separation. The polarization properties and optical cross-sections of individual pairs of DNA linked gold nanoparticles depend not only on the magnitude of the projection of the long dimer axis into the chamber plane and its orientation but also on the interparticle separation (s). The orientation and distance dependence of the scattered light polarization encode important information about the dimer structure and its change as function of time. This information can be accessed using polarization sensitive darkfield microscopy.
The optical set-up used in this study is outlined in Figure 1b. The dimers were imaged in a darkfield microscope[42] using a 1.4 NA oil darkfield condenser with a 60x objective (numerical aperture NA = 0.75) using unpolarized whitelight illumination (100 W Tungsten lamp). The light scattered off the individual dimers was channeled through a polarizing beamsplitter that divides the collected light into two beams with perpendicular polarizations. The beams were translated and reimaged on two separate areas of the same CCD detector (Andor-Ixon). Images were captured at a frame rate of 46 Hz using an active area of 256 × 256 pixels and a hardware 2 × 2 pixel binning. These settings allowed us to monitor all the dimers in an area of 80 by 30 microns simultaneously on two orthogonal polarization channels. The intensities I1 and I2 as obtained through integration of the light collected from each individual dimer on the two polarization channels were used to compute the intensity ratios (P):
| (2) |
where channel 1 was chosen to lie perpendicular and channel 2 to lie parallel to the flow direction in the flow chamber.
P is a sensitive measure for changes in the scattered light polarization due to a rotation of the long dimer axis or due to changes in the interparticle separation. Since I1 and I2 are orthogonal, rotations of the long dimer axis cause anti-correlated changes of I1 and I2 and large changes in P. We point out that while the scattering polarization anisotropy η is defined within the internal coordinate system of the dimers, P is defined in the lab coordinate system. Consequently, |P| corresponds to η only if the long dimer axis is confined to the surface and oriented along one of the detection channels. In this case, the experimental P values can be used to extract interparticle separations if an adequate functional relationship for the distance dependence of η is applied.
The model: dipolar interparticle coupling
Using numerical discrete dipole approximation (DDA)[31] calculations, Grecco and Martinez have shown that the distance dependence of the polarization anisotropy can be used to derive interparticle separations between 20 nm diameter gold nanoparticles.[30] A simplified but general analytical solution for the distance dependence of the polarization anisotropy can be found in the quasistatic approximation assuming that dipolar coupling is the major contribution to the interparticle coupling and taking advantage of the universal scaling behavior of the distance decay of plasmon coupling reported by Jain and El-Sayed[41].
The scattering cross-sections for the longitudinal and vertical plasmon modes in the dimers are obtained from the pair-polarizability α’ of the dimer:[29]
| (3) |
with k being the angular wavenumber k = 2π/λ. The pair polarizabilities of the longitudinal and vertical plasmon modes can be calculated in the dipolar coupling approximation from the polarizabilities of the individual spheres taking into account an orientation factor κ:[41]
| (4) |
where ε0 is the dielectric constant of vacuum and L= s + D is the sum of the surface-to-surface separation (s) and the particle diameter (D). The orientation factor κ depends on the orientation of the dipoles in the dimer; for the longitudinal plasmon mode, the net dipoles in the individual particles are aligned head-to-tail and κ=2. For the vertical plasmon mode, the dipoles are aligned parallel in a side-to-side fashion and κ= −1.[29] We computed the single particle sphere polarizability, α, using the Clausius-Mosotti equation and the experimental gold dielectric function ε reported by Johnson and Christy[43]:
| (5) |
where εm is the dielectric constant of surrounding medium. The value of γ depends on the geometry of the nanoparticles. In DNA linked gold nanoparticle dimers it depends on the interparticle separation; the effective γ can be calculated in the dipolar-coupling limit as a function of the ratio of the surface-to-surface separation (s) and the particle diameter (D) :[28]
| (6) |
With equation (1) and equation (3) – equation (6) the distance dependence of η can be evaluated provided that the wavelength dependence of the gold dielectric function ε(λ) is considered. We computed ε at the resonance wavelengths λres corresponding to specific s/D values.[41] The longitudinal resonances λres(s/D) for different refractive indices (nr) were obtained through fits to previous DDA simulations of λres as function of nr and s/D performed by Jain and El-Sayed[28]. The vertical resonance was assumed to be independent of the separation and was kept constant at the resonance wavelength (λres) of an individual particle.
Assumptions and Limitations
A quantitatively correct conversion of the measured light polarization properties into distances is only possible when the long dimer axis lies in the plane of the chamber surface. As we discuss below in more detail, for most dimers the degree of polarization indicates that the long dimer axis has indeed a strong in-plane component. However, a non-zero out-of-plane angle with the surface plane can in general not be excluded. Another shortcoming of the applied model is its limitation to dipolar interparticle interactions. Especially at very short interparticle separations, modes of higher order angular momenta are expected to contribute significantly to the coupling as well.[44–46] Because of the small size of the particles of interest in this study (D ≤ 40 nm), and for the sake of simplicity, electromagnetic retardation is ignored in our model.[47]
It is clear that – due to the above assumptions – the derived distances can only be approximates. Nevertheless, a plasmon sensitive detection improves the usability of plasmon rulers in most applications. Changes in the optical response of plasmon rulers in solution can be the result of various (synergistic) processes including an increase in the refractive index in the micro-environment, a decrease of the interparticle separation, and potentially the tilt of the long dimer axis into the surface plane. A polarization sensitive detection provides additional information about the cause for the observed optical response and is thus instrumental in dissecting the relative contributions from the individual effects.
Refractive Index vs. Distance Dependence of Polarization Anisotropy and Total Scattering Intensity
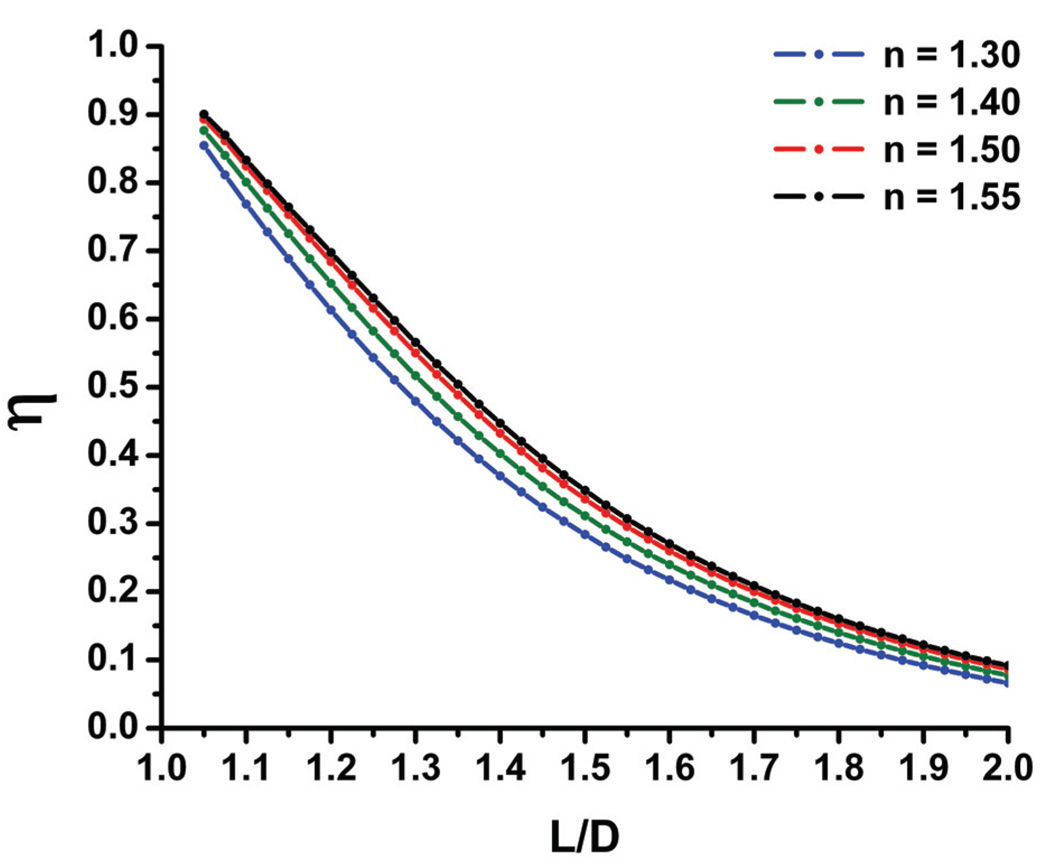
The plasmon resonance wavelength (λres) – and hence the metal dielectric function (ε(λres)) and the polarization anisotropy (η) – depend both on the interparticle separation and the refractive index (nr) of its micro-environment.[25, 27] To estimate the relative contributions from these two factors, η(L/D) relationships for solutions with different refractive indices (nr = ε1/2) were calculated. Figure 2 contains plots of η(L/D) for refractive indices varying from nr = 1.30 to nr = 1.55; this interval was chosen because it covers the refractive index range from pure water nr = 1.33 to pure dendrimers[48] nr≈ 1.50. Figure 2 shows that for any fixed interparticle separation fluctuations in this nr range lead to variations in η on the order of Δη ≈ 0.07. We expect that under our experimental conditions the changes arising from fluctuation in the refractive index are even lower. A PEG passivation layer on the gold surface stabilizes the local refractive index in the immediate vicinity of the gold particles at around nr≈ 1.5. Consequently, we will use the fitted η(L/D) relationship obtained for nr = 1.5 unless otherwise stated in the following: η(L/D) = 2.276exp[−0.841*(L/D)2].
Figure 2.
Simulated distance dependence of the polarization anisotropy (η) in solutions with refractive indices between nr = 1.30 and nr = 1.55 (see legend) for 40 nm gold nanoparticle dimers. The polarization anisotropy is plotted against the ratio of center-to-center distance (L) and diameter (D).
Next, we want to calculate the anticipated change in η during the compaction of the DNA tether through dendrimers at constant refractive index (nr = 1.5). To that end, an estimate of the surface-to-surface separation (s0) of the dimers in the initial state is required. In the case of a single DNA tether between the particles, the average surface-to-surface separation (s) is determined by the end-to-end distance of the polymer linker (Re). The DNA spacers used to tether the two gold nanoparticles contain both single and double stranded regions and the 3’ and 5’ ends are embedded in approximately 3 nm thick PEG monolayers self-assembled on the gold surfaces (see Figure S2). Although the precise end-to-end distance (Re) for such a short but complex tether molecule is difficult to predict quantitatively, an approximation can be obtained in the framework of the worm-like-chain model. The latter has been shown to be applicable to polymers with sections of different flexibilities.[49] We assume that the first 10 nucleotides of the DNA tether on each side bridge the 3 nm thick PEG layers surrounding the gold nanoparticles. The remaining DNA tether is treated as a composite polymer comprising a single stranded part (persistence length PssDNA = 1.3 nm;[50] contour length CssDNA = 26 nm) and a double stranded part (PdsDNA = 53 nm;[49] CdsDNA = 13 nm). This model yields an approximate initial interparticle separation of s0 ≈ 21.5 nm. Based on the simulated distance dependence of the polarization anisotropy in a medium with refractive index of nr = 1.5, we anticipate that the polarization anisotropy for a dimer of 40 nm gold nanoparticles separated by the estimated equilibrium distance is η(L/D = 1.54) ≈ 0.31. The minimum interparticle separation in the compacted state is defined by 3 nm thick PEG monolayers around the gold nanoparticles which are assumed to be incompressible. The corresponding polarization anisotropy for an interparticle separation of 6 nm at nr = 1.5 is η(L/D = 1.15) ≈ 0.75.
The maximum change in η due to refractive index fluctuation of Δη ≈ 0.07 compares with Δη ≈ 0.42 obtained for dimer compaction at constant refractive index. Even if we take into account that the equilibrium interparticle separations are estimates, the quintessence of Figure 2 is that large changes in the interparticle separation affect the polarization anisotropy significantly more than even large fluctuations in the refractive index.
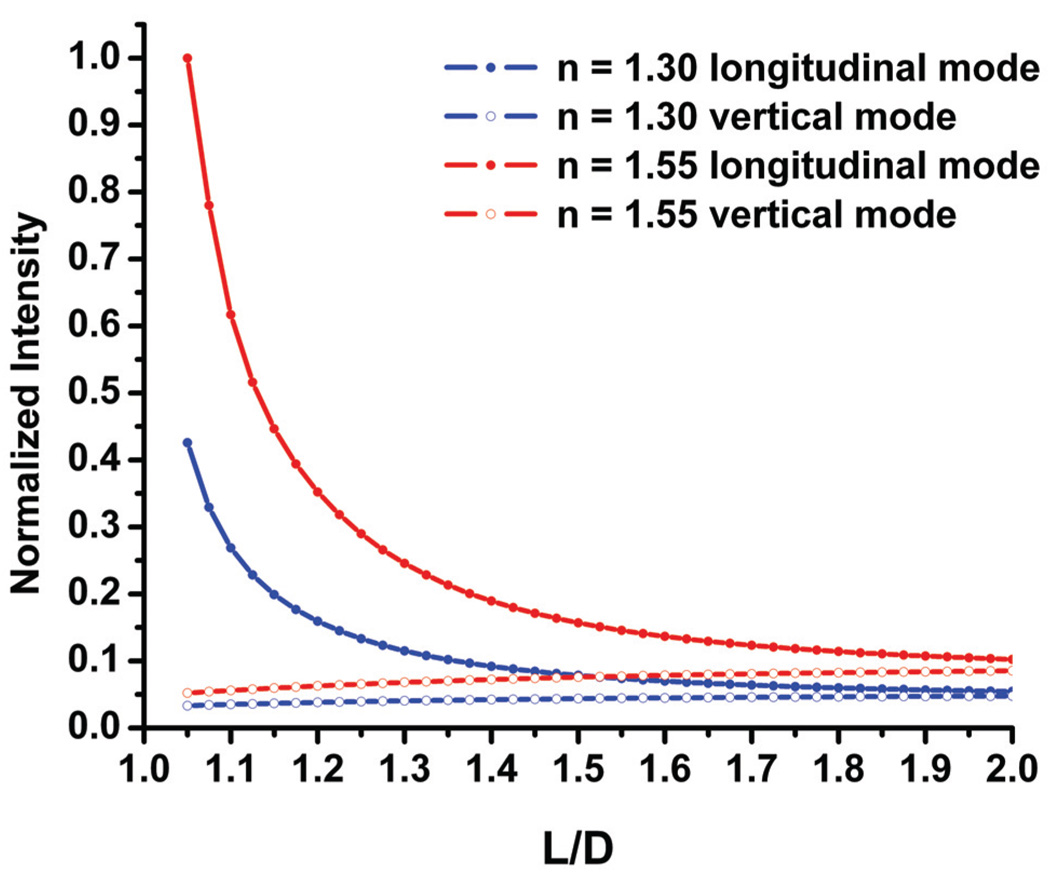
We point out that the polarization anisotropies are less affected by changes in the refractive index than the scattering intensities. In Figure 3, simulated scattering intensities for both the longitudinal and vertical plasmon modes are plotted as a function of L/D for two different refractive indices: nr = 1.30 and nr = 1.55. The plots are normalized by setting the maximum scattering intensity of the nr = 1.55 longitudinal mode plot arbitrarily to 1. The plots of the longitudinal modes are systematically shifted with regard to each other and, most importantly, the difference between them increases notably with decreasing L/D. At short interparticle separations, fluctuations in the refractive index between nr = 1.30 and nr = 1.55 can lead to intensity changes comparable to those obtained upon compaction of the DNA spacer at a constant refractive index. While this behavior can be exploited for designing sensitive plasmonic refractive index sensors, it complicates the use of the scattering intensity for ruler applications. These considerations underline the advantage of the polarization anisotropy as more robust measure for structural and distance changes in gold nanoparticle dimers.
Figure 3.
Simulated scattering intensities as function of the ratio of center-to-center separation (L) and particle diameter (D) for pairs of 40 nm gold nanoparticles in media with refractive indices of nr = 1.30 and nr = 1.55, respectively.
Dendrimer Induced Structural Changes in Single Stranded DNA Tethered 40 nm Gold Nanoparticle Dimers
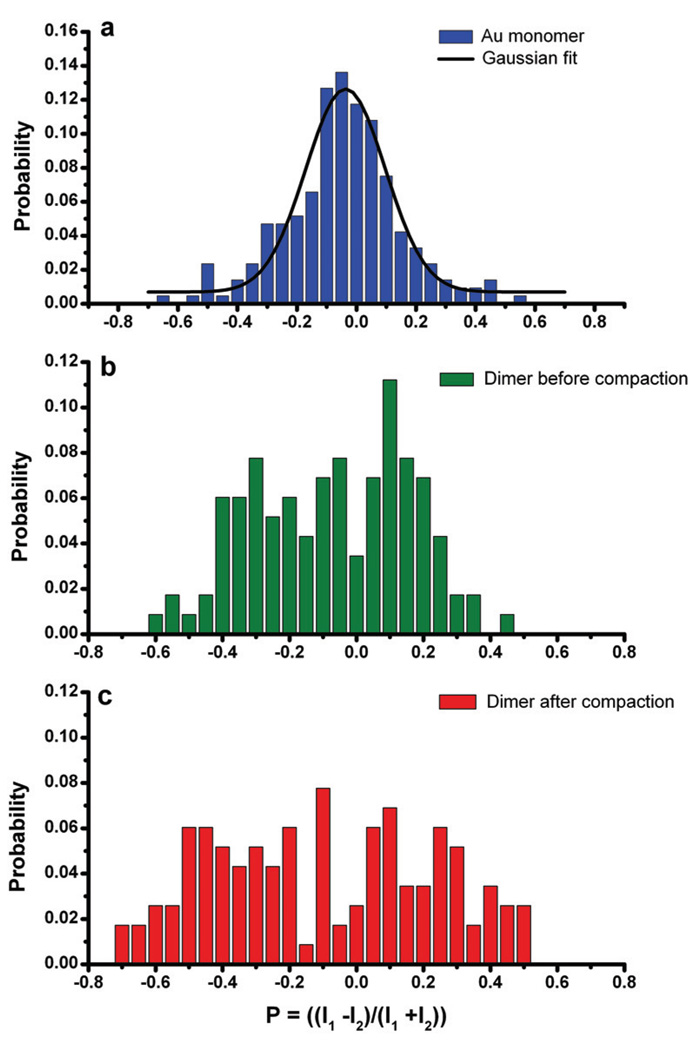
PAMAM dendrimers offer a convenient way of inducing structural changes into individual dimers of DNA tethered nanoparticles. Under physiological conditions the dendrimers are highly positively charged ensuring their efficient binding to the DNA tether and potentially also to the negative surface of the PEG passivated gold particles. In order to obtain a better understanding of the structural response that the dendrimers induce after binding to the dimers, we analyzed the polarization of the light scattered off ~150 gold dimers before and after exposure to the dendrimers. In a control experiment we first determined the P values of ~150 individual 40 nm gold particles. The P value distribution of the individual particles plotted in Figure 4a shows a slight shift to the left. A Gaussian fit to the experimental data shows that the distribution is centered at around P = −0.04. The shift of the distribution to negative P values indicates that the detected light is slightly polarized along channel 2. The optical set-up and the dark-field condenser in particular contain multiple reflective surfaces which induce a weak polarization of the excitation light in the focal plane.
Figure 4.
P-distributions for a) individual gold monomers, b) dimers of DNA linked 40 nm gold nanoparticles before compaction and c) dimers after compaction.
The width of the P value distribution for the monomers is determined by the intrinsic instrument response function and the shape distribution of the gold sol. “Spherical” particle preparations always also contain contaminations of particles with anisotropic shapes such as rods or nanoprisms that polarize the scattered light and thus broaden the P distribution.[12] For dimers that are oriented perpendicular to the surface of the flowchamber, we anticipate a P value distribution similar to that of the monomers. A first inspection of the P value distribution of the dimers before compaction in Figure 4b reveals, however, that the P value distribution of the dimers is significantly broader. The differences in P between monomers and dimers show that i) there is substantial interparticle coupling at the interparticle separation s0 and ii) many dimers are either lying in the surface of the flowchamber or have a significant projection into this plane. For a random distribution of the dimer orientations we expect the P values to lie within the interval [-η:+η]. If the polarization of the incident light is taken into account then this interval is given as [−0.35:0.27] for dimers before compaction. Most of the dimers (84%) lie within this range, but as a whole the distribution is shifted towards the negative values. We ascribe this systematic shift to a preferential alignment of the long dimer axis parallel to the flow direction. The dimers prefer to bind to the surface with the long axis oriented parallel to the flow direction to minimize hydrodynamic resistance.
Addition of dendrimers further broadens the P distribution (see Figure 4c) which is in line with an increase in the polarization anisotropy due to a dendrimer induced decrease in the interparticle separation. After compaction, all of the dimers in Figure 4c lie within the P value range of [−0.79:0.71] predicted for the minimum surface-to-surface separation (s = 6 nm). As observed for the dimers before addition of dendrimers, the distribution demonstrates an overall negative shift. Due to the distance dependence of the polarization anisotropy, this shift to the left is more pronounced for the compacted dimers in Figure 4c.
Polarization Resolved Analysis of Single Plasmon Ruler Trajectories
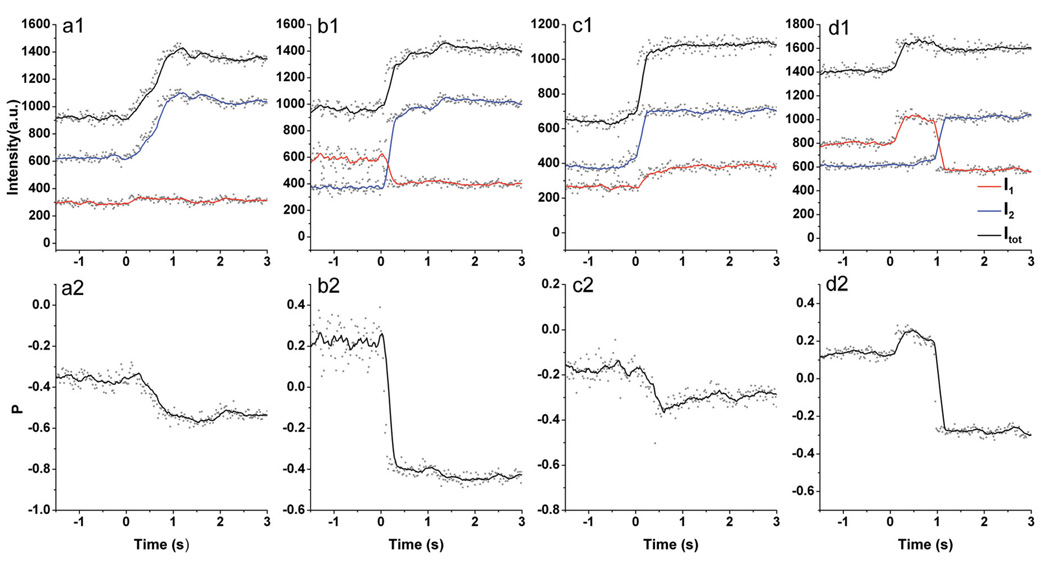
Having analyzed the light polarization properties of individual gold particles and their dimers on the ensemble level, we set out to investigate the interactions between individual dimers and dendrimers in real time next. In a typical experiment we started flushing (flow speed: ~60 mm/s) 10 s after we started recording. We maintained the flow for 5 s and kept recording for another 20 s without flow. In Figure 5 we plot I1, I2, Itot = I1 + I2, and P for four representative DNA tethered dimers during their interaction with dendrimers. One feature that all the trajectories in Figures 5 share is an increase in total scattering intensity shortly after a 500 nM aqueous solution of fourth generation PAMAM dendrimers has been flushed into the chamber. All trajectories in Figure 5 exhibit P changes |ΔP| > 0.2 and are thus larger than can be explained with refractive index fluctuations in the range nr = 1.33 – 1.55 alone. The observed gains in total scattering intensity, the magnitude of the accompanying P value changes, and the irreversibility of these changes are strong indications of dendrimer induced compactions. A closer analysis of the distribution of the light on the two detection channels as a function of time reveals additional details about the structural changes in the individual dimers.
Figure 5.
Experimental scattering intensities and derived P values of individual dimers of DNA linked gold nanoparticles during interactions with dendrimers. Top panel: I1 (red ), I2 (blue) and Itot = I1 + I2 (black) as function of time. Bottom panel: Corresponding P values as function of time. The continuous lines represent 10 point sliding averages of the raw data.
In Figure 5a the intensity on channel 2 increases strongly while channel 1 remains almost constant. This observation implies that the long dimer axis (or its projection into the plane) coincides with the polarization axis of channel 2 and that the compaction of the dimer axis occurs along that direction. Consequently, the orthogonal detection channel records the vertical plasmon mode which is almost independent of the interparticle separation. In this case the computed |P| values correspond to the particle’s polarization anisotropy η provided that the long dimer axis is lying in the chamber plane. In Figure 5a the dendrimers induce a change in P from ~ −0.37 to ~ −0.53. According to our simulations, the polarization anisotropy of a dimer of 40 nm gold nanoparticles separated by the estimated initial distance of s0 ≈ 21.5 nm is η ≈ 0.31. This value defines the |P| value for a 40 nm dimer with interparticle separation s0 whose long axis in the chamber plane is aligned parallel to one of the detection channels. A comparison of the simulated η value with the initial P value in Figure 5a suggests that this dimer fulfills these requirements. The observed change in η can therefore be used to calculate an approximate change in the interparticle separation, which in this case corresponds to Δs = 5.8 nm.
In Figure 5b the anti-correlation of the intensity changes on the two detection channels indicates a rotation of the long dimer axis as part of a concerted dendrimer induced structural change. Refractive index changes can account for gains on one or both of the detection channels, but only a structural change entailing a rotation of the long dimer axis can cause strong anti-correlated intensity changes. In Figure 5b the intensity gain during the dendrimer induced structural response on channel 2 over-compensates the drop in intensity on channel 1, leading to a net gain in total scattering intensity. The starting P value is relatively low with P ≈ 0.23 in this case, and it is feasible that both a further tilting of the long dimer axis into the surface chamber as well as a decrease in the interparticle separation contribute to the observed gain in total scattering intensity. The final |P| value of ~0.42 is higher than η(s0) and is thus indicative of an interparticle separation that is shorter than the initial interparticle distance s0. In Figure 5b the rotation of the long dimer axis is accompanied by its compaction.
In Figure 5c the intensity increases simultaneously on both detection channels, and the P value changes from ~ −0.19 to ~ −0.30. In principle, an irreversible increase in the refractive index of the dimer’s micro-environment can account for a simultaneous increase in the scattering intensity. The magnitude of the total intensity change (factor of ~1.7) requires, however, that the change in the refractive index occurs in a dimer with very short interparticle separation (see Figure 3) which is not supported by the measured initial total scattering intensities. If the interparticle separation was already very short before addition of the dendrimers, the scattering intensity should be high even before any interactions with the dendrimers occur. However, the initial intensity in the trajectory shown in Figure 5c is lower than that in all the other trajectories. Instead, we ascribe the observed simultaneous increase in scattering intensities on channels 1 and 2 to the compaction of a dimer whose long axis is oriented between two orthogonal detection channels. In the example in Figure 5c the long axis remains fixed and compaction is not coupled to a rotation.
All of the trajectories in Figures 5a–c contain one common feature: the dendrimer induced transition between the initial and final P state. In some cases the trajectories exhibited a more complex sequence of fluctuations in I1 and I2, indicating several subsequential structural minima. This is exemplified in the trajectory shown in Figure 5d. After the initial compaction at t ≈ 0, which occurs mainly along channel 1, the intensities on the two polarization channels contain additional strong anti-correlated changes that cancel out. This pattern indicates a dendrimer induced dimer compaction that is followed by an in-plane rotational movement of the long dimer axis which leaves the total scattering intensity essentially unchanged. The latter becomes detectable only through the polarization sensitive detection.
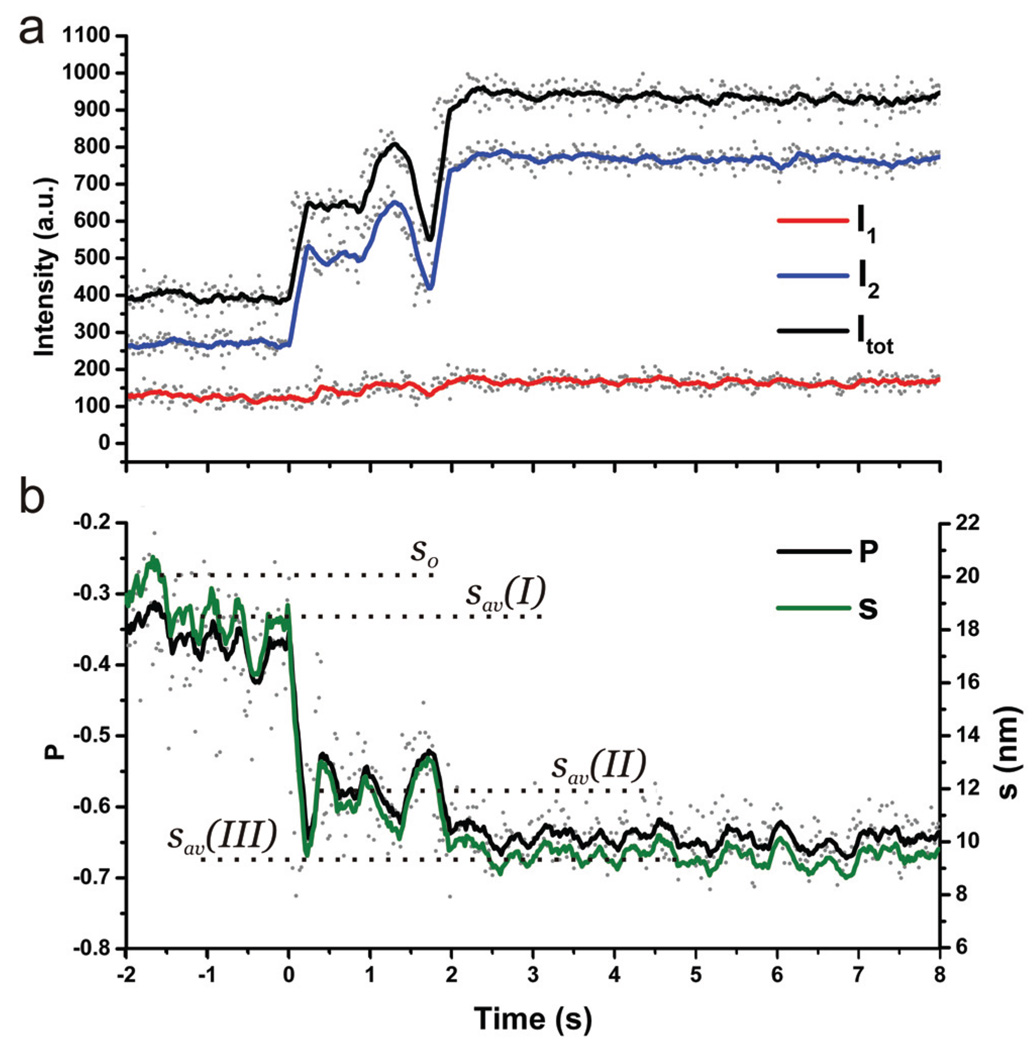
In our discussion so far we have included all dimers that show a change in total scattering intensity and/or change in the P value. Only a subset of these dimers is however amenable to a distance analysis. Only if the dimer’s long axis lies in the chamber plane and points along one detector axis, the measured |P| values correspond to the polarization anisotropy (η). Time resolved η measurements allow then a monitoring of the interparticle separation in real time (see Figure 6). The trajectory in Figure 6a contains changes in I2 at nearly constant I1. As discussed above, this intensity pattern suggests that the dimer compaction occurs along one detection channel. Together with the high initial P value (|P| ≈ 0.35) for the uncompacted dimer these observations justify the assumption that the dimer is lying in the chamber plane with its long axis oriented along detection channel 2.
Figure 6.
a) Scattering intensities I1, I2, and Itot as function of time and b) corresponding P and surface-to-surface separation (s). The continuous lines represent 10 point sliding averages of the raw data.
Figure 6b contains computed P values and corresponding interparticle separations (s); the light was assumed to be essentially unpolarized for this conversion. The average interparticle separation before dendrimer addition fluctuates around s0 ≈ 20 nm, which – taken into account all assumptions – is in good agreement with the theoretically predicted value of 21.5 nm. Upon flushing the chamber with dendrimer containing solution, the average interparticle separation decreases to sav(I) ≈ 19 nm. One possible explanation for this observation is that the liquid flow in the chamber exerts a drag force on the DNA tethered particle in solution. Depending on the orientation of the dimer in the flow chamber this force can slightly increase or decrease the interparticle separation. After approximately 1300 ms of flushing, the buffer in the chamber has been exchanged, and the dendrimers reach the dimers. Binding of the dendrimers to the dimers and subsequent dimer compaction is indicated by a sudden drop in the average interparticle separation to sav(II) ≈ 12nm. The system has, however, not yet found a stable minimum and after a dwell time of 1700 ms, the average interparticle separation further decreases to sav(III) ≈ 9 nm.
Although the computed interparticle separations derived from the polarization anisotropies are currently only approximate, the above analyses show that polarization resolved plasmon coupling microscopy aids the interpretation of single plasmon ruler scattering trajectories. Using this technology we identified a series of different structural modifications in the dimers in Figure 5&Figure 6. The dendrimers induce dimer compactions, rotations, or combinations of both. We recorded a total of ~150 compaction trajectories; the observed changes in P upon compaction are broadly distributed indicating that dimer compaction through dendrimers is a random process.
IV. Conclusions
The need for novel approaches to measure distances below the persistence length of double stranded DNA has recently been underlined by small angle X-ray scattering studies of gold nanocrystal end-labeled DNAs that question the validity of the conventional elastic rod model for short double-stranded DNAs 50 years after the discovery of the double helix structure of DNA.[51] Motivated by this need we have explored here the applicability of discrete pairs of DNA tethered gold nanoparticles to optically probe the structural dynamics in nanostructures using polarization sensitive dark-field microscopy. Individual pairs of DNA tethered gold nanoparticles, so called plasmon rulers, are active nanostructures whose optical responses depend both on the structure and orientation of the dimers. Due to plasmon coupling between the particles, the light scattered off individual DNA linked gold nanoparticle dimers is strongly polarized along the long dimer axis. The degree and direction of the light polarization depend on the interparticle separation and the orientation of the long dimer axis in space. Although fluctuations in the refractive index can influence the interparticle coupling as well, far-field polarization microscopy facilitates the identification and quantification of structural changes. Rotations of the long dimer axis in the chamber plane, for instance, are identified through anti-correlated intensity changes on two orthogonal detection channels. Far-field polarization microscopy greatly enhances the sensitivity for the detection of structural changes in DNA linked gold nanoparticle dimers. We use this approach to investigate the interactions between fourth generation PAMAM dendrimers and DNA linked gold nanoparticles. The structural changes induced by the highly charged dendrimers in individual dimers of DNA linked gold nanoparticles are monitored in real time, and distance and orientation changes are resolved simultaneously. Our studies reveal that the dendrimers can cause rotations, compactions and combined hinge-compaction motions of the long dimer axis. In the framework of a dipolar coupling model, experimental polarization anisotropies of discrete nanoparticle dimers could be converted into approximate distances, enabling the quantification of the interparticle separations during the dimer collapse.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by Grant 1 R21 EB008822–01 by the National Institute of Health. The authors thank L. R. Skewis for proof-reading the manuscript.
REFERENCES
- 1.Ke AL, et al. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429(6988):201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 2.Winkler WC, et al. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428(6980):281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 3.Morgan MJ, et al. Single Molecule Spectroscopic Determination of Lac Repressor-DNA Loop Conformation. Biochemical and Biophysical Research Communications. 2005;89:2588–2696. doi: 10.1529/biophysj.105.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao CD, et al. A nanomechanical device based on the B-Z transition of DNA. Nature. 1999;397(6715):144–146. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- 5.Yurke B, et al. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406(6796):605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 6.Viasnoff V, Meller A, Isambert H. DNA nanomechanical switches under folding kinetics control. Nano Letters. 2006;6(1):101–104. doi: 10.1021/nl052161c. [DOI] [PubMed] [Google Scholar]
- 7.Sherman WB, Seeman NC. A precisely controlled DNA biped walking device2004) Nano Letters. 2004;4(9):1801–1801. [Google Scholar]
- 8.Xiao SJ, et al. Selfassembly of metallic nanoparticle arrays by DNA scaffolding. Journal of Nanoparticle Research. 2002;4(4):313–317. [Google Scholar]
- 9.Le JD, et al. DNA-templated self-assembly of metallic nanocomponent arrays on a surface. Nano Letters. 2004;4(12):2343–2347. [Google Scholar]
- 10.Alivisatos AP, et al. Organization of 'nanocrystal molecules' using DNA. Nature. 1996;382(6592):609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 11.Su KH, et al. Interparticle coupling effects on plasmon resonances of nanogold particles. Nano Letters. 2003;3(8):1087–1090. [Google Scholar]
- 12.Reinhard BM, et al. Calibration of dynamic molecular rule based on plasmon coupling between gold nanoparticles. Nano Letters. 2005;5(11):2246–2252. doi: 10.1021/nl051592s. [DOI] [PubMed] [Google Scholar]
- 13.Xi H, et al. Magneto-elastic coupling and Peierls-like phase transition of one-dimensional magnetic nanoparticle chains. J. Phys. D: Appl. Phys. 2006;39:1333–1336. [Google Scholar]
- 14.Schliwa A, et al. Exciton Level Crossing in Coupled InAs/GaAs Quantum Dot Pairs. phys. stat. sol. (b) 2001;224(2):405–408. [Google Scholar]
- 15.Elghanian R, et al. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277(5329):1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 16.Storhoff JJ, et al. What? controls the optical properties of DNA-linked gold nanoparticle assemblies? Journal of the American Chemical Society. 2000;122(19):4640–4650. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]
- 17.Sonnichsen C, et al. A molecular ruler based on plasmon coupling of single gold and silver nanoparticles. Nature Biotechnology. 2005;23(6):741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 18.Rechberger W, et al. Optical properties of two interacting gold nanoparticles. Optics Communications. 2003;220(1–3):137–141. [Google Scholar]
- 19.Quinten M, Kreibig U. Absorption and Elastic-Scattering of Light by Particle Aggregates. Applied Optics. 1993;32(30):6173–6182. doi: 10.1364/AO.32.006173. [DOI] [PubMed] [Google Scholar]
- 20.Reinhard BM, et al. Use of plasmon coupling to reveal the dynamics of DNA bending and cleavage by single EcoRV restriction enzymes. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(8):2667–2672. doi: 10.1073/pnas.0607826104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skewis LR, Reinhard BM. Spermidine modulated ribonuclease activity probed by RNA plasmon rulers. Nano Letters. 2008;8(1):214–220. doi: 10.1021/nl0725042. [DOI] [PubMed] [Google Scholar]
- 22.Yguerabide J, Yguerabide EE. Resonance light scattering particles as ultrasensitive labels for detection of analytes in a wide range of applications. Journal of Cellular Biochemistry. 2001:71–81. doi: 10.1002/jcb.10077. [DOI] [PubMed] [Google Scholar]
- 23.Yguerabide J, Yguerabide EE. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications - I. Theory. Analytical Biochemistry. 1998;262(2):137–156. doi: 10.1006/abio.1998.2759. [DOI] [PubMed] [Google Scholar]
- 24.Yguerabide J, Yguerabide EE. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications - II. Experimental characterization. Analytical Biochemistry. 1998;262(2):157–176. doi: 10.1006/abio.1998.2760. [DOI] [PubMed] [Google Scholar]
- 25.Novo C, et al. Influence of the medium refractive index on the optical properties of single gold triangular prisms on a substrate. Journal of Physical Chemistry C. 2008;112(1):3–7. [Google Scholar]
- 26.Miller MM, Lazarides AA. Sensitivity of metal nanoparticle surface plasmon resonance to the dielectric environment. Journal of Physical Chemistry B. 2005;109(46):21556–21565. doi: 10.1021/jp054227y. [DOI] [PubMed] [Google Scholar]
- 27.Kelly KL, et al. The optical properties of metal nanoparticles: The influence of size, shape, and dielectric environment. Journal of Physical Chemistry B. 2003;107(3):668–677. [Google Scholar]
- 28.Jain PK, el-Sayed MA. Noble Metal Nanoparticle Pairs: Effect of Medium for Enhanced Nanosensing. Nano Letters. 2008;8:43–47. doi: 10.1021/nl8021835. [DOI] [PubMed] [Google Scholar]
- 29.Kreibig U, Vollmer M. Optical Properties of Metal Clusters. Berlin: Springer-Verlag. 1995:13–193. [Google Scholar]
- 30.Grecco HE, Martinez OE. Distance and orientation measurement in the nanometric scale based on polarization anisotropy of metallic dimers. Optics Express. 2006;14(19):8716–8721. doi: 10.1364/oe.14.008716. [DOI] [PubMed] [Google Scholar]
- 31.Draine BT, Flatau PJ. Discrete-Dipole Approximation for Scattering Calculations. Journal of the Optical Society of America a-Optics Image Science and Vision. 1994;11(4):1491–1499. doi: 10.1364/josaa.25.002693. [DOI] [PubMed] [Google Scholar]
- 32.Rosenberg SA, et al. Rotational motions of macromolecules by single-molecule fluorescence microscopy. Accounts of Chemical Research. 2005;38(7):583–593. doi: 10.1021/ar040137k. [DOI] [PubMed] [Google Scholar]
- 33.Ha T, et al. Polarization spectroscopy of single fluorescent molecules. Journal of Physical Chemistry B. 1999;103(33):6839–6850. [Google Scholar]
- 34.Sase I, et al. Axial rotation of sliding actin filaments revealed by single-fluorophore imaging. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(11):5646–5650. doi: 10.1073/pnas.94.11.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Empedocles SA, Neuhauser R, Bawendi MG. Three-dimensional orientation measurements of symmetric single chromophores using polarization microscopy. Nature. 1999;399(6732):126–130. [Google Scholar]
- 36.Hu JT, et al. Linearly polarized emission from colloidal semiconductor quantum rods. Science. 2001;292(5524):2060–2063. doi: 10.1126/science.1060810. [DOI] [PubMed] [Google Scholar]
- 37.Weston KD, Goldner LS. Orientation imaging and reorientation dynamics of single dye molecules. Journal of Physical Chemistry B. 2001;105(17):3453–3462. [Google Scholar]
- 38.Prummer M, et al. Three-dimensional optical polarization tomography of single molecules. Journal of Chemical Physics. 2003;118(21):9824–9829. [Google Scholar]
- 39.Sonnichsen C, Alivisatos AP. Gold nanorods as novel nonbleaching plasmon-based orientation sensors for polarized single-particle microscopy. Nano Letters. 2005;5(2):301–304. doi: 10.1021/nl048089k. [DOI] [PubMed] [Google Scholar]
- 40.Holmberg A, et al. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis. 2005;26(3):501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- 41.Jain PK, Huang WY, El-Sayed MA. On the universal scaling behavior of the distance decay of plasmon coupling in metal nanoparticle pairs: A plasmon ruler equation. Nano Letters. 2007;7(7):2080–2088. [Google Scholar]
- 42.Sonnichsen C, et al. Spectroscopy of single metallic nanoparticles using total internal reflection microscopy. Applied Physics Letters. 2000;77(19):2949–2951. [Google Scholar]
- 43.Johnson PB, Christy RW. Physical Review B. 1972;6:4370. [Google Scholar]
- 44.Khlebtsov B, et al. Absorption and scattering of light by a dimer of metal nanospheres: comparison of dipole and multipole approaches. Nanotechnology. 2006;17(5):1437–1445. [Google Scholar]
- 45.Atay T, Song JH, Nurmikko AV. Strongly interacting plasmon nanoparticle pairs: From dipole-dipole interaction to conductively coupled regime. Nano Letters. 2004;4(9):1627–1631. [Google Scholar]
- 46.Lassiter JB, et al. Close encounters between two nanoshells. Nano Letters. 2008;8(4):1212–1218. doi: 10.1021/nl080271o. [DOI] [PubMed] [Google Scholar]
- 47.Vallee F. Optical Properties of Metallic Nanoparticles, in Nanomaterials and Nanochemistry. In: Brechignac C, Houdy P, M L, editors. Berlin: Springer; 2008. pp. 197–227. [Google Scholar]
- 48.Tokuhisa H, et al. Preparation and characterization of dendrimer monolayers and dendrimer-alkanethiol mixed monolayers adsorbed to gold. Journal of the American Chemical Society. 1998;120(18):4492–4501. [Google Scholar]
- 49.Rivetti C, Walker C, Bustamante C. Polymer chain statistics and conformational analysis of DNA molecules with bends or sections of different flexibility. Journal of Molecular Biology. 1998;280(1):41–59. doi: 10.1006/jmbi.1998.1830. [DOI] [PubMed] [Google Scholar]
- 50.Smith SB, Cui YJ, Bustamante C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271(5250):795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 51.Matthew-Fenn RS, Das R, B HPA. Remeasuring the Double Helix. Science. 2008;322:446–449. doi: 10.1126/science.1158881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.