Abstract
Cytochrome c oxidase (COX) is composed of 13 subunits, of which COX I, II, and III are encoded by a mitochondrial gene. COX I and II function as the main catalytic components, but the function of COX III is unclear. Because myocardial ischemia affects mitochondrial oxidative metabolism, we hypothesized that COX activity and expression would be affected during postischemic cardiomyopathy. This hypothesis was tested in a monkey model following myocardial infarction (MI) and subsequent pacing-induced heart failure (HF). In this model, COX I protein expression was decreased threefold after MI and fourfold after HF (P < 0.05 vs. sham), whereas COX II expression remained unchanged. COX III protein expression increased 5-fold after MI and further increased 10-fold after HF compared with sham (P < 0.05 vs. sham). The physiological impact of COX III regulation was examined in vitro. Overexpression of COX III in mitochondria of HL-1 cells resulted in an 80% decrease in COX I, 60% decrease in global COX activity, 60% decrease in cell viability, and threefold increase in apoptosis (P < 0.05). Oxidative stress induced by H2O2 significantly (P < 0.05) increased COX III expression. H2O2 decreased cell viability by 47 ± 3% upon overexpression of COX III, but only by 12 ± 5% in control conditions (P < 0.05). We conclude that ischemic stress in vivo and oxidative stress in vitro lead to upregulation of COX III, followed by downregulation of COX I expression, impaired COX oxidative activity, and increased apoptosis. Therefore, upregulation of COX III may contribute to the increased susceptibility to apoptosis following MI and subsequent HF.
Keywords: cytochrome oxidase, mitochondria, myocardial ischemia, oxidative stress
heart failure (HF) following myocardial infarction (MI) is characterized by extensive remodeling, including increased apoptosis and cell death. However, the molecular mechanisms that induce apoptosis and cell death in MI and HF are not fully elucidated. In previous studies, we showed that such mechanisms include regulation of the expression and activity of stress-responsive chaperones, autophagy pathway, and cell cycle regulators (7, 29, 35). In addition to these mechanisms, it was shown previously that the production of reactive oxygen species (ROS) during ischemia impairs the expression of genes encoded by the mitochondrial DNA and that a downregulation of expression and activity of enzymes composing the respiratory chain complexes plays an important role in the progression of left ventricular remodeling and failure after MI (14). For example, the enzyme activity of electron transport chain complex I (NADH-ubiquinone oxidoreductase), III (ubiquinol-cytochrome c oxidoreductase), and IV [cytochrome c oxidase (COX)] decreased in mice subjected to MI (14). However, the role of individual subunits in such conditions remains largely unknown.
In this study, we tested the hypothesis that MI followed by HF would affect the expression and activity of the COX complex. COX, the terminal oxidase of the mitochondrial electron transport chain, is composed of 13 subunits. The subunits COX I, II, and III are encoded by a single mitochondrial gene. COX I and II belong to the catalytic core, which is key for the assembly and the function of the complex. However, the function of COX III is not clear. It has been suggested that COX III, which is a hydrophobic protein, may be involved in the folding and/or stability of COX (22, 33), but it is not an essential element of the proton pump (10) and is not involved in proton translocation (17, 27). It has been demonstrated that COX III is involved in oxidative stress (36). For example, homocysteine-induced production of ROS in endothelial cells significantly increases COX III expression (26), although the functional consequences of such regulation were not determined in that condition. In addition, the role and regulation of COX III in cardiac disease states are not understood. Therefore, we specifically tested whether COX III participates in stress-induced apoptosis and, thereby, contributes to myocardial remodeling and dysfunction following MI and HF. This hypothesis was tested first in a monkey model of MI followed by HF and, subsequently, more mechanistically by manipulation of COX III expression in HL-1 myocytes. Our results demonstrate that mitochondrial overexpression of COX III occurs after MI and HF and results in decreased cell viability, which suggests that COX III plays an important role in cardiac apoptosis.
MATERIALS AND METHODS
Animal model.
MI was induced in 14 young male monkeys (Macaca fascicularis, 6.3 ± 0.4 yr old, 5.5 ± 0.4 kg body wt) by ligation of the left anterior descending coronary artery in the heart. At 2 mo after MI, nine of these monkeys were subjected to 3 wk of pacing-induced (270 beats/min) HF. Five sham monkeys were used as controls. After anesthesia with pentobarbital sodium (50 mg/kg), the left ventricular tissue was excised and dissected into three portions representing infarcted, adjacent, and remote areas, respectively, on the basis of visual inspection of the tissue, as we described previously (18). Samples were frozen rapidly in liquid nitrogen and stored at −80°C. This investigation conforms to the National Institutes of Health Guide for the Care and Use of Laboratory Animals [Department of Health and Human Services Publication (NIH) No. 83-23, revised 1996]. All animal care and use protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Medicine and Dentistry of New Jersey, New Jersey Medical School.
Plasmid construction.
Site-directed mutagenesis was performed with the QuikChange multisite-directed mutagenesis kit (Stratagene, La Jolla, CA) in 12 sites of the mouse COX III coding sequence. A 25-amino acid-long NH2-terminal mitochondrial target peptide was fused to the mutated COX III sequence, together with a COOH-terminal FLAG tag. This sequence was ligated into the pEF-BOS vector (a gift from Dr. Nagata) after digestion of the Xba I site according to the method of Mizushima and Nagata (23).
Cell culture.
All the experiments were performed in triplicate from at least three different cultures. The mouse myocyte cell line HL-1 (a gift from Dr. Claycomb) was cultured as previously described (6). Transfection was conducted using Lipofectamine 2000 (Invitrogen). At 24 h after transfection, the cells were treated with different doses of H2O2 (Sigma-Aldrich, St. Louis, MO) for 24 h. COX activity was measured from cell extracts by the single-wavelength spectrophotometric assay using Complex IV Microplate kits (MitoScience) according to the manufacturer's instructions. Cell viability was assessed by trypan blue staining. Caspase-3 activity was measured with the caspase-3 activity detection kit (Molecular Probe) according to the manufacturer's instruction. Briefly, cells were harvested and washed with PBS, protein was extracted, and 50 μl of 2× substrate working solution were added in a 96-well microplate. Fluorescence (∼342-nm excitation and 441-nm emission) was measured after incubation at room temperature for 30 min. For detection of ROS, the cells were loaded with 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA, 100 μmol/l; Molecular Probes) for 30 min at 37°C in the dark. Cells were trypsinized for 2 min, and the reaction was stopped after addition of PBS-10% FCS. After centrifugation, cells were washed twice with PBS. Formation of ROS was detected by fluorescence (485-nm excitation and 530-nm emission). DNA fragmentation of cells was detected in situ by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) with the ApopTag peroxidase in situ apoptosis detection kit (Chemicon International). Cells grown on coverslips (1 × 105 cells/well of an 8-well plate) were washed with PBS, fixed in paraformaldehyde solution (4% in PBS) for 30 min at room temperature, and postfixed in precooled ethanol-acetic acid (2:1) for 5 min at −20°C. After they were washed with PBS, cells were quenched in 3% H2O2 for 5 min at room temperature and incubated with the terminal deoxynucleotidyl transferase for 1 h at 37°C. After addition of anti-digoxigenin peroxidase conjugate, the coverslips were washed with PBS and counterstained in 0.5% methyl green.
Except for the dose-response experiments, all data were obtained after transfection with the lowest dose of plasmid that showed a significant difference in viability (4 μg). The higher doses of plasmid require an increased amount of the transfection reagent Lipofectamine, which would potentially be toxic to the cells (2).
Detection of COX III transcript.
The expression of COX III in HL-1 myocytes was determined by RT-PCR and real-time quantitative PCR (qPCR). All the experiments were performed in triplicate from at least three different cultures. Total RNA was extracted using the phenol-chloroform method (5). After reverse transcription, PCR analysis of COX III was performed using the forward primer 5′-caaggccaccacactcctat-3′ and the reverse primer 5′-attcctgttggaggtcagca-3′, with 20 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 45 s, and extension at 72°C for 1 min. RT-PCR products were analyzed in a 1.5% agarose gel. qPCR was performed on a real-time PCR system (model 7300, Applied Biosystems, Foster City, CA) after reverse transcription of the RNA of interest with the TaqMan RT kit (Applied Biosystems) followed by qPCR with the SYBR Green PCR Master Mix (Applied Biosystems). Expression of β-actin was measured as an internal loading control.
Immunocytochemistry.
The localization of COX III was detected using MitoTracker Red (Molecular Probes) and an anti-FLAG-FITC antibody (Sigma). Briefly, cells were incubated with 100 nM MitoTracker Red for 30 min at 37°C and washed with PBS. Cell fixation and permeabilization were performed by incubation in cooled methanol for 10 min followed by cooled acetone for 1 min; then the cells were washed with PBS. Cells were blocked by incubation with 1% BSA for 10 min at room temperature. Then the cells were incubated with a monoclonal anti-FLAG-FITC antibody at room temperature for 1 h. Images were acquired with a fluorescent microscope (Olympus).
Western blotting.
Proteins were extracted with a buffer [20 mM Tris·HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, and 1 mM β-glycerophosphate] supplemented with protease and phosphatase inhibitors. Protein concentration was measured with the bicinchoninic acid reagent (Pierce Biotechnology). Proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, incubated with primary anti-COX I, II, or III antibody (all at 1:1,000 dilution; Invitrogen, Carlsbad, CA) or anti-cleaved (Asp175) caspase-3 antibody (1:1,000 dilution; Cell Signaling Technology, Danvers, MA) as recommended by the manufacturer, and detected by chemiluminescence. Protein abundance was determined with Quantity One software after the films were scanned on a densitometer (model GS-800, Bio-Rad, Hercules, CA).
Statistical analysis.
Values are means ± SE. Statistical analysis was performed using a two-tailed unpaired Student's t-test. A multigroup analysis was also performed by ANOVA with post hoc Tukey's correction. For Fig. 3, repeated-measures designs were implemented, using SAS, with the three separate times treated as the repeated measure. In each case, treatment as class variable was defined as either vector or COX III overexpressed (pEF-BOS-C8C3F). In each case, appropriate tests for equality of variance showed no evidence of the fact that the variances are unequal over the time-by-treatment classes. The true variances were therefore assumed to be equal in each case over the time-by-treatment classes. The parsimonious generalized linear models were obtained with COX I expression, COX II expression, or COX activity as the dependent variable and time and treatment with interaction as the independent variables. When two variables changed at different doses, e.g., Fig. 7, a two-way ANOVA was used. Differences were considered significant for P < 0.05.
Fig. 3.
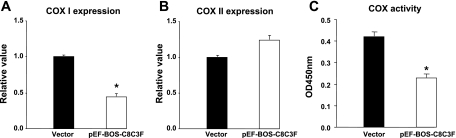
Effects of COX III overexpression on level of COX I and II and total activity of COX. Compared with cells transfected with empty vector, overexpression of COX III (4 μg) resulted in a decrease (P < 0.05) in COX I expression (A), a slight, but not significant (P = 0.09), increase in COX II expression (B), and a significant decrease in total COX activity (C). Values are means ± SE for experiments performed in triplicate. *P < 0.05 vs. vector.
Fig. 7.
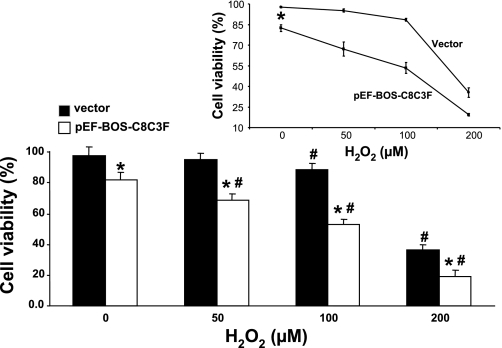
Decreased cell viability upon COX III overexpression in the presence of H2O2 in HL-1 cells. Viability of cells transfected with the empty vector or with 4 μg of pEF-BOS-C8C3F plasmid was evaluated by trypan blue in response to oxidative stress induced by increasing concentrations of H2O2 (0, 50, 100 and 200 μM). Values are means ± SE for experiments performed in triplicate. *P < 0.05 vs. corresponding vector. #P < 0.05 vs. corresponding group at 0 μM H2O2. Inset: average H2O2-induced decrease in cell viability was greater with COX III overexpression than in the presence of the empty vector (*P < 0.05 vs. vector).
RESULTS
Expression of COX I, II, and III in monkey hearts with chronic MI and HF following MI.
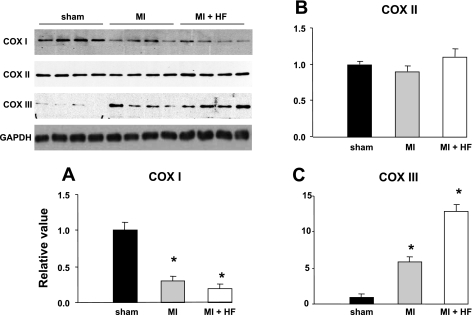
We investigated the alterations in expression of mitochondrion-encoded COX I, II, and III in five monkeys after chronic (2 mo) MI and in nine additional monkeys with superimposed HF induced by 3 wk of regional ventricular pacing following 2 mo of MI. The physiological characteristics of this model have been described previously (29). We investigated specifically the area adjacent to MI to avoid necrotic tissue. In that area, apoptosis, assessed by TUNEL, was increased from 0.11 ± 0.13 (sham, n = 4) to 0.62 ± 0.04 cells/μm2 in MI and further increased to 3.95 ± 1.6 cells/μm2 in HF (both P < 0.05 vs. sham). By Western blotting, COX I protein expression was significantly (P < 0.05) decreased (Fig. 1A), whereas COX II expression remained unchanged (Fig. 1B), both after MI and after HF. Reciprocally, COX III protein expression increased 5-fold after MI and further increased 10-fold after HF (Fig. 1C) compared with sham (both P < 0.05 vs. sham).
Fig. 1.
Regulation of cytochrome c oxidase (COX) subunits during ischemic cardiomyopathy. COX I, II, and III proteins from the heart of sham-treated monkeys (n = 4), a monkey model of myocardial infarction (MI, n = 6), and a monkey model of myocardial infarction and subsequent heart failure (MI + HF, n = 9) were detected by immunoblotting, and protein abundance was calculated. A: COX I was significantly (P < 0.05) decreased in MI and HF monkeys. B: COX II remained unchanged in MI and HF monkeys. C: COX III was increased significantly, by 5-fold in MI vs. sham and further increased 2-fold in HF. Values are means ± SE. *P < 0.05 vs. sham.
Overexpression of COX III impairs COX activity.
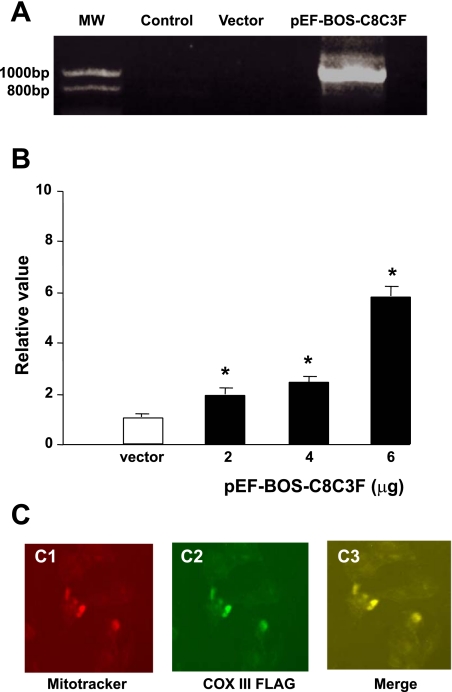
For further evaluation of the impact of COX III upregulation, a plasmid containing a tagged COX III sequence with a mitochondrial target peptide was transfected in mitochondria of HL-1 myocytes. Mitochondrial expression of the plasmid was confirmed by RT-PCR and immunocytochemistry. RT-PCR results demonstrated that the plasmid was successfully expressed in HL-1 cells (Fig. 2A). In addition, COX III expression increased dose dependently according to the amount of the plasmid transfected as determined by qPCR (Fig. 2B). Using the mitochondrial probe MitoTracker Red and an anti-FLAG-FITC antibody, we confirmed that overexpression of COX III was localized in the mitochondria (Fig. 2C).
Fig. 2.
Overexpression and localization of COX III in mitochondria of HL-1 cells. A: agarose gel showing that the pEF-BOS-COX VIII-COX III-FLAG plasmid (pEF-BOS-C8C3F, 4 μg) was successfully transfected and overexpressed in HL-1 cells as detected by RT-PCR. Total length of the DNA sequence is 880 bp, which consists of 150 bp of COX VIII (mitochondrial targeted peptide), 780 bp of site-directed mutated COX III genes, and 30 bp of FLAG genes. MW, molecular weight. B: dose-dependent overexpression of COX III as a function of plasmid amount was detected by quantitative real-time PCR (qPCR). Different amounts of PEF-BOS-C8C3F plasmid (2, 4 and 6 μg) were transfected into 6-cm plates and cultured for 48 h. β-Actin was used as an internal control. Each condition was measured in triplicate. Values are means ± SE for the triplicates. *P < 0.05 vs. vector. C: localization of overexpressed COX III (4 μg) was confirmed using a mitochondrial probe, MitoTracker Red (C1), and anti-FLAG-FITC antibody (C2) followed by merging of both images (C3), which demonstrates that overexpressed COX III was localized in mitochondria.
Compared with the cells transfected with the empty vector, overexpression of COX III with 4 μg of plasmid resulted in a 50% decrease (P < 0.05) in the level of COX I transcript expression (Fig. 3A). Expression of COX II showed a slight, but not significant (P = 0.09), increase (Fig. 3B). The total catalytic activity of the COX complex was decreased by ∼50% (P < 0.05) in cells overexpressing COX III compared with controls (Fig. 3C), suggesting that COX III is a negative regulator of COX.
COX III overexpression promotes cell death.
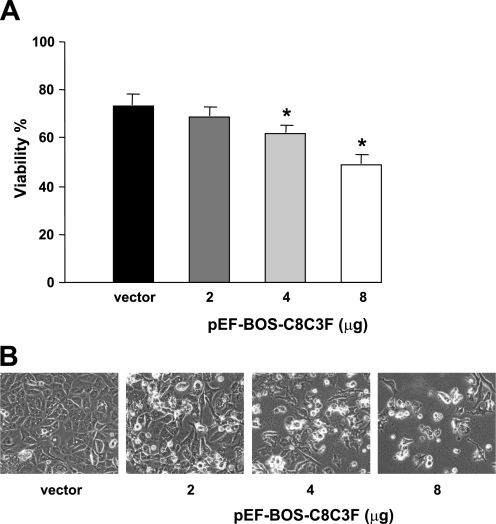
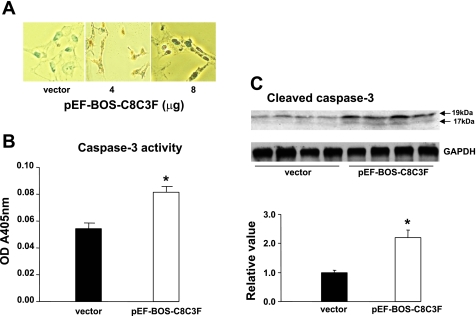
We determined next how the decrease in COX activity with COX III overexpression would affect HL-1 cell viability and susceptibility to apoptosis. Cell viability was measured by exclusion of trypan blue (Fig. 4A). On exposure to different amounts of the COX III plasmid, cell viability showed a dose-dependent decrease compared with control conditions in the presence of the empty vector (Fig. 4B). We determined whether such a decrease in cell viability would be associated with increased susceptibility to apoptosis, which was measured by TUNEL and by activation of caspase-3. TUNEL-positive cells were detected upon overexpression of COX III but not in control conditions (Fig. 5A). Caspase-3 activity, as measured by ELISA, significantly increased by 40% in COX III-overexpressing cells compared with the control group (Fig. 5B). Similarly, caspase-3 cleavage, which reflects the activation of the apoptotic effector, was doubled upon COX III overexpression (Fig. 5C).
Fig. 4.
Dose-dependent decrease in cell viability induced by COX III overexpression. A: cell viability, as measured by trypan blue, progressively decreased in response to increased concentrations (2, 4, and 8 μg) of pEF-BOS-C8C3F plasmid. Values are means ± SE for experiments performed in triplicate. *P < 0.05 vs. vector. B: changes in cellular morphology were induced by COX III overexpression as a consequence of decreased viability.
Fig. 5.
Overexpression of COX III induces apoptosis in HL-1 cardiac myocytes. A: terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) analysis of HL-1 cells exposed to increased concentrations (4 and 8 μg) of pEF-BOS-C8C3F plasmid. B: activity of caspase-3, as measured by ELISA, in HL-1 cells exposed to empty vector or 4 μg of pEF-BOS-C8C3F plasmid. OD A450nm, optical density absorbance at 450 nm. Values are means ± SE for experiments performed in triplicate. *P < 0.05 vs. vector. C: caspase-3 cleavage, as measured by Western blotting, in HL-1 cells exposed to empty vector or 4 μg of pEF-BOS-C8C3F plasmid. Values are means ± SE for experiments performed in triplicate. *P < 0.05 vs. vector.
COX III increases oxidative stress.
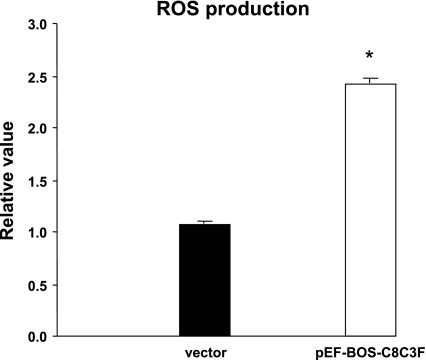
Dysfunction of COX catalytic activity impairs the overall activity of the oxidative phosphorylation chain, which results in increased production of ROS. We therefore determined whether COX III overexpression would result in increased ROS production in HL-1 cells. ROS production was quantified using the fluorescent probe H2DCF-DA. As demonstrated in Fig. 6, ROS production was more than doubled in cells infected with the COX III plasmid compared with the empty vector. To determine whether COX III would alter the cellular response to oxidative stress, HL-1 cells were transfected with the COX III construct or the empty vector and incubated for 24 h in the presence of different concentrations of H2O2 (0, 50, 100, and 200 μM). As shown in Fig. 7, without H2O2 treatment, a greater (P < 0.05) decrease in cell viability was observed in the presence of COX III overexpression than in the presence of the empty vector. A dose-dependent, significant (P < 0.05) decrease in cell viability with H2O2 treatment was also observed with COX III overexpression, whereas in the vector group, the decrease in cell viability was significant only after exposure to 100 μM H2O2. The average H2O2-induced decrease in cell viability was greater with COX III overexpression than in the vector group (P < 0.05, by 2-way ANOVA). These data indicate that COX III overexpression decreases the cell's tolerance to oxidative stress.
Fig. 6.
Reactive oxygen species (ROS) production in HL-1 cells exposed to COX III overexpression. ROS production was detected using the fluorescent marker 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA). ROS production was increased 2.4-fold by transfection with 4 μg of pEF-BOS-C8C3F plasmid compared with the empty vector. Values are means ± SE for experiments performed in triplicate. *P < 0.05 vs. vector.
COX III is a sensitive marker of oxidative stress.
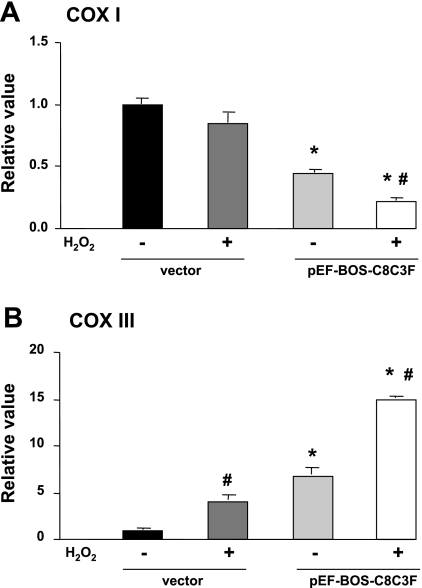
We have shown that COX III overexpression in HL-1 cells increases the sensitivity to oxidative stress, which is accompanied by decreased cell survival. Next, we tested the reciprocal situation, i.e., whether COX III expression is regulated by oxidative stress. HL-1 cells were exposed to 200 μM H2O2 for 24 h; then the transcript expression of COX I and III was measured by qPCR in the residual viable cells. In these conditions, H2O2 led to a threefold increase in COX III transcript and a trend to a decrease in COX I expression (Fig. 8). When this experiment was repeated with overexpression of COX III, the transcript level of COX III was increased sixfold compared with control (Fig. 8B); this increase was accompanied by a significant decrease in COX I expression (Fig. 8A). Addition of H2O2 upon COX III overexpression resulted in a further doubling of the COX III transcript (Fig. 8B), which was accompanied by a further and significant reduction of COX I mRNA expression (Fig. 8A). Taken together, these results suggest that COX III transcript expression is directly increased by oxidative stress, whereas the decreased COX I expression results from COX III overexpression, rather than the oxidative stress itself.
Fig. 8.
Expression of COX I and III in COX III transcripts in HL-1 cells treated with or without 200 μM H2O2 and in the presence or absence of COX III overexpression. A: transcription of COX I, as measured by quantitative real-time PCR, was not changed in the vector group treated with H2O2 but was significantly decreased upon COX III overexpression (4 μg of plasmid). B: COX III transcription was increased significantly after treatment with H2O2 in transfected (4 μg of plasmid) and nontransfected (vector) controls compared with the corresponding groups without H2O2 treatment. Values are means ± SE for experiments performed in triplicate. *P < 0.05 vs. corresponding vector. #P < 0.05 vs. corresponding group without H2O2.
DISCUSSION
Our study demonstrates a specific regulation of COX III in the monkey hearts following MI and HF. In particular, we demonstrate for the first time a reciprocal regulation of COX III and I in the context of myocardial ischemic stress. Previous studies indicated that COX I is the core subunit regulating the function of COX, whereas COX III negatively regulates the activity of COX. Because of its structure and complexity, COX is an excellent model for the study of holoenzyme assembly and mitochondrial biogenesis and function in response to physiological stimuli (30). The stoichiometry of COX subunits dictates the activity of the COX complex. A differential regulation pattern of COX subunits has been shown in a previous study, in which thyroid hormone decreased COX III mRNA but increased COX I transcripts and overall COX activity in the heart (30). We show the reciprocal condition, i.e., an increase in COX III and a simultaneous decrease in COX I resulting in decreased COX activity. Therefore, COX activity seems to be dictated by the COX I-to-COX III ratio.
To detect the function of COX III and its effect on COX activity, we overexpressed COX III in mitochondria of HL-1 myocytes. Reciprocally, examination of the effects of COX I overexpression on COX III level is complicated by the fact that the COX I sequence contains 18 stop codons (vs. 12 stop codons for the COX III sequence), which need to be mutated before an RNA that can be translated into a functional protein can be produced. An alternative approach to our experiments of overexpression would be the silencing techniques; however, there are no practical methods to knock down COX III in mitochondria, because small interfering RNA, which is an effective method to downregulate the gene expression of nuclear encoded genes, has not been successful in knocking down the expression of mitochondrial genes. Therefore, we could not block the effects of COX III.
We also demonstrate that COX III overexpression results in a decreased abundance of COX I and a decrease in COX activity, which is accompanied by decreased cell viability, increased apoptosis, and a greater sensitivity to oxidative stress in transfected cells. In addition, overexpression of COX III induced ROS production. HL-1 cardiac myocytes treated with a proapoptotic agent showed an increased expression of COX III, followed by increased ROS production and increased apoptosis. Together, our results suggest that COX III plays an important role in ROS-mediated apoptosis in cardiac myocytes. It is not known whether the mitochondrial dysfunction observed here directly results from an accumulation of COX III per se or, rather, from an imbalance between COX I and COX III expression. It seems that the latter hypothesis might prevail, because mutations impairing COX III function actually decrease proton flux through the COX complex and lead to a “functional suicide” of the complex (13). It is therefore likely that a correct stoichiometry between the different components of the complex is paramount to its optimal function. Although COX II shows a nonsignificant trend of upregulation upon COX III overexpression, this subunit is certainly not downregulated and, therefore, cannot explain the loss of COX activity.
Our experiments suggest that COX III expression level, rather than oxidative stress per se, is responsible for regulating the expression level of COX I. Indeed, we show that COX III transcript expression is directly increased by oxidative stress induced by H2O2, whereas COX I expression remains stable under oxidative stress. Instead, decreased COX I expression is observed upon COX III overexpression, whether oxidative stress is applied or not. Mitochondria are a primary source of ROS in the cell and also a major target for ROS-mediated damage, e.g., apoptosis. The mitochondrial electron transport chain is the key site for the production of intracellular ROS, and inhibition of one or more of the complexes of the electron transport chain may increase the rate of mitochondrial ROS generation (15). Complexes I, II, and III in the electron transport chain are reported to be the main source of ROS production in mitochondria (1, 3, 8, 12, 19, 20, 32, 34, 37). However, there are few studies regarding the relationship between complex IV (COX) and the production of ROS. This relationship is, however, of primary importance, because oxidative stress and mitochondrial oxidative damage are directly involved in several forms of heart disease. ROS in the heart are produced by different sources under various pathophysiological conditions, including ischemia-reperfusion injury, MI, and cardiomyopathy (4, 11, 16, 21). Potential sources for the generation of ROS include not only the dysfunctional mitochondrial electron transport chain, but also arachidonic acid metabolism, neutrophil infiltration and activation, xanthine-xanthine oxidase, and catecholamines (4). The increased production of ROS activates the Bcl-2 family proteins and the caspases of the mitochondrial pathway of apoptosis and various other intracellular signaling pathways that lead to cellular hypertrophy, apoptosis, necrosis, and, eventually, heart dysfunction (11, 21, 24). In reperfused myocardium following ischemia, an increase in oxidative stress and a decrease in antioxidant defense have been shown to be responsible for cardiomyocyte death and cardiac dysfunction (9, 25, 28, 31). Reciprocally, the cardioprotection conferred by ischemic preconditioning is associated with lower ROS production from mitochondria (38). Whether the COX I-to-COX III ratio is also preserved in that condition remains to be determined.
In conclusion, the ischemic stress induced by MI in vivo leads to specific upregulation of COX III, which can be reproduced in vitro in the presence of oxidative stress. Overexpression of COX III is sufficient to downregulate the catalytic subunit of COX I, impair COX enzyme activity, and increase cell death by apoptosis. Thus upregulation of COX III may contribute to the increased apoptosis following chronic MI and HF.
GRANTS
This work was supported by National Institutes of Health Grants AG-027211, HL-033107, HL-059139, HL-069752, HL-095888, HL-069020, AG-023137, and AG-014121.
ACKNOWLEDGMENTS
We are grateful to Dr. D. Pain (Department of Pharmacology and Physiology, University of Medicine and Dentistry of New Jersey, New Jersey Medical School) for helpful comments.
REFERENCES
- 1.Barja G. Mitochondrial oxygen radical generation and leak: sites of production in states 4 and 3, organ specificity, and relation to aging and longevity. J Bioenerg Biomembr 31: 347–366, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Bauer M, Kristensen BW, Meyer M, Gasser T, Widmer HR, Zimmer J, Ueffing M. Toxic effects of lipid-mediated gene transfer in ventral mesencephalic explant cultures. Basic Clin Pharmacol Toxicol 98: 395–400, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Boveris A. Mitochondrial production of hydrogen peroxide in Saccharomyces cerevisiae. Acta Physiol Latinoam 26: 303–309, 1976 [PubMed] [Google Scholar]
- 4.Cai H, Harrison DG. Endothelial dysfunction in cardiovascular diseases: the role of oxidant stress. Circ Res 87: 840–844, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159, 1987 [DOI] [PubMed] [Google Scholar]
- 6.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ., Jr HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci USA 95: 2979–2984, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danan IJ, Rashed ER, Depre C. Therapeutic potential of H11 kinase for the ischemic heart. Cardiovasc Drug Rev 25: 14–29, 2007 [DOI] [PubMed] [Google Scholar]
- 8.Dionisi O, Galeotti T, Terranova T, Azzi A. Superoxide radicals and hydrogen peroxide formation in mitochondria from normal and neoplastic tissues. Biochim Biophys Acta 403: 292–300, 1975 [DOI] [PubMed] [Google Scholar]
- 9.Elsasser A, Suzuki K, Lorenz-Meyer S, Bode C, Schaper J. The role of apoptosis in myocardial ischemia: a critical appraisal. Basic Res Cardiol 96: 219–226, 2001 [DOI] [PubMed] [Google Scholar]
- 10.Haltia T, Saraste M, Wikstrom M. Subunit III of cytochrome c oxidase is not involved in proton translocation: a site-directed mutagenesis study. EMBO J 10: 2015–2021, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haunstetter A, Izumo S. Apoptosis: basic mechanisms and implications for cardiovascular disease. Circ Res 82: 1111–1129, 1998 [DOI] [PubMed] [Google Scholar]
- 12.Herrero A, Barja G. Localization of the site of oxygen radical generation inside the complex I of heart and nonsynaptic brain mammalian mitochondria. J Bioenerg Biomembr 32: 609–615, 2000 [DOI] [PubMed] [Google Scholar]
- 13.Hosler JP. The influence of subunit III of cytochrome c oxidase on the D pathway, the proton exit pathway and mechanism-based inactivation in subunit I. Biochim Biophys Acta 1655: 332–339, 2004 [DOI] [PubMed] [Google Scholar]
- 14.Ide T, Tsutsui H, Hayashidani S, Kang D, Suematsu N, Nakamura K, Utsumi H, Hamasaki N, Takeshita A. Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ Res 88: 529–535, 2001 [DOI] [PubMed] [Google Scholar]
- 15.Jacobson J, Duchen MR, Hothersall J, Clark JB, Heales SJ. Induction of mitochondrial oxidative stress in astrocytes by nitric oxide precedes disruption of energy metabolism. J Neurochem 95: 388–395, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Kaul N, Siveski-Iliskovic N, Hill M, Slezak J, Singal PK. Free radicals and the heart. J Pharmacol Toxicol Methods 30: 55–67, 1993 [DOI] [PubMed] [Google Scholar]
- 17.Kennaway NG, Carrero-Valenzuela RD, Ewart G, Balan VK, Lightowlers R, Zhang YZ, Powell BR, Capaldi RA, Buist NR. Isoforms of mammalian cytochrome c oxidase: correlation with human cytochrome c oxidase deficiency. Pediatr Res 28: 529–535, 1990 [DOI] [PubMed] [Google Scholar]
- 18.Kim YK, Kim SJ, Kramer CM, Yatani A, Takagi G, Mankad S, Szigeti GP, Singh D, Bishop SP, Shannon RP, Vatner DE, Vatner SF. Altered excitation-contraction coupling in myocytes from remodeled myocardium after chronic myocardial infarction. J Mol Cell Cardiol 34: 63–73, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Lenaz G. The mitochondrial production of reactive oxygen species: mechanisms and implications in human pathology. IUBMB Life 52: 159–164, 2001 [DOI] [PubMed] [Google Scholar]
- 20.Loschen G, Azzi A, Richter C, Flohe L. Superoxide radicals as precursors of mitochondrial hydrogen peroxide. FEBS Lett 42: 68–72, 1974 [DOI] [PubMed] [Google Scholar]
- 21.MacLellan WR, Schneider MD. Death by design. Programmed cell death in cardiovascular biology and disease. Circ Res 81: 137–144, 1997 [DOI] [PubMed] [Google Scholar]
- 22.Meunier B, Taanman JW. Mutations of cytochrome c oxidase subunits 1 and 3 in Saccharomyces cerevisiae: assembly defect and compensation. Biochim Biophys Acta 1554: 101–107, 2002 [DOI] [PubMed] [Google Scholar]
- 23.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res 18: 5322, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moudgil R, Menon V, Xu Y, Musat-Marcu S, Kumar D, Jugdutt BI. Postischemic apoptosis and functional recovery after angiotensin II type 1 receptor blockade in isolated working rat hearts. J Hypertens 19: 1121–1129, 2001 [DOI] [PubMed] [Google Scholar]
- 25.Palace V, Kumar D, Hill MF, Khaper N, Singal PK. Regional differences in non-enzymatic antioxidants in the heart under control and oxidative stress conditions. J Mol Cell Cardiol 31: 193–202, 1999 [DOI] [PubMed] [Google Scholar]
- 26.Perez-de-Arce K, Foncea R, Leighton F. Reactive oxygen species mediates homocysteine-induced mitochondrial biogenesis in human endothelial cells: modulation by antioxidants. Biochem Biophys Res Commun 338: 1103–1109, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem 65: 563–607, 1996 [DOI] [PubMed] [Google Scholar]
- 28.Qin F, Shite J, Mao W, Liang CS. Selegiline attenuates cardiac oxidative stress and apoptosis in heart failure: association with improvement of cardiac function. Eur J Pharmacol 461: 149–158, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Qiu H, Dai H, Jain K, Shah R, Hong C, Pain J, Tian B, Vatner DE, Vatner SF, Depre C. Characterization of a novel cardiac isoform of the cell cycle-related kinase that is regulated during heart failure. J Biol Chem 283: 22157–22165, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheehan TE, Kumar PA, Hood DA. Tissue-specific regulation of cytochrome c oxidase subunit expression by thyroid hormone. Am J Physiol Endocrinol Metab 286: E968–E974, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Singal PK, Khaper N, Palace V, Kumar D. The role of oxidative stress in the genesis of heart disease. Cardiovasc Res 40: 426–432, 1998 [DOI] [PubMed] [Google Scholar]
- 32.Sipos I, Tretter L, Adam-Vizi V. The production of reactive oxygen species in intact isolated nerve terminals is independent of the mitochondrial membrane potential. Neurochem Res 28: 1575–1581, 2003 [DOI] [PubMed] [Google Scholar]
- 33.Taanman JW, Kateeb I, Muntau AC, Jaksch M, Cohen N, Mandel H. A novel mutation in the deoxyguanosine kinase gene causing depletion of mitochondrial DNA. Ann Neurol 52: 237–239, 2002 [DOI] [PubMed] [Google Scholar]
- 34.Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J 191: 421–427, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan L, Sadoshima J, Vatner DE, Vatner SF. Autophagy: a novel protective mechanism in chronic ischemia. Cell Cycle 5: 1175–1177, 2006 [DOI] [PubMed] [Google Scholar]
- 36.You KR, Wen J, Lee ST, Kim DG. Cytochrome c oxidase subunit III: a molecular marker for N-(4-hydroxyphenyl)retinamise-induced oxidative stress in hepatoma cells. J Biol Chem 277: 3870–3877, 2002 [DOI] [PubMed] [Google Scholar]
- 37.Zhang L, Yu L, Yu CA. Generation of superoxide anion by succinate-cytochrome c reductase from bovine heart mitochondria. J Biol Chem 273: 33972–33976, 1998 [DOI] [PubMed] [Google Scholar]
- 38.Zhu BH, Ueno M, Matsushita T, Fujisawa H, Seriu N, Nishikawa T, Nishimura Y, Hosokawa M. Effects of aging and blood pressure on the structure of the thoracic aorta in SAM mice: a model of age-associated degenerative vascular changes. Exp Gerontol 36: 111–124, 2001 [DOI] [PubMed] [Google Scholar]