Abstract
IGF-I increases skeletal muscle mass, but whether IGF-I increases type IIb myosin heavy chain (MyHC) transcriptional activity is not known. C2C12 myotubes were cultured with or without IGF-I to determine whether IGF-I increases type IIb MyHC promoter activity, and if so, what region of the promoter might IGF-I signaling regulate. At differentiation days 3 and 4, IGF-I increased type IIb MyHC mRNA and mouse 3.0-kb type IIb MyHC promoter activity. Deletion construct studies identified a potential IGF-I-responsive region between 1.25 and 1.2 kb of the type IIb MyHC promoter, which contained an exact 6-bp T-cell factor/lymphoid enhancer factor (Tcf/Lef) binding site at position −1206 to −1201. Site-specific mutation of the putative Tcf/Lef binding site reduced IGF-I-induced 1.3-kb type IIb MyHC promoter activity. To identify potential IGF-I signaling molecules, the phosphatidylinositol 3-kinase (PI3K) inhibitors wortmannin and LY-294002 were both found to markedly attenuate IGF-I activation of the 1.3-kb type IIb MyHC promoter. Downstream signaling of IGF-I can phosphorylate and inactivate GSK-3β, thereby enhancing β-catenin protein. The GSK-3β inhibitor, LiCl, dramatically enhanced IGF-I induction of the 1.3-kb type IIb MyHC promoter, and constitutively active GSK-3β attenuated IGF-I-induced 1.3-kb type IIb MyHC promoter activity. Finally, IGF-I increased nuclear β-catenin protein, and small interfering RNA knockdown of β-catenin attenuated IGF-I-induced 1.3-kb type IIb MyHC promoter activity and type IIb MyHC mRNA. In summary, IGF-I stimulation of C2C12 myotubes increases mouse type IIb MyHC promoter activity, likely through signaling of PI3K, GSK-3β, β-catenin, and a Tcf/Lef binding site at −1,206 to −1,201 bp in the promoter.
Keywords: insulin-like growth factor I, C2C12 myocytes, β-catenin
exogenous igf-i delivered either through osmotic pumps (1) or genetic overexpression (20) results in greater skeletal muscle mass in rodents. A similar response has been noted on application of IGF-I to cultured myotubes, where larger myotube diameters and higher protein contents have been reported (22, 33). To increase muscle mass, IGF-I would have to increase myosin heavy chain (MyHC) expression, because it is a major contractile protein in skeletal muscle. MyHC has multiple isoforms in limb skeletal muscle (MyHC I, IIa, IIx/d, and IIb), each with its own gene (24). Barton-Davis et al. (3) demonstrated that overexpression of IGF-I for 9 mo in extensor digitorum longus (EDL) muscles (primarily composed of type IIb fibers) prevented muscle atrophy and the loss of type IIb fibers in old mice. In transgenic mice that overexpress IGF-I in skeletal muscle, the cross-sectional diameter of type IIb fibers was increased (20). In addition, Flint et al. (10) demonstrated that IGF-I overexpression in denervated muscle prevents the loss of type IIb MyHC protein. We therefore chose to study whether IGF-I signaling increases type IIb MyHC promoter activity in mouse skeletal muscle. Our notion is that an IGF-I downstream target, such as type IIb MyHC, could serve as a source to identify a potential regulatory site for IGF-I interactions with other genes.
Under steady-state conditions, most agree that MyHC genes are regulated at the transcriptional/pretranslational level (2). Nuclear run-on assays indicate that the type IIb MyHC gene is transcriptionally activated as myoblasts fuse and mature to myotubes in tissue culture (7) and during whole skeletal muscle development (6). These observations have been confirmed and extended by others. Swoap (28) and Wheeler et al. (34) determined that the type IIb MyHC promoter has ∼35 times greater in vivo transcriptional activity in the rat tibialis anterior muscle [26–46% IIb fibers (9)] than in the slow-twitch soleus [0% IIb fibers (9)]. Furthermore, changes in contractile activity, e.g., muscle training or hindlimb unloading, can alter MyHC phenotype, and these changes at the protein level are preceded by changes at the mRNA level (4, 13, 15). Nonetheless, how IGF-I modulates type IIb MyHC and many other skeletal muscle genes remains unknown. To our knowledge, there are no data at the level of the type IIb MyHC promoter that support a role for IGF-I increasing type IIb MyHC mRNA. Therefore, the purpose of the current study was to investigate whether IGF-I increases type IIb MyHC promoter activity and potential signaling mechanism(s) associated with its increase.
METHODS
Cell culture.
All cell culture experiments were performed with C2C12 myoblasts (American Type Culture Collection CRL-1722) under standard conditions (37°C and 10% CO2) and maintained at a subconfluent density in growth medium (GM). GM consisted of Dulbecco's modified Eagle's medium (DMEM; Gibco), 20% (vol/vol) fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 40 μg/ml gentamicin. Transient transfections were performed using 0.078 pmol of plasmid DNA and Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations in antibiotic-free GM for 6 h. Differentiation was induced 18 h after transfection by replacing the GM with differentiation media [DM; DMEM, 2% (vol/vol) horse serum] or DM + IGF-I (250 ng/ml; PeproTech). DM and DM + IGF-I were replaced every 24 h. Dimethyl sulfoxide (vehicle), lithium chloride [10 mM (21)], wortmannin [100 nM (8)], LY-294002 [10 μM (32)], and rapamycin [1 μg/ml (25)] were purchased from Sigma Chemicals. The constitutively active glycogen synthase kinase-3β plasmid, pGSK-3A9 (caGSK-3β), was a gift from James R. Woodgett (University of Toronto, Toronto, ON, Canada) (26). The β-catenin antibody was purchased from Cell Signaling (no. 9581).
Generation of promoter reporter, 5′-deletion constructs, and site-directed mutagenesis.
The 2,567-base pair (bp) mouse type IIb MyHC promoter sequence published by Takeda et al. (29), accession number M92099, was used to generate the ∼3,000-bp (3.0-kb) type IIb MyHC promoter and subsequent constructs as follows. The published sequence was input into the University of California Santa Cruz Genome Bioinformatics database (http://genome.ucsc.edu) (17), and 414 additional bases at the 5′-end were retrieved. Thus, 2,981 bp of the 5′-flanking region (−2,968 bp − +13 bp relative to the transcription start site) of the mouse type IIb MyHC gene were polymerase chain reaction (PCR) amplified from purified bacterial artificial chromosome DNA. Briefly, searching the National Center for Biotechnology Information (NCBI) nucleotide data base revealed that clone RP23-294E23 (accession no. AL596129.27) contains the region of chromosome 11 with the MyHC IIb gene (myh4, GeneID: 17884) and its 5′-flanking region (M92099). The clone and the promoter were aligned via the NCBI Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov) for confirmation. The bacterial artificial chromosome containing clone RP23-294E23 was purchased from Invitrogen. PCR-based methods using AccuPrime Pfx DNA polymerase (Invitrogen) and primers with engineered Mlu I (5′-primers) and Sma I (3′-primers) restriction endonuclease sites for subsequent ligation were used to amplify the 3,000 bp type IIb MyHC promoter segment. Following restriction endonuclease digestion, the promoter clone was ligated into the multiple cloning site of the pGL3 Basic (Promega) reporter vector driving firefly luciferase expression, termed the 3.0-kb type IIb MyHC promoter reporter. Restriction enzyme digest and automated bidirectional sequencing (University of Missouri-Columbia DNA Core Facility) were used to confirm insert size, PCR fidelity, and insert orientation of the entire 2,981 bp. Nineteen 5′-deletion constructs were made using the 3.0-kb type IIb MyHC promoter construct as the template using primers with engineered Mlu I and Sma I restriction endonuclease sites and annealed to the reporter vector. Each 5′-deletion construct has a numerical designation referring to the 5′-promoter sequence most relative to the transcription start site, and the 3′-end of all constructs ends at +13 relative to the transcription start site.
Site-specific mutations on the 1.3-kb type IIb MyHC promoter were accomplished using the QuickChange II site-directed mutagenesis kit (Stratagene). Complementary DNA oligos were created changing three bases (−1206, −1204, −1202) from the target sequence in the center of the oligos (underlined in each sequence); forward primer sequence: GAACACTTTTCTTTCCGGTTCTTAGCCTAACACTTGGGG; reverse primer sequence: CCCCAAGTGTTAGGCTAAGAACCGGAAAGAAAAGTGTTC. Mutated plasmids were amplified by PCR, followed by the digestion of template 1.3-kb type IIb MyHC plasmid with Dpn I. Plasmids were transformed and grown up in One Shot Top10 competent cells (Invitrogen) and purified with the Qiafilter Plasmid Midi Kit (Qiagen).
Small interfering RNA transfections.
To optimize transfection conditions, C2C12 myoblasts were cotransfected with the wild-type 1.3-kb type IIb MyHC promoter (same concentration as previous experiments; 0.078 pmol) and increasing concentrations of either three different small interfering RNA (siRNA) constructs against mouse β-catenin (s63417; no. 1, s63418; no. 2, and s63419; no. 3), nontargeting, negative control siRNA (NT siRNA; 4390843), or GAPDH-positive control siRNA (4390849), all from Applied Biosystems (AB). Preliminary experiments on β-catenin siRNA constructs revealed that siRNA construct no. 3 (s63419) was the most effective of the three. Optimization experiments on β-catenin siRNA construct no. 3 revealed ∼80% knockdown of β-catenin mRNA with transfection of 3–9 nM (Fig. 7B); therefore 3 nM β-catenin siRNA construct no. 3 was used for the remaining siRNA experiments. For type IIb MyHC promoter activity and mRNA experiments with β-catenin siRNA, C2C12 myoblasts were cotransfected with either the wild-type or mutated 1.3-kb type IIb MyHC promoter and/or either 3 nM β-catenin siRNA or 25 nM NT siRNA using Lipofectamine 2000. At 24 h posttransfection, cells were washed with PBS and changed to DM or DM + IGF-I (250 ng/ml). Cells were harvested after 3 days of differentiation and analyzed for type IIb MyHC promoter luciferase activity or type IIb MyHC mRNA.
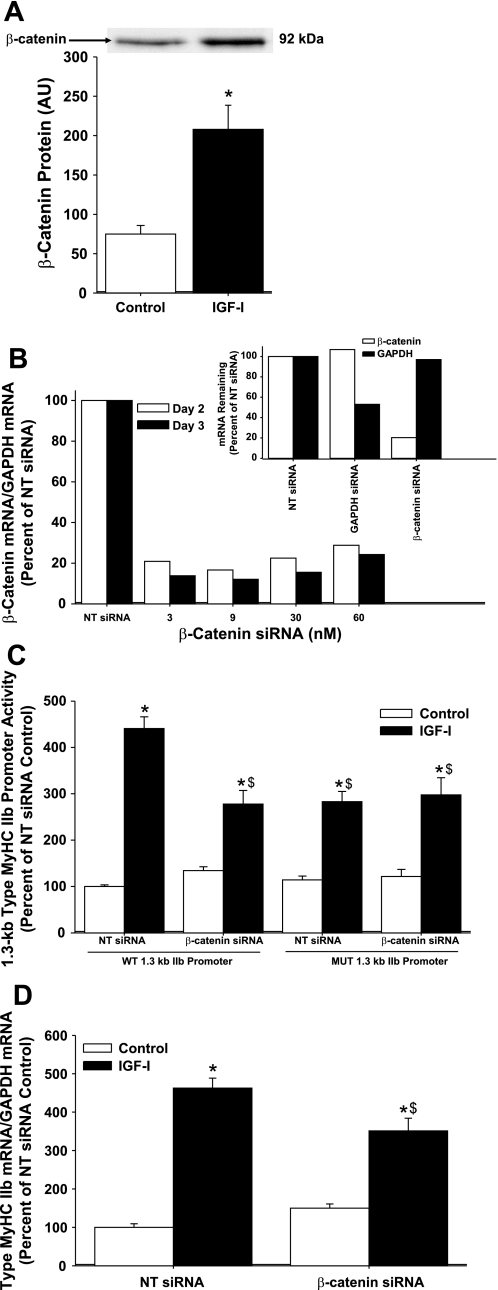
Fig. 7.
Role of β-catenin in IGF-I-induced type IIb MyHC transcriptional activity. A: enriched nuclear extracts from differentiating C2C12 myocytes were analyzed after 3 days of differentiation in DM (control) or DM with 250 ng/ml of IGF-I (IGF-I). Enriched nuclear extract protein was prepared, and equal amounts (10 μg each) were separated by SDS-PAGE; nuclear β-catenin protein levels were determined by Western blot analysis. IGF-I increased nuclear β-catenin 2.8-fold, compared with control. Representative immunoblots are depicted above data. Band intensity was quantified and is expressed in arbitrary units (AU) and reported as average ± SE, n = 4 per group. *P < 0.05 from control. B: optimization of β-catenin small interfering RNA (siRNA) transfection in differentiating C2C12 myocytes. β-Catenin mRNA and GAPDH mRNA were analyzed at 2 and 3 days after initiation of differentiation in C2C12 myocytes transfected with increasing concentrations of β-catenin siRNA (s63419; no. 3) or 25 nM nontargeting siRNA (NT siRNA; negative control). Inset: C2C12 myocytes were transfected with 25 nM NT siRNA, 5 nM GAPDH siRNA (positive control), or 3 nM β-catenin siRNA, and β-catenin and GAPDH mRNA were analyzed 2 days after initiation of differentiation. C: C2C12 myocytes were cotransfected with either 3 nM β-catenin siRNA or 25 nM NT siRNA and either the WT 1.3-kb type IIb MyHC promoter or the mutated 1.3-kb type IIb MyHC promoter and harvested after 3 days of differentiation in DM (control) or DM with 250 ng/ml IGF-I (IGF-I) for the analysis of type IIb MyHC promoter activity. Knockdown of β-catenin with siRNA attenuates IGF-I-induced increases in 1.3-kb WT type IIb MyHC promoter activity. Values are expressed as a percentage of control NT siRNA WT and are reported as the average ± SE of n = 4 wells/group. *P < 0.05 from control NT siRNA, $P < 0.05 from IGF-I NT siRNA. D: C2C12 myocytes were transfected with either 3 nM β-catenin siRNA or 25 nM NT siRNA and harvested after 3 days of differentiation in DM (control) or DM with 250 ng/ml IGF-I (IGF-I) for the analysis of type IIb MyHC mRNA. Knockdown of β-catenin with siRNA attenuates IGF-I-induced increases in type IIb MyHC mRNA. Values are relative to control NT siRNA and are reported as the average ± SE of n = 4 wells/group. *P < 0.05 from control NT siRNA, $P < 0.05 from IGF-I NT siRNA.
Real-time PCR.
Total cellular RNA was isolated from C2C12 myotubes at specified time points (QIAshredder and RNeasy Micro; Qiagen) and reverse transcribed (Superscript III First-Strand Synthesis System; Invitrogen) using random hexamers. Each reaction, performed in duplicate, contained 25 ng cDNA, 250 nM MGB probe, 900 nM primers, and Taqman Universal PCR Master Mix (AB) in a 25-μl volume. An AB Prism 7000 sequence detection system was used for all real-time PCR reactions. Type IIb MyHC primers and probe were designed using Primer Express (AB); forward primer sequence: GAGAGGTGACAGGAGAAATCACAA; reverse primer sequence: TGCAGAATTTATTTCCGTGATATACAC; probe sequence: TGTGACGTTCTTTGTCACTGT. Relative changes in type IIb MyHC mRNA levels were determined using the ΔΔCt method (where Ct is threshold cycle; AB User Bulletin no. 2) and normalized to 18S rRNA (18S was not different over time or with IGF treatment). TaqMan Ribosomal RNA control reagents (AB) were used to determine 18S levels. β-Catenin mRNA levels were determined using the AB TaqMan Gene Expression Assay mix (AB; Mm00483033_m1) and normalized to GAPDH mRNA (AB, Endogenous Control mix; 4352339E) with Universal PCR Master Mix.
Reporter gene analysis.
At the specified time points, cells were harvested with Passive Lysis Buffer (Promega) and stored at −80°C until analysis. Firefly luciferase activity was determined using Luciferase Assay Reagent II according to the manufacturer's instructions (Promega) and measured using a Veritas Microplate Luminometer (Turner BioSystems). Firefly luciferase activity was normalized to control (non-IGF-I) 3.0-kb or 1.3-kb type IIb MyHC promoter activity.
Nuclear isolation and Western blot.
Myotubes were harvested after 3 days of differentiation with or without 250 ng/ml of IGF-I, and nuclei were isolated using the NE-PER Kit (Pierce; Rockford, IL) with protease and phosphatase inhibitors (Sigma) per the manufacturer's instructions. Enriched nuclear extract protein concentrations were determined using the BCA protein assay kit (Pierce). Equal amounts of enriched nuclear extract protein were separated by SDS-PAGE and subsequently transferred to nitrocellulose membranes. Membranes were stained with Ponceau S (Sigma), and the image was quantified to verify equal loading [P = 0.576; data not shown (19)]. β-Catenin protein was analyzed by Western blot and detected with a primary antibody from Cell Signaling Technology (no. 9562; Beverly, MA) and horseradish peroxidase-conjugated secondary antibody and SuperSignal West Dura chemiluminescence reagent from Pierce. Immunoblots were developed and analyzed using the Kodak 4000R Molecular Imaging System (Rochester, NY).
Statistical analysis.
Type IIb MyHC and β-catenin mRNA and all promoter data were analyzed by analysis of variance, and where significant differences existed, a Newman-Keuls test was used post hoc. Enriched nuclear extract β-catenin protein was analyzed by Student's t-test. Significance was established at P < 0.05. Data are reported as means ± SE.
RESULTS AND DISCUSSION
Barton-Davis et al. (3) previously reported that overexpression of IGF-I completely prevents the age-related loss of type IIb muscle fibers in old mouse EDL muscle. In addition, IGF-I has been shown to increase type IIb MyHC protein in denervated skeletal muscle (3, 10). However, mechanisms that might regulate type IIb MyHC expression in response to IGF-I are largely unknown. Since type IIb MyHC expression is transcriptionally regulated in response to thyroid hormones and mechanical stimuli (2), it seemed logical to hypothesize that IGF-I might increase type IIb MyHC promoter activity. Therefore, the purpose of this study was to investigate whether IGF-I increases type IIb MyHC promoter activity using reporter gene assays and, if so, to identify a regulatory element and potential upstream signaling to this element. Using C2C12 muscle cells, we demonstrate, for the first time, that IGF-I increases type IIb MyHC mRNA levels and activity of the type IIb MyHC promoter. Furthermore, these novel findings suggest that IGF-I-induced promoter activity of type IIb MyHC involves GSK-3β, β-catenin, and a putative Tcf/Lef binding site in the promoter region of the type IIb MyHC gene.
IGF-I induces type IIb MyHC promoter activity.
To investigate whether IGF-I increases type IIb MyHC mRNA, we differentiated C2C12 muscle cells with or without IGF-I for 4 days (Fig. 1). Type IIb MyHC mRNA was not detectable in undifferentiated myocytes (data not shown), nor was it detectable after 1 day of differentiation under control or IGF-I conditions (data not shown). In the absence of IGF-I, type IIb MyHC mRNA levels increased 2- and 5-fold at differentiation days 3 and 4, respectively, as compared with day 2 (Fig. 2A). These data are in accordance with a previous study reporting increased levels of type IIb MyHC mRNA during differentiation without added IGF-I (30). However, when IGF-I was added to the differentiation media, type IIb MyHC mRNA levels increased to a greater degree than without IGF-I (12- and 7-fold at days 3 and 4 of differentiation, respectively) (Fig. 2A). Based on our finding that IGF increases type IIb MyHC mRNA and on the fact that mRNA levels are determined by the rates of transcription and/or mRNA stabilization and MyHC gene transcription is a primary regulatory point for MyHC expression (see Ref. 2 for reference), promoter analysis studies were pursued.
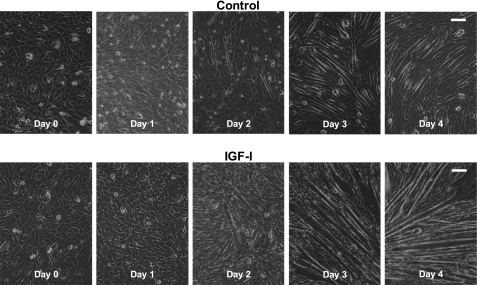
Fig. 1.
C2C12 differentiation time course. Representative photomicrographs of C2C12 myocytes at the time differentiation was initiated (day 0) and after 1–4 days of differentiation in differentiation media (DM; control) or DM with 250 ng/ml of insulin-like growth factor I (IGF-I). Bar in top right corner = 200 μm.
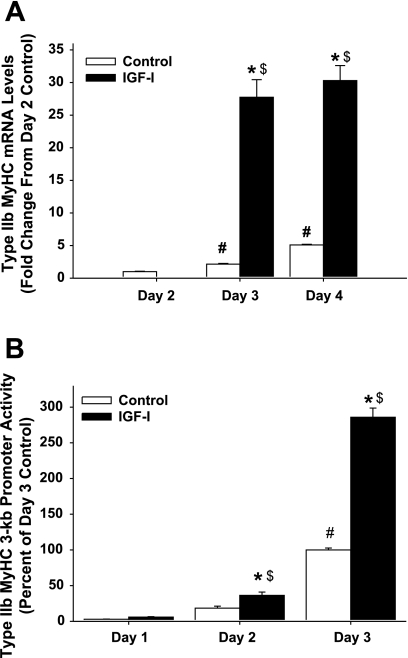
Fig. 2.
Time course of type IIb myosin heavy chain (MyHC) mRNA levels (A) and type IIb MyHC promoter activity (B) in differentiating C2C12 myocytes. C2C12 myocytes were harvested after 1–4 days of differentiation in DM (control) or DM with 250 ng/ml IGF-I (IGF-I). Type IIb MyHC mRNA was not detectable at the time differentiation was initiated (day 0) nor after 1 day of differentiation under control or IGF-I conditions (data not shown). Type IIb MyHC mRNA was detectable after 2 days under IGF-I conditions, but because of the scale of the graph the data are not visible. Data are reported as the average ± SE of n = 4 wells/group. *P < 0.05 from time-matched control, #P < 0.05 from earliest day control, $P < 0.05 from earliest day IGF-I.
To examine whether the IGF-I-induced increases in type IIb MyHC mRNA were due to increased activity of the type IIb MyHC promoter, we constructed a promoter reporter containing ∼3,000 bp (3.0-kb) of the mouse type IIb MyHC promoter region. C2C12 muscle cells transfected with the 3.0-kb type IIb MyHC promoter reporter were differentiated with or without IGF-I for 3 days, and luciferase promoter activity assays were performed. Type IIb MyHC promoter activity was detectable, albeit low, in the presence and absence of IGF-I after 1 day of differentiation. However, by days 2 and 3 of differentiation, IGF-I increased type IIb MyHC promoter activity 2- and ∼3-fold, respectively, compared with no added IGF-I (Fig. 2B). Together, these data show that IGF-I enhances type IIb MyHC promoter activity in differentiating C2C12 myocytes.
Deletion analysis reveals an IGF-I responsive region between −1,250 and −1,200 bp on the type IIb MyHC promoter.
To determine the region(s) of IGF-I-induced transcriptional activity in the type IIb MyHC promoter, 5′-deletion constructs were generated from the 3.0-kb type IIb MyHC promoter reporter. Luciferase promoter activity assays revealed three potential IGF-I-responsive regions in the type IIb MyHC promoter: −1,300 to −1,200; −1,100 to −1,000; and −500 to −400 bp (Fig. 3A). Of the three potential IGF-I-responsive regions in the type IIb MyHC promoter, the −1,300- to −1,200-bp deletion produced the largest decrement in IGF-I activation, and therefore the −1,300- to −1,200-bp region was pursued. When an additional 50-bp deletion construct was generated between −1,300 and −1,200 bp (1.25-kb), a significant decrease in IGF-I-induced promoter activity was detected between the 1.25-kb to 1.2-kb constructs, but not between 1.3-kb and 1.25-kb (Fig. 3B), suggesting a potential IGF-I-responsive region between −1,250 and −1,200 bp in the type IIb MyHC promoter.
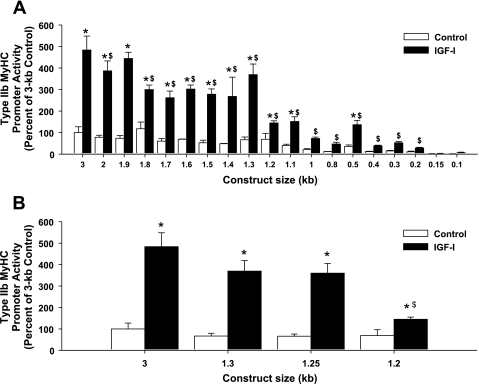
Fig. 3.
IGF-I responsiveness in type IIb MyHC promoter activity is lost with 5′-deletions. C2C12 myocytes were harvested after 3 days of differentiation in DM (control) or DM with 250 ng/ml IGF-I (IGF-I), and type IIb MyHC promoter activity was assessed. A: successive 5′-deletions were made to the 3-kb type IIb MyHC promoter. Values are expressed as a percentage of control 3-kb type IIb MyHC promoter activity and are reported as the average ± SE of n = 4 wells/group. *P < 0.05 from control within same promoter length, $P < 0.05 from IGF-I 3-kb activity. B: an additional 50-bp 5′-deletion between 1.3 kb and 1.2 kb revealed a potential IGF-I-responsive region in the type IIb MyHC promoter. Values are expressed as a percentage of control 3-kb type IIb MyHC promoter activity and are reported as the average ± SE of n = 4 wells/group. *P < 0.05 from control within same promoter length, $P < 0.05 from IGF-I 1.3-kb and IGF-I 1.25-kb activity.
IGF-I-PI3K/Akt-GSK-3β signaling to the type IIb MyHC promoter.
IGF-I signaling activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway in skeletal muscle hypertrophy (for review see Ref. 12). Downstream signaling from the IGF-I receptor and PI3K involves Akt-directed phosphorylation and inactivation of glycogen synthase kinase-3β (GSK-3β) in skeletal muscle cells (8, 32, 33).
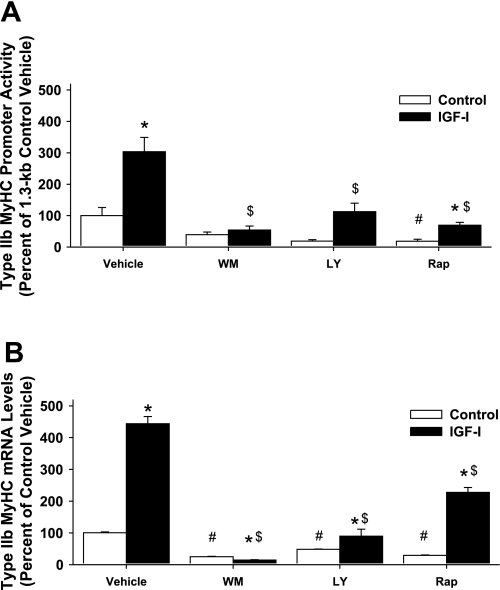
To gain insight into potential IGF-I signaling mechanism(s) to the −1,300 to −1,200 bp region of the type IIb MyHC promoter, a pharmacological approach was first undertaken. C2C12 muscle cells were differentiated with or without IGF-I for 3 days; the pharmacological inhibitors for molecules involved in the PI3K signaling cascade were added during the final 24 h. Inhibiting PI3K with wortmannin completely abolished 1.3-kb type IIb MyHC promoter activity (Fig. 4A) and type IIb MyHC mRNA (Fig. 4B) in response to IGF-I. The PI3K inhibitor, LY-294002, and the inhibitor of mammalian target of rapamycin (mTOR; a downstream target of PI3K/Akt signaling), rapamycin, both significantly attenuated type IIb MyHC promoter activity (Fig. 4A) and type IIb MyHC mRNA levels (Fig. 4B) in response to IGF-I, implying that multiple pathways may be involved in signaling to the type IIb MyHC promoter. A possible limitation to our pharmacological approach is that wortmannin, LY-294002, and rapamycin can specifically inhibit transcription and/or translation of the luciferase reporter gene, or have nonspecific effects on general transcription and/or translation. Additionally, it should be emphasized that inhibitors were present in the differentiation media for only the last 24 h; thus, normal expression of trans-activating factors of IGF-I signaling molecules were not impacted by the inhibitors during the initial 48 h of differentiation. These data (Fig. 4, A and B) suggest that the significant increases in type IIb MyHC mRNA (Fig. 2A) and promoter activity by IGF-I (Fig. 2B) that occur between 48 and 72 hrs of differentiation likely require IGF-I activation of PI3K/Akt signaling.
Fig. 4.
Chemical inhibition of the 1.3-kb type IIb MyHC promoter and type IIb MyHC mRNA levels. C2C12 myocytes were harvested after 3 days of differentiation in DM (control) or DM with 250 ng/ml IGF-I (IGF-I). Vehicle (DMSO) or the inhibitors wortmannin (WM, 100 nM), LY-294002 (LY, 10 μM), and rapamycin (Rap, 1 μg/ml) were added during the final 24 h of 3 days of differentiation. A: 1.3-kb type IIb MyHC promoter activity. Values are expressed as a percentage of control vehicle. B: type IIb MyHC mRNA levels. Values are expressed as a percentage of control vehicle and are reported as the average ± SE of n = 4 wells/group. *P < 0.05 from control within same treatment, #P < 0.05 from control vehicle, $P < 0.05 from IGF-I vehicle.
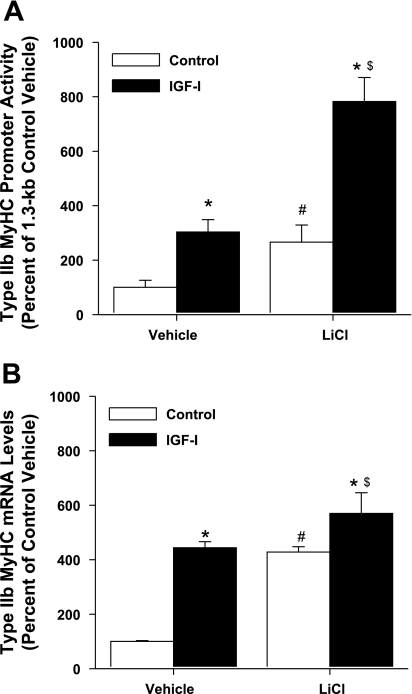
Genetic (22) or chemical (33) inhibition of GSK-3β has been demonstrated to induce significant myotube hypertrophy. Thus, we sought to determine whether modulating GSK-3β activity by chemical inhibition or genetic manipulation would alter type IIb MyHC promoter activity. We hypothesized that lithium chloride (LiCl), a noncompetitive inhibitor of GSK-3β, would enhance IGF-I-induced activation of the 1.3-kb type IIb MyHC promoter. Indeed, addition of LiCl significantly increased both 1.3-kb type IIb MyHC promoter activity (Fig. 5A) and the levels of type IIb MyHC mRNA (Fig. 5B). Furthermore, when IGF-I and LiCl treatments were combined, the effects on 1.3-kb type IIb MyHC promoter activity were additive, reminiscent of the additive effects of GSK-3β inhibition observed with the insulin-induced hypertrophic response in C2.7 myogenic cells reported by Rochat et al. (21).
Fig. 5.
Lithium chloride (LiCl) modulation of the 1.3-kb type IIb MyHC promoter and type IIb MyHC mRNA levels. C2C12 myocytes were harvested after 3 days of differentiation in DM (control) or DM with 250 ng/ml IGF-I (IGF-I). DMSO (vehicle) or LiCl (10 mM) was added during the final 24 h of 3 days of differentiation. A: 1.3-kb type IIb MyHC promoter activity. Values are expressed as a percentage of control vehicle. B: type IIb MyHC mRNA levels. Values are expressed as a percentage of control vehicle and are reported as the average ± SE of n = 4 wells/group. *P < 0.05 from control within same treatment, #P < 0.05 from control vehicle, $P < 0.05 from IGF-I vehicle.
Promoter analysis reveals a putative Tcf/Lef binding element and β-catenin as a potential link between IGF-I/GSK-3β signaling and type IIb MyHC promoter activity.
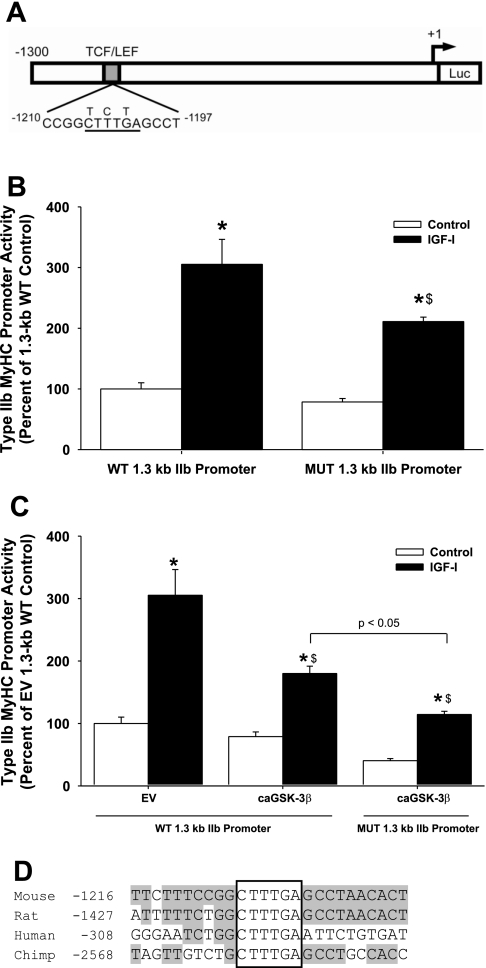
To further investigate the IGF-I-responsive region in the type IIb MyHC promoter at −1,300 to −1,200 bp, MatInspector (http://www.genomatix.de/) (5) was utilized to search this region for possible transcription factor binding sites. The search revealed an exact 6-bp T-cell factor/lymphoid enhancer factor (Tcf/Lef) DNA binding site located between −1,206 and −1,201 bp of the mouse type IIb MyHC promoter. β-Catenin, a transcriptional coactivator, binds to an NH2-terminal domain of Tcf/Lef facilitating the assembly of multimeric complexes containing transcriptional coactivators, such as CBP/p300, BCL9/LGS and Pygo, which can activate transcription of target genes. Tcf/Lef proteins bind to a conserved DNA sequence, classically known as the Wnt-response element (WRE: C/T-C-T-T-T-G-A/T-A/T) or as referred to here as the Tcf/Lef binding site (14). When canonical Wnt signaling is absent, GSK-3β phosphorylates β-catenin, targeting it for ubiquitination and degradation by the proteasome, thereby disrupting the formation of these transcriptional complexes (see Refs. 11 and 35 for references). Previously, we reported that IGF-I phosphorylates and inactivates GSK-3β (33). Taken together, these data suggest a possible link between IGF-I/GSK-3β signaling, β-catenin, and the Tcf/Lef binding element within this region of the type IIb MyHC promoter.
To verify the putative Tcf/Lef binding site's IGF-I responsiveness, we next performed site-directed mutagenesis on the −1,206 to −1,201 bp region of the 1.3-kb type IIb MyHC promoter (Fig. 6A). Site-directed mutagenesis of three bases between −1206 and −1201 resulted in a 31% decrease in IGF-I-induced type IIb MyHC promoter activity (Fig. 6B). These data suggest that a Tcf/Lef binding site at −1206 to −1201 on the type IIb MyHC promoter is functional and, for the first time, we demonstrate an IGF-I-responsive region on the type IIb MyHC promoter.
Fig. 6.
IGF-I activation of the wild-type (WT) and mutant (MUT) 1.3-kb type IIb MyHC promoter with or without constitutively active GSK-3β. A: schematic of the putative T-cell factor/lymphoid enhancer factor (Tcf/Lef) binding site at −1,206 to −1,201 bp in the WT 1.3-kb type IIb MyHC construct. The conserved Tcf/Lef binding site is underlined, and site-directed mutations are denoted above the original sequence. Luc, luciferase. B: C2C12 myocytes were transfected with the 1.3-kb WT type IIb MyHC promoter or the mutated 1.3-kb type IIb MyHC promoter and harvested after 3 days of differentiation in DM (control) or DM with 250 ng/ml IGF-I (IGF-I). Mutation of the conserved Tcf/Lef binding site attenuates IGF-I-induced 1.3-kb type IIb MyHC promoter activity. Values are expressed as a percentage of control WT 1.3-kb type IIb MyHC promoter activity and are reported as the average ± SE of n = 4 wells/group. *P < 0.05 from control WT 1.3-kb promoter, $P < 0.05 from IGF-I WT 1.3-kb promoter. C: C2C12 myocytes were cotransfected with the WT 1.3-kb type IIb MyHC promoter and the pGL3 basic empty vector (EV), the WT 1.3-kb type IIb MyHC promoter and constitutively active glycogen synthase kinase-3β (caGSK-3β), or the MUT 1.3-kb type IIb MyHC promoter and caGSK-3β, then harvested after 3 days of differentiation in DM (control) or DM with 250 ng/ml IGF-I (IGF-I). caGSK-3β attenuates IGF-I-induced 1.3-kb type IIb MyHC promoter activity. Values are expressed as a percentage of control EV WT 1.3-kb type IIb MyHC promoter activity and are reported as the average ± SE of n = 4 wells/group. *P < 0.05 from control WT 1.3-kb promoter; $P < 0.05 from IGF-I WT 1.3-kb promoter. D: nucleotide sequence comparison of Tcf/Lef binding sites in mouse, rat, human, and chimp type IIb MyHC promoters. The published mRNA sequence for type IIb MyHC for each respective species was input into the University of California, Santa Cruz Genome Bioinformatics database, and 3000 bases 5′ to the transcription start site were retrieved and analyzed for Tcf/Lef binding site and flanking region homology. The following genome assemblies were used for retrieval of genomic DNA sequences: mouse - July 2007 (mm9); rat - November 2004 (rn4); human - March 2006 (hg18); and chimp - March 2006 (panTro2). The core Tcf/Lef binding site (boxed) is conserved across these species, and the flanking region sequence homology is indicated by gray shading.
Next, to further examine the potential linkage of GSK-3β to the type IIb MyHC promoter, we employed a genetic approach to modulate GSK-3β activity in the opposite direction as LiCl. We tested whether a constitutively active GSK-3β (caGSK-3β) would attenuate the increase in type IIb MyHC promoter activity in response to IGF-I. Overexpression of caGSK-3β, in the absence of IGF-I, did not affect 1.3-kb type IIb MyHC promoter activity. However, in the presence of IGF-I, the IGF-I-induced increase in type IIb MyHC promoter activity was significantly attenuated by 41% with caGSK-3β (Fig. 6C), suggesting that IGF-I-induced inactivation of GSK-3β may play an important role in transcriptional activation of type IIb MyHC.
To this point, we have reported that both site-directed mutagenesis of the −1206 to −1201 Tcf/Lef binding site and overexpression of caGSK-3β (Fig. 6, B and C, respectively) each attenuate, but do not completely abolish IGF-I-induced increases in 1.3-kb type IIb MyHC promoter activity. These data suggest that mutation of the Tcf/Lef binding site and overexpression of caGSK-3β have similar effects on inhibiting IGF-I signaling to the type IIb MyHC promoter. Therefore, we were surprised with the finding that combining both the mutant 1.3-kb type IIb MyHC promoter and caGSK-3β together induced further attenuation of 1.3-kb type IIb MyHC promoter activity in response to IGF-I (Fig. 6C).
Cross-species nucleotide sequence comparisons showed 100% conservation of the core nucleotide sequence for Tcf/Lef binding sites in the mouse, rat, human, and chimp type IIb MyHC promoters, despite differences in location across species (Fig. 6D). The 5′- and 3′-flanking regions, however, showed less than perfect sequence homology across species. Divergence in core element location and 5′- and 3′-flanking region sequence is thought to allow for different binding factors to interact within the transcriptional complex and is likely to confer developmental-, tissue-, or perturbation-specific responsiveness (18). For example, the proximal MCAT binding site on the β-MyHC promoter is highly conserved between human and rat; however, sequence divergence of the 5′-flanking region of this MCAT element exerts profound effects on TEAD-protein binding specificity, affinity, and function (16, 31).
Nonetheless, these data demonstrate for the first time that transcriptional activation of the mouse type IIb MyHC promoter in response to IGF-I occurs, in part, through components of a Tcf/Lef signaling network. Recently, Schakman et al. (23) reported that Akt, GSK-3β, and β-catenin signaling plays a critical role in the anti-atrophic action of IGF-I in skeletal muscle atrophy caused by glucocorticoids. Concurrently, we were investigating whether the Tcf/Lef binding site and β-catenin play a role in IGF-I-induced type IIb MyHC transcriptional activity. We hypothesized that if IGF-I signaling occurs via the Tcf/Lef binding site on the type IIb MyHC promoter, IGF-I treatment would increase nuclear β-catenin protein and thus provide further insight into the mechanism by which IGF-I promoted transcriptional activity of type IIb MyHC. Here, for the first time, we show that IGF-I increases nuclear β -catenin protein 2.8-fold in differentiating C2C12 myocytes, compared with differentiating C2C12 myocytes without IGF-I treatment (Fig. 7A). Recently, Sun and Jin (27) demonstrated that insulin increases nuclear β-catenin content and induces the nuclear translocation and binding of β-catenin to two Tcf-binding sites in the human c-Myc gene promoter in intestinal cell lines. Therefore, the possibility exists that increased nuclear β-catenin protein could interact with Tcf/Lef binding sites on the type IIb MyHC promoter to increase promoter activity of type IIb MyHC in skeletal muscle cells.
Knockdown of β-catenin blunts IGF-I-induced type IIb MyHC promoter activity and type IIb MyHC mRNA.
Initially, we inhibited GSK-3β using LiCl, which is known to result in stabilization and nuclear translocation of β-catenin (21), and chronically activated GSK-3β by genetic mutation in C2C12 myocytes, resulting in opposing effects on IGF-I-induced transcriptional activity of type IIb MyHC. In addition, we showed that mutation of the Tcf/Lef binding site attenuated 1.3-kb type IIb MyHC promoter activity. These experiments demonstrate that GSK-3β and the Tcf/Lef binding site at −1,206 to −1,201 bp in the type IIb MyHC promoter play important roles in IGF-I-induced increases in type IIb MyHC transcriptional activity. However, the link that connects these two separate findings has yet to be described. Therefore, we sought to determine whether β-catenin is involved in IGF-I-induced increases in type IIb MyHC transcriptional activity.
On that note, we pursued attenuation of β-catenin expression using siRNA technology. Optimization experiments on β-catenin siRNA revealed ∼80% knockdown of β-catenin mRNA with transfection of 3–30 nM β-catenin siRNA (Fig. 7B). Furthermore, transfection of positive control GAPDH siRNA attenuated GAPDH mRNA levels by 50% but did not affect β-catenin mRNA and vice versa (Fig. 7B, inset). Moreover, when β-catenin expression was knocked down using siRNA, IGF-I-induced 1.3-kb type IIb MyHC promoter activity was attenuated ∼50% (Fig. 7C). Moreover, no further attenuation of IGF-I-induced 1.3-kb type IIb MyHC promoter activity occurred with either mutation of the Tcf/Lef binding site on the type IIb MyHC promoter, or with a combination of β-catenin siRNA and mutation of the Tcf/Lef binding site. Furthermore, knockdown of β-catenin using siRNA showed similar attenuation of IGF-I-induced increases in type IIb MyHC mRNA (Fig. 7D). These results confirm the involvement of β-catenin in IGF-I signaling to the Tcf/Lef binding site at −1,206 to −1,201 bp in the mouse type IIb MyHC promoter in differentiating C2C12 myocytes.
In summary, our data are the first to demonstrate that IGF-I increases 1) type IIb MyHC mRNA levels, 2) transcriptional activity of the mouse type IIb MyHC promoter, and 3) nuclear levels of β-catenin protein in differentiating C2C12 myocytes. We also utilized the conserved Tcf/Lef binding site at −1,206 to −1,201 bp in the type IIb MyHC promoter as a tool to explore potential signaling mechanism(s) involved in IGF-I-induced type IIb MyHC transcriptional activation. Our data suggest that transcriptional activation of type IIb MyHC in response to IGF-I occurs, in part, through GSK-3β inactivation, nuclear translocation of β-catenin protein, and an intact Tcf/Lef binding site on the mouse type IIb MyHC promoter. These data provide valuable insight into how IGF-I modulates the complex regulatory mechanisms of type IIb MyHC gene expression and present a novel mechanism by which IGF-I may regulate transcriptional activity of other genes involved in muscle hypertrophy. Furthermore, these findings could prove valuable as a guide in searching for additional IGF-I-induced Tcf/Lef-dependent genes in skeletal muscle, other than type IIb myosin heavy chain.
GRANTS
This work was supported by National Institutes of Health (NIH) Grant AR-051640 (to R. A. Shanely), Fellow of the Health and Activity Rehabilitation Research and Training Center (to K. A. Zwetsloot), NIH Grant AG-18780 (to F. W. Booth), and generous gifts (to F. W. Booth).
REFERENCES
- 1.Adams GR, McCue SA. Localized infusion of IGF-I results in skeletal muscle hypertrophy in rats. J Appl Physiol 84: 1716–1722, 1998 [DOI] [PubMed] [Google Scholar]
- 2.Baldwin KM, Haddad F. Effects of different activity and inactivity paradigms on myosin heavy chain gene expression in striated muscle. J Appl Physiol 90: 345–357, 2001 [DOI] [PubMed] [Google Scholar]
- 3.Barton-Davis ER, Shoturma DI, Musaro A, Rosenthal N, Sweeney HL. Viral mediated expression of insulin-like growth factor I blocks the aging-related loss of skeletal muscle function. Proc Natl Acad Sci USA 95: 15603–15607, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caiozzo VJ, Haddad F, Baker MJ, Baldwin KM. Influence of mechanical loading on myosin heavy-chain protein and mRNA isoform expression. J Appl Physiol 80: 1503–1512, 1996 [DOI] [PubMed] [Google Scholar]
- 5.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Cox RD, Buckingham ME. Actin and myosin genes are transcriptionally regulated during mouse skeletal muscle development. Dev Biol 149: 228–234, 1992 [DOI] [PubMed] [Google Scholar]
- 7.Cox RD, Weydert A, Barlow D, Buckingham ME. Three linked myosin heavy chain genes clustered within 370 kb of each other show independent transcriptional and post-transcriptional regulation during differentiation of a mouse muscle cell line. Dev Biol 143: 36–43, 1991 [DOI] [PubMed] [Google Scholar]
- 8.Cross DA, Alessi DR, Vandenheede JR, McDowell HE, Hundal HS, Cohen P. The inhibition of glycogen synthase kinase-3 by insulin or insulin-like growth factor 1 in the rat skeletal muscle cell line L6 is blocked by wortmannin, but not by rapamycin: evidence that wortmannin blocks activation of the mitogen-activated protein kinase pathway in L6 cells between Ras and Raf. Biochem J 303: 21–26, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delp MD, Duan C. Composition and size of type I, IIA, IID/X, and IIB fibers and citrate synthase activity of rat muscle. J Appl Physiol 80: 261–270, 1996 [DOI] [PubMed] [Google Scholar]
- 10.Flint PW, Nakagawa H, Shiotani A, Coleman ME, O'Malley BW., Jr Effects of insulin-like growth factor-1 gene transfer on myosin heavy chains in denervated rat laryngeal muscle. Laryngoscope 114: 368–371, 2004 [DOI] [PubMed] [Google Scholar]
- 11.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta 1653: 1–24, 2003 [DOI] [PubMed] [Google Scholar]
- 12.Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol 37: 1974–1984, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Haddad F, Qin AX, Zeng M, McCue SA, Baldwin KM. Interaction of hyperthyroidism and hindlimb suspension on skeletal myosin heavy chain expression. J Appl Physiol 85: 2227–2236, 1998 [DOI] [PubMed] [Google Scholar]
- 14.Hoppler S, Kavanagh CL. Wnt signalling: variety at the core. J Cell Sci 120: 385–393, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Huey KA, Roy RR, Baldwin KM, Edgerton VR. Temporal effects of inactivity on myosin heavy chain gene expression in rat slow muscle. Muscle Nerve 24: 517–526, 2001 [DOI] [PubMed] [Google Scholar]
- 16.Karasseva N, Tsika G, Ji J, Zhang A, Mao X, Tsika R. Transcription enhancer factor 1 binds multiple muscle MEF2 and A/T-rich elements during fast-to-slow skeletal muscle fiber type transitions. Mol Cell Biol 23: 5143–5164, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ. The UCSC Genome Browser Database. Nucleic Acids Res 31: 51–54, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin SB, Ordahl CP. Multiple layers of control in transcriptional regulation by MCAT elements and the TEF-1 protein family. In: Heart Development, edited by Harvey RP, Rosenthal N. San Diego, CA: Academic, 1999, p. 307–329 [Google Scholar]
- 19.Machida S, Booth FW. Increased nuclear proteins in muscle satellite cells in aged animals as compared to young growing animals. Exp Gerontol 39: 1521–1525, 2004 [DOI] [PubMed] [Google Scholar]
- 20.Musaro A, McCullagh K, Paul A, Houghton L, Dobrowolny G, Molinaro M, Barton ER, Sweeney HL, Rosenthal N. Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat Genet 27: 195–200, 2001 [DOI] [PubMed] [Google Scholar]
- 21.Rochat A, Fernandez A, Vandromme M, Moles JP, Bouschet T, Carnac G, Lamb NJ. Insulin and wnt1 pathways cooperate to induce reserve cell activation in differentiation and myotube hypertrophy. Mol Biol Cell 15: 4544–4555, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol 3: 1009–1013, 2001 [DOI] [PubMed] [Google Scholar]
- 23.Schakman O, Kalista S, Bertrand L, Lause P, Verniers J, Ketelslegers JM, Thissen JP. Role of Akt/GSK-3beta/beta-catenin transduction pathway in the muscle anti-atrophy action of insulin-like growth factor (IGF)-I in glucocorticoid-treated rats. Endocrinology 149: 3900–3908, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev 76: 371–423, 1996 [DOI] [PubMed] [Google Scholar]
- 25.Semsarian C, Wu MJ, Ju YK, Marciniec T, Yeoh T, Allen DG, Harvey RP, Graham RM. Skeletal muscle hypertrophy is mediated by a Ca2+-dependent calcineurin signalling pathway. Nature 400: 576–581, 1999 [DOI] [PubMed] [Google Scholar]
- 26.Stambolic V, Woodgett JR. Mitogen inactivation of glycogen synthase kinase-3 beta in intact cells via serine 9 phosphorylation. Biochem J 303: 701–704, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J, Jin T. Both Wnt and mTOR signaling pathways are involved in insulin-stimulated proto-oncogene expression in intestinal cells. Cell Signal 20: 219–229, 2008 [DOI] [PubMed] [Google Scholar]
- 28.Swoap SJ. In vivo analysis of the myosin heavy chain IIB promoter region. Am J Physiol Cell Physiol 274: C681–C687, 1998 [DOI] [PubMed] [Google Scholar]
- 29.Takeda S, North DL, Lakich MM, Russell SD, Whalen RG. A possible regulatory role for conserved promoter motifs in an adult-specific muscle myosin gene from mouse. J Biol Chem 267: 16957–16967, 1992 [PubMed] [Google Scholar]
- 30.Tomczak KK, Marinescu VD, Ramoni MF, Sanoudou D, Montanaro F, Han M, Kunkel LM, Kohane IS, Beggs AH. Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J 18: 403–405, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Tsika RW, McCarthy J, Karasseva N, Ou Y, Tsika GL. Divergence in species and regulatory role of β-myosin heavy chain proximal promoter muscle-CAT elements. Am J Physiol Cell Physiol 283: C1761–C1775, 2002 [DOI] [PubMed] [Google Scholar]
- 32.Van der Velden JLJ, Langen RC, Kelders MC, Wouters EF, Janssen-Heininger YM, Schols AM. Inhibition of glycogen synthase kinase-3β activity is sufficient to stimulate myogenic differentiation. Am J Physiol Cell Physiol 290: C453–C462, 2006 [DOI] [PubMed] [Google Scholar]
- 33.Vyas DR, Spangenburg EE, Abraha TW, Childs TE, Booth FW. GSK-3β negatively regulates skeletal myotube hypertrophy. Am J Physiol Cell Physiol 283: C545–C551, 2002 [DOI] [PubMed] [Google Scholar]
- 34.Wheeler MT, Snyder EC, Patterson MN, Swoap SJ. An E-box within the MHC IIB gene is bound by MyoD and is required for gene expression in fast muscle. Am J Physiol Cell Physiol 276: C1069–C1078, 1999 [DOI] [PubMed] [Google Scholar]
- 35.Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev 20: 1394–1404, 2006 [DOI] [PubMed] [Google Scholar]