Abstract
The SCN5A-encoded cardiac sodium channel underlies excitability in the heart, and dysfunction of sodium current (INa) can cause fatal ventricular arrhythmia in maladies such as long QT syndrome, Brugada syndrome (BrS), and sudden infant death syndrome (SIDS). The gene GPD1L encodes the glycerol phosphate dehydrogenase 1-like protein with homology to glycerol phosphate dehydrogenase (GPD1), but the function for this enzyme is unknown. Mutations in GPD1L have been associated with BrS and SIDS and decrease INa through an unknown mechanism. Using a heterologous expression system, we show that GPD1L associated with SCN5A and that the BrS- and SIDS-related mutations in GPD1L caused a loss of enzymatic function resulting in glycerol-3-phosphate PKC-dependent phosphorylation of SCN5A at serine 1503 (S1503) through a GPD1L-dependent pathway. The direct phosphorylation of S1503 markedly decreased INa. These results show a function for GPD1L in cell physiology and a mechanism linking mutations in GPD1L to sudden cardiac arrest. Because the enzymatic step catalyzed by GPD1L depends upon nicotinamide adenine dinucleotide, this GPD1L pathway links the metabolic state of the cell to INa and excitability and may be important more generally in cardiac ischemia and heart failure.
Keywords: cardiac arrhythmia, sodium current, ion channel, cell metabolism, glycerol 3-phosphate dehydrogenase 1-like, protein kinase C, cardiac sodium channel α-subunit
the scn5a gene encodes the cardiac sodium channel α-subunit (SCN5A; also called Nav1.5) that carries the sodium current (INa) that underlies excitation and conduction of the cardiac impulse. Loss-of-function and gain-of-function mutations in SCN5A or other genes that encode sodium channel interacting proteins (ChIPs) cause potentially lethal inheritable arrhythmia syndromes or cardiac channelopathies including long QT syndrome (LQTS) (10, 19, 23, 24), Brugada syndrome (BrS) (3, 9, 25), and sudden infant death syndrome (SIDS) (1, 4). The gene GPD1L on chromosome 3p22.3 encodes the protein glycerol 3-phosphate dehydrogenase 1-like (GPD1L); the letters stand for glycerol 3-phosphate dehydrogenase 1-like because it shares 84% homology with glycerol 3-phosphate dehydrogenase 1 (GPD1) (13). GPD1L was first noted as part of mammalian gene sequence collection program (18) in 2002, but the function or importance of GPD1L, if any, was unknown. Mutations in GPD1L were linked to BrS in a large family (26), and in 2007 the mutation (GPD1L-A280V) (9) and the novel SIDS-associated mutation (GPD1L-E83K) (22) were both shown to decrease cardiac INa amplitude. This decrease could account for their arrhythomogenesis, but the mechanism by which GPD1L mutations decreased INa was unknown.
The related gene GPD1 encodes a NAD-dependent cytosolic enzyme that is an important link between the glycolytic pathway and triglyceride synthesis. GPD1 catalyzes the reversible conversion of glycerol-3-phosphate (G3P) to dihydroxyacetone phosphate (DHAP). GPD1 has direct and indirect interactions with genes and proteins involved in the insulin pathway and cellular metabolism, and GPD1 dysfunction has been linked to obesity, diabetes, and other diseases (14). We hypothesized that like its namesake GPD1, GPD1L catalyzes the reaction of G3P to DHAP. A loss of function of the enzymatic activity of GPD1L would be expected to increase levels of G3P as has been observed in a mouse GPD1-knockout model (2). By a GPD1L-dependent SCN5A phosphorylation pathway (Fig. 2B), decreased GPD1L activity would then increase the substrate G3P and feed the PKC-mediated phosphorylation of SCN5A at serine residue 1503 (S1503) where such phosphorylation is known to decrease INa (11). To investigate this hypothesis, we performed experiments with wild-type (WT) and mutant SCN5A and GPD1L constructs coexpressed in heterologous cell system [human embryonic kidney (HEK)293 cells].
Fig. 2.
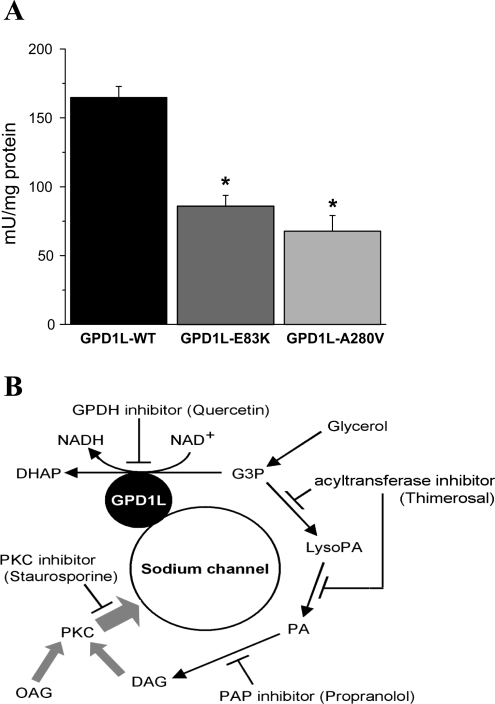
Enzymatic activity of GPD1L decreased by the GPD1L missense mutations E83K and A280V. A: G3P oxidation was measured spectrophotometrically at 37°C from the WT and glutathione S-transferase (GST)-labeled GPD1L proteins cleaved by factor Xa to be separated from GST-tag (materials and methods; n = 6 in each group). B: diagram of proposed GPD1L-PKC pathway with substrates and pharmacological blockers (indicated by T-shaped lines) that affect SCN5A-GPD1L association via PKC activation. DAG, diacylglycerol; PA, phosphatidic acid; LysoPA, lsophoshpatidic acid; DHAP, dihydroxyacetone phosphate. *P < 0.05 vs. GPD1L WT.
MATERIALS AND METHODS
Reagents and antibodies.
Cell culture media were Opti-MEM, RPMI 1640, and fetal calf serum from Invitrogen (Carlsbad, CA) and FuGENE6 from Roche Molecular Biochemicals (Basel, Switzerland). Staurosporine, 1-oleoyl-2-acetylglycerol (OAG), thimerosal, and propranolol were from Sigma-Aldrich (St. Louis, MO). Rabbit FITC-conjugated anti-FLAG antibody, rabbit anti-SCN5A antibody, and goat horseradish peroxidase-conjugated anti-rabbit IgG (H+L) antibody were from Immunology Consultants Laboratory (Newberg, OR), Upstate (Charlottesville, VA), and Bio-Rad laboratories (Hercules, CA), respectively.
Plasmids construction.
The cDNA of GPD1L (GenBank Accession No. BC028726) was subcloned into pIRES2EGFP (Clontech, Palo Alto, CA) to generate pGPD1L-IRES2EGFP. E83K and A280V mutants in GPD1L were incorporated into WT clones by the site-directed mutagenesis. The cDNAs of SCN5A (GenBank Accession No. AB158469), and β1-subunit gene, SCN1B (GenBank Accession No. BC112922) were subcloned into pcDNA3, and the FLAG peptide of DYKDDDDK was incorporated at R45 in SCN1B and confirmed by sequencing. S1503A in SCN5A was incorporated by the site-directed mutagenesis. For glutathione S-transferase (GST)-fusion GPD1L constructs, the coding region of GPD1L was subcloned pGEX-5X-2 (Amersham Biosciences, Piscataway, NJ). All clones were sequenced and confirmed.
Cell culture and transfection.
Cells were maintained in MEM supplemented with 10% heat-inactivated fetal calf serum. For transfections, cells were grown in 60-mm dishes and then transfected with 1.5 μg DNA of SCN5A and 0.3 μg DNA of GPD1L-IRES-GFP per dish using FuGENE6.
Electrophysiological study.
Macroscopic voltage-gated sodium current was recorded using the whole-cell method of the patch clamp technique in the HEK293 cells 48 h after transfection as described previously (12) at room temperature with cells continuously perfused with solution containing (in mM) 140 NaCl, 4 KCl, 1.8 CaCl2, 0.75 MgCl2, and 5 HEPES (pH 7.4 set with NaOH). The pipette (intracellular) solution contained (in mM) 120 CsF, 20 CsCl2, 5 EGTA, and 5 HEPES (pH 7.4 set with CsOH). Except where indicated otherwise, drugs and substrates were added to the culture medium and incubated for 3–5 h and then washed before the electrophysiological experiment. INa density was measured at −20 mV from holding potential −140 mV and normalized to cell capacitance.
Flowcytometry.
The transfected HEK293 cells were harvested by incubation with 0.5 mM EDTA-PBS for 10 min at 37°C and washed with RPMI 1640 supplemented with 1 mM EDTA (pH 7.4), 3% FCS, and 0.02% azide (staining medium). The FITC-conjugated anti-FLAG antibody (Sigma-Aldrich) incubations were performed in staining medium at 4°C and then washed by PBS supplemented with 1 mM EDTA (pH 7.4) and 1% FCS. The stained cells were examined for surface expression with FACSCalibur (BD Biosciences, San Jose, CA).
GST-pulldown assay.
Rosetta gami 2 bacteria (Novagen, Madison, WI) expressing GST-labeled GPD1L induced by isopropyl-β-d-thiogalactopyranoside (IPTG) were sonicated in PBS with 1% Triton X-100, purified and fixed on MagneGST particles (Promega, Madison, WI) per manufacturer instructions. Purity and concentration of fusion proteins were determined by SDS-PAGE followed by Coomasie Blue staining. GST-bait was incubated in protein-binding buffer overnight at 4°C with cell lysates obtained from stably expressing SCN5A cells that were precleared for 1 h at 4°C with MagneGST particles without GPD1L. Samples were washed four times in binding buffer. Bound proteins were liberated by boiling in Laemmli buffer with 50 mM DTT and subjected to SDS-PAGE and immunoblotted with anti-SCN5A antibody on 7.5% SDS-polyacrylamide gels (Bio-Rad).
G3P oxidation activity assay.
WT and mutated GPD1L with GST label were obtained from bacterial expression induced by lactose analog IPTG and purified by MagenGST particles. The purified proteins were cleaved by factor Xa (Novagen). G3P oxidation activity of the GPD1L WT and mutants were measured spectrophotometrically at 25°C by following the disappearance of the oxidation of G3P and formation of NADH at 340 nm (28). The assay mixture contained 1 mM EDTA and 1 mM 2-mercaptoethanol, 5 mM NAD + and 5 mM G3P, buffered by 50 mM Tris·HCl (for pH 6.5–7.4).
Statistics.
Summary data are presented as means ± SE and statistical significance for the difference between mean values in GPD1L-WT versus GPD1L mutants or control versus treatment and tested by the Student's t-test.
RESULTS
Stimulation of the GPD1L-WT PKC pathway reduces INa density.
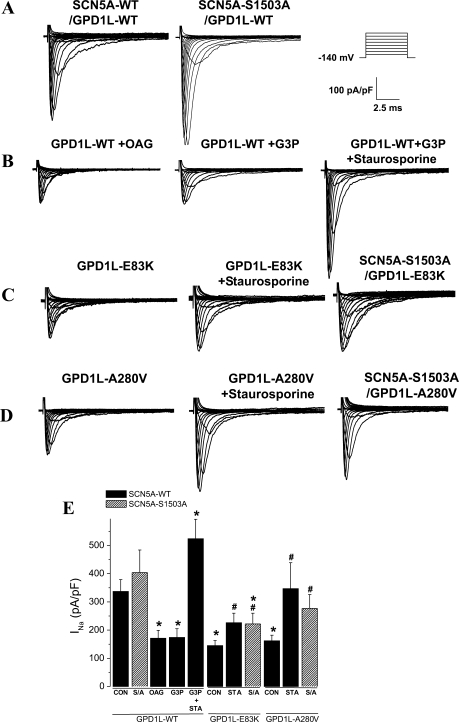
GPD1L-WT was coexpressed with SCN5A-WT, or with SCN5A-S1503A, a channel with an engineered mutation at serine 1503 to an alanine at a consensus PKC phosphorylation site (11). The INa density from whole-cell voltage clamp showed a small but insignificant increase with SCN5A-S1503A compared with SCN5A-WT (Fig. 1, A and E). Activation of the GPD1L-PKC pathway using the direct PKC agonist OAG and the substrate G3P both significantly decreased INa density, and this decrease was abrogated by the PKC blocker staurosporine (Fig. 1, B and E). As reported previously (15), OAG shifted inactivation in the negative direction (by 7 mV; data not shown), which would tend to further reduce INa from holding potentials more positive than those used in this study. The SIDS mutation GPD1L-E83K (22) (Fig. 1, C and E) and the BrS mutation GPD1L-A280V (9) (Fig. 1, D and E) significantly decreased INa density compared with GPD1L-WT, and the pathway blocker staurosporine abrogated this decrease. In addition, the decreased INa density with the GPD1L mutations was not observed when coexpressed with the phosphorylation-deficient mutant channel SCN5A-S1503A (Fig. 1, C–E). Together these data support the hypothesis that GPD1L plays a role in regulating INa by a GPD1L-PKC phosphorylation pathway depicted in Fig. 2B.
Fig. 1.
Glycerol 3-phosphate dehydrogenase 1-like (GPD1L) mutations decreased sodium current (INa) density mediated by PKC phosphorylation at serine 1503 (S1503). A–D: representative traces for families of macroscopic INa measured from human embryonic kidney (HEK293) cells coexpressing wild-type (WT) and mutant constructs of cardiac sodium channel α-subunit (SCN5A) and GPD1L as indicated. The cells were depolarized for 24 ms to different potentials in increments of 10 mV from a holding potential of −140 mV (see protocol inset) in control conditions and after 3–5 h incubation with 1-oleoyl-2-acetylglycerol (OAG; 20 μM), glycerol-3-phosphate (G3P; 10 μM), and G3P plus staurosporine (Sta; 1 μM) before patch-clamp recordings. E: summary data for INa density measurements at −20 mV from a holding potential of −140 mV for SCN5A with GPD1L-WT and mutants (n = 5–32). S/A, SCN5A-S1503A *P < 0.05 vs. GPDlL-WT control (Con) and vs. GPD1L-WT/SCN5A-S1503A; #P < 0.05 vs. GPD1L-mutant/SCN5A-WT in Con.
GPD1L enzymatic activity decreased by mutations.
A G3P oxidation assay with GPD1L-WT and mutant GPD1L showed that both E83K and A280V exhibited significantly less enzymatic activity than GPD1L-WT (Fig. 2A). These results support the hypothesis that these mutations affect INa by a direct decrease in enzymatic activity, although other conformational effects of the mutations on the complex such as chaperoning functions cannot be excluded.
Manipulation of the GPD1L-PKC pathway.
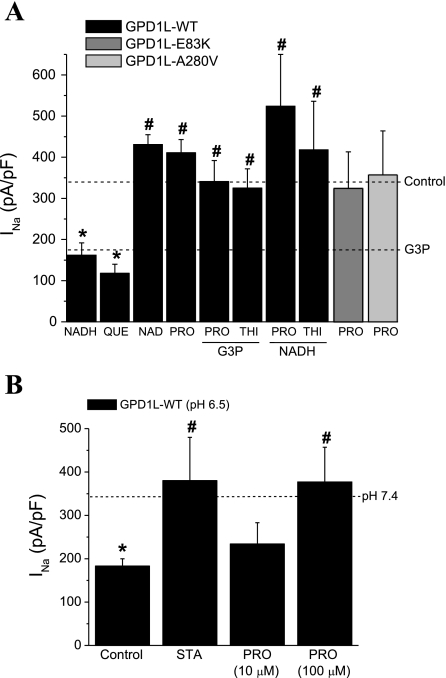
The proposed pathway (Fig. 2B) makes a number of predictions regarding the effects of various substrates and blockers on INa density, which we tested (Fig. 2A). As predicted, NADH and the glycerol phosphate dehydrogenase (GPDH) inhibitor quercetin both significantly decreased INa density compared with the control GPD1-L WT. NAD+ did not significantly increase INa density above control levels (Fig. 3A), suggesting the relatively low basal state of phosphorylation of SCN5A under our study conditions, also consistent with the nonsignificant increase with SCN5A-S1503A compared with SCN5A-WT (Fig. 1, A and E). The NADH effect to decrease INa was nearly completely abrogated by the GPD1L-dependent SCN5A phosphorylation pathway blockers the phosphatidic acid phosphatase inhibitor propranolol and the acelytransferase inhibitor thimerosal (Fig. 3A), supporting the hypothesis that this pathway is a predominant mechanism for NADH modulation of INa. As with the mutations, propanolol abrogated the effects of GPD1L-E83K and GPD1L-A280V mutants. These data (Fig. 3A) were obtained after 3–5 h incubation with drugs and reagents, and the reagents were removed before testing for INa. In more acute experiments, inclusion of 10 μM G3P in the pipette solution significantly reduced peak INa to 69 ± 79 pA/pF (n = 4) compared with peak INa of 379 ± 68 pA/pF (n = 6), and thimerosol significantly increased peak INa to 309 ± 58 pA/pF (n = 5) from 248 ± 53 pA/pF (n = 6) in cells coexpressing SCN5A and GPD1L-WT (P < 0.05). Together, these results support the hypothesis that GPD1L-E83K and GPD1L-A280V decrease INa by the pathway indicated in Fig. 2B, where loss of enzymatic function and accumulation of G3P leads to increased PKC-dependent phosphorylation of SCN5A.
Fig. 3.
GPD1L-PKC stimulation through the G3P pathway decreased INa density for SCN5A with GPD1L-WT. A: summary data for INa density at −20 mV from a holding potential of −140 mV for SCN5A with GPD1L-WT after treatment with various pathway substrates and inhibitors: 10 μM G3P, 300 μM NADH, 10 μM OAG, 100 μM quecertin (Que), 300 μM NAD, 10 μM propranolol (Pro), 1 μM staurosporine (Sta), and 10 μM thimerosol (Thi). For columns 5–8, the treatment was with 2 compounds as indicated by the labels with a column, with G3P present or NADH present. For GPD1L-E83K (dark gray) and GPD1L-A280V (light gray), recordings were performed after incubation with 100 μM Pro added 3–5 h before patch-clamp recording. The dotted lines represent the INa density in GPD1L-WT for control conditions and in G3P from Fig. 1E (n = 5–23). *P < 0.05 vs. control; #P < 0.05 vs. G3P, Que, or NADH alone. B: acidosis decreased INa density through a PKC-dependent mechanism. HEK293 cells were transfected with SCN5A-WT and GPD1L-WT and incubated overnight at pH of 7.4 or pH of 6.5 before transfer and recording of INa in control (normal pH) bath solution. The dotted line represents the value for SCN5A-WT + GPD1L-WT at pH 7.4 from Fig. 1E. The INa-attenuating effects of acidic pH with GPD1L-WT were prevented by both Sta and Pro (n = 6–12). *P < 0.05 vs. control at pH 7.4; #P < 0.05 vs. control at pH 6.5.
Acidosis is another important component of metabolic state, and the enzymatic activity of GPD1L was reduced under more acidic conditions (data not shown). INa density was also significantly reduced after incubation at pH 6.5 compared with pH 7.4 (Fig. 3B), and this reduction was abrogated by the PKC inhibitor staurosporine and by propranalol. Protons have long been known to affect INa by both pore-blocking and surface charge effects (7, 8), but the voltage clamp data were obtained after transfer to control pH, suggesting additional important longer term pH effects on INa may be mediated through G3P metabolism and the GPD1L-dependent SCN5A phosphorylation pathway.
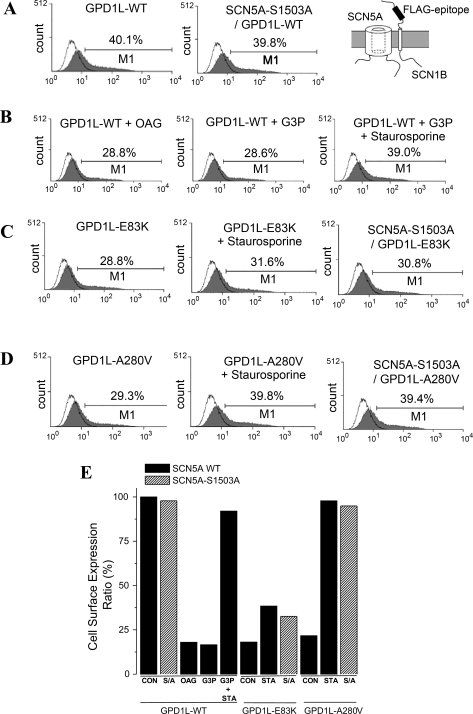
GPD1L mutants decrease SCN5A cell surface expression and INa through a GPD1L-SCN5A phosphorylation pathway.
Previously, the mutations GPD1L-A280V (9) and GPD1L-E83K (22) were shown to decrease INa and A280V was noted to decrease cell surface expression of SCN5A by quantitative immunostaining and confocal microscopy (9). We used flowcytometry and INa measurements to confirm that A280V as well as E83K decreased INa and SCN5A cell surface expression compared with GPD1L-WT (Fig. 4, A–E). When the E83K and A280V mutations were expressed with the channel lacking the PKC-dependent phosphorylation site at S1503 (SCN5A-S1503A), cell surface expression and the INa density were partially restored compared with when they were expressed with SCN5A-WT (Fig. 4, C–E). The decrease in cell surface expression and INa density caused by the GPD1L mutants was reversed (partially for E83K, completely for A280V) by the PKC inhibitor staurosporine or by the S1503A SCN5A mutant lacking the PKC consensus site (Fig. 4, C–E), further supporting the hypothesis that these GPD1L mutations decrease INa through PKC phosphorylation, but the less than complete reversal by PKC block for E83K suggests other mechanisms may contribute to the effect of this mutation on INa. Gating properties of SCN5A such as activation and inactivation were not affected by the GPD1L mutations (data not shown) in agreement with previous reports (9, 22) suggesting the major effect is a reduction in the cell surface expression of Nav1.5.
Fig. 4.
Flowcytometric analysis of cell surface expression of the sodium channel complex. HEK293 cells transiently transfected with SCN5A, GPD1L, and FLAG-SCN1B were harvested (see materials and methods), and cell surface staining was performed by FITC-conjugated anti-FLAG M2 antibody, without permeabilization. Gray areas reflect signals from cells expressing SCN5A, GPD1L, and FLAG-SCN1B constructs, and the black line indicates signals from SCN5A, GPD1L, and SCN1B without FLAG as negative control. In this method, the count of cells with surface expression is reflected in the long tail rather than the peak. Marker 1 (M1) was established at a point containing 5% of negative control. The numbers above M1 represent the percentage of positive events falling within the M1 region and reflect cell surface expression. A and B: cell surface expression of the SCN5A. GPD1L complex was measured from HEK cells coexpressing SCN5A, SCN1B, and GPD1L-WT under control conditions (A) and after 3–5 h incubation with OAG (20 μM), or G3P (10 μM) and G3P plus staurosporine (1 μM) 1 μM stausporin (B), or with GPD1L-E83K or GPD1L-A280V (C and D). GPD1L-E83K and -A280V decreased cell surface expression, and this effect was reversed by staurosporine and by expression of SCN5A with a mutated PKC phosphorylation site (SCN5A-S1503A; C and D). E: summary data for INa density measurements shown in A–D. The data represent the percent ratio with the minimal (26.4%) surface expression obtained by testing SCN5A-G1743R, an established trafficking defective mutation (Ref. 20) (data not shown) and maximal (40.1%) surface expression seen in control conditions.
SCN5A and GPD1L associate.
GPD1L was used as bait to pull-down SCN5A in a GST-fusion protein assay to show that GPD1L was associated with SCN5A. Neither the SIDS-associated (E83K) nor the BrS-associated (A280V) GPD1L mutation disrupted the association between GPD1L and SCN5A since the GST-GPD1L fusion protein containing these mutations also pulled down SCN5A (Fig. 5). This suggests that the hypothesized loss of GPD1L function is not caused by disruption of the complex and is consistent with our data that the mutations interfere directly with the enzymatic activity (Fig. 2A).
Fig. 5.
GPD1L associated with SCN5A and GPD1L mutations do not disrupt this association. HEK cells stably expressing SCN5A were transfected with GST constructs of GPD1L-WT, E83K, or A280V and lysed 48 h after transfection. Cell lysates were examined for protein interaction using the pull-down approach. GST-GPD1L-bound proteins were analyzed by SDS-PAGE and immunoblotted with anti-cardiac sodium channel antibody.
DISCUSSION
These results provide a first putative functional role for GPD1L in general cell physiology. The link of GPD1L mutations to sudden death predisposing heritable arrhythmia syndromes such as BrS and SIDS suggests the physiological role of GPD1L may be cardiac specific, but whether GPD1L plays regulatory roles in other excitable tissues remains to be determined. RNA for GPD1L was present in the heart (9), but additional information for the tissue distribution of GPD1L is lacking. Even within the heart, whether GPD1L has a regulatory role for proteins other than the cardiac sodium channel is unknown.
We have shown that GPD1L has a direct or closely coupled association with the pore-forming α-subunit of the cardiac sodium channel (SCN5A, Nav1.5). The location of this association is unknown, and it is also possible that the association is indirect and mediated by ChIPs such as syntrophin (6). The association of GPD1L with SCN5A could confer specificity of GPD1L regulation of SCN5A by substrate localization, perhaps analogous to that shown for nitrosylation of SCN5A by the syntrophin complex (19). In that study, mutations in syntrophin were shown to cause SCN5A dysfunction by disrupting the association of important enzymes in the complex (19). Here, however, neither the SIDS-associated (E83K) nor the BrS-associated (A280V) GPD1L mutation disrupted the association between GPD1L and SCN5A since the GST-GPD1L fusion protein containing these mutations also pulled down SCN5A (Fig. 5). Our results suggest that the mutated GPD1L remains associated with SCN5A but has reduced or lost its enzymatic function (Fig. 2A).
The results support the idea that GPD1L regulates SCN5A primarily through direct phosphorylation of SCN5A primarily at S1503 by a GPD1L-dependent SCN5A phosphorylation pathway proposed in Fig. 2B. The GPD1L mutants, however, also increased phosphorylation of SCN5A-S1503A compared with GPD1L-WT (data not shown), suggesting the possible presence of additional PKC phosphorylation sites in addition to S1503 on SCN5A, perhaps analogous to PKC sites on the domain I-II linker of the brain sodium channel (16). We also cannot exclude the possibility that other proteins that regulate SCN5A may be phosphorylated by the GPD1L mechanism. For example, PKC activates Ras GTPase, Ras GTPase phosphorylation activates ERK, and ERK has been shown to reduce INa density (27). It is also possible that NAD+ and NADH may have other direct or indirect effects on SCN5A function, but the complete block of NADH effects on INa density by pathway inhibitors (Fig. 3A) suggests the primary mechanism for NADH effects on INa is mediated through this GPD1L-dependent SCN5A phosphorylation pathway. In addition, other substrates and inhibitors used in the pathway have many actions in addition to the putative action on the pathway and may have additional effects or exert their action primarily outside the pathway. For example, thimerosol has been used as a thiol oxidant and postulated to have a direct effect on L-type calcium channels (5). Propranalol also has many additional effects, and the concentration used in this study (10 μM) is supratherapeutic so implications for therapy must be considered with caution. A limitation of our study is that the experiments were performed completely in heterologous expression systems and await confirmation in native myocytes. Finally, it is important to note that other pathways, such as ANG II downregulation of INa by redox-sensitive NF-κβ (17) may also play a role in redox regulation of INa amplitude.
Despite these limitations, the GPD1L-dependent SCN5A phosphorylation pathway we describe provides a plausible mechanism for the decrease in INa and the pathogenicity of the BrS2-associated mutation GPD1L-A280V and the SIDS-associated mutation GPD1L-E83K. In addition to suggesting possible therapeutic approaches to loss of SCN5A function, the elucidation of this pathway suggests that the other genes that encode the various proteins responsible for the metabolism of glycerol-3-phosphate may deserve consideration as sudden death-susceptibility candidate genes.
Although the rare heritable arrhythmia syndrome mutations in GPD1L are likely to account for a relatively small proportion of sudden cardiac arrests, these results have more general implications for cardiac excitability and arrhythmia. Because metabolism of G3P to DHAP by the enzymatic activity of GPD1L depends upon the ratio of oxidized NAD+ and reduced NADH (ratio NAD+-to-NADH; Fig. 2B), the GPD1L-dependent SCN5A phosphorylation pathway may be a general regulatory mechanism linking the metabolic state of the cardiocyte to cellular excitability by modulating INa density. This metabolic pathway may link both acidosis and the oxidation-reduction state of the cardiac cell to INa and excitability that may play important regulatory roles in arrhythmia vulnerability seen in common diseases such as cardiac ischemia and heart failure.
GRANTS
Funding was provided by the University of Wisconsin Cellular and Molecular Arrhythmia Research Program (to J. C. Makielski) and the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program (to M. J. Ackerman), the Established Investigator Award from the American Heart Association (to M. J. Ackerman), and the National Institutes of Health Grants HD-42569 (to M. J. Ackerman) and HL-71092 (to J. C. Makielski).
ACKNOWLEDGMENTS
The current address for K. Ueda is with Research and Development, Bristol-Myers K.K., Tokyo 163-1328, Japan. We thank Masayasu Hiraoka for reading of the manuscript.
REFERENCES
- 1.Ackerman MJ, Siu BL, Sturner WQ, Tester DJ, Valdivia CR, Makielski JC, Towbin JA. Postmortem molecular analysis of SCN5A defects in sudden infant death syndrome. JAMA 286: 2264–2269, 2001 [DOI] [PubMed] [Google Scholar]
- 2.Brown LJ, Koza RA, Marshall L, Kozak LP, MacDonald MJ. Lethal hypoglycemic ketosis and glyceroluria in mice lacking both the mitochondrial and the cytosolic glycerol phosphate dehydrogenases. J Biol Chem 277: 32899–32904, 2002 [DOI] [PubMed] [Google Scholar]
- 3.Chen Q, Kirsch GE, Zhang D, Brugada R, Brugada J, Brugada P, Potenza D, Moya A, Borggrefe M, Breithardt G, Ortiz-Lopez R, Wang Z, Antzelevitch C, O′ Brien RE, Schulze-Bahr E, Keating MT, Towbin JA, Wang Q. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature 392: 293–296, 1998 [DOI] [PubMed] [Google Scholar]
- 4.Cronk LB, Ye B, Kaku T, Tester DJ, Vatta M, Makielski JC, Ackerman MJ. Novel mechanism for sudden infant death syndrome: persistent late sodium current secondary to mutations in caveolin-3. Heart Rhythm 4: 161–166, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fearon IM, Palmer AC, Balmforth AJ, Ball SG, Varadi G, Peers C. Modulation of recombinant human cardiac L-type Ca2+ channel alpha1C subunits by redox agents and hypoxia. J Physiol 514: 629–637, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gavillet B, Rougier JS, Domenighetti AA, Behar R, Boixel C, Ruchat P, Lehr HA, Pedrazzini T, Abriel H. Cardiac sodium channel Nav1.5 is regulated by a multiprotein complex composed of syntrophins and dystrophin. Circ Res 99: 407–414, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Hille B. Charges and potentials at the nerve surface. Divalent ions and pH. J Gen Physiol 51: 221–236, 1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hille B, Woodhull AM, Shapiro BI. Negative surface charge near sodium channels of nerve: divalent ions, monovalent ions, and pH. Philos Trans R Soc Lond B Biol Sci 270: 301–318, 1975 [DOI] [PubMed] [Google Scholar]
- 9.London B, Michalec M, Mehdi H, Zhu X, Kerchner L, Sanyal S, Viswanathan PC, Pfahnl AE, Shang LL, Madhusudanan M, Baty CJ, Lagana S, Aleong R, Gutmann R, Ackerman MJ, McNamara DM, Weiss R, Dudley SC., Jr Mutation in glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) decreases cardiac Na+ current and causes inherited arrhythmias. Circulation 116: 2260–2268, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medeiros-Domingo A, Kaku T, Tester DJ, Iturralde-Torres P, Itty A, Ye B, Valdivia C, Ueda K, Canizales-Quinteros S, Tusie-Luna MT, Makielski JC, Ackerman MJ. SCN4B-encoded sodium channel β4 subunit in congenital long-QT syndrome. Circulation 116: 136–142, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray KT, Hu NN, Daw JR, Shin HG, Watson MT, Mashburn AB, George ALJ. Functional effects of protein kinase C activation on the human cardiac Na+ channel. Circ Res 80: 370–376, 1997 [DOI] [PubMed] [Google Scholar]
- 12.Nagatomo T, Fan Z, Ye B, Tonkovich GS, January CT, Kyle JW, Makielski JC. Temperature dependence of early and late currents in human cardiac wild-type and long QT Δ KPQ Na+ channels. Am J Physiol Heart Circ Physiol 275: H2016–H2024, 1998 [DOI] [PubMed] [Google Scholar]
- 13.Ou X, Ji C, Han X, Zhao X, Li X, Mao Y, Wong LL, Bartlam M, Rao Z. Crystal structures of human glycerol 3-phosphate dehydrogenase 1 (GPD1). J Mol Biol 357: 858–869, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Park JJ, Berggren JR, Hulver MW, Houmard JA, Hoffman EP. GRB14, GPD1, and GDF8 as potential network collaborators in weight loss-induced improvements in insulin action in human skeletal muscle. Physiol Genomics 27: 114–121, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Qu Y, Rogers J, Tanada T, Scheuer T, Catterall WA. Modulation of cardiac Na+ channels expressed in a mammalian cell line and in ventricular myocytes by protein kinase C. Proc Natl Acad Sci USA 91: 3289–3293, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheuer T, Catterall WA. Control of neuronal excitability by phosphorylation and dephosphorylation of sodium channels. Biochem Soc Trans 34: 1299–1302, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Shang LL, Sanyal S, Pfahnl AE, Jiao Z, Allen J, Liu H, Dudley SC., Jr NF-κB-dependent transcriptional regulation of the cardiac SCN5A sodium channel by angiotensin II. Am J Physiol Cell Physiol 294: C372–C379, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA 99: 16899–16903, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueda K, Valdivia C, Medeiros-Domingo A, Tester DJ, Vatta M, Farrugia G, Ackerman MJ, Makielski JC. Syntrophin mutation associated with long QT syndrome through activation of the nNOS-SCN5A macromolecular complex. Proc Natl Acad Sci USA 105: 9355–9360, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valdivia CR, Tester DJ, Rok BA, Porter CB, Munger TM, Jahangir A, Makielski JC, Ackerman MJ. A trafficking defective, Brugada syndrome-causing SCN5A mutation rescued by drugs. Cardiovasc Res 62: 53–62, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Van Norstrand DW, Valdivia CR, Tester DJ, Ueda K, London B, Makielski JC, Ackerman MJ. Molecular and functional characterization of novel glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) mutations in sudden infant death syndrome. Circulation 116: 2253–2259, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vatta M, Ackerman MJ, Ye B, Makielski JC, Ughanze EE, Taylor EW, Tester DJ, Balijepalli RC, Foell JD, Li Z, Kamp TJ, Towbin JA. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation 114: 2104–2112, 2006 [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell 80: 805–811, 1995 [DOI] [PubMed] [Google Scholar]
- 25.Watanabe H, Koopmann TT, Le Scouarnec S, Yang T, Ingram CR, Schott JJ, Demolombe S, Probst V, Anselme F, Escande D, Wiesfeld AC, Pfeufer A, Kaab S, Wichmann HE, Hasdemir C, Aizawa Y, Wilde AA, Roden DM, Bezzina CR. Sodium channel beta1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J Clin Invest 118: 2260–2268, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss R, Barmada MM, Nguyen T, Seibel JS, Cavlovich D, Kornblit CA, Angelilli A, Villanueva F, McNamara DM, London B. Clinical and molecular heterogeneity in the Brugada syndrome: a novel gene locus on chromosome 3. Circulation 105: 707–713, 2002 [DOI] [PubMed] [Google Scholar]
- 27.Yanagita T, Kobayashi H, Yamamoto R, Kataoka H, Yokoo H, Shiraishi S, Minami S, Koono M, Wada A. Protein kinase C-alpha and -epsilon down-regulate cell surface sodium channels via differential mechanisms in adrenal chromaffin cells. J Neurochem 74: 1674–1684, 2000 [DOI] [PubMed] [Google Scholar]
- 28.Zolnierowicz S, Swierczynski J, Zelewski L. Isolation and properties of glycerol-3-phosphate oxidoreductase from human placenta. Eur J Biochem 154: 161–166, 1986 [DOI] [PubMed] [Google Scholar]