Abstract
The Zfhx1a gene expresses two different isoforms; the full length Zfhx1a-1 and a truncated isoform termed Zfhx1a-2 lacking the first exon. Deletion analysis of the Zfhx1a-1 promoter localized cell-specific repressors, and a proximal G-string that is critically required for transactivation. Transfection of Zfhx1a-1 cDNA, but not Zfhx1a -2, downregulates Zfhx1a-1 promoter activity. Mutation of an E2-box disrupted the binding of both Zfhx1a isoforms. Consistent with this, transfected Zfhx1a-1 does not regulate the transcriptional activity of the E-box mutated Zfhx1a-1 promoter. Competitive EMSAs and transfection assays show that Zfhx1a-2 can function as a dominant negative isoform since it is able to compete and displace Zfhx1a-1 from its binding site and overcome Zfhx1a-1 induced repression of the Zfhx1a-1 promoter in cells. Hence, the Zfhx1a-1 gene is autoregulated in part by negative feedback on its own promoter which is, in turn, modified by the availability of the negative dominant isoform Zfhx1a-2.
Keywords: Zfhep/ZEB1/δEF1, autoregulation, gene regulation, promoter characterization
INTRODUCTION
Zfhx1a (δEF1, ZEB, Zfhep1) is a member of the ZFH family, containing zinc finger domains widely flanking a central homeodomain-like sequence [1, 2]. Zfhx1a is involved in smooth muscle differentiation, neurodifferentiation, skeletal patterning, lymphopoiesis and cancer metastasis [3–6]. Null mice have defects of craniofacial development, skeletal patterning, and severe T cell deficiency, and die at birth due to a failure to breathe, however, the cellular and molecular basis for this is unknown [5]. In humans, heterozygous mutations cause metaplasia of the cornea [7]. Zfhx1a is expressed as two different isoforms, Zfhx1a-1 which encodes the full-length protein and an N-terminal-deleted isoform termed Zfhx1a-2 [8]. Zfhx1a-2 mRNA does not contain the first exon of the gene, and therefore the protein is missing a PCAF/p300 interaction domain near the N-terminus, but contains the CtBP co-repressor and the SMAD binding sites [9]. Both Zfhx1a isoforms strongly bind the E2-box consensus sequence CACCTG [1 and references there in]. Binding and repression by Zfhx1a-1 has been shown with the GATA3, CD4, E-cadherin, p73 and IL2 target genes [10, 11]. However, little is known about how the Zfhx1a gene is regulated to bring about the observed developmental roles of Zfhx1a.
We initially carried out deletion analysis of the human Zfhx1a-1 promoter in order to identify the DNA elements that account for transcriptional activity in different cell types. Sequence analysis of the proximal promoter with TRANSFAC [12] identified consensus binding sites for the transcription factors Runx1, Sp1 and MZF1 and two putative Zfhx1a binding sites. We demonstrate that Zfhx1a-1 is able to repress expression of its own gene by binding to one of these E2-box sites. Surprisingly, the alternative isoform Zfhx1a-2 competes for binding and acts as a dominant negative of Zfhx1a-1, antagonizing auto-repression of the gene.
MATERIALS AND METHODS
Cell lines
CHO-K1 (Chinese Hamster Ovary), C2C12 (mouse myoblast), and Jurkat (human lymphoblastic T-leukemia) were obtained from the American Type Culture Collection (ATCC, Rockville, MD).
Reporter and expression plasmids
Full-length Zfhx1a-1 cDNA (3407 bp) and Zfhx1a-2 cDNA (2900 bp) were subcloned into the mammalian cell expression vector pCDNA4/HisMaxB (Invitrogen) and referred to as HisMax/Zfhx1a-1 and HisMax/Zfhx1a-2, respectively. The human Zfhx1a gene promoter was isolated by PCR amplification of human genomic DNA. Z1p.1000Luc contains from −913 to +44 of the human Zfhx1a gene inserted into a promoterless luciferase reporter vector (pGL3-basic, Promega, Madison, WI). Subclones of this promoter were prepared by standard cloning methods. The 5′-deletions are: Z1p.359Luc (−359 to +44 of the Zfhx1a promoter), Z1p.219Luc (−219 to +44), Z1p.133Luc (−133 to +44), Z1p.133ΔG (deleting bases −57 to −13 from Z1p.133Luc), and Z1p.12Luc (−12 to + 44). In addition, a 3′-deletion was prepared, Z1p.786ΔEagI, containing the fragment from −913 to −129 of the Zfhx1a promoter inserted into pGL3-promoter (Promega). All clones were confirmed by sequence analysis.
In vitro mutagenesis
A mutation of the E2-box at −120/−125 (Zfhx1a-binding site 2; BS2) was generated by using a sequential PCR strategy according to Cornack [13]. The following primers were employed: 5′-CTAGCAAAATAGGCTGTCCC-3′, 5′-TTCCTCAtaTGTGGTGGG-3′, 5′-CCCACCACAtaTGAGGAA-3′ and 5′-GGAACCAGGGCGTATCTC-3′. The PCR amplicons were subcloned into pGL3-basic, and verified by DNA sequencing.
Transfection Studies
C2C12 cells were transfected by the calcium-phosphate method [13]; CHO-K1 and Jurkat cells were lipofected with Fugene6 (Roche Diagnostics, IN). C2C12 cells were plated at a density of 1.5 × 105 cells and CHO-K1 cells at 2 × 105 per well in 6-well plates. CHO-K1 and C2C12 were transfected with 1.0 μg of the Zfhx1a-1 promoter construct or pGL3-basic and with 0.5 μg of a β-galactosidase vector, CMVβ, as a control for transfection efficiency. Jurkat cells were seeded in 12-well plates (1.5 × 106 cells/well) and transiently transfected with 0.7 μg of the Zfhx1a promoter construct and 0.3 μg of pCMVβ. For co-transfections, CHO-K1 cells were transfected with 1.0 μg of Zfhx1a-1 promoter constructs and 0.8 μg of either HisMax/Zfhx1a-1 or HisMax/Zfhx1a-2, and 0.6 μg CMVβ. Luciferase and β-galactosidase activity were evaluated as described (Cabanillas, 2001 #42). Results were expressed as luciferase/β-galactosidase activity. Means were compared by one-way ANOVA followed by Student-Newman-Keuls (SNK) tests or t-tests. P<0.05 was considered significant. All experiments were done in triplicate and were repeated at least 3 times.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from Jurkat cells as described [14]. Zfhx1a-2 protein was synthesized by in vitro translation in rabbit reticulocyte lysates (RRL) according to the manufacturer’s instructions (T7-TnT Quick Coupled Transcription/Translation System, Promega) using HisMax/Zfhx1a-2. Three different DNA probes were used and designated as: Binding Site 1 (BS1), an NheI/XhoI fragment of Z1p133ΔGLuc which contains the promoter sequence from −132 to −58; Binding Site 2 (BS2), an oligonucleotide spanning from −197 to −236 of the Zfhx1a-1 promoter (5′-TCTCCCCACCACACCTGAGGAAAACTTTTCCCTCGCCCCT-3′); and BS1+2, an NheI/XhoI fragment of Z1p268Luc with the DNA sequence from –268 to +7 of the Zfhx1a-1 promoter. In addition, a mutated version of BS2 (BS2 mut) was synthesized with the following sequence: 5′-TCTCCCCACCACAtaTGAGGAAAACTTTTCCCTCGCCCCT-3′. DNA fragments were labelled with [α-32P]dATP by fill-in with Klenow. One μl of the nuclear extract (3 μg) or 3 μl of Zfhx1a-2 programmed RRL were incubated with [32P]-labeled DNA probe (50,000 cpm) in binding buffer (20 mM Hepes, pH 7.9, 50 mM KCl, 2 mM MgCl2, 100 μM ZnCl2, 1 mM DTT, 0.6 mg/ml BSA, 0.6 mg/ml poly dI/dC) in a total volume of 15 μl and incubated at room temperature for 60 minutes. For supershift analysis, 0.2 μg of antibody was incubated with the protein 20 min before adding the DNA probe. Antibodies for supershifts were anti-Zfhx1a-1 antibodies directed against N-terminus (SC-E20) or C-terminus (SC-C20) peptides (Santa Cruz Biotech). Samples were loaded onto a 5% polyacrylamide gel and electrophoresed in 0.25X TBE buffer. The gel was exposed to Rx film overnight.
Potential Zfhx1a binding sites into the Zfhx1a-1 promoter were identified by comparison with the TRANSFAC transcription factor database [12] using MOTIF sequence motif search.
Western Blots
Nuclear extracts from Jurkat, SupT1, and C33a were used for western immunoblots with our polyclonal anti-Zfhx1a antibody as described [4, 14].
RESULTS
Regulatory elements in the Zfhx1a-1 promoter
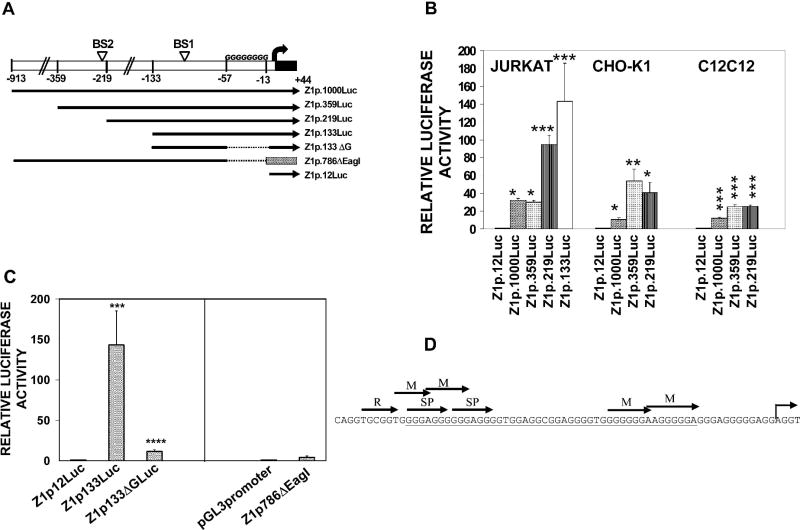
The human Zfhx1a-1 promoter was isolated to define the major regulatory elements in different cell types. Constructs containing 913 bp (Z1p.1000Luc), 359 bp (Z1p.359Luc), 219 bp (Z1p.219Luc), or 12 bp (Z1p.12Luc) of the promoter in a luciferase reporter (Fig. 1A) were transfected into CHO-K1, C2C12, or Jurkat cells. Data are expressed relative to Z1p.12Luc (control), which did not differ from the pGL3 vector. The highest activity was obtained in Jurkat cells, where Z1p.1000Luc was 32.4-fold higher (P<0.05) than Z1p.12Luc (Fig. 1B). Deletion of nucleotides −913 to −360 (creating Z1p.359Luc) had no effect on activity in Jurkat cells, however, the other cell types had increased activity with the smaller construct, indicating the presence of a cell-specific repressor element. Further deletion to −219 significantly increased (P< 0.001) Zfhx1a-1 promoter activity in Jurkat cells, indicating another repressor element (Fig. 1B). This element is not observed in CHO-K1, C2C12 (Fig. 1B) or P19 cells (not shown) suggesting that Zfhx1a-1 promoter has two different cell-specific regulatory regions.
Figure 1. Zfhx1a-1 promoter activity in Jurkat, CHO-K1 and C2C12 cells and localization of an essential Zfhx1a-1 promoter enhancer to the G-string.
(A) Schematic of DNA constructs containing the 5′-flanking region of Zfhx1a-1 gene in the pGL3 vector. Z1p.786ΔEag1 contains from −913 to −129 of the Zfhx1a promoter inserted into pGL3-promoter vector having the SV40 promoter (stripped box). The bent arrow is the transcriptional start site.
(B) Luciferase activity of 5′-deletions of Zfhx1a-1 promoter in Jurkat (immature lymphoblast), CHO-K1 (ovary) and C2C12 (myoblast) cells. Clones were transfected into triplicate wells and the results were expressed as luciferase/β-galactosidase activity. Values are the fold activation compared to Z1p.12Luc, expressed as the mean±SEM of three to six experiments. Means were compared by one-way ANOVA followed by Student-Newman-Keuls. *P<0.05 vs. Z1p.12Luc; **P<0.01 vs. Z1p.12Luc, ***P<0.001 vs. Z1p.12Luc.
(C) Deletions of the proximal promoter were transfected into Jurkat cells. Values are the fold activation compared to the appropriate controls (either Z1p.12Luc, or pGL3-promoter), expressed as the mean±SEM of three experiments. ****P<0.0001 vs. Z1p.133Luc. (D) The sequence of the human Zfhx1a promoter analyzed by TFSEARCH (at http://www.cbrc.jp/research/db/TFSEARCH.html) identifies candidate regulatory transcription factors; Runx (R), MZF1 (M), and SP1 (SP) binding sites. The underlined sequence is deleted in the Z1p.133ΔG construct.
In order to characterize the minimal Zfhx1a-1 promoter, additional clones were transfected into Jurkat cells. Expression of Z1p.133Luc was not significantly different from the 219 bp construct, whereas Z1p.786ΔEagI (containing bases −913 to −129 of the Zfhx1a gene driving an SV40 proximal promoter) had little activity, confirming that the strong activity of the Zfhx1a-1 promoter is within 128 bases of the start site (Fig. 1C).
The proximal promoter contains 77% guanosine nucleotides from −66 to −1 relative to the transcriptional start site, and lacks canonical TATA or CAAT boxes (Fig. 1D). To evaluate the importance of this G-string, a deletion of bases −57 to −13 was created in the context of Z1p.133Luc (termed Z1p.133ΔG). Luciferase expression of Z1p.133ΔG was strongly reduced (12-fold) as compared to Z1p.133Luc (P<0.0001, Fig. 1C), demonstrating the importance of the G-string in Jurkat cells. In addition, Z1p.133ΔG retained basal activity above the Z1p.12Luc control plasmid.
Zfhx1a-1 autoregulates its promoter
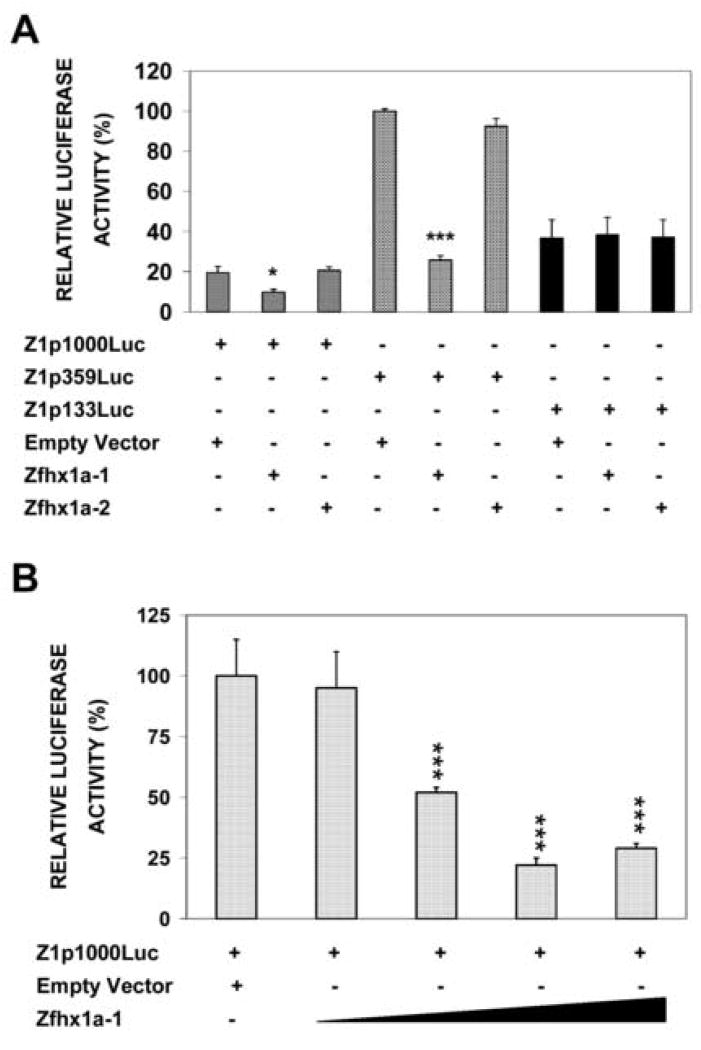
Analysis of the Zfhx1a-1 promoter sequence with the TRANSFAC program revealed two potential binding sites for Zfhx1a protein. Binding site 1 (BS1) is on the lower strand at −63 to − 68, and is preceded by two imperfect sites between −97 and −112. BS2 is at nucleotide −220 to −225 of the Zfhx1a-1 promoter. These binding sites have perfect E2-box sequences (CACCTG) for Zfhx1a binding [15]. To investigate the functional importance of these elements we first performed transient transfection assays. Zfhx1a-1 promoter-luciferase constructs were co-transfected with Zfhx1a-1 or Zfhx1a-2 expression clones or with the empty expression vector (HisMax) into CHO-K1 cells. Zfhx1a-1 cotransfection repressed the basal activity of Z1p1000Luc to 50 % and repressed Z1p359Luc to 25 % (Fig 2A lanes 2 and 5). Co-transfection of increased amounts of Zfhx1a-1 caused increased repression (Fig. 2B). However, Zfhx1a-2 did not repress the activity of the promoter (Fig 2A lanes 3 and 6). Interestingly, neither of the Zfhx1a isoforms were able to regulate the activity of Z1p133Luc (Fig 2A lanes 7–9). These results indicate that Zfhx1a-1 represses its own gene, however, the proximal binding site, BS1, is not sufficient for regulation. Autoregulation requires sequences between −359 and −133, which contains BS2.
Figure 2. Zfhx1a-1 represses its own promoter activity, but Zfhx1a-2 does not.
(A) Co-transfection of Zfhx1a promoter clones with Zfhx1a-1 or -2 expression plasmids. Cells were co-transfected with the indicated reporter constructs and Zfhx1a-1 or Zfhx1a-2 expression vectors, or the empty expression vector (pcDNA4/HisMax). The promoter activity of each construct was normalized to the condition with the highest activity. (B) CHO-K1 cells were co-transfected with 1.0 μg of Z1p1000Luc and with 0.5 μg, 0.8 μg, 1.0 μg and 1.3 μg of the Zfhx1a-1 expression vector, or with 1.0 μg of the empty vector. Results are mean±SEM in both figures (n=4–6).
Both Zfhx1a isoforms bind to the Zfhx1a-1 promoter
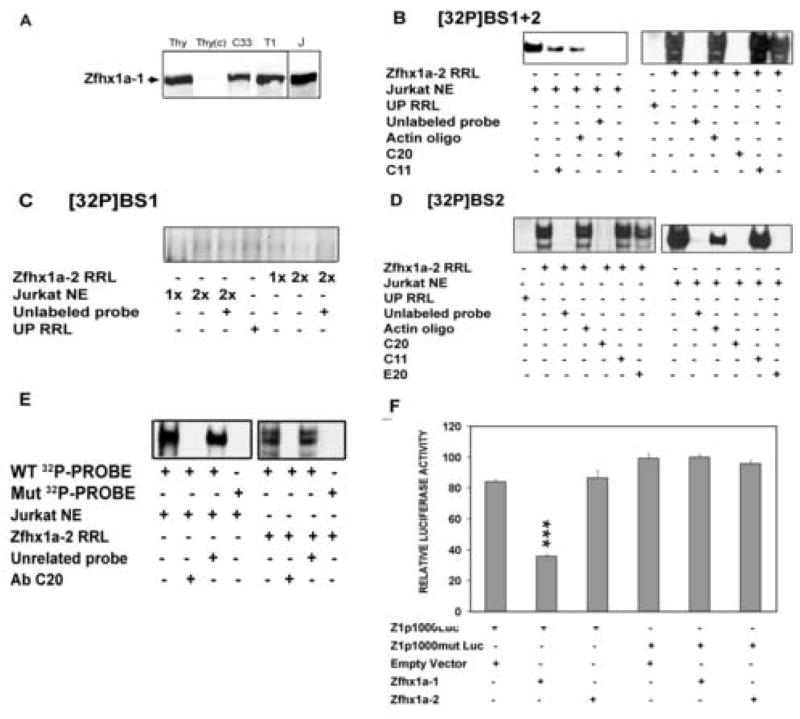
EMSAs were used to examine the formation of specific Zfhx1a complexes at each binding site. We used nuclear extracts from Jurkat cells as a source of Zfhx1a-1 protein because of the high endogenous expression of Zfhx1a-1 (Fig 3A). Zfhx1a-2 protein was synthesized in vitro in rabbit reticulocyte lysates. In vitro translation performed in the presence of [35S] methionine showed three major bands specific for Zfhx1a–2, which correlate with three well defined Zfhx1a-2-specific bands on EMSA analysis.
Figure 3. BS2 sequence is necessary for each Zfhx1a isoform binding to Zfhx1a-1 promoter.
(A) Zfhx1a is strongly expressed in Jurkat and mouse thymocytes. Western analysis of Zfhx1a protein expression in thymocyte nuclear extract (Thy), and cytosol (Thy(c)) and nuclear extracts from C33a (C33), SupT1 (T1), and Jurkat (J) cells. (B) EMSAs were performed with Jurkat nuclear extracts (NE), Zfhx1a-2 programmed reticulocyte lysate (Zfhx1a-2 RRL) or unprogrammed reticulocyte lysate (UP RRL) plus [32P]BS 1+2 oligonucleotide. (C) EMSA using the same protein sources and the [32P]BS1 probe, showing no binding of protein. (D) EMSA using the [32P]BS2 probe. 32P-probes were competed by 100-fold molar excess of cold specific oligonucleotides (Unlabeled probe) or cold actin oligonucleotide (Actin oligo). Antibodies used for supershift are a) SC-E20 Zfhx1a N-terminus antibody (E20), b) SC-C20 Zfhx1a C-terminus antibody (C20), and c) anti-Actin antibody (C11). (E) EMSAs with Jurkat NE or Zfhx1a-2 RRL were incubated with the wild type [32P]BS2 probe (WT 32P-PROBE) or its mutant (Mut 32P-PROBE). (F) CHO-K1 cells were co-transfected with 0.8 μg of the Zfhx1a-1, Zfhx1a-2 expression vector or the empty vector and 1.0 μg of the Z1p1000Luc or Z1p1000mut Luc. Results are expressed as mean±SD and are representative of 3 experiments with same results. ***P<0.001 vs. control (first column on the left).
Both Zfhx1a-1 and Zfhx1a-2 bind specifically to the BS1+2 probe, which contains both putative binding sites (Fig 3B). The complex observed with Jurkat nuclear extract was disrupted by anti-Zfhx1a-1 antibodies directed against either the N- (not shown) or C-terminus of the protein. The band containing Zfhx1a-2 and BS1+2 was supershifted only by the C20 antibody (raised against C-terminal region) but not by the N-terminus-directed antibody (E20) since Zfhx1a-2 lacks the N-terminus region of the full-length isoform. The addition of an unrelated antibody (anti-Actin) did not inhibit either Zfhx1a-1 or Zfhx1a-2 complex formation (Fig 3B).
We performed EMSAs using separate BS1 or BS2 probes. Neither Zfhx1a-1 nor Zfhx1a-2 bound the BS1 probe (Fig. 3C). On the contrary, Zfhx1a-1 and Zfhx1a-2 each showed strong complex formation with [32P]-BS2 (Fig. 3D lanes 2 and 8) that could be competed by an excess of the same cold oligonucleotide but not by the unrelated oligonucleotide. The Zfhx1a-1 complexes were supershifted by C20 and E20 antibodies but not by anti-actin antibody showing the specificity of the complex (Fig 3D). As expected Zfhx1a-2 complex formation was only disrupted by the C20 antibody to the C terminal sequence (Fig 3D, lane 5).
In order to ascertain whether the E-box in BS2 was involved in the Zfhx1a binding, we generated the BS2mut probe with a double mutation (CACCTG →CAtaTG) in the E2-box sequence, which disrupts the binding of Zfhx1a-1 [15]. As expected, neither of the Zfhx1a isoforms bound to this mutated probe (Fig 3E).
Functional studies using a Zfhx1a-1 promoter mutated in BS2 (Z1p1000mutLuc) showed no repression by either Zfhx1a-1 or Zfhx1a-2 expression vectors (Fig. 3F). Therefore, disruption of BS2 was sufficient to disrupt Zfhx1a-1 autoregulation.
Zfhx1a-1 and Zfhx1a-2 compete to bind and regulate the Zfhx1a-1 promoter
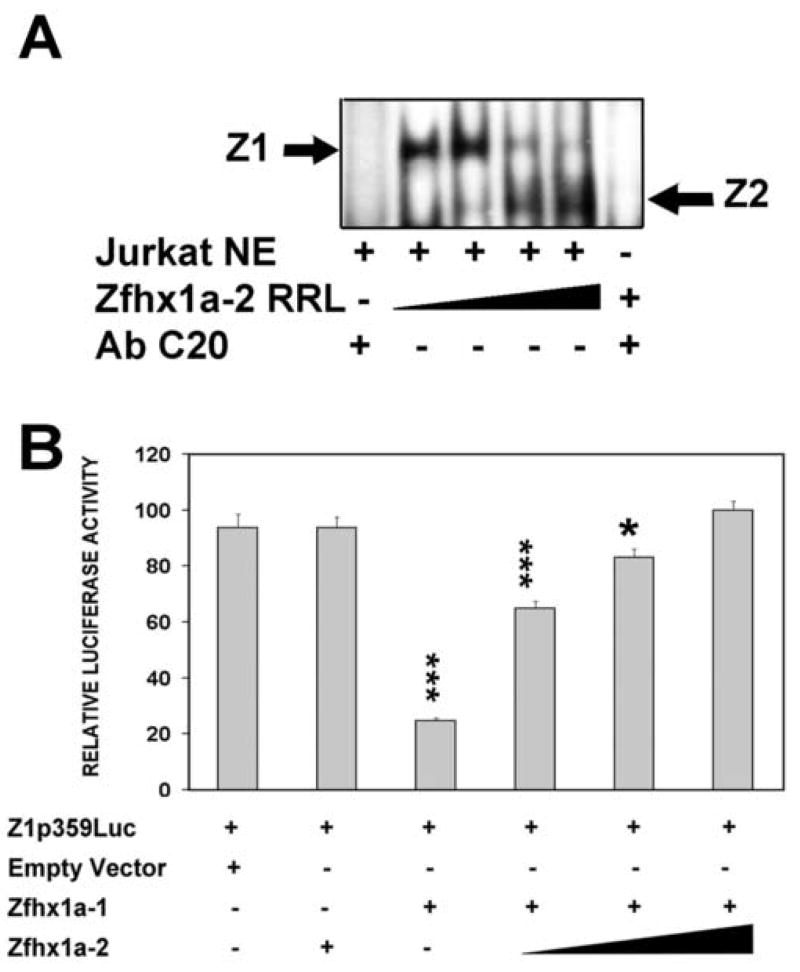
In spite of the strong binding of the Zfhx1a-2 isoform to the binding site 2, the protein did not regulate the Zfhx1a-1 promoter (Fig 2A). In order to investigate competition between the isoforms, we incubated 32P-BS2 with 1μl of Jurkat nuclear extract (containing Zfhx1a-1) and increasing amounts of RRL programmed with Zfhx1a-2. The resulting complexes were resolved by EMSA. As shown in Figure 4A, increased amounts of Zfhx1a-2 resulted in a reduction of the signal of the Zfhx1a-1 complex, while increasing the signal of the lower Zfhx1a-2 complex. Specificity of both complexes was determined by addition of an anti-Zfhx1a antibody that disrupted both complexes (Fig. 4 lanes 1 and 6). Binding of the isoforms to the BS2 site seems to be mutually exclusive. The function of Zfhx1a-2 was also studied by transfections. While Zfhx1a-2 alone does not regulate its promoter, Zfhx1a-1 alone repressed the promoter, and addition of increasing amounts of Zfhx1a-2 progressively relieved that repression (Fig 4B). Hence, Zfhx1a-2 disrupts auto-repression of the gene by Zfhx1a-1, apparently by competition at the DNA-binding site.
Figure 4. Zfhx1a-2 blocks the silencer activity of Zfhx1a-1 and displaces Zfhx1a-1 from its binding site.
(A) EMSA where the [32P]BS2 probe was incubated with 1.0 μl of Jurkat NE plus increasing amounts of Zfhx1a-2 RRL (1–9 μl). (B) CHO-K1 cells were co-transfected with 0.8 μg of Z1p359Luc, 0.8 μg of the Zfhx1a-1 expression vector and increasing amounts (0.25 μg, 0.5 μg and 0.8 μg) of the Zfhx1a-2 expression vector. Total DNA concentrations were equalized using the empty expression vector. Results are expressed as mean±SD and they are representative of 3 experiments with same results. *P<0.05, ***P<0.001 vs. control (first column on the left).
DISCUSSION
Functional analysis of the Zfhx1a-1 promoter shows cell specific positive and negative regulatory elements. However, the proximal promoter region is sufficient to support much of the transcriptional activity of the Zfhx1a-1 gene. Analysis of the human Zfhx1a-1 promoter in Jurkat cells, a human immature single positive lymphoblast cell line, identified a key guanosine-rich region at positions −1 to −66 relative to the transcriptional start site. The basal activity of this TATA-less promoter appears to be concentrated in this region. Sequence analysis with Transfac 5.0 (using 100% core factor identity and matrix identity >90%) identified consensus binding sites for the transcription factors Runx1 (AML-1/CBFA2), SP1, and MZF1 (Znf42), all of which are conserved within the mouse and human G-string sequence (Fig. 1D). These three factors are involved in hematopoiesis [16] [17], and Runx1 is absolutely required for normal differentiation of the T and B cell lineages [18]. Runx1 mRNA is highly expressed in double negative (DN) thymocytes [19] where it participates in differentiation of DN1-4 populations [18]. Runx1 may act in part by activating the Zfhx1a gene through the G-string sequence since differentiation of DN cells is disrupted in Zfhx1a-null mice [3]. Further studies are needed to characterize the expression of Zfhx1a-1 during lymphopoiesis.
Our studies indicate that Zfhx1a-1 protein is capable of repressing its own promoter by binding directly to one of two consensus sites. Transfection and DNA-binding assays identified BS2 as the active repressor element, and demonstrated that BS1 was not active despite having a perfect E2-box sequence. The functionally important BS2 site is located − 220 to −225 relative to the transcriptional start site and may contribute to the repressor element observed at −220 to −359 in Jurkat cells (Fig. 1B). Multiple Zfhx1a-binding motifs do not seem to be critical for Zfhx1a-1 autoregulation as described previously for other Zfhx1a target genes [15, 20, 21].
Zfhx1a-2 isoform lacks the N-terminal p300 and P/CAF interaction domain [9], and most of the N-terminal zinc finger cluster [8]. However, Zfhx1a-2 does contain the CtBP-interaction domain, which mediates repression by binding CtBP1/2. The P/CAF-interaction domain allows P/CAF to acetylate the CtBP binding site, thereby disrupting the ability of Zfhx1a to repress transcription [9]. Since Zfhx1a-2 lacks the P/CAF interacting domain, it might be anticipated to have constitutive repressor activity. However, our functional data indicate that Zfhx1a-2 is not able to repress the Zfhx1a-1 promoter in spite of strong binding to the E2-box present in BS2. Competition experiments demonstrate that Zfhx1a-2 competes with the full length Zfhx1a-1 protein for the BS2 site, behaving as a dominant negative isoform of Zfhx1a. We have previously shown tissue-specific expression of the Zfhx1a-2 (Zfhep-2) mRNA [8], suggesting that this isoform modifies the activity of Zfhx1a-1 in a cell-specific manner.
In conclusion, we show that the Zfhx1a gene is autoregulated at the promoter level and also through the expression of a dominant negative isoform, Zfhx1a-2. In addition to this transcriptional regulation, separate results from our laboratory indicate that the activity of Zfhx1a protein is regulated by changes of phosphorylation [14]; Arroyo et al, submitted).
Acknowledgments
This work was supported by NIH DE13614 (to DSD), CONICET and SECYT-UNC grants (to AMC). AMC is an Associate Researcher from CONICET, and was supported partially by the FULBRIGHT SCHOLAR PROGRAM.
Footnotes
We have previously termed this gene ‘Zfhep,’ however, will now refer to it as ‘Zfhx1a’. Accordingly, Zfhep-1 is Zfhx1a-1 and Zfhep-2 is Zfhx1a-2. Other names for this gene in vertebrates include TCF8, ZEB, δEF1, BZP, Nil2a, and AREB6.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Darling D, Gaur N, Zhu B. A zinc finger homeodomain transcription factor binds specific thyroid hormone response elements. Mol Cell Endocrinol. 1998;139:25–35. doi: 10.1016/s0303-7207(98)00076-8. [DOI] [PubMed] [Google Scholar]
- 2.Williams T, Moolten D, Burlein J, Romano J, Bhaerman R, Godillot A, Mellon M, Rauscher F, 3rd, Kant J. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science. 1991;254:1791–1794. doi: 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- 3.Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, Kikutani H, Kondoh H. Impairment of T Cell Development in delta EF1 Mutant Mice. J Exp Med. 1997;185:1467–1480. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darling D, Stearman R, Qi Y, Qiu M, Feller J. Expression of Zfhep/deltaEF1 protein in palate, neural progenitors, and differentiated neurons. Gene Expr Patterns. 2003;3:709–717. doi: 10.1016/s1567-133x(03)00147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takagi T, Moribe H, Kondoh H, Higashi Y. DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development. 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- 6.Nishimura G, Manabe I, Tsushima K, Fujiu K, Oishi Y, Imai Y, Maemura K, Miyagishi M, Higashi Y, Kondoh H, Nagai R. DeltaEF1 mediates TGF-beta signaling in vascular smooth muscle cell differentiation. Dev Cell. 2006;11:93–104. doi: 10.1016/j.devcel.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Krafchak CM, Pawar H, Moroi SE, Sugar A, Lichter PR, Mackey DA, Mian S, Nairus T, Elner V, Schteingart MT, Downs CA, Kijek TG, Johnson JM, Trager EH, Rozsa FW, Mandal MN, Epstein MP, Vollrath D, Ayyagari R, Boehnke M, Richards JE. Mutations in TCF8 cause posterior polymorphous corneal dystrophy and ectopic expression of COL4A3 by corneal endothelial cells. Am J Hum Genet. 2005;77:694–708. doi: 10.1086/497348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabanillas A, Darling D. Alternative splicing gives rise to two isoforms of Zfhep, a zinc finger/homeodomain protein that binds T3-response elements. DNA Cell Biol. 1996;15:643–651. doi: 10.1089/dna.1996.15.643. [DOI] [PubMed] [Google Scholar]
- 9.Postigo A, Depp J, Taylor J, Kroll K. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003;22:2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Grunsven L, Schellens A, Huylebroeck D, Verschueren K. SIP1 (Smad Interacting Protein 1) and {delta}EF1 ({delta}-Crystallin Enhancer Binding Factor) are Structurally Similar Transcriptional Repressors: A Current Survey of Their Functions and Mechanisms of Action in Transforming Growth Factor-{beta} Signalling. J Bone Joint Surg Am. 2001;83:S40–47. [PubMed] [Google Scholar]
- 11.Fontemaggi G, Gurtner A, Strano S, Higashi Y, Sacchi A, Piaggio G, Blandino G. The Transcriptional Repressor ZEB Regulates p73 Expression at the Crossroad between Proliferation and Differentiation. Mol Cell Biol. 2001;21:8461–8470. doi: 10.1128/MCB.21.24.8461-8470.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, Krull M, Matys V, Michael H, Ohnhauser R, Pru{beta} M, Schacherer F, Thiele S, Urbach S. The TRANSFAC system on gene expression regulation. Nucl Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current Protocols in Molecular Biology. Wiley & Sons, Inc; New York: 2001. [Google Scholar]
- 14.Costantino M, Stearman R, Smith G, Darling D. Cell-specific phosphorylation of Zfhep transcription factor. Biochemical and Biophysical Research Communications. 2002;296:368–373. doi: 10.1016/s0006-291x(02)00880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, Verschueren K, Smith JC, Huylebroeck D. New mode of DNA binding of multi-zinc finger transcription factors: delta EF1 family members bind with two hands to two target sites. EMBO J. 1999;18:5073–5084. doi: 10.1093/emboj/18.18.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen QY, Jackson N. Human CD1D Gene Has TATA Boxless Dual Promoters: An SP1-Binding Element Determines the Function of the Proximal Promoter. J Immunol. 2004;172:5512–5521. doi: 10.4049/jimmunol.172.9.5512. [DOI] [PubMed] [Google Scholar]
- 17.Hromas R, Collins S, Hickstein D, Raskind W, Deaven L, O’Hara P, Hagen F, Kaushansky K. A retinoic acid-responsive human zinc finger gene, MZF-1, preferentially expressed in myeloid cells. J Biol Chem. 1991;266:14183–14187. [PubMed] [Google Scholar]
- 18.Taniuchi I, Osato M, Egawa T, Sunshine M, Bae S, Komori T, Ito Y, Littman D. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 19.Levanon D, Groner Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23:4211–4219. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- 20.Postigo A, Dean D. Independent Repressor Domains in ZEB Regulate Muscle and T-Cell Differentiation. Mol Cell Biol. 1999;19:7961–7971. doi: 10.1128/mcb.19.12.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabanillas A, Smith G, Darling D. T3-activation of the rat growth hormone gene is inhibited by a zinc finger/homeodomain protein. Mol Cell Endocrinol. 2001;181:131–137. doi: 10.1016/s0303-7207(01)00531-7. [DOI] [PubMed] [Google Scholar]