Summary
Regulated activity of Src kinases is critical for cell growth. Src kinases can be activated by trans-phosphorylation of a tyrosine located in the central activation loop of the catalytic domain. However, because the required exposure of this tyrosine is not observed in the down-regulated X-ray structures of Src kinases, transient partial opening of the activation loop appears to be necessary for such processes. Umbrella sampling molecular dynamics simulations are used to characterize the free energy landscape of opening of the hydrophilic part of the activation loop in the Src kinase Hck. The loop prefers a partially open conformation where Tyr416 has increased accessibility, but remains partly shielded. An asymmetric distribution of the charged residues in the sequence near Tyr416, which contributes to shielding, is found to be conserved in Src family members. A conformational equilibrium involving exchange of electrostatic interactions between the conserved residues Glu310 and Arg385 or Arg409 affects activation loop opening. A mechanism for access of unphosphorylated Tyr416 into an external catalytic site is suggested based on these observations.
Keywords: RMSD restraint, phosphorylation, molecular dynamics, potential of mean force, umbrella sampling
Introduction
The Src-family tyrosine kinases are highly conserved allosteric enzymes playing a key role in cellular signaling. They catalyze the transfer of the terminal phosphate from an ATP molecule to a tyrosine residue in a substrate polypeptide sequence. Because their catalytic activity can be modulated in response to specific cellular signals, they are critical control points in signal transduction pathways governing cell growth and proliferation.1 Regulating their ability to phosphorylate tyrosine residues in their substrate proteins yields control over these pathways, whereas loss of such control results in the unchecked activation of downstream signaling pathways involved in diseases like cancer.
All nine members of the family (Src, Yes, Fyn, Lyn, Lck, Blk, Hck, Fgr, and Yrk) share a common structural topology comprising a catalytic tyrosine kinase domain preceded by two peptide-binding modules, the Src-homology domains SH2 and SH3. The SH2 and SH3 binding modules cooperate in the autoinhibitory interactions regulating the activity of the catalytic domain responsible for the phosphorylation.2 Downregulation of catalytic domain activity requires three simultaneous occurrences: Tyr416 (chicken c-Src numbering) in the activation loop remaining unphosphorylated, the SH3 module binding to the proline-containing linker between the SH2 module and the SH2 module at the N-terminus of the catalytic domain bound to the phosphorylated Tyr527 residue in the C-terminal tail of the catalytic domain.3 Conversely, disruption in any one of these three interactions may result in the upregulation of kinase activity. Amongst these three factors, phosphorylation of Tyr416 is the only activating factor localized in the immediate vicinity of the kinase active site in the catalytic domain.4
Different members of the Src family have been characterized structurally using X-ray crystallography: Hemopoetic cell kinase (Hck) has been crystallized in the inactive state 5,6 and recently in the active state 7; c-Src has been crystallized in the inactive state 8,9 and active states 10,11; and Lymphocytic cell kinase (Lck) has been crystallized in its active state with the activation loop tyrosine phosphorylated.12 Structures of these and other protein kinases such as c-AMP activated protein kinase A (PKA)13 and the insulin receptor kinase (IRK)14 allow clear structural distinctions to be drawn between the inactive and active states. The inactive state of the catalytic domain has the activation loop with Tyr416 buried into the catalytic site, and the α-C 4 helix (residues 304–316, shown in green in Figure 1B) rotated outward into the solvent with the catalytically critical Lys295-Glu310 interaction 5,8 disrupted. In the active state, there is an outward movement of the activation loop and an inward rotation of the α-C helix, which permits the formation of the Lys295-Glu310 pair. Structural studies on other protein kinases have shown that their catalytic domains adopt active state conformations similar to PKA, but may vary widely in their inactive state conformations.15
Figure 1.
(A) The reference states used for the umbrella sampling MD simulations and the equilibrated states that are obtained after unrestrained equilibration of these reference states; (B) Relevant structural motifs and residues in the Src kinase catalytic domain with the DFG motif comprised of Asp404-Phe405-Gly406 shown in tan and the α-C helix shown in green. 25
Trans-phosphorylation occurring via a bimolecular encounter with a second kinase (presumably of the same type) in its activated state is believed to be the dominant activation mechanism in Src.16, 17 The combined information from the crystallographic studies shows the conformational change occurring between the inactivated auto-inhibitory state to the fully activated state. The process clearly involves an outward movement of the activation loop with Tyr416, but also a significant rotation of helix α-C and a displacement of the N-terminus of the catalytic domain. It is unclear, however, whether presenting Tyr416 to the active site of an incoming kinase in a transient bimolecular encounter event would require the complete conformational change or could be restricted to a limited segment of the activation loop. There is no apparent reason why the molecule that is being phosphorylated needs, itself, to assume the fully active conformation observed in the active state crystal structure. This view is supported by the fact that two distinctive regions of the activation loop can be clearly identified 18, a buried hydrophobic part comprising residues 402–411 and a flexible hydrophilic part comprising residues 412 to 424 (shown in red in Figure 1B). The buried part of the loop, including the highly conserved DFG motif 19 (shown in tan in Figure 1B), interacts with hydrophobic residues in the β-sheets of the N-lobe as well as residues in the α-C helix.18 This suggests that its motion might be tightly coupled to the larger conformational changes (such as the rotation of the α-C helix) that are required for attaining the fully active state. In contrast, the hydrophilic part of the activation loop including the Tyr416 residue is more readily solvent-accessible and relatively unencumbered. The likelihood that this region would show greater flexibility is also supported by the fact that it was partly disordered and unresolved in crystal structures of Hck and c-Src. 5,8 Therefore, the notion that Tyr416 could become accessible for trans-phosphorylation via a limited conformational change involving mainly the hydrophilic flexible region of the activation loop seems plausible on structural grounds.
In the present study, extensive umbrella sampling Molecular Dynamics (MD) simulations of an isolated and unphosphorylated Hck catalytic domain in explicit solvent are carried out to characterize the free energy landscape governing the opening and closing of the flexible segment of the activation loop (residues 412–424). The Root Mean Square Distance (RMSD) is used here as a generic “order parameter” to map the potential of mean force (PMF) between the two end-point reference states corresponding to the buried and partially accessible conformations of the activation loop, respectively. Umbrella sampling using similar RMSD biasing restraints have been used successfully in previous computational studies on Src kinases 18, 20, 21, 22, 23, 24 to study dynamic correlations, hinge-bending motions, bidirectional allostery, domain assembly, and structural details of activation in these proteins. Here, two conformational minima are identified for the activation loop, and the relationship between Tyr416 availability and interactions between residues on adjacent sides of the activation loop is delineated through the statistical analysis of conformations obtained during the umbrella sampling MD simulations. A proposed model, supported by sequence analysis, discusses the implications of interacting residue asymmetry in the activation loops of Src kinases.
Materials and Methods
The umbrella sampling calculations were carried out using the c33a2 academic version of the CHARMM program 25 and the all22 CHARMM force field for proteins 26. The starting “Y416-buried” state conformation of the Hck catalytic domain (residues 253–523) lacking the C-terminal tail was the 2.0 Å resolution crystal structure (PDB ID: 1QCF) in the presence of the inhibitor PP1. Since only the opening of the hydrophilic part of the activation loop was being studied, the “Y416-exposed” state structure was artificially modeled based on refinement of the open loop conformation of the 1.7 Å resolution Lck active form structure (PDB ID: 3LCK) 12 for Hck using sequence alignment through CLUSTALW 27 and homology modeling through MODELLER.28 The rest of the catalytic domain, including the buried part of the activation loop and the α-C helix, were in their inactive state conformation in this structure. These Y416-buried and Y416-exposed configurations were solvated using a 150 mM KCl box with dimensions 77.6 Å × 62.1 Å × 52.4 Å such that the edge of the box from any protein non-hydrogen atom was at least 8 Å. The systems were minimized with 5 kcal/mol harmonic restraints on non-hydrogen solute atoms for 500 Steepest Descent steps, followed by 100 Adopted Basis Newton Raphson steps. Periodic boundary conditions were created by generating images using the CRYSTAL module in CHARMM.29 Covalent bonds with hydrogen atoms were constrained using SHAKE 30 to allow an integration time step of 2 fs. The Particle Mesh Ewald (PME) approach 31 with a B-spline order of 4 and a Fast Fourier Transform grid of one point per Å and a real-space gaussian-width kappa of 0.3 Å−1 was used to treat long-range electrostatic interactions. Real space and Lennard-Jones (LJ) interaction cutoffs of 10 Å were used with non-bond interaction lists maintained and heuristically updated out to 16 Å. The migration of the solute protein outside the primary solvent box was discouraged by weak (0.5 kcal/mol) center-of-mass translational and rotational restraints using the MMFP module of CHARMM.32 Constant pressure and temperature (NPT) MD simulation 33 at 300 K was carried out with harmonic restraints on non-hydrogen atoms that were released gradually over the first 100 ps. For the Y416-buried state, the minimized crystal structure was used as the inactive state reference. For the Y416-exposed state, the artificial model prior to equilibration was expected to provide the widest separation from the inactive state crystal conformation and was therefore used as the reference structure. The Y416-buried and Y416-exposed starting reference states were subjected to 4 ns and 8 ns equilibration MD simulations, and as shown in Figure 1A, the inactive state (left) remains quite stable whereas the active state (right) relaxes to a conformation intermediate between the two reference states. Unrestrained simulations probing the influence of Arg409 motion on activation loop opening were carried out using NAMD version 2.6b2 34 using the same CHARMM force field and simulation setup and settings as described above. For these calculations, 12 crystal structures (1FMK, 1AD5, 1QCF, 1Y57, 1YI6, 2C0I, 2C0O, 2C0T, 2H8H, 2HCK, 2HK5, 2SRC) were used to generate initial models. The loop regions for structures in which they were disordered were modeled using the 2SRC 9 and 1QCF 6 structures that had these loop regions resolved. The resulting trajectories were analyzed using the CHARMM program.
For the umbrella sampling calculations, the initial structures for the intermediate windows bridging the Y416-buried and Y416-exposed conformations were generated by 5 separate MD simulations with different random seeds for assigning initial velocities and slightly different temperatures using a 1D RMSD restraint pulling the structure gradually (0.1 Å RMSD intervals with 10 ps of relaxation at each interval) from the inactive to the active state and back, yielding 10 sets of initial structures for each window with a total of 67 windows. The atoms included in the restraint were all non-hydrogen atoms of residues 412–424 of the activation loop except atoms involving terminal sidechain atoms with multiple interaction possibilities (e.g. carboxyl oxygens of glutamate and aspartate residues). All 10 starting configurations for each window were then subjected to umbrella sampling MD simulations with the restraint wj on the ΔDRMSD order parameter implemented in CHARMM,35
where Dmin is the desired minimum of the restraint potential. The ΔDRMSD order parameter is the subtraction between the RMSD values of each structure from the two reference states:
where Xt is the instantaneous structure, XY416-buried and XY416-exposed are the Y416-buried and Y416-exposed reference structures, respectively. For this coordinate, each window is separated by 0.2 Å. The force constant for this harmonic restraint was gradually reduced from 500 kcal/mol/Å2 to 10 kcal/mol/Å2 over a period of 80 ps NPT MD simulation. The final system in each window was then allowed to evolve with the final weak relative 1D RMSD restraint of 10 kcal/mol/Å2 using NPT MD simulations for variable durations. In total, 5 independent forward and 5 backward umbrella sampling MD simulations ranging from 0.2 ns to 2 ns per window were performed. Sampling for one forward simulation was extended to 5 ns sampling per window to test the convergence of the calculation. The total aggregate sampling time for the 10 umbrella sampling calculations for all the sets of windows combined (about 25000 atoms each) was > 0.9 µs. The ΔDRMSD values saved at each timestep of the production MD simulations were used to create a biased histogram along the ΔDRMSD reaction coordinate that was unbiased using the Weighted Histogram Analysis Method (WHAM) algorithm 36,37 to get the PMF. All molecular pictures were produced using DINO (http://www.dino3d.org) or VMD (www.ks.uiuc.edu/Research/vmd), all graphs were made using the OPENDX program version 4.4.4 (http://www.opendx.org) or gnuplot version 4.0 (http://www.gnuplot.info) and the Gnu Image Manipulation Program (GIMP) version 1.2 (http://www.gimp.org).
Results
Choice of reference conformations
Umbrella sampling is a method that uses a biasing potential in Monte Carlo or MD simulations to enforce the occurrence of rare events along a given reaction coordinate.38 Unbiasing the resulting probability distributions allows computation of the potential of mean force exerted (or the free energy profile) along that reaction coordinate. Since the equilibrium opening and closing of the activation loop of Src kinases likely occurs on a very slow timescale for MD simulations, it is precisely the type of rare event amenable to umbrella sampling calculations. The first requirement for these calculations is choosing the “end-point” references states, which will enable to accurately monitor the conformational transformation via changes in the order parameter. The reference structures are important, though their details do not limit our ability to discover and sample of stable novel conformations that differs from both end-points. For example, in a previous umbrella sampling study on the relative stability of A-DNA and B-DNA in aqueous solution using “canonical” A- and B-forms as reference end-points, the global free energy minimum in the PMF of the CTCGAG hexamer did not match either of the two canonical end state conformations but rather corresponded almost exactly to its x-ray structure. 35
The reference structure chosen to represent the closed loop inactive state in which Tyr416 is partially buried in the active site, called “Y416-buried”, is the Hck crystal structure with all its residues resolved crystallographically, and Arg409 from the activation loop interacting with Glu310 in the α-C helix. 6,9 The relative position of the various important residues (Glu310, Arg385, Arg409, and Tyr416) and conformational motifs (DFG motif and α-C helix) near the active site for this Y416-buried reference are shown in Figure 1B. The reference structure on the opposite end of the reaction coordinate, called “Y416-exposed”, is a model in which only the relatively flexible segment of the activation loop (residues 412–424) has been modified to adopt the conformation it assumes in the active state structures. 7,12 It is shown on the upper right in Figure 1A (note that the active state crystal structures of Lck 12 and Hck 7 are essentially identical). The reference model is not expected to be a genuine stable state, which is confirmed by the relaxation of the protein conformation during an 8 ns simulation (see Figure 1A, lower right), but as stated above, it does not need to be stable to serve as an end-point reference for the RMSD order parameter in the umbrella sampling.
Free energy landscape of activation loop
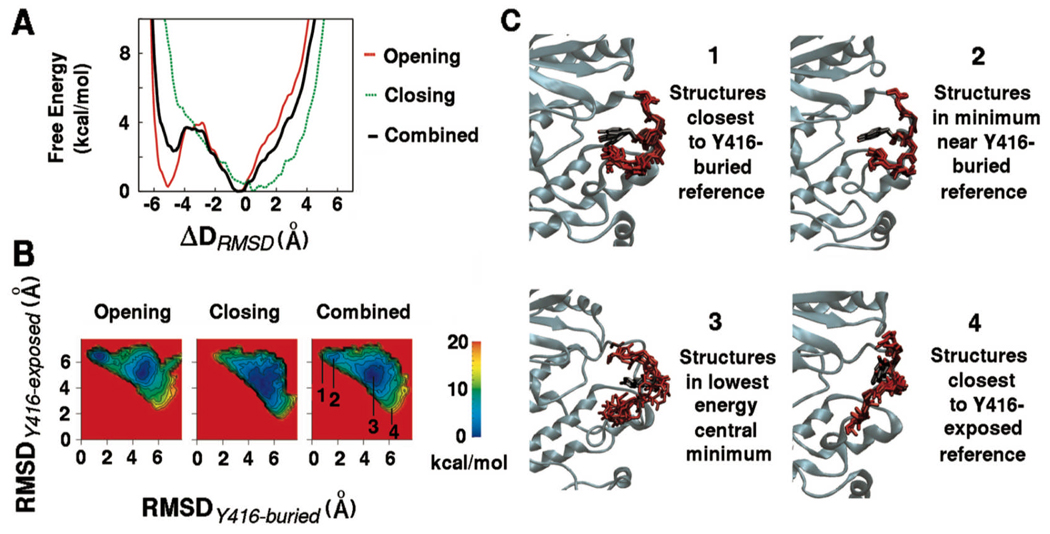
The potential of mean force (PMF) profiles obtained by consolidation of the five opening (Y416-buried to Y416-exposed) and five closing (Y416-exposed to Y416-buried) umbrella sampling MD simulations for the hydrophilic part of the activation loop are shown in Figure 2. It is clear from the one-dimensional (1D) profiles in Figure 2A and the two-dimensional (2D) profiles in Figure 2B that some hysteresis remains between the opening and closing directions used to generate initial window coordinates. Such hysteresis is consistent within a particular direction in that the five umbrella sampling calculations initiated from opening trajectories resemble one another in having their most stable minimum near the original Y416-buried state. A few of the calculations generated in the opening direction also show emergence of lower energy regions in states intermediate between the two references. The five umbrella sampling calculations initiated from closing trajectories also resemble one another (see supplementary information for more details). The combined PMFs from all 10 umbrella sampling MD simulations, which incorporate both pathways and are likely to have greatest accuracy 39,40, indicate the presence of a minimum energy configuration near the Y416-buried reference and a more stable configuration centrally located between the Y416-buried and the Y416-exposed reference structures. Extension of the sampling in one of the opening PMFs up to 5 ns per window leads to a free energy profile progressively resembling the combined PMF profile (see supplementary information), which indicates that the qualitative features of the free energy landscape are unlikely to change much upon further sampling.
Figure 2.
The potential of mean force (PMF) describing opening and closing of the hydrophilic part of the activation loop (residues 412–424). The PMFs for opening (5 simulations from Y416-buried to Y416-exposed states), closing (5 simulations from Y417-exposed to Y416-buried states), and combined (all 10 simulations) along (A) the 1D-ΔRMSD reaction coordinate; (B) the 2D of RMSD from the Y416-buried reference and RMSD from the Y416-exposed reference. The numeric labels indicate regions of the 2D RMSD space from which corresponding instantaneous snapshots of the activation loop were extracted; (C) Instantaneous snapshots of the activation loop extracted from specific regions of the 2D RMSD space. The snapshots labeled 1 are those closest to the starting crystal structure or the Y416-buried reference while the snapshots labeled 2 are those in the proximal minimum into which this starting crystal structure relaxes. Similarly, snapshots labeled 4 are those closest to the Y416-exposed reference and shapshots labeled 3 are from the central lowest energy minimum of the free energy landscape.
Figure 2C shows some instantaneous snapshots extracted from the umbrella sampling simulations corresponding to specific regions 2D space of the two RMSD variables. The snapshots were extracted from both opening and closing trajectories for loop residues 412–424. Snapshots chosen to represent the reference states (labels 1 and 4) are extracted from 2D RMSD regions that are within 20 kcal/mol of the global minimum. Snapshots labeled 1 are those closest to the Y416-buried reference while snapshots labeled 2 are those from the proximal minimum that this initial Y416-buried reference structure relaxes to. It is clear that these two sets of structures are very similar and are correspondingly very close in 2D RMSD space. The centrally located lowest free energy basin between the two reference structures covers a broad area of 2D RMSD space (label 3). The broader area indicates that a larger number of structures belong to this minimum, which suggests that conformational entropy might be contributing to its stability. The backbone conformations of the structures extracted from this minimum are similar. The helical nature adopted by residues 415–418 in the Y416-buried reference is not preserved, and Tyr416 adopts a wide range of sidechain conformations, which are all relatively unexposed. In contrast, the minimum located very close to the Y416-buried reference structure covers a smaller area of 2D RMSD space, confirming that it corresponds to an ordered structure similar to the Y416-buried reference crystal structure.6,9 The estimated barrier between these two stable states is close to 3 kcal/mol, suggesting that the transitions back and forth would be feasible (though the present calculations do not allow an estimation of the actual transition rate).
Loop opening and Glu310 exchange between Arg385 and Arg409
The crystal structures for c-Src and Hck Src kinases in the inactive state can be divided into two categories. The 1997 inactive structures have the central portion of the activation loop disordered with a salt bridge formed between Glu310 and Arg385 in the HRD motif.5,8 The 1999 inactive structures (again for both c-Src and Hck) have the activation loop fully ordered but the ion-pair formed by Glu310 is with Arg409 in the activation loop.6,9 The present study is focused only on the movement of the most flexible region around Tyr416 (residues 412–424). The inactive crystal structure, with all residues resolved and the interaction between Glu310 and Arg409 present, was therefore chosen as the Y416-buried reference state.6,9 The RMSD restraint is applied only to residues 412 to 424 in the umbrella sampling calculations and the salt bridges involving Glu310, Arg385 and Arg409 are not directly affected by the biasing restraint during the simulations. Nevertheless, it is of interest to correlate the present results with the two possible Glu-Arg salt bridge organizations observed experimentally.
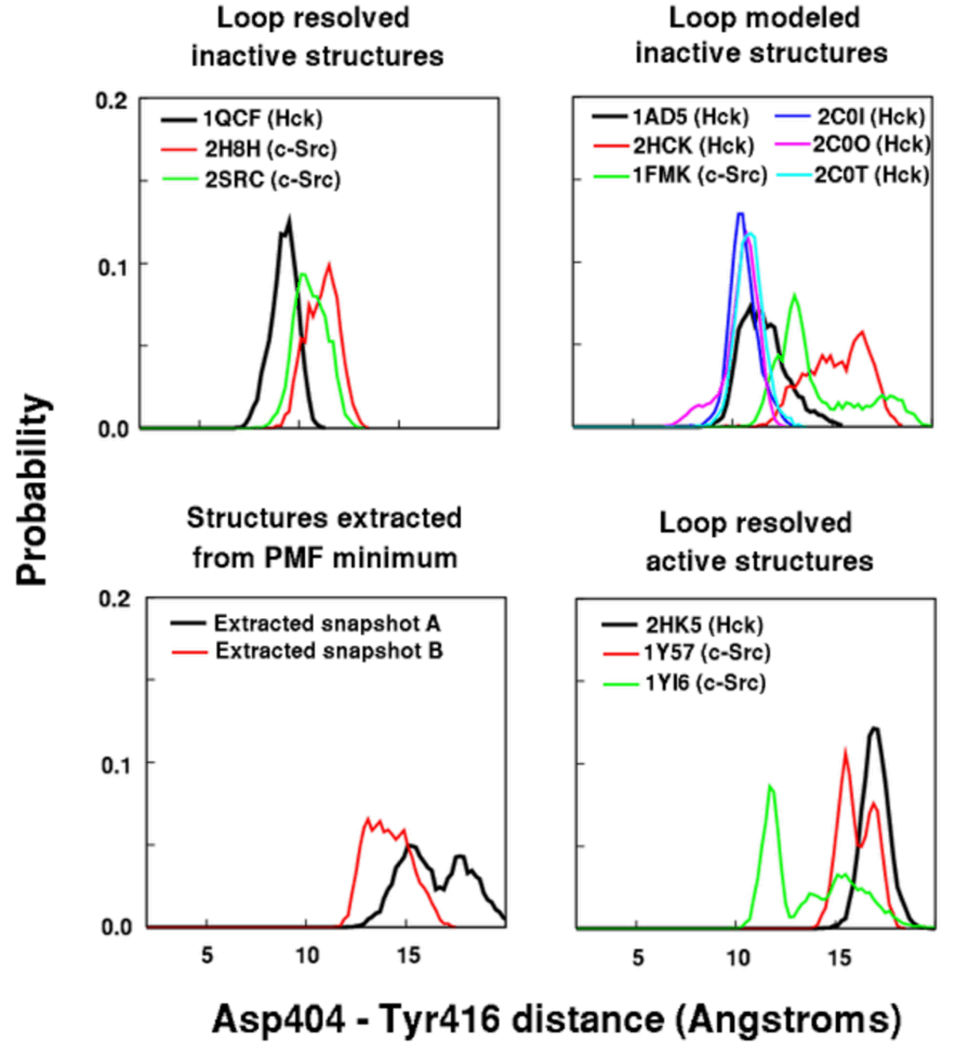
To further assess the loop opening movements around Tyr416 allowed by the two different salt bridge organizations, we carried out additional unrestrained explicit solvent MD simulations on all available structures of either Hck or c-Src kinase with PDB IDs: 1FMK 8, 1AD5 5, 1QCF 6, 1Y57 10, 1YI6 11, 2C0I, 2C0O, 2C0T 41, 2H8H 42, 2HCK 5, 2HK5 7, 2SRC 9, and two structures extracted from the most stable minimum in the computed energy landscape. For structures that had loop residues unresolved, these residues were modeled based on their conformation in the corresponding structures where they are resolved.6,9 Figure 3 shows the probability distributions of the distance between the active site residue Asp404 (sidechain Cγ atom) and Tyr416 (sidechain O atom) for all these systems. This distance monitors the movement of Tyr416 away from the active site since Asp404 is relatively immobile. As expected, the structures where the entire loop adopts its active state structure (2HK5, 1Y57, and 1YI6), the distance shows Tyr416 remaining stable away from the active site. In the structures that have Arg409 dissociated from Glu310 and flipped into the active site (1AD5, 2HCK, and 1FMK), Tyr416 shows a greater propensity to move away from the Asp404. In the structures that maintain the Arg409-Glu310 interaction (1QCF and 2SRC), Tyr416 tends to remain in its original buried position closer to the active site. The two structures extracted from the global free energy minimum (#3 in Figure 2B and 2C) have Tyr416 populating states away from the active site.
Figure 3.
The probability distributions of the distance between the active site (represented by Asp404) and the Tyr416 residue in unrestrained simulations of 12 c-Src and Hck crystal structures labeled by their PDB IDs. The structures that have Arg409 displaced from its interaction with Glu310 by Arg385 (1AD5, 2HCK, 1FMK) show a spontaneous tendency for Tyr416 to move away from its starting conformation near the active site.
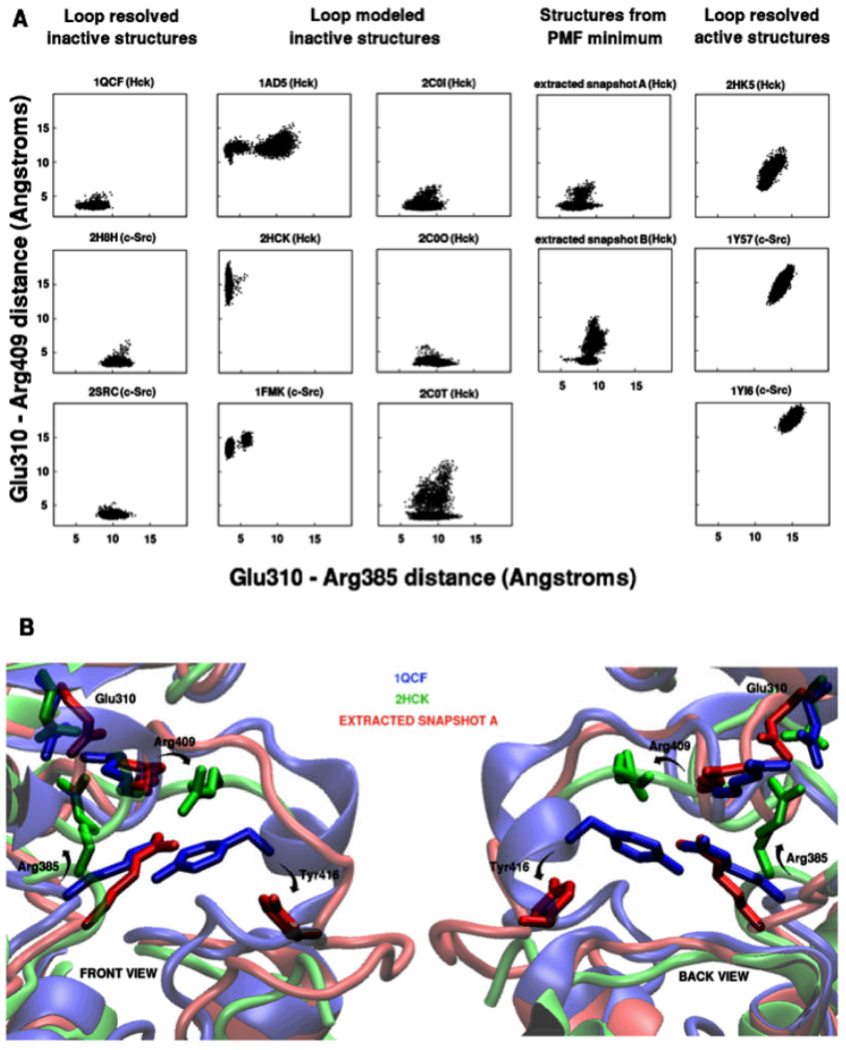
An important issue is whether the Glu-Arg salt bridge organization is affected indirectly by the opening up of the flexible part of the loop. Figure 4A shows the scatter plots of interaction distances between Glu310 (Cδ atom) and NH1 or NH2 (whichever is closer) atoms of Arg385 (X-axis) or Arg409 (Y-axis) for all the unrestrained simulations. Of the structures that start with the Glu310-Arg385 interaction present (1FMK, 1AD5, and 2HCK), 1FMK and 1AD5 show some tendency of Arg385 to dissociate and re-associate with Glu310, but this interaction is very stable in 2HCK. Dissociation of the Glu310-Arg385 pair is not accompanied by corresponding association of the Glu310-Arg409 pair in these systems. Active state structures (1Y57, 1YI6, and 2HK5), where neither Arg385 nor Arg409 interact with Glu310, maintain the separation between these residue pairs. Of the structures that start with the Glu310-Arg409 interaction present, all Hck systems except 1QCF show some dissociation and reassociation of this interaction. In 1QCF and the corresponding c-Src systems (2H8H and 2SRC), the interaction seems to be more stable. In the 5 ns timescale of the simulations, no exchange occurs between the Glu310-Arg385 and the Glu310-Arg409 interactions.
Figure 4.
A. Scatter plots showing distribution of Glu310-Arg385 and Glu310-Arg409 distances in simulations of all available crystal structures of c-Src and Hck (1FMK, 1AD5, 1QCF, 1Y57, 1YI6, 2C0I, 2C0O, 2C0T, 2H8H, 2HCK, 2HK5, and 2SRC), along with 2 extracted structures from the global minimum of the computed free energy landscape (labeled as extracted snapshots A and B); B. Final structures from the equilibration simulations of 1QCF (blue), 2HCK (green), and extracted snapshot A (red). Residue rearrangements distinguishing 2HCK and 1QCF are change of orientation of Arg385 to interact with Glu310 and flipping of Arg409. Extracted snapshot A shows only displacement of Tyr416 without Arg409 or Arg385 rearrangements. The section of 2HCK unresolved in the crystal structure (residues 412–421) is not shown for clarity.
The umbrella sampling simulations for activation loop opening indicate that the opening of the stretch of residues 412–424 is compatible with leaving the ion-pair Glu310-Arg409 intact. However, unrestrained simulations do show that Arg385 can approach Glu310 in the Y416-exposed conformation corresponding to the umbrella sampling PMF free energy minimum within a 5 ns timescale. Figure 4B shows these rearrangements of the residues Arg385, Arg409, and Tyr416 in the equilibrated structures of 1QCF, 2HCK, and a snapshot extracted from the PMF minimum. The rearrangement of Arg409 and Arg385 that occurs in 2HCK, such that Arg385 replaces Arg409 in interacting with Glu310, does not occur in the PMF minimum where Arg409 still maintains its interaction with Glu310, even though the loop is partially open and Tyr416 is relatively exposed. Based on the distributions in Figure 3 and the scatter plots in Figure 4A, it is expected that flipping of Arg409 as seen in 2HCK is likely to stabilize the partially open loop conformation seen in the lowest energy PMF minimum by destabilizing the closed loop conformation of Tyr416 and its surrounding residues seen in 1QCF. Thus, even if an exchange of the Glu310-Arg409 interaction for the Glu310-Arg385 interaction may not necessary for reaching the Y416-exposed state (#3 in Figure 2B and C), the system seems poised to allow such an exchange once this state is reached.
Overall, these results suggest that Tyr416 displays a propensity to move into more exposed positions when Arg385 displaces Arg409 as the residue interacting with Glu310, and Arg409 flips into the active site. This is consistent with the fact that the loop residues surrounding Tyr416 are disordered only in the crystal structures where the Glu310-Arg385 interaction is present, the Glu310-Arg409 interaction is absent. These results also suggest that formation of the Glu310-Arg385 interaction could lead to a further stabilization of the Y416-exposed free energy minimum state detected in the umbrella sampling calculations.
Asymmetric interactions between activation loop residues
Activation of the kinase involves the reorganization of complex networks of electrostatic interactions.18,23 In particular, the presence of multiple charged residues in the neighborhood of Tyr416 in the activation loop allow the possibility of a number of interactions, which could be especially relevant in stabilizing intermediate states during loop opening and closing. Due to the overall conformation of the activation loop and its orientation with respect to the catalytic domain, interactions between oppositely charged residues that are on alternate sides of Tyr416 in the primary sequence have the potential to shield the tyrosine sidechain from the environment. In the inactive state crystal structure 6,9, there is a good example of just such an electrostatic interaction between Asp413 and Lys423. The conformations obtained during the umbrella sampling MD simulations provide the opportunity to detect the transient formation of other such electrostatic interactions within the activation loop.
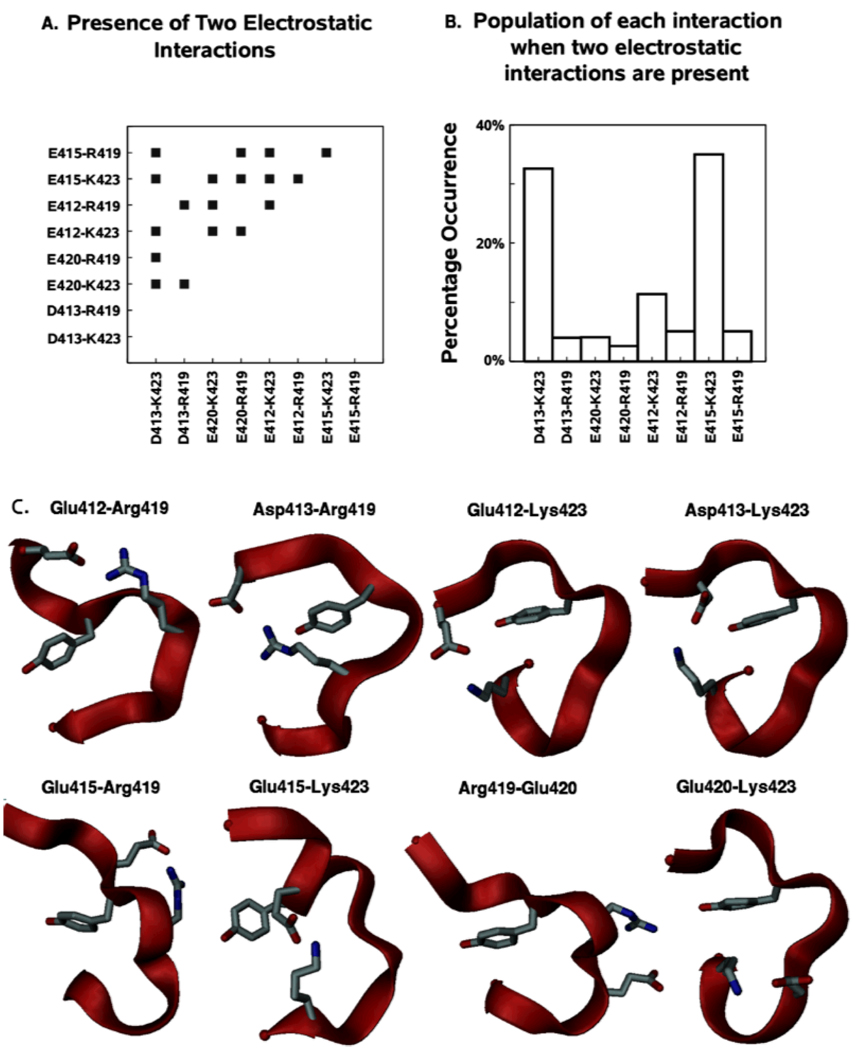
Figure 5 shows the presence and relative frequency of electrostatic interactions in the hydrophilic region of the Hck activation loop during opening and closing. It is clear that all 8 possible salt links between the four negatively charged (Glu412, Asp413, Glu415, and Glu420) and the two positively charged residues (Arg419 and Lys423) occur individually during activation loop motion. There is also an intrinsic asymmetry in the distribution of these charged residues, such that six of these interactions actually occur across the Tyr416 residue. Only two occur where both residues are on the same side (N-terminal or C-terminal) with respect to Tyr416. In addition, there is a substantial population of two or more salt bridges occurring simultaneously. Amongst these, the two most frequently occurring salt links (Asp413-Lys423 and Glu415-Lys423) between segments of the activation loop on opposite sides of Tyr416 in the primary sequence. Such salt links acts as “clips” that staple the two bordering regions of Tyr416 to each other, thereby decreasing the freedom of motion and accessibility of Tyr416. Each salt link has its own local association and dissociation kinetics. Since different salt links are seen in the Y416-buried and the Y-416 exposed states, any lag in association/dissociation of individual salt links while transitioning between the two structures, would appear as hystereses between the forward and backward umbrella sampling trajectories. The multiplicity of possible salt links also raises the possibility that various backbone conformations of the loop could be stabilized by different combinations of sidechain salt links. Such multiple salt links could therefore be a contributing factor in the structural plasticity observed in protein kinase activation loop regions.15
Figure 5.
Electrostatic interactions in the Hck activation loop opening and closing umbrella sampling MD simulations. (A) Shows the presence of specific pair-wise interactions between all charged residues in the hydrophilic region using a distance criteria, (B) shows the percentage occurrence of each interaction when two or more interactions occur simultaneously, (C) Structures extracted from the umbrella sampling MD simulations showing presence of each of the charged residue interactions by indicating interacting sidechains along with the Tyr416 sidechain in stick form.
Possible role of charge asymmetry in trans-phosphorylation
Trans-phosphorylation requires one Src kinase to present its Tyr416 residue to the active site of a second Src kinase molecule in the activated state. The activation loop charged residue asymmetry detected in the umbrella sampling MD simulations is suggestive of a possible mechanism by which such access may be facilitated (illustrated in Figure 6). Let us consider two Src kinase molecules, A and B. If phosphorylation of its Tyr416 residue has previously activated kinase molecule B, then some of its activation loop charged residues are effectively released from their intramolecular interactions. This active kinase molecule B can then bind to kinase molecule A and its released charged residues can then seek complementary interactions with oppositely charged residues in the activation loop of kinase molecule A. This association can then allow Tyr416 from kinase molecule a greater access to the active site of kinase molecule B leading to the phosphorylation of Tyr416 from kinase molecule A in the active site of kinase molecule B.
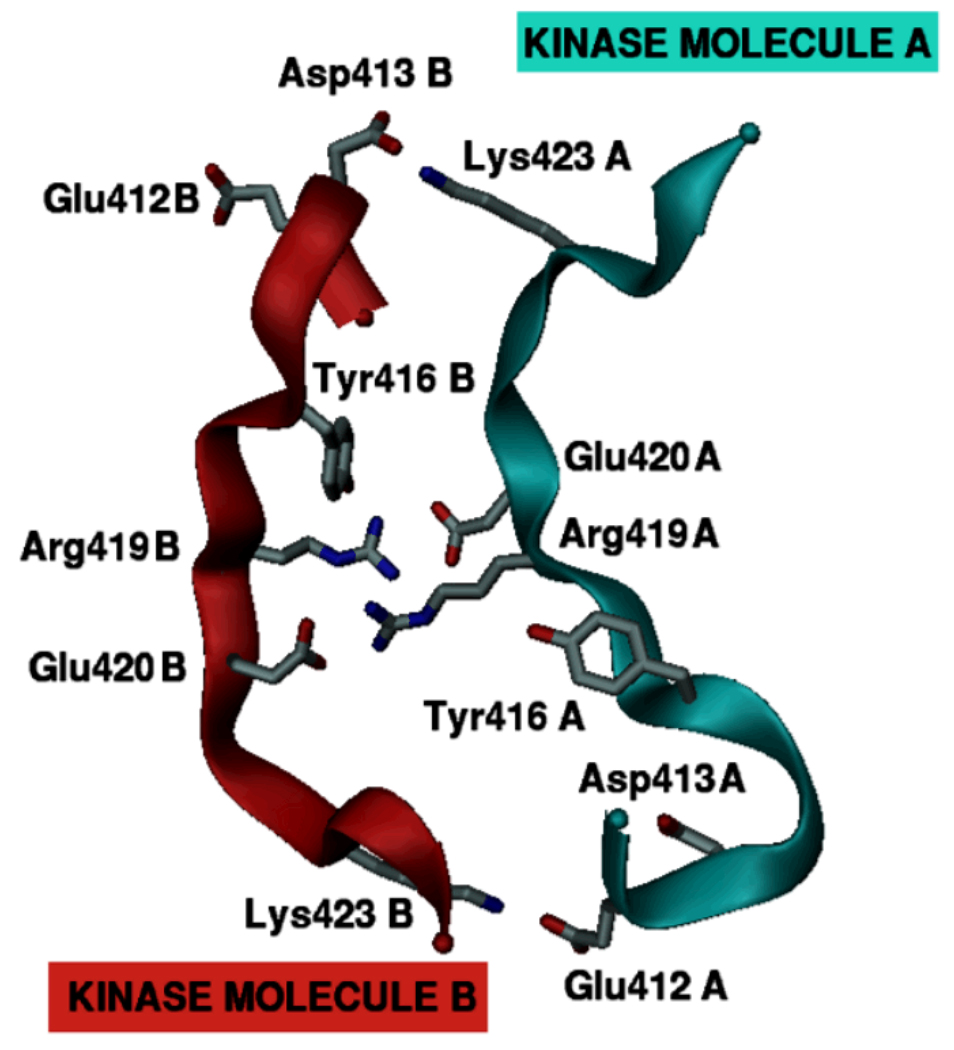
Figure 6.
A proposed mechanism for the role of activation loop charge asymmetry in Src kinase. The three-dimensional model of two interacting activation loop motifs (from Hck) shown here is generated artificially to schematically illustrate the main concept. Though presumably one of the two kinases should be in an activated state, this is left unspecified here for the sake of simplicity. The activation loops of both kinase molecules utilize their intrinsic asymmetry in the distribution of charged residues to facilitate the interaction. Charged residues initially interacting across the Tyr416 within the activation loop instead interact with oppositely charged residues belonging to the second kinase molecule.
For this mechanism to be feasible, it is necessary for the spatial distribution of the charged residues to be complementary between the two Src molecules. If the charged residues are distributed asymmetrically around Tyr416, then even identical kinase molecules possess, by construction, such activation loop complementarity as shown in Figure 6. Three observations should be noted. First, transient kinase dimer formation could occur in variable orientations of the overall kinase domains with charged residue complementarity achieved within the loop regions through local three-dimensional (3D) spatial adjustments. Second, the complementarity need not be only of the charged electrostatic type, the same principle should hold to a lesser capacity for other weaker polar or non-polar interactions. Third, differing activation loop sequences and resulting mismatches in complementarity with substrate kinase activation loops provide a straightforward mechanism by which differential substrate specificity can be achieved through modulation of the activation loop dynamics in these kinases. The detailed mechanics of such trans-phosphorylation still has to be a function of the complex free energy landscape that includes interactions between all atoms of the solute and solvent environment. The mechanism proposed here reduces the complexity of trans-phosphorylation to patterns in primary protein sequence. As explained in the next section, it has the advantage of being easy to test by standard experimental assays that can judge the validity of its inherent simplifications.
Sequence conservation of charge asymmetry
The high propensity of electrostatic interactions between the charged residues disposed asymmetrically with respect to Tyr416 in the activation loop of Hck is clearly observed from the present calculations. However, it is not possible to determine from the umbrella sampling simulation data alone whether this is a general feature of Src kinases since only one sequence has been studied. In the context of the simple model proposed above, however, perusal of primary sequences is sufficient to detect the recurring presence of such charge asymmetry. For this purpose, nine primary Src kinase sequences: hck human (P08631), lck human (P06239), lyn human (P07948), src human (P12931), blk human (P51451), fgr human (P09769), fyn human (P06241), yes human (P07947), and yrk chick (Q02977); were independently used in PSI-BLAST 43 searches to identify homologous sequences. The first 50 hits for each sequence were selected and pooled together. The high homology between the original nine sequences resulted in a lot of redundancy in the pooled sequences. Removing redundant sequences and sequences not belonging to the Src kinase family left a total of 47 sequences that included the original nine Src family members (the list is given in the supplementary material). These were used to analyze the possibility of salt links within the activation loop in Src kinases.
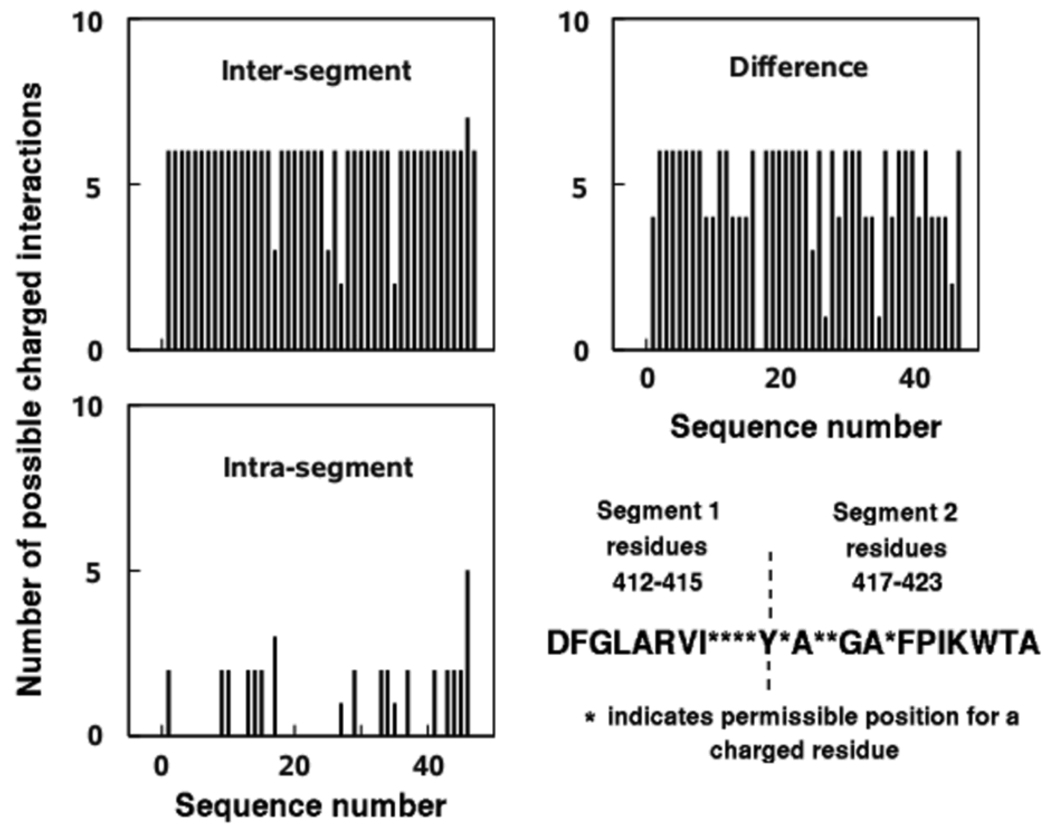
For the purposes of this analysis, the activation loop is divided into two segments based on their position in the primary sequence with respect to Tyr416. The two segments comprise residues 412–415 and 417–423, preceding and following Tyr416, respectively. If the total number of positively charged residues is m and the total number of negatively charged residues is n, then the maximum number of possible salt links is m × n. The salt links can be classified as intrasegment or inter-segment, depending on whether the two oppositely charged residues are in the same segment or not. As shown in Figure 7, a majority of the non-redundant Src kinase sequences show asymmetry in their hydrophilic activation loop regions, and show greater likelihood of inter-segment salt links than intra-segment salt links. The differences between the number of possible inter- and intra-segment salt links mostly vary between +4 and +6, indicating the greater propensity of inter-segment interactions. While the number of inter- and intra-segment salt links does cancel out for one of the sequences, none of them show greater possibility for intra-segment salt links. According to the proposed model, more inter-segment salt links satisfy two needs. First, maintenance of an autoinhibited activation loop conformation where Tyr416 is at least partially occluded from the environment, and second, susceptibility to loop opening in the presence of another kinase molecule with complementary charged residue distribution in its activation loop.
Figure 7.
The pattern of distribution of charged residues and their possible interactions in the activation loops of 47 Src kinase sequences. The classification of residues into specific segments is based on their primary sequence position (either before or after Tyr416). Intrasegment refers to possible salt links between charged residues within each segment, intersegment refers to possible salt links between charged residues in separate segments. It should be noted that the difference in number of salt links (Intersegment – Intrasegment) is always positive, indicating that the activation loop sequences mostly tend towards forming intersegment salt links.
Analysis of the multiple sequence alignment of these redundant Src kinase sequences yields 8 positions within the activation loop that may display polar or charged residues. Starting from the conserved DFG motif with the tyrosine to be phosphorylated underlined, a sequence of the activation loop indicating these 8 positions by “x” is DFGLARVIxxxxYxAxxGAxFPI. Since there are four charged amino acids (Asp, Glu, Lys, Arg) at physiological pH, and there are 8 possible positions where they can be placed, a total of 65536 sequences (48) are possible. A reduction in the number of sequences is achieved by considering just oppositely charged residues as the polarizing differences. Ignoring the differences between Lys and Arg or between Asp and Glu, would result in 28 or 256 sequences. If the hypothesized model is correct, sequences that have greater inter-segment interactions as compared to intra-segment interactions would be resistant to activation, except through trans-phosphorylation by kinases with complementary sequences. High-throughput kinase assays of all or part of this charged residue sequence space composed of up to 65536 possibilities should be able to rank the kinase mutants according to their substrate specificity and resistance to activation, thereby providing a straightforward basis for validation of the present model.
A simpler way to scan this sequence space could be to do it for peptide substrates instead of entire kinase domains. Prior studies have worked on identifying peptide sequences that are good substrates or inhibitors for specific Src kinases.44,45,46,47 In fact, one of the commercially available Src kinase optimal peptide substrates AEEEIYGEFEAKKKK (ANASPEC catalog number 60222-1) does show a strongly asymmetric charged residue distribution around the tyrosine residue to be phosphorylated. But since a peptide substrate has much greater conformational flexibility and less steric hindrance than kinase activation loops, it is difficult to predict whether the influence of charged residue complementarity is larger or smaller than that for the same peptide sequence in the context of a kinase activation loop.
Conclusion
Different conformations are accessible to the activation loop of protein kinases in inactive states.15 Presumably, the specific conformation that dominates depends on the amino acid sequence of the loop region, its phosphorylation state, the presence of substrates or inhibitors, and the environment into which it folds. Prediction of the preferred conformation of the loop is a complex task requiring an accurate description of the multidimensional free energy landscape of the protein in its environment. Here, extensive umbrella sampling MD simulations were used to characterize the energetic costs of opening a specific part of the activation loop of Hck kinase, rendering Tyr416 accessible for phosphorylation by another kinase in a bimolecular encounter.
The free energy profile obtained indicates that the most stable state of the hydrophilic part of the activation loop is an intermediate conformation between the canonical closed inactive and open active forms. Charged residues on either side of Tyr416 interacting with each other occlude Tyr416 from the external environment and contribute to the plasticity of the hydrophilic part of the activation loop. All possible electrostatic interactions between residues of opposite charges occur during the opening and closing MD simulations, with two or more interactions sometimes occurring simultaneously, but interactions formed across Tyr416 are the most frequent. The opening of the activation loop is likely to be affected by the conformational equilibrium between the Glu310-Arg385 and Glu310-Arg409 interactions. A model attributing a functional role to the charge asymmetry in activation loop for trans-phosphorylation is proposed. Sequence analysis confirms that such asymmetry is prevalent in Src kinases and also suggests experimental analysis that can corroborate the proposed model.
Supplementary Material
Acknowledgments
We would like to thank José Faraldo-Gómez and Sichun Yang for helpful discussions. This work was supported financially by grant CA93577-01 from the National Institutes of Health and the Keck Postdoctoral Fellowship for NKB. Computational support from the Pittsburgh Supercomputing Center (PSC) obtained through the National Resource Allocation Committee (NRAC) was used for the calculations.
References
- 1.Brown M, Cooper J. Regulation, substrates and functions of Src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 2.Sicheri F, Kuriyan J. Structures of Src-family tyrosine kinases. Curr Opin Struct Biol. 1997;7:777–785. doi: 10.1016/s0959-440x(97)80146-7. [DOI] [PubMed] [Google Scholar]
- 3.Porter M, Schindler T, Kuriyan J, Miller W. Reciprocal regulation of Hck activity by phosphorylation of Tyr(527) and Tyr(416) Effect of introducing a high affinity intramolecular SH2 ligand. J Biol Chem. 2000;275:2721–2726. doi: 10.1074/jbc.275.4.2721. [DOI] [PubMed] [Google Scholar]
- 4.Azarnia R, Reddy S, Kmiecik TE, Shalloway D, Loewenstein WR. The cellular src gene product regulates junctional cell-to-cell communication. Science. 1988;239:398–401. doi: 10.1126/science.2447651. [DOI] [PubMed] [Google Scholar]
- 5.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 6.Schindler T, Sicheri F, Pico A, Gazit A, Levitzki A, Kuriyan J. Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol Cell. 1999;3:639–648. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 7.Sabat M, Van Rens JC, Laufersweiler MJ, Brugel TA, Maier J, Golebiowski A, De B, Easwaran V, Hsieh LC, Walter RL, Mekel MJ, Evdokimov A, Janusz MJ. The development of 2-benzimidazole substituted pyrimidine based inhibitors of lymphocyte specific kinase (Lck) Bioorg Med Chem Lett. 2006;16:5973–5977. doi: 10.1016/j.bmcl.2006.08.132. [DOI] [PubMed] [Google Scholar]
- 8.Xu W, Doshi A, Lei M, Eck M, Harrison S. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 9.Xu W, Harrison S, Eck M. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 10.Cowan-Jacob SW, Fendrich G, Manley PW, Jahnke W, Fabbro D, Liebetanz J, Meyer T. The Crystal Structure of a c-Src Complex in an active conformation suggests possible steps in c-Src Activation. Structure. 2005;13:861–871. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Fleury D, Sarubbi E, Courjaud A, Guitton JD, Ducruix A. Structure of the unphosphorylated C-terminal tail segment of the Src kinase and its role in Src activity regulation. (Unpublished, but structure deposited in PDB under PDB ID: 1YI6) [Google Scholar]
- 12.Yamaguchi H, Hendrickson W. Structural basis for activation of human lymphocyte kinase Lck upon tyrosine phosphorylation. Nature. 1996;384:484–489. doi: 10.1038/384484a0. [DOI] [PubMed] [Google Scholar]
- 13.Taylor SS, Radzio-Andzelm E. 3 Protein-kinase structures define a common motif. Structure. 1994;2:345–355. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 14.Hubbard S. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO Journal. 1997;16:5573–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 16.Hubbard S, Mohammadi M, Schlessinger J. Autoregulatory mechanisms in protein-tyrosine kinases. J Biol Chem. 1998;273:11987–11990. doi: 10.1074/jbc.273.20.11987. [DOI] [PubMed] [Google Scholar]
- 17.Hubbard S, Till J. Protein tyrosine kinase structure and function. Ann Rev Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- 18.Banavali NK, Roux B. Anatomy of a structural pathway for activation of the catalytic domain of Src kinase Hck. Proteins: Structure, Function, and Bioinformatics. 2007;67:1096–1112. doi: 10.1002/prot.21334. [DOI] [PubMed] [Google Scholar]
- 19.Johnson L, Lewis RJ. Structural basis for control by phosphorylation. Chem Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 20.Young M, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J. Dynamic coupling between the SH2 and SH3 domains of c-Src and Hck underlies their inactivation by C-terminal tyrosine phosphorylation. Cell. 2001;105:115–126. doi: 10.1016/s0092-8674(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 21.Mendieta J, Gago F. In Silico activation of Src tyrosine kinase reveals the molecular basis for intromolecular phosphorylation. J. Mol. Graph. Model. 2004;23:189–198. doi: 10.1016/j.jmgm.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Banavali NK, Roux B. The N-terminal end of the Catalytic Domain of Src kinase Hck is a conformational switch implicated in long-range allosteric regulation. Structure. 2005;13:1715–1723. doi: 10.1016/j.str.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Ozkirimli E, Post CB. Src kinase activation: A switched electrostatic network. Protein Science. 2006;15:1051–1062. doi: 10.1110/ps.051999206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faraldo-Gomez JD, Roux B. On the importance of a funneled energy landscape for the assembly and regulation of multidomain Src tyrosine kinases. Proc Natl Acad Sci USA. 2007;104:13643–13648. doi: 10.1073/pnas.0704041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brooks B, Bruccoleri R, Olafson B, States D, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy minimization and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 26.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marti-Renom MA, Stuart A, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Ann Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 29.Field MJ, Karplus M. Ph.D. thesis. Cambridge, MA: Harvard University; 1992. CRYSTAL: program for crystal calculations in CHARMM. [Google Scholar]
- 30.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-alkanes. J Comp Phys. 1977;23:327–341. [Google Scholar]
- 31.Darden T, York D, Pedersen L. Particle Mesh Ewald - an {N.log(N)} method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 32.Beglov D, Roux B. Finite representation of an infinite bulk system: Solvent boundary potential for computer simulations. J Chem Phys. 1994;100:9050–9063. [Google Scholar]
- 33.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation - the Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 34.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banavali NK, Roux B. Free Energy Landscape of A-DNA to B-DNA Conversion in Aqueous Solution. J Am Chem Soc. 2005;127:6866–6876. doi: 10.1021/ja050482k. [DOI] [PubMed] [Google Scholar]
- 36.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules .1. The method. J Comp Chem. 1992;13:1011–1021. [Google Scholar]
- 37.Souaille M, Roux B. Extension to the weighted histogram analysis method: combining umbrella sampling with free energy calculations. Comput Phys Comm. 2001;135:40–57. [Google Scholar]
- 38.Torrie GM, Valleau JP. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J Comp Phys. 1977;23:187–199. [Google Scholar]
- 39.Jarzinsky C. Non-equilibrium equality for free energy differences. Phys Rev Lett. 1997;87:2690–2693. [Google Scholar]
- 40.Shirts MR, Pande VS. Comparison of efficiency and bias of free energies computed by exponential averaging, the Bennett acceptance ratio, and thermodynamic integration. J Chem Phys. 2005;122:144107. doi: 10.1063/1.1873592. [DOI] [PubMed] [Google Scholar]
- 41.Burchat A, Borhani DW, Calderwood DJ, Hirst GC, Li B, Stachlewitz RF. Discovery of A-770041, a src-family selective orally active lck inhibitor that prevents organ allograft rejection. Bioorg Med Chem Lett. 2006;16:118–122. doi: 10.1016/j.bmcl.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 42.Hennequin LF, Allen J, Breed J, Curwen J, Fennell M, Green TP, Lambert-van der Brempt C, Morgentin R, Norman RA, Olivier A, Otterbein L, Ple PA, Warin N, Costello G. N-(5-chloro-1,3-benzodioxol-4-yl)-7-[2-(4-methylpiperazin-1-yl)ethoxy]-5-(tetrahydro-2 H p y r a n-4-yloxy)quinazolin-4-amine, a novel, highly selective, orally available, dual-specific c-Src/Abl kinase inhibitor. J Med Chem. 2006;49:6465–6488. doi: 10.1021/jm060434q. [DOI] [PubMed] [Google Scholar]
- 43.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen K, Hines AC, Schwarzer D, Pickin KA, Cole PA. Protein kinase structure and function analysis with chemical tools. Biochim Biophys Acta. 2005;1754:65–78. doi: 10.1016/j.bbapap.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Pellicena P, Stowell KR, Miller WT. Enhanced Phosphorylation of Src Family Kinase Substrates Containing SH2 Domain Binding Sites. J Biol Chem. 1998;273:15325–15328. doi: 10.1074/jbc.273.25.15325. [DOI] [PubMed] [Google Scholar]
- 46.Nair SA, Warren SD, Choi S, Songyang Z, Cantley LC, Hangauer DG. Identification of efficient pentapeptide substrates for the tyrosine kinase pp60c src. J Med Chem. 1995;13:4276–4283. doi: 10.1021/jm00021a017. [DOI] [PubMed] [Google Scholar]
- 47.McMurray JS, Budde RJ, Dyckes DF. Cyclic peptide substrates of pp60c-src. Synthesis and evaluation. Int J Pept Protein Res. 1993;42:209–215. doi: 10.1111/j.1399-3011.1993.tb00134.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.