Abstract
The internal ribosome entry site (IRES), a highly conserved structured element of the hepatitis C virus genomic RNA, is an attractive target for antiviral drugs. Here we show that benzimidazole inhibitors of the HCV replicon act by conformational induction of a widened interhelical angle in the IRES subdomain IIa which facilitates the undocking of subdomain IIb from the ribosome and ultimately leads to inhibition of IRES-driven translation in HCV-infected cells.
Hepatitis C virus (HCV) infection affects ~170 million people worldwide and is a major cause of chronic hepatitis as well as hepatocellular carcinoma.1 Current therapies suffer from low efficiency and serious side effects. Therefore, there is an urgent need for novel antiviral agents for the treatment of HCV infection. Among the potential targets for HCV inhibitors is the highly conserved 5′ untranslated region (UTR) of the viral RNA genome which harbors an internal ribosome entry site (IRES). The HCV IRES binds with high affinity2 to host cell 40S ribosomal subunits and initiates translation in a cap-independent fashion.3,4 The IRES element adopts an ordered structure5 which is dominated by independently-folding RNA domains.6,7 The subdomain IIa is the target for benzimidazole inhibitors (1, 2) that reduce viral RNA levels in the HCV replicon at micromolar concentrations (Fig. 1).8 Here, we have used fluorescence labeling guided by structure information to study the mechanics of target interaction of the benzimidazole inhibitors.
Figure 1.
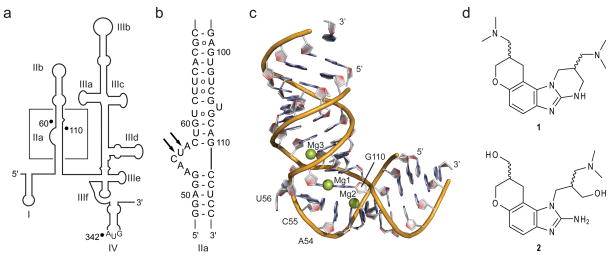
The HCV IRES RNA target. (a) Secondary structure of the HCV 5′ NTR (nucleotides 1–341 of HCV genotype 1b) that contains the IRES element. In addition to residues of the NTR, the IRES includes 26 nt of the reading frame in the hairpin loop of domain IV. (b) Secondary structure of the IRES subdomain IIa. Arrows indicate protection from RNase A digestion at an internal loop of IIa in the presence of benzimidazole 1.8 (c) Three-dimensional structure of the IIa-1 RNA (Supplementary Fig. 1a) corresponding to the subdomain IIa.9 Mg2+ ions are shown as spheres. (d) Benzimidazole inhibitors of the HCV replicon. Compound 1 has a binding affinity for the IRES subdomain IIa of KD=0.72μM, as determined by mass spectrometry, and inhibits HCV replicon at EC50=5.4μM.8 Compound 2 is a precursor to 1.
The three-dimensional structure of the IRES subdomain IIa was previously determined in our laboratory by X-ray crystallography revealing an overall bent architecture (Fig. 1c),9 in agreement with NMR studies of the full domain II10 and cryo-electron microscopy (cryo-EM) investigations of IRES-40S complexes.11,12 The cryo-EM work revealed that the L-shaped conformation of subdomain IIa directs the apical hairpin loop IIb towards the ribosomal E site in proximity of the active site.11 Ribosomal association of domain II induces a conformational change in the 40S head12 and closes the mRNA binding cleft.11 Correct binding of the viral mRNA at the ribosome depends critically on the L-shaped architecture of the domain II.7
Guided by the crystal structure, we have previously identified a key adenine (A54) in the subdomain IIa for replacement by the fluorescent nucleobase analog 2-aminopurine (2AP, 3) to monitor metal ion binding as well as RNA folding (see Supplementary Results, Supplementary Fig. 1a).9 The fluorescence response of 2AP54 renders it a sensitive reporter of the IIa conformation. Upon addition of Mg2+, dose-dependent quenching of 2AP fluorescence ocurred, indicating the stacking of base 54 in the interior of the structure during metal-induced RNA folding.
To study the effect of benzimidazole (1) binding on the conformation of the IIa target, fluorescence of the 2AP54-labeled RNA was recorded in the presence of Mg2+ and increasing concentrations of the ligand (see Supplementary Methods and Supplementary Results, Supplementary Fig. 1b). Binding of 1 resulted in a dose-dependent fluorescence increase, partially reverting the Mg2+-induced quenching. The benzimidazole-triggered fluorescence increase was modulated by the presence of Mg2+, suggesting at least partial competition between binding of the ligand and the metal and supporting the proposed binding site in proximity of structural Mg2+ sites (Fig. 1c). We concluded that 1 induced a conformational change either of the residue 2AP54 or of the entire internal loop region, both of which might result in increased exposure of the 2AP and, hence, increased fluorescence. We further hypothesized that the ligand-induced conformational change in the IIa domain might lead to an incorrect orientation of the apical hairpin loop IIb at the ribosome, effectively preventing the IRES function.
To investigate the molecular mechanism of the ligand-induced conformational change in IIa and to distinguish between a local effect on residue 54 versus a global structural rearrangement, an assay was conceived based on fluorescence resonance energy transfer (FRET) (see Supplementary Methods and Supplementary Fig. 2). The assay allowed the monitoring of the interhelical angle between the stems flanking the internal loop in IIa via measurement of the distance between the stem termini. Labeling of the 5′ termini in IIa with a pair of fluorescent cyanine dyes resulted in the IIa-2 RNA construct. Using the crystal structure of the IIa-1 RNA as a guide,9 the length of the stems was adjusted such that the distance between the dyes would be ~7Å shorter than the Forster radius of the dye pair. This distance gives a maximized sensitivity of FRET to widening of the bend in the native RNA structure while maintaining dynamic range to monitor events that constrict the interhelical angle. At the chosen distance of the dyes even small changes in the overall conformation of the native IIa structure translate into decrease or increase of FRET via, respectively, widening or closing of the interhelical angle.
The FRET assay was validated by titration of IIa-2 RNA, which was initially free of divalent metal ions, with increasing amounts of Mg2+ and monitoring of FRET (see Supplementary Methods and Supplementary Results, Supplementary Fig. 3). As expected, FRET was not detectable in the absence of Mg2+ when the internal loop of subdomain IIa does not stably fold9,10 and the RNA adopts an extended conformation which places the dyes beyond the Forster radius. Upon addition of Mg2+, but not monovalent cations, FRET appeared in a dose-dependent fashion, reflecting the formation of folded RNA-Mg2+ complex. The distance between the helix termini, calculated from the FRET efficiency, was in agreement with the IIa crystal structure.
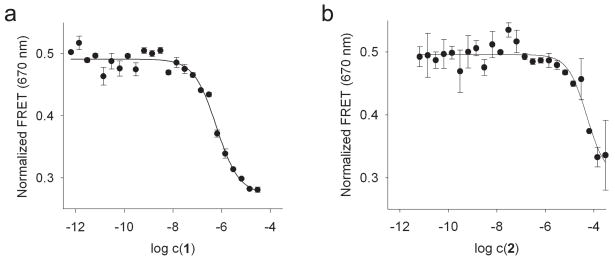
The FRET assay was then used to investigate ligand binding to the subdomain IIa. Therefore, IIa-2 RNA was titrated with 1 and the precursor 2 in the presence of 2mM Mg2+ (Fig. 2). Addition of the benzimidazoles resulted in dose-dependent quenching of FRET, suggesting that ligand binding at the internal loop induced a conformational change which led to a 23° widening of the interhelical angle of the IIa target. The EC50 value of the FRET quenching for 1 (600±80nM) corresponded well with the reported KD (720nM).8 The precursor 2 required higher concentrations for FRET quenching, suggesting weaker target binding in agreement with published data.8
Figure 2.
Normalized FRET signal for titrations of Cy3/Cy5-labeled IIa-2 RNA with benzimidazole ligands (a, compound 1; b, precursor 2) in the presence of 2mM Mg2+. Fitting of dose-response curves resulted in EC50 values for ligand binding of 600±80nM for compound 1 and 90±75μM for precursor 2. An interhelical angle of 112±5° was calculated from asymptotic FRET efficiencies for the ligand-bound state of the IIa RNA. Error bars represent ± s.d. calculated from three independent titrations.
To assess the target specificity of 1, FRET titrations were performed in the presence of competitor tRNA (Supplementary Fig. 4). Binding of 1 to IIa was not affected by 50-fold excess of eukaryotic tRNA which contains bent loop structures of similar complexity as the IIa RNA. The specific nature of the interaction of the IIa subdomain with 1 was further supported by titrations with promiscuous RNA binders which did not elicit FRET quenching (Supplementary Fig. 5).
To narrow down the interaction of 1 with the RNA target, we studied the impact of mutations in the IIa subdomain on ligand binding. Five single and one double base exchange were chosen corresponding to mutations occurring in HCV clinical isolates (Supplementary Fig. 6), which were selected to obtain changes in the benzimidazole binding site that would result in a still functional IRES. As expected, all mutants were competent for Mg2+-induced folding as indicated by FRET which translated into interhelical angles between 82–96° (see Supplementary Results, Supplementary Table 1). Benzimidazole 1 was found to bind and widen the angle of IIa in all cases, albeit to a slightly lesser extent than in the wild type (Supplementary Table 1). Mutation of A57 decreased affinity for 1 by 7-fold, suggesting that the base at position 57 might be close to the ligand binding site. While the affinity of the other mutants for 1 was comparable to wild type RNA, the A57U change promised to provide a useful tool for the study of the inhibitor mechanism in the cellular context.
The dose-dependent FRET quenching upon binding of 1 and 2 to the IIa target provided a rationale for the mechanism of action for these HCV inhibitors. Initiation of viral translation depends critically on the correct positioning of the IRES element on the host cell ribosome. The bent architecture of the subdomain IIa ensures the docking of the apical hairpin of domain IIb into the mRNA exit site on the 40S subunit.7 Consequently, we propose that widening of the angle at the internal loop of IIa facilitates undocking of the IIb hairpin from the ribosome which impedes IRES-driven translation. To test this hypothesis, we investigated the impact of 1 on HCV translation and replication in human cells (see Supplementary Methods). To determine translation effects, compound was added to cells 4h after electroporation with replicon RNA. Replication effects were studied by compound addition to cells stably transfected with replicon. The BM4-5 FEO replicon was used in both experiments, which carries a luciferase reporter under the control of the HCV IRES.13 In the cells electroporated with replicon RNA only translation is occurring in the first 4–8 hours.14 Thus, the impact of 1 on HCV translation can be tested using freshly electroporated cells and compared to reduction of overall replication in the stably transfected line.
In cells electroporated with replicon, the benzimidazole 1 inhibited reporter expression, corresponding to IRES-driven translation, at low micromolar concentrations (Fig. 3a,b). The A57U mutation, which lowers affinity of 1 for IIa, reduced reporter expression as well (Fig. 3c), consistent with weaker binding of 1 to the mutant in the cellular context which ultimately affects the extent of conformational change of the RNA target.
Figure 3.
HCV translation inhibition by benzimidazole 1 in human Huh-7.5 cells. (a, b) Inhibition of luciferase reporter expressed 4h (a) and 8h (b) after transfection with the BM4-5 FEO HCV replicon RNA. Fitting of dose-response curves results in IC50 values for translation inhibition by compound 1 of 4.0μM (4h) and 9.5μM (8h). (c) Comparison of luciferase reporter expressed 4h after transfection with wild type or A57U IRES mutant RNA in the absence and presence of inhibitor 1. In the control (left, no inhibitor) reporter expression was normalized to wild type. Expression in the presence of 25 μM inhibitor (right) was normalized to control for each wild type and mutant. While the A57U change originates from an HCV clinical isolate, the efficiency of IRES-driven translation is drastically reduced in this mutant (15% of wild type activity). (d) Inhibition of luciferase reporter expression after 48h in cells stably transfected with HCV replicon. Fitting of a dose-response curve results in an IC50 value for replicon inhibition by compound 1 of 2.0μM. Assessment of cell viability in the presence of compound ruled out that signal decrease was due to cytotoxicity (Supplementary Fig 8). Error bars represent ± s.d. calculated from triplicate experiments.
In cells stably transfected with the wild type replicon, reporter expression was inhibited at a slightly lower IC50 value of 2μM (Fig. 3d). Testing of 1 for effects on NS5B proved that the compound did not inhibit the HCV polymerase (see Supplementary Methdos and Supplementary Fig. 7). The higher potency of 1 against replicating HCV might indicate an additional effect of ligand binding to the IIa subdomain on the interaction between the 5′ IRES and 3′ UTR, which presumably plays a role in the switch between translation and replication.15
Replicon inhibition in both transiently and stably transfected cells in combination with FRET data thus suggest that benzimidazole 1 acts as a HCV-specific translation inhibitor by conformational induction of a widened RNA interhelical angle at the IRES subdomain IIa. This is the first example of such a conformational mechanism proven for a biologically active small molecule that targets an RNA structure outside the bacterial ribosome. The concept of interference with the conformational state of structured RNA has been recognized before as an important principle for the inhibitory mechanism of RNA-directed ligands,16 most prominently exploited by antibiotics that target the bacterial ribosome.17
Supplementary Material
Figure 4.
Acknowledgments
We thank Bao Ho and Cody Higginson for help with compound synthesis. A kind gift of NS5BΔ21 protein by Gilead Sciences, Foster City, is acknowledged. This work was supported in part by the National Institutes of Health, grants R01 AI72012 (TH) and K08 AI069989 (DLW).
Footnotes
Author Contributions
M.P.C. synthesized the IRES-binding benzimidazole inhibitors. S.D. synthesized NS5B inhibitors. S.M.D. participated in the design of the RNA FRET construct. J.P. performed biochemical experiments and was involved in concept development of the FRET assay. D.L.W. conceived and performed cell-based assays. T.H. conceived and supervised the project and wrote the manuscript. J.P., D.L.W. and T.H. were involved in the discussion of the experimental data and its interpretation.
Competing Financial Interests
None
References
- 1.Maheshwari A, Ray S, Thuluvath PJ. Lancet. 2008;372:321–332. doi: 10.1016/S0140-6736(08)61116-2. [DOI] [PubMed] [Google Scholar]
- 2.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Proc Natl Acad Sci U S A. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pestova TV, et al. Proc Natl Acad Sci U S A. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto GA, Puglisi JD. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 5.Brown EA, Zhang H, Ping LH, Lemon SM. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kieft JS, et al. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 7.Lukavsky PJ. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seth PP, et al. J Med Chem. 2005;48:7099–7102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- 9.Dibrov SM, Johnston-Cox H, Weng YH, Hermann T. Angew Chem Int Ed Engl. 2007;46:226–229. doi: 10.1002/anie.200603807. [DOI] [PubMed] [Google Scholar]
- 10.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Nat Struct Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 11.Spahn CM, et al. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 12.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure (Camb) 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Wyles DL, Kaihara KA, Vaida F, Schooley RT. J Virol. 2007;81:3005–3008. doi: 10.1128/JVI.02083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blight KJ, McKeating JA, Marcotrigiano J, Rice CM. J Virol. 2003;77:3181–3190. doi: 10.1128/JVI.77.5.3181-3190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Y, et al. J Virol. 2006;80:11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermann T. Angew Chem Int Ed Engl. 2000;39:1890–1904. doi: 10.1002/1521-3773(20000602)39:11<1890::aid-anie1890>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 17.Hermann T. Curr Opin Struct Biol. 2005;15:355–366. doi: 10.1016/j.sbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.