Abstract
MAPK (mitogen-activated protein kinase) kinase 4 (MKK4) is a pivotal upstream activator of c-Jun-N-terminal kinase (JNK) and p38MAPK. Here, we report that MKK4 is increased in senescent human diploid fibroblasts (HDFs) through enhanced translation. We identified four microRNAs (miR-15b, miR-24, miR-25, and miR-141) that target the MKK4 mRNA; the abundance of these microRNAs decreased during replicative senescence. Individually modulating the amount of each microRNA did not modify MKK4 abundance, but their concomitant overexpression decreased MKK4, whereas their joint reduction increased MKK4. Reporter analyses indicated that these microRNAs acted through the MKK4 5′ and 3′ untranslated regions. Elevated MKK4 inhibited cell proliferation and increased the phosphorylation of p38 and PRAK (p38-regulated/activated protein kinase), thereby contributing to the senescent phenotype of HDFs. Thus, multiple microRNAs acting on a single target, the MKK4 mRNA, collectively influence MKK4 abundance during replicative senescence.
INTRODUCTION
Replicative senescenceis a process whereby cells halt proliferation irreversibly after completing a finite number of divisions but remain metabolically active (1,2). It is believed to represent a tumor-suppressive mechanism and a contributing factor in aging (3,4). Although several transcription factors are responsible for modulating protein abundance during replicative senescence, the contribution of posttranscriptional events, including altered mRNA stability and translation, is increasingly recognized (5). These processes are efficiently modulated by specific RNA-binding proteins (RBPs) and microRNAs (miRNAs) that bind to specific mRNA sequences (6,7). miRNAs are ~22-nucleotide (nt) long non-coding RNAs that associate with a target mRNA in a sequence-specific manner and typically repress gene expression by lowering the stability of the mRNA, decreasing its translation rate, or performing both functions (8). miRNAs interact with target mRNAs through the ‘seed region’, which spans residues 2–9 of miRNAs and typically complement the target mRNA at 7 or 8 nucleotides (8). They are involved in the posttranscriptional regulation of genes implicated in complex cellular processes such as aging, cancer, proliferation, differentiation, and apoptosis (9,10).
The global patterns of mRNA and protein abundance change during replicative senescence. Although proliferative and stress-response proteins are typically reduced in senescent cells, several proteins are elevated [such as p16, p21, p27, p53, and cyclin D1 (11,12)] and play important biological roles in senescence and aging. While investigating age-related changes in gene expression, we discovered that the abundance of the signaling protein MKK4 increased with age in several human tissues. In response to external stimuli such as environmental stress or inflammation, the dual-specificity kinase MKK4 [also known as JNKK1, MEK4, and SEK (stress-activated protein kinase/extracellular signal-regulated kinase kinase 1)] phosphorylates and thereby activates JNK and p38, which in turn elicit biological responses such as cell proliferation, differentiation, and apoptosis (13,14). Clinical correlative studies have identified a putative tumor suppressor function for MKK4 in various tumor types (15). Functional in vivo studies have identified a metastasis suppressor activity for MKK4 in prostate and ovarian cancers (16,17), and other clinical and experimental studies suggest that MKK4 may have pro-oncogenic properties, indicating that the role of MKK4 is dependent upon cellular and environmental contexts (18,19).
Here, we demonstrate in an in vitro model of human replicative senescence that MKK4 abundance is regulated by the joint action of four microRNAs. The increase in MKK4 abundance with replicative senescence is elicited by the combined decline in multiple microRNAs – miR-15b, miR-24, miR-25, and miR-141– that suppress MKK4 translation by binding to sites in its 3′-and 5′-untranslated regions (UTRs).
RESULTS
MKK4 abundance increases with replicative senescence
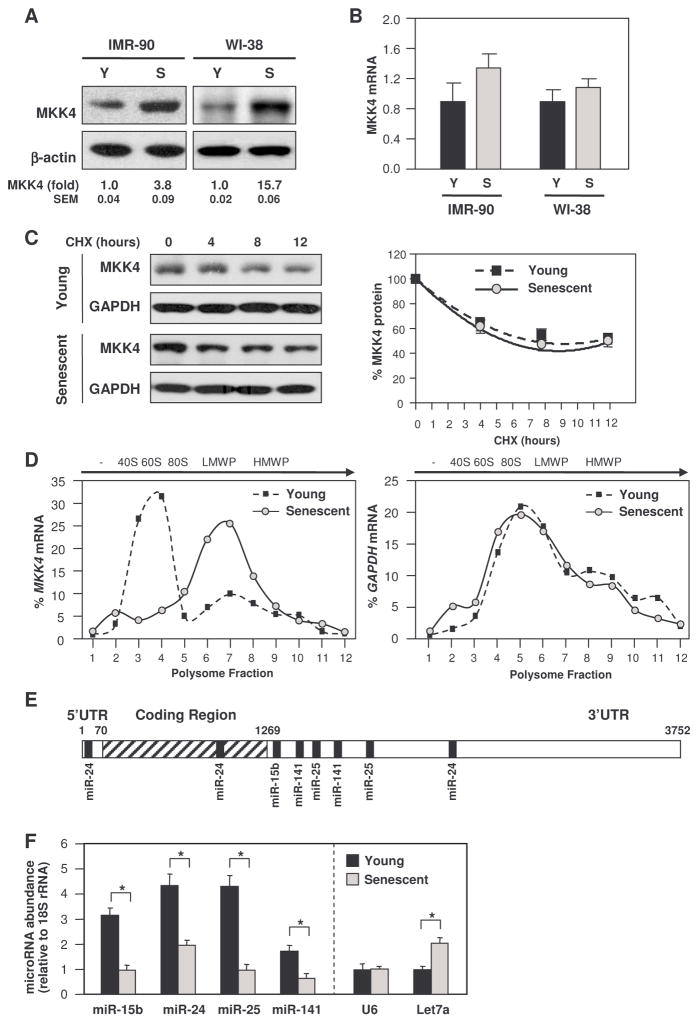
In several human tissues, including pancreas and ovary, MKK4 abundance increased with age, as determined using a panel of tissue biopsies from human donors (fig. S1). We used human fetal lung diploid fibroblasts (HDFs) WI-38 and IMR-90 as in vitro models of aging. We defined late passage [50–55 population doublings (Pdl)], non-proliferating HDFs as senescent, and early passage (typically 20–30 Pdl), proliferating HDFs as “young”. Senescent HDFs contained 3- to 15-fold more MKK4 than young HDFs (Fig. 1A). Lack of correlation between MKK4 abundance and the amount of MKK4 mRNA was previously shown in prostate cancer cells, suggesting a role for altered translation rates (19). Accordingly, MKK4 mRNA abundance (Fig. 1B) and the stability of MKK4 protein (Fig. 1C) did not differ between senescent and young HDFs. Instead, the amount of MKK4 protein in senescent cells appeared to increase by translational mechanisms. As shown in Fig. 1D, analysis of the association of MKK4 mRNA with translating ribosomes (polysomes) indicated that MKK4 mRNA was more actively translated in senescent HDFs compared with young HDFs. After fractionation of the cytoplasm through sucrose gradients and isolation of RNA in each fraction, MKK4 mRNA in young cells was found mostly in light fractions of the gradient, where mRNAs are not translated or are minimally translated (fractions 1–5) (Fig. 1D). By contrast, in senescent cells the MKK4 mRNA was found in heavier parts of the gradient where the polysomes are located (low- and high-molecular weight polysomes, LMWP and HMWP, respectively), supporting the notion that MKK4 was more actively translated in senescent cells. The distribution of individual transcripts (MKK4 and GAPDH mRNAs) was determined by reverse transcription (RT) followed by quantitative polymerase chain reaction (qPCR) analysis of RNA prepared from each fraction of the sucrose gradients (Fig. 1D) [see reference (20) for details of polysome analysis]. A survey of RBPs did not identify specific regulators of MKK4 translation in prostate cancer or WI-38 cells (fig. S2). Thus, we hypothesized that miRNAs might contribute to the differential abundance of MKK4 during replicative senescence.
Fig. 1. Senescent human diploid fibroblasts (HDFs) show elevated MKK4 and reduced MKK4-directed miRNAs.
(A) Representative Western blot analysis of MKK4 abundance in young (Y, Pdl 11) and senescent (S, Pdl 51) IMR-90 fibroblasts, as well as in young (Pdl 24) and senescent (Pdl 54) WI-38 fibroblasts. β-actin was included as loading control. MKK4 signals from at least three Western blots were quantified by densitometry and normalized to β-actin signals. (B) MKK4 mRNA, as measured by RT-qPCR analysis of total RNA isolated from IMR-90 and WI-38 fibroblasts. (C) MKK4 protein stability was determined in young (Pdl 24) and senescent (Pdl 54) WI-38 cells that had been treated with cycloheximide (CHX, 50 μg/ml) and immunoblotting for MKK4 and GAPDH as a loading control. (D) Association of MKK4 mRNA (and GAPDH mRNA as a control) with cellular polysomes. After centrifugation of cytoplasmic components through linear 10–50% sucrose gradients, mRNA amounts in each fraction were measured by RT-qPCR and plotted as a percentage of the total mRNA in the sample. Arrow, direction of sedimentation; -, fractions lacking ribosome components; 40S and 60S, small and large ribosomal subunits, respectively; 80S, monosome; LMWP and HMWP, low- and high-molecular weight polysomes, respectively. (E) Schematic of the MKK4 mRNA, including the predicted target sites for miR-15b, miR-24, miR-25, and miR-141. (F) miR-15b, miR-24, miR-25, and miR-141 in young (Pdl 24) and senescent (Pdl 54) WI-38 cells were quantified by RT-qPCR analysis, normalized to 18S rRNA. As controls, U6 (which was unchanged between young and senescent cells) and let7a miRNA (which was elevated in senescent cells) were also measured. Data represent standard error of the mean (SEM) from 3 independent experiments; *, p<0.05, paired t test.
We previously found differences in miRNA profiles of young and senescent WI-38 fibroblasts (20). Among the miRNAs predicted to associate with the MKK4 mRNA, four miRNAs – miR-15b, miR-24, miR-25, and miR-141 – were decreased in senescent HDFs, as measured by RT-qPCR of each individual miRNA (Fig. 1, E and F) (20). Moreover, the abundance of these four miRNAs was generally reduced in tissues from older compared to those from younger human donors (fig. S3). Only miR-15 was predicted to target the MKK4 3′UTR at one site; miR-25 and miR-141 each were predicted to target the MKK4 3′UTR at two sites, and miR-24 was predicted to target at one site in the 5′UTR, one in the coding region, and one in the 3′UTR (Fig. 1, E and F, and fig. S4). Therefore, we tested the ability of miR-15b, miR-24, miR-25, and miR-141 to regulate the translation of MKK4.
MicroRNAs miR-15b, miR-24, miR-25, and miR-141 jointly, but not individually, repress MKK4 translation
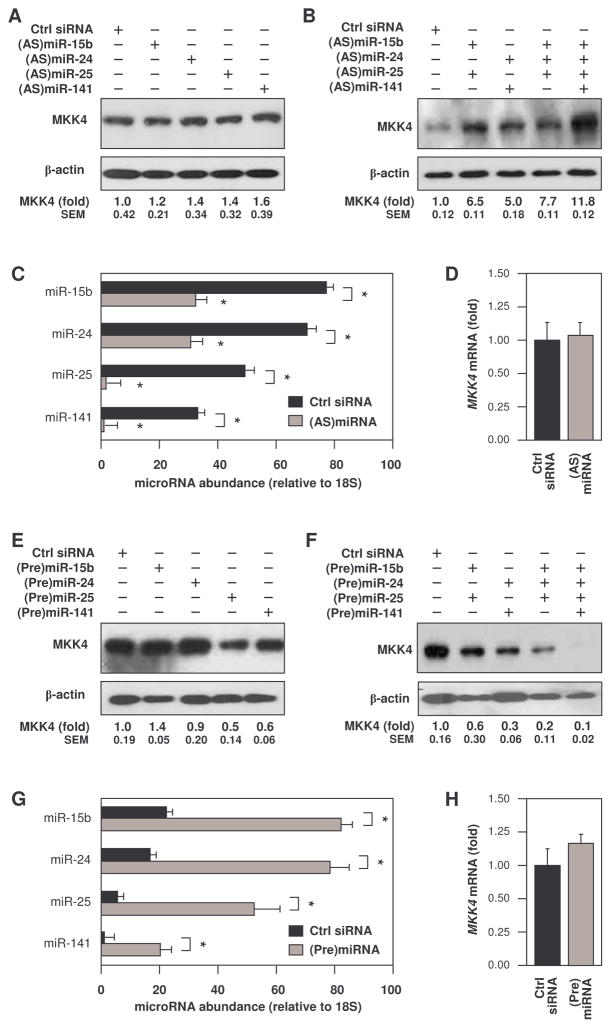
Individual reduction of each miRNA was accomplished by transfecting young WI-38 cells with antagomirs, short transcripts complementary (antisense; AS) to each miRNA. Individual knockdown of each miRNA had negligible influence on MKK4 protein amounts, as assessed by Western blot analysis (Fig. 2A). However, concomitant knockdown of the 4 miRNAs elevated MKK4 protein abundance (Fig. 2B). These interventions lowered each miRNA between two-fold and >20-fold over basal miRNA concentrations (Fig. 2C). MKK4 mRNA abundance was unchanged after transfecting all four (AS)miRNAs (Fig. 2D), indicating that the increase in miRNAs directly affected the MKK4 protein abundance. To test if miRNA overexpression had the opposite effect, precursor (Pre)miRNAs were transfected into senescent WI-38 cells. Transfection of (Pre)miRNAs individually had little or no effect on MKK4 abundance (Fig. 2E), but combined transfections of (Pre)miRNAs lowered MKK4 abundance, especially when all four (Pre)miRNAs were transfected together (Fig. 2, E and F, and fig. S5). Transfection of senescent WI-38 cells with precursor (Pre)miRNAs increased the abundance of each miRNA by 3- to >10-fold (Fig. 2G). MKK4 mRNA remained elevated after transfection of the four (Pre)miRNAs (Fig. 2H), indicating that the (Pre)miRNAs specifically reduced MKK4 at the protein level.
Fig. 2. miRNAs directed against MKK4 cooperatively reduce MKK4 protein abundance.
(A to D) Young WI-38 cells were transfected with control (Ctrl) siRNA or with different combinations of antisense (AS)miRNAs (100 nM final). 48 hours later, the individual (A) or combined (B) effects of the (AS)miRNAs on MKK4 abundance was determined by Western blot analysis, using β-actin as a loading control. RT-qPCR was used to quantify each miRNA individually (C) as well as MKK4 mRNA in young WI-38 cells transfected with control siRNA or with all four (AS)miRNAs, using 18S rRNA for normalization (D). (E to H) Senescent WI-38 cells were transfected with either control (Ctrl) siRNA or with different combinations of precursor (Pre)miRNAs (100 nM final). The individual (E) or combined (F) effect of the (Pre)miRNAs on MKK4 abundance was determined by Western blot analysis, using β-actin as a loading control. RT-qPCR was used to quantify each miRNA (G), and MKK4 mRNA in S WI-38 cells transfected with Ctrl siRNA or all four (Pre)miRNAs, using 18S rRNA for normalization (H). MKK4 signals were quantified by densitometry from 3 independent Western blots (A, B, E, and F). Data are shown as SEM from 3 independent experiments (C, D, G, and H); *, p<0.05, paired t test..
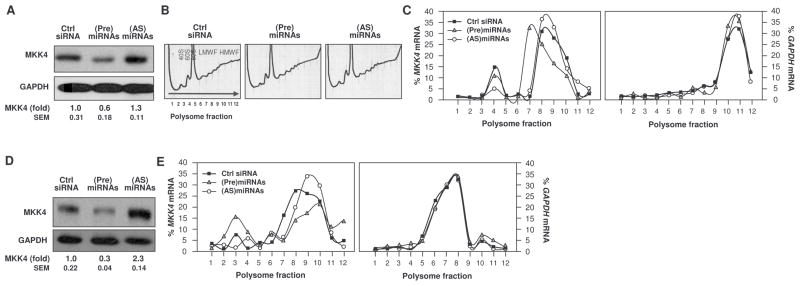
To determine the action of these miRNAs, we employed HeLa (human cervical carcinoma) cells, because subsequent analyses required larger volumes of cells, which were easier to obtain from this cell system. In HeLa cells, transfection of all four (Pre)miRNAs also reduced MKK4 abundance, although transfection of all four (AS)miRNAs only modestly elevated MKK4 abundance compared with the control transfection group (Fig. 3A). Sucrose gradient fractionation of cytoplasmic lysates of transfected cells, followed by RT-qPCR analysis of MKK4 mRNA (and housekeeping GAPDH mRNA), showed that transfection of all four (Pre)miRNAs moderately shifted MKK4 mRNA distribution to fractions with less active translation (lower molecular-weight polysomes) of the gradient compared with the control transfection group (Fig. 3, B and C). In contrast, transfection of all four (AS)miRNAs only slightly increased the percentage of transcripts in the actively translating (higher molecular-weight) polysomes (Fig. 3, B and C).
Fig. 3. miRNAs directed against MKK4 cooperatively repress MKK4 association with polysomes.
(A) HeLa cells were transfected with Ctrl siRNA, with four pooled (Pre)miRNAs [ (Pre)miR-15b, (Pre)miR-24, (Pre)miR-25, and (Pre)miR-141] or with four pooled (AS)miRNAs [(AS)miR-15b, (AS)miR-24, (AS)miR-25, and (AS)miR-141]. 48 hours later, MKK4 and GAPDH as a loading control were assessed by Western blot analysis. (B) Analysis of polysome profiles in cells transfected as described in (A). (C) Distribution of the mRNAs for MKK4 (left) and GAPDH (right) on polysome gradients prepared from the transfection groups described in panel A. (D and E) WI-38 cells of intermediate passage (Pdl 39) were transfected as described in (A). 48 hours later, MKK4 was assessed by Western blot analysis (D) and MKK4 mRNA distribution on polysome gradients was determined (E). MKK4 signals were quantified by densitometry from 3 Western blots (A and D).
We also used WI-38 cells that had undergone 39 population doublings (Pdl 39) and were thus intermediate in age between young and senescent cells in order to study both increases and decreases in the polysomal association of MKK4 mRNA. Transfection of (Pre)miRNAs into Pdl 39 WI-38 cells reduced MKK4 protein abundance and the association of MKK4 mRNA with actively translating polysomes compared with the control transfection group (Fig. 3, D and E). Conversely, transfection of Pdl-39 WI-38 cells with (AS)miRNAs elevated MKK4 abundance and moderately increased the association of MKK4 mRNA with actively translating polysomes (Fig. 3, D and E). It is important to note that small but consistent shifts in distribution of a given mRNA by one or two gradient fractions can have a strong effect upon the translation of the mRNA, as seen previously for p16 mRNA (20).
Both the 3′UTR and 5′UTR of MKK4 contribute to the translational repression by miRNAs
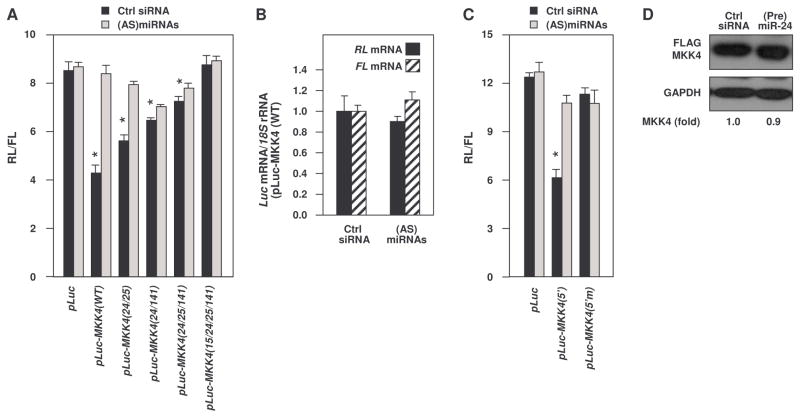
To assess the contribution of the individual miRNA sites within the MKK4 mRNA, we prepared reporter constructs. For the analysis of 3′UTR sites, the 1.3 kb region spanning the predicted miRNA hit sites was cloned at the 3′ end of the Renilla luciferase (RL) reporter cDNA in the psiCHECK-2 plasmid. A second reporter protein (firefly luciferase, FL) expressed from the same plasmid using a separate promoter served as internal control for transfection. Several reporters were prepared, including an ‘empty’ plasmid (pLuc), a plasmid expressing the intact MKK4 3′UTR region, pLuc-MKK4(WT), and additional reporters in which 2 or 3 nt within the seed region of the miRNA sites were mutated (fig. S6 and fig. S7). The ratios of RL/FL activities in transfected WI-38 cells were measured in order to quantify the contribution of the MKK4 3′UTR (wild-type or with mutations in various miRNA sites) to miRNA-mediated translation repression of MKK4. Insertion of the MKK4(WT) 3′UTR region reduced the activity of reporter pLuc-MKK4(WT) to 45% of that seen for pLuc, whereas the sequential addition of mutations in the miRNA sites (first 24, 25, 141, and finally 15; fig. S6 and fig. S7) progressively elevated RL/FL ratios, eventually reaching those seen with pLuc (Fig. 4A). Moreover, reduction of miRNAs by transfection of (AS)miRNAs increased the expression of all of the reporters (Fig. 4A, gray bars), indicating that the decrease in RL/FL activity in reporters bearing the MKK4 3′UTR (wild-type and mutated at 2 or 3 sites) required the presence of the miRNAs. The loss in RL/FL was not due to a selective reduction in RL mRNA, because the relative abundance of RL and FL mRNAs was unchanged, as shown for the pLuc-MKK4(WT) plasmid under conditions of normal and reduced miRNA concentrations (Fig. 4B). A similar analysis in HeLa cells yielded comparable results (fig. S7).
Fig. 4. MKK4 reporter analysis.
(A) psiCHECK-2-derived reporter constructs used are shown in fig. S7. pLuc, control vector expressing Renilla luciferase (RL) and firefly luciferase (FL). pLuc-MKK4(WT) is a fusion of RL and the 1.3 kb proximal MKK4 3′UTR segment bearing all six predicted miRNA sites. pLuc-MKK4(24/25) was derived from pLuc-MKK4(WT) by introducing mutations in the seed regions of miR-24 and miR-25 sites. In pLuc-MKK4(24/141), the mutations were introduced in miR-24 and miR-141 sites. In pLuc-MKK4(24/25/141), the mutations were introduced in miR-24, miR-25, and miR-141 sites. In pLuc-MKK4(15b/24/25/141), all six miRNA sites on the MKK4 3′ UTR were mutated (fig. S6). WI-38 cells that were transfected with either control siRNA or pooled (AS)miRNAs were subsequently transfected with reporter plasmids (fig. S7), and RL activity and FL activity were measured 24 hours later. RL/FL values are shown. (B) RL and FL mRNAs were measured in WI-38 cells (with normal or reduced amounts of miRNAs) by RT-qPCR analysis 24 hours after transfection of pLuc-MKK4(WT). (C) Cells were transfected with pLuc, pLuc-MKK4(5′) (which expresses an RL chimeric RNA with the wild-type MKK4 5′UTR), and pLuc-MKK4(5′m) (which expresses an RL chimeric RNA with the mutated MKK4 5′UTR (fig. S7). 24 hours later, RL and FL activities were measured. (D) The plasmid pcDNA-FLAG-MKK4 and the miR-24 site within the MKK4 coding region are shown in fig. S7. WI-38 cells were cotransfected with either control siRNA or (Pre)miR-24 along with pFLAG-MKK4. 24 hours later, MKK4 was quantified by Western blot and densitometry analysis of the FLAG signals. Data are representative of 3 experiments. Data represent SEM from 3 independent experiments (A, B, and C). *, p < 0.05. Statistical significance was determined by ANOVA followed by Tukey multiple comparisons test.
Although most miRNAs are believed to function through 3′UTR sites, examples are emerging of miRNAs acting through the 5′UTR and coding region (10,20–22). Because putative miR-24 target sites were also found in the MKK4 5′UTR and coding region, the contribution of these sites was also tested. pLuc-MKK4(5′) contained the wild-type MKK4 5′UTR (5′), and pLuc-MKK4(5′m) had a mutation that disrupted miR-24 binding (5′m) to the 5′ end of RL (fig. S7). pLuc-MKK4(5′) showed 49% RL/FL activity compared with pLuc activity, whereas the 5′ mutation in pLuc-MKK4(5′m) abrogated this repression (Fig. 4C), indicating that the miR-24 site on the MKK4 5′UTR could independently repress the RL reporter. In each case, transfection with (AS)miRNAs blocked this effect (Fig. 4C). The contribution of the miR-24 site in the coding region to the translation repression of MKK4 was assessed by measuring the abundance of a FLAG-tagged MKK4 protein, expressed from a transcript devoid of MKK4 3′ or 5′UTR sequences (fig. S7). Unlike the endogenous MKK4 protein, the abundance of which was reduced by (Pre)miRNAs (Fig. 3A), FLAG-MKK4 was refractory to the presence of transfected (Pre)miR-24, suggesting that the site in the coding region did not contribute to modulating MKK4 abundance (Fig. 4D). Together, these findings support the notion that the 3′UTR-targeting miRNAs function jointly with those targeting the 5′UTR to suppress MKK4 production.
MKK4 mRNA-directed miRNAs influence proliferation, senescence, and senescence-associated cell signaling
DNA replication, as assessed by 3H-thymidine incorporation, was used as a measure of proliferation. 3H-thymidine incorporation was higher in HeLa cells transfected with (Pre)miRNAs to reduce MKK4 abundance compared to control siRNA-transfected HeLa cells (Fig. 5, A to C). Both transfection groups showed reduced 3H-thymidine incorporation when MKK4 was overexpressed and increased 3H-thymidine incorporation when MKK4 was silenced by small interfering RNA (siRNA) (Fig. 5, A to C). The growth inhibitory influence of MKK4 was also investigated in HDFs. In young WI-38 cells, MKK4 abundance was lowered by siRNA, an effect that was reversed by cotransfection of (AS)miRNAs. As observed in HeLa cells, modulation of MKK4 in WI-38 cells was accompanied by changes in DNA replication, with 3H-thymidine incorporation increasing when MKK4 was reduced and decreasing when MKK4 was elevated (Fig. 5D). The increase in cell proliferation seen in WI-38 cells with lower MKK4 amounts contrasts with the slight reduction in proliferation reported for MKK4-null mouse embryo fibroblasts (23). Differences in the degree of reduction of MKK4 abundance, the species studied (human versus mouse), and the time periods of MKK4 loss or silencing could explain the distinct effect of MKK4 on proliferation seen in our study and that of Tournier et al. (23). WI-38 cell morphology was altered, with high MKK4-expressing cells becoming enlarged, a characteristic of pre-senescent and senescent cells (Fig. 5E). No apoptosis was detected after short-term silencing of MKK4 (fig. S8).
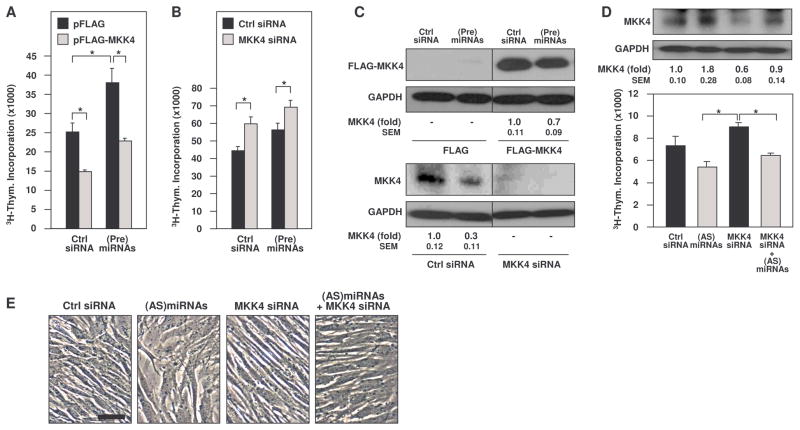
Fig. 5. MKK4 reduces proliferation.
(A to C) 3H-thymidine incorporation was measured 48 hours after cotransfection of HeLa cells with control siRNA or Pre(miRNAs) and (A) pFLAG or pFLAG-MKK4 plasmids or (B) control or MKK4 siRNA. The abundance of MKK4, FLAG-MKK4, and the loading control GAPDH was determined by Western blot analysis and densitometry (C). (D) Quantification of MKK4 by Western blot analysis and densitometry (top) and 3H-thymidine incorporation (graph) 48 hours after transfection of WI-38 cells of intermediate Pdl (Pdl 39) with the small RNAs shown. (E) Phase-contrast micrographs of cells from the transfection groups described in (D). Scale bars, 50 μm (A and E). Data represent SEM from 3 independent experiments (A, B, and D). Western blots are representative of 3 independent experiments (C and D). *, p<0.05. Statistical significance was determined by ANOVA followed by Tukey multiple comparisons test.
Overexpression of FLAG-MKK4 triggered a senescent phenotype in young WI-38 cells, as determined by the increased activity of a broadly used marker, senescence-associated β-galactosidase [SA-β-gal; (24)]; in comparison, cells expressing the empty vector (FLAG) were SA-β-gal negative (Fig. 6A). Likewise, the senescence-associated proteins p21 and cyclin D1 were more abundant in MKK4-overexpressing cells (Fig. 6B). To study the influence of miRNAs on the senescence phenotype, MKK4-directed (Pre)miRNAs and (AS)miRNAs were transfected every 4 days for 5 weeks. At the end of this period, the cells in the (AS)miRNA-transfected group contained higher amounts of MKK4, were larger, and divided more slowly than control cells, were positive for SA-β-gal activity, and showed elevated abundance of the senescence marker p16. In contrast, the cells transfected with the (Pre)miRNAs were smaller, proliferated more rapidly, and showed lower abundance of p16 and MKK4 than control cells (Fig. 6, C and D).
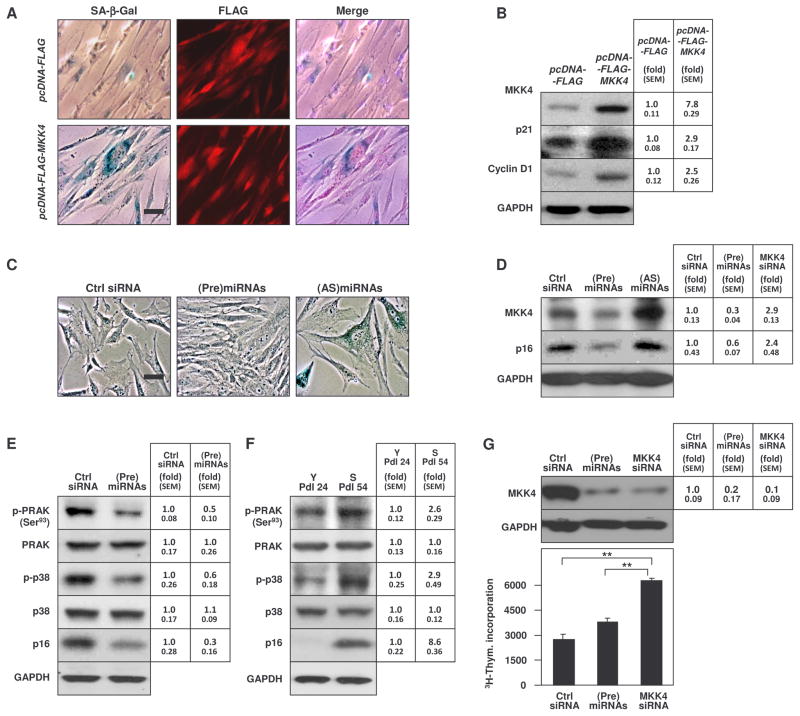
Fig. 6. Altering MKK4 abundance changes markers of replicative senescence.
(A and B) 48 hours after transfection of young WI-38 cells with pcDNA-FLAG or pcDNA-FLAG-MKK4, SA-β-gal activity and FLAG signals were detected by phase-contrast microscopy and fluorescence microscopy, respectively (A). The abundance of MKK4, the loading control GAPDH, and the senescence-associated proteins p21 and cyclin D1 was assessed by Western blot analysis (B). (C) Young WI-38 cells (Pdl 24) were sequentially transfected (every 4 days for 5 weeks) with the small RNAs indicated. Cellular morphology and SA-β-gal activity were assessed by phase-contrast microscopy. Images are representative of N=2 independent experiments. (D) Western blot analysis of the abundance of MKK4, the senescence marker p16, and the loading control GAPDH in the populations described in panel C. (E and F) Western blot analysis of total PRAK and phosphorylated PRAK (p-PRAK), total and phosphorylated p38 (p-p38), p16, and the loading control GAPDH, as assessed 48 hours after transfection of WI-38 cells (Pdl 39) with control siRNA or (Pre)miRNAs. Protein abundance was also assessed in untransfected young (Pdl 24) and senescent (Pdl 54) WI-38 cells. (G) MKK4 was quantified by Western blot analysis and densitometry (top) and 3H-thymidine incorporation (graph) in S WI-38 (Pdl 54) cells 48 hours after transfection of the small RNAs shown. Data are shown as SEM. The signals in 3 independent Western blot analyses were quantified by densitometry (B, D, E, F, and G). **, p<0.001. Statistical significance was determined by ANOVA followed by Tukey multiple comparisons test.
Sun and coworkers showed that the p38 substrate PRAK is critical for the induction of senescence (25). Accordingly, cells transfected with (Pre)miRNAs, and hence containing less MKK4, also showed reduced p16 and phosphorylated PRAK at Ser93 (Fig. 6E). Conversely, phosphorylated PRAK, phosphorylated p38, and p18 were higher overall in senescent compared to young WI-38 cells (Fig. 6F). In contrast, the JNK pathway did not appear to be affected by alterations in MKK4 abundance (fig. S9). Finally, in senescent WI-38 cells, transfection of MKK4 siRNA or (Pre)miRNAs reduced MKK4 abundance and promoted 3H-thymidine incorporation (Fig. 6G).
DISCUSSION
Together, our data indicate that MKK4 abundance is increased during replicative senescence, largely because of the reduced abundance of microRNAs miR-15b, miR-24, miR-25, and miR-141. Furthermore, increased MKK4 contributes to decreased cell proliferation, a hallmark characteristic of senescent cells. Ongoing studies are aimed at elucidating the mechanisms by which the abundance of miR-15b, miR-24, miR-25, and miR-141 are decreased during replicative senescence and how increased MKK4 affects the responsiveness of senescent cells to stress-causing agents.
In contrast to MKK4, evidence suggests that MKK7, which can also phosphorylate JNK, plays a role in delaying senescence (26). The premature senescence observed in MKK7−/− mouse embryo fibroblasts (MEFs) could not be rescued by MKK4, which is likely due to differences in the substrate specificity of these two MAPK kinases. Unlike MKK7, MKK4 can phosphorylate both JNK and p38 MAPKs, the latter of which promotes the induction of replicative senescence (27). Sun et al. reported that the p38 substrate PRAK was critical for inducing senescence (25). PRAK−/− mice displayed enhanced tumorigenesis and PRAK−/− cells exhibited impaired senescence; tumorigenesis and senescence were linked to the phosphorylation of the transcription factor p53, which is also a tumor suppressor and senescence-associated protein. p53 was proposed to be a key effector of the Ras pathway that leads to PRAK activation (25). According to our analysis of the senescence markers SA-β-gal and p16, reducing MKK4 abundance by either siRNA or (Pre)miRNAs inhibited the senescence phenotype, whereas elevating MKK4 enhanced cellular senescence (Fig. 6, A to D).
MKK4 abundance in HDFs was dependent on the combinatorial influence of at least four miRNAs acting through the MKK4 3′ and 5′ UTRs. The elevated amounts of miR-15b, miR-24, miR-25, and miR-141 contributed to the reduced abundance of MKK4 in proliferating fibroblasts, whereas their reduction led to increased MKK4 production in senescent fibroblasts. A similar pattern was observed in tissues from human donors (fig. S1 and fig. S3), suggesting that increased MKK4 may also occur in vivo in senescent cells.
Together, these findings support a model in which individual miRNAs may only have an incremental effect upon target mRNA abundance, whereas the combined action of several miRNAs on a given target mRNA affects protein abundance. The combined effect of miRNAs affords the cell with a versatile means of increasing and decreasing the amount of a protein with a high degree of accuracy. This ability to fine-tune protein abundance by collections of miRNAs is increasingly recognized in pathophysiological processes that require adaptive gene expression patterns (6, 8). As shown here for MKK4, microRNAs ensure a flexible, potent, and precise control of protein abundance during key cellular processes – such as replicative senescence – underlying cancer and aging.
MATERIALS AND METHODS
Cell culture and transfections
Early-passage, proliferating (‘young’, ~20–30 Pdl) and late-passage, senescent (50–55 Pdl) human WI-38 diploid fibroblasts (HDFs, Coriell Cell Repositories) were cultured as described (20). HeLa cells were cultured in DMEM supplemented with 10% FBS. Control (Ctrl) siRNA, 2′-O-methyl antisense (AS)miR-15b, (AS)miR-24, (AS)miR-25, (AS)miR-141, precursor (Pre)miR-15b, (Pre)miR-24, (Pre)miR-25, and (Pre)miR-141, were from Ambion. RNAs were transfected at a final concentration of 100 nM; oligonucleotides and plasmids were transfected with Lipofectamine 2000 (Invitrogen). Plasmids pcDNA3.1-FLAG-MKK4 and pcDNA3.1-FLAG were transfected at 2 μg per 60 mm plate. siRNA directed again MKK4 (siMKK4, Santa Cruz) was transfected into cells at a final concentration of 100 nM using Oligofectamine (Invitrogen).
Plasmids for luciferase analysis were derived from the dual luciferase reporter vector psiCHEK-2 (Promega, Madison, WI, referred to as pLuc). Plasmid pLuc-MKK4(WT) comprised the MKK4 3′UTR spanning the first 1.3 kb where all six predicted miRNA sites are located. Additional plasmids were derived from pLuc-MKK4(WT) by mutating individually and in combination (fig. S7) using a site-directed mutagenesis kit (Stratagene) (fig. S6). The mutated miRNA seed recognition sequences for each miRNA recognition site are as follows: GC to TA for miR-15b, GG to TT for miR-24, GGC to TTA and GC to TA for miR-25, and CG to AT, and GG to TT for miR-141. HeLa cells (and WI-38 or IMR-90 cells, fig. S7) were transfected with 200 ng of plasmid DNA using Lipofectamine-2000. Cells were lysed 24 hours after transfection and the ratios of RL and FL activities were measured with a dual-luciferase assay (Promega).
RNA and protein analysis
Total cellular RNA was isolated directly from cells or from gradient fractions using Trizol (Invitrogen). After reverse transcription (RT) using random hexamers and SSII reverse transcriptase (Invitrogen), real-time quantitative (q)PCR analysis was performed using gene-specific primer pairs (below) and SYBR Green PCR master mix (Applied Biosystems), as described (20). The following primers were used: CTCTTCACTCCCAACAAT and ACATGGGAGAGCTGGGAGTA for MKK4 mRNA; TGCACCACCAACTGCTTA and GGCATGGACTGTGGTCATGAG for GAPDH mRNA; CCCTGATCAAGAGCGAAGAG and GTCTAACCTCGCCCTTCTCC for RL mRNA; TTCGCTAAGAGCACCCTGAT and GTAATCAGAATGGCGCTGGT for FL mRNA; CCCTATCAACTTTCGATGGTAGTCG and CCAATGGATCCTCGTTAAAGGATTT for 18S rRNA.
Mature miR-15b, miR-24, miR-25, and miR-141 were quantified using the Taqman microRNA Detection Assay (Applied Biosystems).
Prediction of MKK4 mRNA as a target of miR-15b, miR-24, miR-25, and miR-141
The MKK4 mRNA was predicted to be target of miR-15b, miR-24, miR-25, and miR-141 at several sites within the 3′UTR after performing computational analyses with prediction algorithms that included Pictar, TargetScan, RNAhybrid, and RNA22. Using RNA22 (http://cbcsrv.watson.ibm.com/rna22.html) and the sequences of MKK4 mRNA and miR-15b, miR-24, miR-25, and miR-141, the program returned the hits shown in fig. S4.
Immunohistochemistry, immunofluorescence, and SA-β-galactosidase activity
For immunofluorescence analysis, cells were fixed with 4% paraformaldehyde, then permeabilized with 0.2% Triton X-100 in PBS. For FLAG detection, samples were blocked in 5% BSA for 1 hour, incubated with a 1:1000 dilution of the rabbit monoclonal antibody against FLAG (Sigma Aldrich) in the background reducing antibody diluent (DAKO) for 16 hours at 4°C. Slides were washed with PBS-T and then incubated with a 1:1000 dilution of the FITC-labeled anti-rabbit monoclonal antibody for 1 hour at room temperature. The cell preparations were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) in Prolong GOLD antifade (Invitrogen-Molecular Probes, Carlsbad, CA). SA-β-gal activity was detected with a kit (Cell Signaling) following the manufacturer’s recommendations. SA-β-gal images were acquired using an inverted AX10 Observer D1 microscope (Carl Zeiss, Inc.) and images were analyzed using Axiovision software. Images of cells that exhibited SA-β-gal and overexpressed pCDNA-FLAG-MKK4 were acquired sequentially using the multi-dimensional acquisition platform and analyzed by Axiovision software.
Western blot analysis
Whole-cell lysates were prepared using RIPA buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS, and 1 mM dithiothreitol) and proteins were resolved by SDS-PAGE and transferred to PVDF membranes (Invitrogen). After incubation with primary antibodies recognizing MKK4, GAPDH, p-p38, PRAK, p-PRAK, p21, p16 or α-Tubulin (Santa-Cruz Biotech), p38 or cyclin D1 (Cell Signaling), β-actin (Abcam), or FLAG (Sigma-Aldrich), blots were incubated with the appropriate secondary antibodies and signals were detected by ECL Plus (Amersham). Western blotting signals were quantified by densitometry using Image J software.
Polyribosome analysis
Cells were incubated with cycloheximide (100 μg/ml; Calbiochem) for 15 min. Cytoplasmic lysates (500 μl) were centrifuged through 10–50% linear sucrose gradients and divided into 10 fractions for analysis RT-qPCR analysis, as previously described (20).
Supplementary Material
Fig. S1. MKK4 abundance increases with aging in a panel of tissue biopsies from young and old individual donors.
Fig. S2. Survey of putative MKK4 mRNA-interacting RNA-binding proteins.
Fig. S3. Higher expression of predicted human MKK4 mRNA-directed microRNAs in fetal tissues compared with adult tissues.
Fig. S4. Predicted microRNA-MKK4 mRNA hybrids.
Fig. S5. Unchanged MKK7 and Hsp27 abundance despite changes in MKK4.
Fig. S6. Point mutations introduced at each predicted microRNA site.
Fig. S7. MKK4 reporter analysis in HeLa cells recapitulates MKK4 reporter analysis in WI-38 HDFs.
Fig. S8. Activities of caspase-3 and caspase-7 remain unaltered after modulating MKK4.
Fig. S9. Analysis of cells treated with (Pre)miRNAs and (AS)miRNAs to modulate MKK4 reveal that MKK4 does not appear to activate the JNK pathway in S cells.
REFERENCES AND NOTES
- 1.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 2.Cristofalo VJ, Lorenzini A, Allen RG, Torres C, Tresini M. Replicative senescence: a critical review. Mech Ageing Dev. 2004;125:827–848. doi: 10.1016/j.mad.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 3.Smith JR, Pereira-Smith OM. Replicative senescence: implications for in vivo aging and tumor suppression. Science. 1996;273:63–67. doi: 10.1126/science.273.5271.63. [DOI] [PubMed] [Google Scholar]
- 4.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 7.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 8.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shelton DN, Chang E, Whittier PS, Choi D, Funk WD. Microarray analysis of replicative senescence. Curr Biol. 1999;17:939–945. doi: 10.1016/s0960-9822(99)80420-5. [DOI] [PubMed] [Google Scholar]
- 12.Trougakos IP, Saridaki A, Panayotou G, Gonos ES. Identification of differentially expressed proteins in senescent human embryonic fibroblasts. Mech Ageing Dev. 2006;127:88–92. doi: 10.1016/j.mad.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001:410, 37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 14.Cuenda A. Mitogen activated protein kinase kinase 4 (MKK4) Int J Biochem Cell Biol. 2000;32:581–587. doi: 10.1016/s1357-2725(00)00003-0. [DOI] [PubMed] [Google Scholar]
- 15.Taylor J, Hickson J, Lotan T, Yamada DS, Rinker-Schaeffer CW. Using metastasis suppressor proteins to dissect interactions among cancer cells and their microenvironment. Cancer Metastasis Rev. 2008;27:67–73. doi: 10.1007/s10555-007-9106-7. [DOI] [PubMed] [Google Scholar]
- 16.Teng DH, Perry WL, 3rd, Hogan JK, Baumgard M, Bell R, Berry S, Davis T, Frank D, Frye C, Hattier T, et al. Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor. Cancer Res. 1997;57:4177–4182. [PubMed] [Google Scholar]
- 17.Robinson VL, Hickson JA, Vander Griend DJ, Dubauskas Z, Rinker-Schaeffer CW. Societal interactions in ovarian cancer metastasis: a quorum-sensing hypothesis. Clin Exp Metastasis. 2003:20, 25–30. doi: 10.1007/s10585-008-9177-z. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy NJ, Davis RJ. Role of JNK in tumor development. Cell Cycle. 2003;2:199–201. [PubMed] [Google Scholar]
- 19.Robinson VL, Shalhav O, Otto K, Kawai T, Gorospe M, Rinker-Schaeffer CW. Mitogen-activated protein kinase kinase 4/c-Jun NH2-terminal kinase kinase 1 protein expression is subject to translational regulation in prostate cancer cell lines. Mol Cancer Res. 2008;6:501–508. doi: 10.1158/1541-7786.MCR-07-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R, Jr, Srikantan S, Subrahmanyam R, Martindale JL, Yang X, Ahmed F, et al. p16(INK4a) translation suppressed by miR-24. PLoS ONE. 2008:3, e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci USA. 2008;105:20297–20302. doi: 10.1073/pnas.0809376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henke JI, Goergen D, Zheng J, Song Y, Schüttler CG, Jünemann C, Niepmann M. MicroRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tournier C, Dong C, Turner TK, Jones SN, Flavell RA, Davis RJ. MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes Dev. 2001;15:1419–1426. doi: 10.1101/gad.888501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, Xie C, Chen J, Deng Q, Yamout M, et al. PRAK is essential for ras-induced senescence and tumor suppression. Cell. 2007;128:295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 26.Wada T, Joza N, Cheng HY, Sasaki T, Kozieradzki I, Bachmaier K, Katada T, Schreiber M, Wagner EF, Nishina H, Penninger JM. MKK7 couples stress signalling to G2/M cell-cycle progression and cellular senescence. Nat Cell Biol. 2004;6:215–226. doi: 10.1038/ncb1098. [DOI] [PubMed] [Google Scholar]
- 27.Han J, Sun P. The pathways to tumor suppression via route p38. Trends Biochem Sci. 2007;32:364–371. doi: 10.1016/j.tibs.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 28.We thank M. Bernier, B. Damdinsuren, and M. Kaileh for providing reagents and N. N. Hooten for help with the preparation of the manuscript. This research was supported by the National Institute on Aging-Intramural Research Program, National Institutes of Health.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. MKK4 abundance increases with aging in a panel of tissue biopsies from young and old individual donors.
Fig. S2. Survey of putative MKK4 mRNA-interacting RNA-binding proteins.
Fig. S3. Higher expression of predicted human MKK4 mRNA-directed microRNAs in fetal tissues compared with adult tissues.
Fig. S4. Predicted microRNA-MKK4 mRNA hybrids.
Fig. S5. Unchanged MKK7 and Hsp27 abundance despite changes in MKK4.
Fig. S6. Point mutations introduced at each predicted microRNA site.
Fig. S7. MKK4 reporter analysis in HeLa cells recapitulates MKK4 reporter analysis in WI-38 HDFs.
Fig. S8. Activities of caspase-3 and caspase-7 remain unaltered after modulating MKK4.
Fig. S9. Analysis of cells treated with (Pre)miRNAs and (AS)miRNAs to modulate MKK4 reveal that MKK4 does not appear to activate the JNK pathway in S cells.