Abstract
Pooling clinical specimens reduces the number of assays needed when screening for infectious diseases. Polymerase chain reaction (PCR)-based assays are the most sensitive tests to diagnose malaria, but its high cost limits its use. We adapted a pooling platform that could reduce the number of assays needed to detect malaria infection. To evaluate this platform, two sets of 100 serum samples, with 1% and 5% malaria prevalence, were tested. DNA, extracted from pooled samples, was amplified by malaria-specific PCR. Additional validation was performed by determining the level of PCR detection based on 1:10 and 1:100 dilution. The platform correctly detected all malaria samples in the two test matrices. The use of stored serum samples also has important implications for studies investigating malaria prevalence rates retrospectively. Field studies, using serum and whole blood specimens, are needed to validate this technique for the adaptation of these methods for clinical utility.
Although microscopy remains the gold standard diagnostic test for malaria in clinical settings,1,2 polymerase chain reaction (PCR)-based assays can have 100-fold greater sensitivity,3-5 especially in the setting of low parasitemia6 or subclinical infections.7 Despite this advantage, the high cost of reagents, equipment, and quality assurance have limited PCR-based diagnostic assays primarily to research settings. Gal and others8 have shown that malaria DNA can be successfully detected in serum rather than the commonly used whole blood samples when using PCR-based assays. Previously, we showed that this applies to stored serum samples as well.9 This allows testing banked sera samples, a commonly stored specimen in clinical studies, to determine malaria infection rates in a sensitive manner. To further advance the usefulness of this technique, we sought to increase efficiency by pooling specimens.
Methods that pool clinical specimens before performing diagnostic tests have been shown to be an efficient way of screening for infectious diseases. This technique was first proposed by Dorfman10 to screen for syphilis in military recruits. Modifications to Dorfman's original pooling algorithm by Finucan11 and Phatarfod and Sudbury12 have further improved the technique by reducing the number of individual tests needed to identify positive samples. Nucleic acid amplification tests using pooled specimens are now used for detecting viral infections such as acute HIV,13 hepatitis B and C,14 and West Nile virus,15 but not malaria.
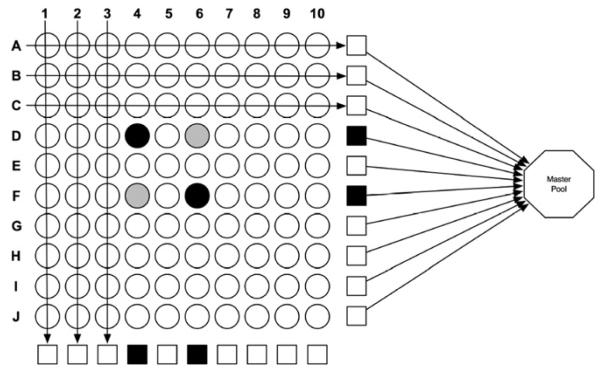
For detecting malaria infection, we propose adapting the pooling platform described by Westreich and others.16 One hundred individual clinical samples are arranged in rows and columns in a 10 × 10 matrix format (Figure 1). Ten individual samples are pooled by row and column such that each sample is represented in 2 of the 20 mini-pools, and each pool is assayed. Negative results of all 20 pools would exclude malaria infection in the 100 samples and preclude further testing. For a matrix with positive pools, additional assays may be needed to determine which sample(s) were positive, depending on the number and location of the individual positive samples. Sample selection for additional assays is guided by the 10 × 10 matrix platform. In our example (Figure 1), the four positive pools (black squares in rows D and F and columns 4 and 6) can result from various combinations of positive samples in the locations: D4, D6, F4, and F6 (gray and black circles). These four samples would require further testing; therefore, in this example, the pooling method would identify the two positive samples (black circles D4 and F6) using only 24 PCR assays (20 row and column pools + 4 individual) instead of the 100 needed by individual testing. In regions of even lower malaria prevalence, a master -pool comprised of all 100 samples by pooling the row or column pools (Figure 1), resulting in 1:100 dilution for a single positive sample, can also be used.
Figure 1.
Example of using a 10 × 10 matrix to screen for malaria infection. Mini-pools (squares) are made up of 10 samples (circles) each. The octagon represents a master-pool made from 10 mini-pools. A negative master-pool would indicate that all 100 samples constituting the pool are negative, precluding further testing. The black squares and circles represent PCR-positive samples. The gray circles represent negative samples that need to be tested individually to rule out infection.
Our goals for this study were to (1) propose a strategy for pooling specimens and (2) show the feasibility of malaria detection by PCR-based methods using pooled, field-collected serum samples.
The details of specimen selection and cohort enrollment have been reported earlier.17 This study was approved by the Institutional Review Boards/Ethical Committees of the University of California San Diego, Universidad Peruana Cayetano Heredia, AB PRISMA, and US Department of Defense. Permission to conduct the study was provided by the Loreto Director of Health, Iquitos, Peru. Informed consent was obtained from all participants before enrollment. Blood (2–4 mL) was collected from participants with acute malaria infection and uninfected controls from Iquitos, Peru, who had Plasmodium vivax or P. falciparum parasitemia confirmed by microscopy. Serum was separated and stored at −20°C for an average period of 6 weeks. The samples were then stored at −80°C for an average of 4.5 years before use in this study. Sufficient serum was available from 15 participants with malaria diagnosed by light microscopy and 5 uninfected individuals.18
The following semi-quantitative system was used: < 1+, < 1 parasite/100 high power fields (HPF); +, 1 to < 2 parasites/HPF; ++, 2–20 parasites/HPF; +++, 21–200 parasites/HPF; ++++, > 200 parasites/HPF. Parasite concentration was also measured by determining the number of parasites per 200 white blood cells (WBCs). The parasitemia, as parasites per microliter, was calculated using a standard WBC count of 8,000/μL.
Two representative sets of 100 specimens each were prepared using malaria positive and negative sera. Because of limited serum samples, sufficient serum for testing in the two matrices was available for 6/15 microscopy-confirmed malaria patients. Commercially available serum (Gemini Bio-Products Catalog 100–512) was used as a substitute for malaria-negative sera. The first set was comprised of serum from one malaria-positive sample (1% prevalence) and the second set contained five malaria-positive samples (5% prevalence) with the remainder made up of 99 and 95 malaria-negative serum samples, respectively. Malaria-positive samples were randomly placed in the two matrices in a blinded fashion. Row and column pools were prepared using 100 μL serum from each sample, resulting in 20 pools of 1 mL each. From each pool, DNA was extracted from 400 μL of pooled sera using the DNeasy Blood and Tissue Kit (Catalog 69506; Qiagen) according to the manufacturer's blood and body fluid protocol. DNA extracted from 200 μL of each undiluted malaria-positive serum sample was used as positive control. Similarly, DNA from 200 μL serum from cases that did not have malaria based on light microscopy and PCR was used as negative control.
The feasibility of detecting parasite DNA if there was only one positive sample in a pool was tested in pools of 10 (1:10 dilution) and 100 (1:100 dilution) serum samples. Row and column pools containing serum (100 μL) from single malaria positive case also included nine malaria-negative cases (900 μL pooled serum; Gemini Bio-Products). A master pool of 100 specimens was prepared by pooling 100 μL of serum from either the 10 row or column pools.
PCR amplification was performed using a modification of the technique originally described by Snounou and others4 with primers targeting the Plasmodium spp. 18S small subunit ribosomal RNA genes. The PCR reaction was performed in a total volume of 50 μL containing 20 μL extracted DNA, 25 μL of HotStar Taq Master Mix (Catalog 203443; Qiagen), and forward and reverse primers (0.2 μmol/L). Cycling conditions were incubation at 95°C for 15 minutes, followed by 35 cycles of 95°C for 30 seconds, and 58°C for 1 minute, with a final incubation at 72°C for 1 minute. Primer sequences were as follows: 5′-TTAAAATTGTTGCAGTTAAAACG-3′ (sense) and 5′-CCTGTTGTTGCCTTAAACTTC-3′ (antisense). The presence of amplification products was detected by ethidium bromide staining after agarose gel (1.8%) electrophoresis.
Plasmodium vivax was the predominant infecting species in our study (12/15), with P. falciparum accounting for the remainder. The parasite counts, as measured by microscopy, were < 1+ to +++ (mean, 2,704 parasites/μL; range, 1,000–7,407 parasites/μL) for subjects with P. vivax malaria and 1+ to ++ (mean, 7,778 parasites/μL; range, 2,210–16,730 parasites/μL) for subjects with P. falciparum malaria.
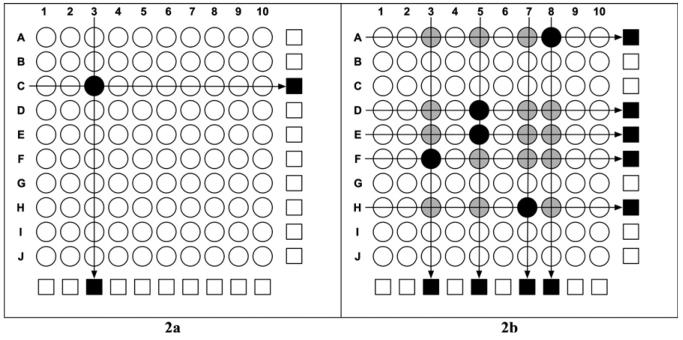
In the first set of 100 specimens, malaria PCR identified two positive pools (Figure 2A). The 10 × 10 matrix identified the individual sample (P. vivax parasitemia 2,694 parasites/μL or ++) that resulted in the positive tests without any further testing. In the second set, there were nine positive pools (Figure 2B). The 10 × 10 matrix guided selection of individual samples for further PCR assays that identified the five malaria-positive samples (P. vivax parasitemia range, 234–7,407 parasites/μL or < 1+ to +++).
Figure 2.
Specimen pooling strategy using malaria-positive samples to represent 1% and 5% malaria prevalence rates. A, Two pools (black squares) tested positive, and the matrix identified the single malaria positive sample (black circle). B, The positive pools (black squares) led to further testing of individual samples (black and gray circles) and identification of five malaria-positive samples (black circles).
The PCR-based assay detected parasite DNA in all 15 (100%) undiluted samples and in mini-pools with a 1:10 dilution. This implies that all 15 samples would have tested positive in a 10 × 10 matrix. In master pools with a 1:100 dilution, 12/15 (80%) samples had detectable DNA, with a mean parasite count of 4,083 parasites/μL (range, 926–16,730 parasites/μL or + to +++). In the three negative samples, the mean parasitemia was 2,260 parasites/μL (range, 234–4,395 parasites/μL or < 1+ to ++). Results from the second matrix using DNA extracted from malaria-positive vertical and horizontal mini-pools and individual serum samples are shown in Figure 3. These PCRs were designed to maximize sensitivity for qualitative purposes (detection of presence or absence of malaria template), and quantitative conclusions cannot be made. All PCR assays were carried out in triplicate.
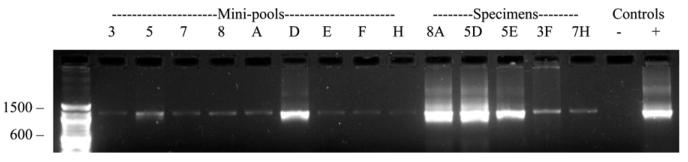
Figure 3.
PCR amplification products using Plasmodium genus-specific primers after DNA extraction from pooled and individual serum samples from matrix shown in Figure 2B. Lane 1, 100-bp DNA size marker ladder; Lanes 2–10, DNA extracted from malaria-positive vertical and horizontal mini-pools; Lanes 11–15, DNA extracted from malaria-positive individual specimens that constituted the mini-pools; Lane 16, DNA extracted from serum from a malaria-negative control; Lane 17, DNA extracted from P. falciparum (3D7) strain.
In this study, we evaluated, for the first time, a strategy that pools clinical samples to improve the efficiency of screening for malaria using a PCR-based assay. We also showed the feasibility of such methods for P. vivax and P. falciparum by using long-term cryostored serum samples (> 4 years), which could be important for the retrospective investigation of specimen banks of cohort studies.
The success of the proposed pooling strategy depends on the ability of PCR to detect parasite DNA despite the dilutional effect of pooling. The detection limit of malaria PCR is ≥ 5 parasites/μL,6,19 and in our experiments, the mini-pools with 1:10 dilution had parasite levels well above this (> 20 parasites/μL). Detectable parasite DNA was also present in all except 3 of 15 master pools with 1:100 dilution. Low parasitemia by microscopy may explain the false-negative PCR in one sample (Sample 14) that would have had a parasite level of 2.34 parasites/μL after dilution in a master pool of 100 samples. The other two samples should have had parasite levels within the detection limit of PCR even after dilution, so the negative results are unclear. We speculate that there could have been a discrepancy between parasitemia quantified by microscopy that measures intra-erythrocytic parasites and amount of parasite DNA present in serum or the degradation of parasite DNA during long-term cryostorage. Future studies should test a wider range of clinical samples with more stringent criteria for measuring parasitemia, such as quantitative PCR. Species-specific PCR may enhance sensitivity of the method and remains to be evaluated. Also, there are no published data on how long parasite DNA persists in serum after malaria treatment, and this needs to be studied.
To detect parasite DNA, malaria PCR must overcome the dilutional effect of pooling, which has implications when selecting appropriate target population. Because of differences in infection rates, we believe that this strategy is better suited for hypoendemic malaria regions than hyperendemic ones. Hypoendemic regions have infection rates that do not exceed 10%.20 The pooling strategy in these settings may have significant cost-savings depending on matrix size. However, the optimal number of specimens in the pool will be limited by the sensitivity of malaria PCR and vary from region to region depending on parasite density. For example, in our setting, mini-pools of 10 specimens in a 10 × 10 matrix were best suited for pooling, but the master pool of 100 specimens had a low sensitivity (80%). On the other hand, in hyperendemic areas, the infection rates are > 50%,20 which would require testing of all individual specimens in the 10 × 10 matrix, thereby eliminating its benefit. Smaller pools will have to be tested until an optimal size is determined. Another potential use of the pooling method in hyperendemic regions could be to evaluate control measures once they have successfully reduced malaria prevalence to make this strategy cost effective.
We recognize that there may be other clinical scenarios in which parasitemia may be so low as to give false-negative results during PCR-based screening with pooled samples, as in asymptomatic malaria infection and inadequate response to antimalarials caused by malarial drug resistance. However, the pooling strategy should still have increased sensitivity over microscopy, because experienced microscopists can detect as low as 20 parasites/μL,21 but routine diagnostic laboratories often have a much inferior sensitivity of detection (500 parasites/μL or 0.01% RBC infected).3
Given the exploratory nature of our study, we would like to address its limitations. First, because of the small sample size, we cannot accurately assess the sensitivity and predictive values of the pooling strategy. These test characteristics will need to be determined in much larger studies and more appropriately in various clinical populations. Second, although this technique should be relatively easy to adapt for clinical care, it will need to be evaluated in the field with both serum and whole blood samples. In fact, the yield of parasite DNA from whole blood or dried blood spots is most likely to be greater than in serum, and these specimens may be better suited to pooling. Third, the cost effectiveness of the proposed methods will need to be formally evaluated. In hypoendemic malaria regions (< 10% prevalence), the mini-pooling strategy may provide significant cost-savings over screening samples using PCR-based assays individually or for screening asymptomatic individuals. In regions of even lower malaria prevalence, a master pool comprised of all 100 samples by pooling the row and column pools might be useful with considerable cost-savings. The optimal size of the pool would, therefore, depend on malaria prevalence in the region and the group of patients of interest, such as symptomatic versus asymptomatic.
In these studies, screening for malaria infection with PCR-based assays using pooling platforms is sensitive and efficient, although field studies using a variety of biological samples and in various malaria endemic populations are needed for validation and to evaluate its cost effectiveness.
Table 1.
Results of microscopy and PCR detection of Plasmodium DNA using undiluted and pooled stored serum
| Microscopy |
PCR result |
||||
|---|---|---|---|---|---|
| Serial no. | Species | Parasites/μL | Undiluted | Mini-pool (1:10 dilution) |
Master-pool (1:100 dilution) |
| 1 | Pf | 16,730 | + | + | + |
| 2 | Pv | 1,942 | + | + | + |
| 3 | Pf | 4,395 | + | + | − |
| 4 | Pv | 1,000 | + | + | + |
| 5 | Pv | 2,684 | + | + | + |
| 6 | Pv | 2,150 | + | + | − |
| 7 | Pv | 3,540 | + | + | + |
| 8 | Pv | 7,407 | + | + | + |
| 9 | Pv | 2,694 | + | + | + |
| 10 | Pv | 4,210 | + | + | + |
| 11 | Pv | 3,042 | + | + | + |
| 12 | Pv | 2,614 | + | + | + |
| 13 | Pv | 926 | + | + | + |
| 14 | Pv | 234 | + | + | − |
| 15 | Pf | 2,210 | + | + | + |
Parasitemia quantification: < 1+, < 1 parasite/100 HPF; +, 1 to < 2 parasites/HPF; ++, 2–20 parasites/HPF; +++, 21–200 parasites/HPF; ++++, > 200 parasites/HPF.
Pf = Plasmodium falciparum; Pv = Plasmodium vivax.
Acknowledgments
The authors thank the patients from the city of Iquitos and the surrounding villages for their participation and our field staff for their assistance.
Financial support: A.R.B. is supported by National Institute of Mental Health Grant R25 MH81482 and the REACH (Research and Education in HIV/AIDS for Resource-Poor Countries) Initiative of Tibotec; J.M.V. is supported by a Doris Duke Charitable Foundation Innovations in Clinical Research Program grant, NIH/NIAID Grant K24AI068903, and NIH Fogarty International Center Global Infectious Diseases Training Grant 5D43TW007120; D.M.S. is supported by National Institutes of Health Grants MH083552, AI077304, AI69432, AI38858, AI43638, AI43752, AI29164, AI47745, MH625N12, AI047745, and AI57167 and the UCSD Center for AIDS Research (AI36214); and the HIV Neurobehavioral Research Center (HNRC) is supported by Center Award P30 MH62512 from the National Institute of Mental Health.
REFERENCES
- 1.Makler MT, Palmer CJ, Ager AL. A review of practical techniques for the diagnosis of malaria. Ann Trop Med Parasitol. 1998;92:419–433. doi: 10.1080/00034989859401. [DOI] [PubMed] [Google Scholar]
- 2.Warhurst DC, Williams JE. ACP broadsheet no 148. July 1996. Laboratory diagnosis of malaria. J Clin Pathol. 1996;49:533–538. doi: 10.1136/jcp.49.7.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milne LM, Kyi MS, Chiodini PL, Warhurst DC. Accuracy of routine laboratory diagnosis of malaria in the United Kingdom. J Clin Pathol. 1994;47:740–742. doi: 10.1136/jcp.47.8.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 5.Hermsen CC, Telgt DS, Linders EH, van de Locht LA, Eling WM, Mensink EJ, Sauerwein RW. Detection of Plasmodium falciparum malaria parasites in vivo by real-time quantitative PCR. Mol Biochem Parasitol. 2001;118:247–251. doi: 10.1016/s0166-6851(01)00379-6. [DOI] [PubMed] [Google Scholar]
- 6.Coleman RE, Sattabongkot J, Promstaporm S, Maneechai N, Tippayachai B, Kengluecha A, Rachapaew N, Zollner G, Miller RS, Vaughan JA, Thimasarn K, Khuntirat B. Comparison of PCR and microscopy for the detection of asymptomatic malaria in a Plasmodium falciparum/vivax endemic area in Thailand. Malar J. 2006;5:121. doi: 10.1186/1475-2875-5-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roshanravan B, Kari E, Gilman RH, Cabrera L, Lee E, Metcalfe J, Calderon M, Lescano AG, Montenegro SH, Calampa C, Vinetz JM. Endemic malaria in the Peruvian Amazon region of Iquitos. Am J Trop Med Hyg. 2003;69:45–52. [PubMed] [Google Scholar]
- 8.Gal S, Fidler C, Turner S, Lo YM, Roberts DJ, Wainscoat JS. Detection of Plasmodium falciparum DNA in plasma. Ann N Y Acad Sci. 2001;945:234–238. doi: 10.1111/j.1749-6632.2001.tb03891.x. [DOI] [PubMed] [Google Scholar]
- 9.Bharti AR, Patra KP, Chuquiyauri R, Kosek M, Gilman RH, Llanos-Cuentas A, Vinetz JM. Polymerase chain reaction detection of Plasmodium vivax and Plasmodium falciparum DNA from stored serum samples: implications for retrospective diagnosis of malaria. Am J Trop Med Hyg. 2007;77:444–446. [PubMed] [Google Scholar]
- 10.Dorfman R. The detection of defective numbers of large populations. Ann Math Stat. 1943;XXX:436–440. [Google Scholar]
- 11.Finucan HM. The blood testing problem. Appl Stat. 1964;13:43–50. [Google Scholar]
- 12.Phatarfod RM, Sudbury A. The use of a square array scheme in blood testing. Stat Med. 1994;13:2337–2343. doi: 10.1002/sim.4780132205. [DOI] [PubMed] [Google Scholar]
- 13.Stramer SL, Glynn SA, Kleinman SH, Strong DM, Caglioti S, Wright DJ, Dodd RY, Busch MP. Detection of HIV-1 and HCV infections among antibody-negative blood donors by nucleic acid-amplification testing. N Engl J Med. 2004;351:760–768. doi: 10.1056/NEJMoa040085. [DOI] [PubMed] [Google Scholar]
- 14.Mine H, Emura H, Miyamoto M, Tomono T, Minegishi K, Murokawa H, Yamanaka R, Yoshikawa A, Nishioka K. High throughput screening of 16 million serologically negative blood donors for hepatitis B virus, hepatitis C virus and human immunodeficiency virus type-1 by nucleic acid amplification testing with specific and sensitive multiplex reagent in Japan. J Virol Methods. 2003;112:145–151. doi: 10.1016/s0166-0934(03)00215-5. [DOI] [PubMed] [Google Scholar]
- 15.Busch MP, Caglioti S, Robertson EF, McAuley JD, Tobler LH, Kamel H, Linnen JM, Shyamala V, Tomasulo P, Kleinman SH. Screening the blood supply for West Nile virus RNA by nucleic acid amplification testing. N Engl J Med. 2005;353:460–467. doi: 10.1056/NEJMoa044029. [DOI] [PubMed] [Google Scholar]
- 16.Westreich DJ, Hudgens MG, Fiscus SA, Pilcher CD. Optimizing screening for acute human immunodeficiency virus infection with pooled nucleic acid amplification tests. J Clin Microbiol. 2008;46:1785–1792. doi: 10.1128/JCM.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bharti AR, Chuquiyauri R, Brouwer KC, Stancil J, Lin J, Llanos-Cuentas A, Vinetz JM. Experimental infection of the neo-tropical malaria vector Anopheles darlingi by human patient-derived Plasmodium vivax in the Peruvian Amazon. Am J Trop Med Hyg. 2006;75:610–616. [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization . Basic Malaria Microscopy, Parts I and II. World Health Organization; Geneva: [Google Scholar]
- 19.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemio-logic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 20.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonkman A, Chibwe RA, Khoromana CO, Liabunya UL, Chaponda ME, Kandiero GE, Molyneux ME, Taylor TE. Cost-saving through microscopy-based versus presumptive diagnosis of malaria in adult outpatients in Malawi. Bull World Health Organ. 1995;73:223–227. [PMC free article] [PubMed] [Google Scholar]