Abstract
Cofilin is a key player in actin dynamics during cell migration. Its activity is regulated by (de)phosphorylation, pH, and binding to phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2]. Here, we here use a human cofilin-1 (D122K) mutant with increased binding affinity for PI(4,5)P2 and slower release from the plasma membrane to study the role of the PI(4,5)P2–cofilin interaction in migrating cells. In fibroblasts in a background of endogenous cofilin, D122K cofilin expression negatively affects cell turning frequency. In carcinoma cells with down-regulated endogenous cofilin, D122K cofilin neither rescues the drastic morphological defects nor restores the effects in cell turning capacity, unlike what has been reported for wild-type cofilin. In cofilin knockdown cells, D122K cofilin expression promotes outgrowth of an existing lamellipod in response to epidermal growth factor (EGF) but does not result in initiation of new lamellipodia. This indicates that, next to phospho- and pH regulation, the normal release kinetics of cofilin from PI(4,5)P2 is crucial as a local activation switch for lamellipodia initiation and as a signal for migrating cells to change direction in response to external stimuli. Our results demonstrate that the PI(4,5)P2 regulatory mechanism, that is governed by EGF-dependent phospholipase C activation, is a determinant for the spatial and temporal control of cofilin activation required for lamellipodia initiation.
INTRODUCTION
The actin depolymerizing factor (ADF)/cofilin family is ubiquitously present in all eukaryotes and involved in various cellular processes that depend on actin dynamics (Bamburg, 1999; Ono, 2007; Van Troys et al., 2008). In nonmuscle cells, two isoforms are expressed: cofilin-1 and ADF (Hotulainen et al., 2005). ADF/cofilins are multipotent actin modulators. They interact both with monomeric (G)- and filamentous (F)-actin and are reported to nucleate, sever, and depolymerize filaments (Carlier et al., 1997; Chan et al., 2000; Andrianantoandro and Pollard, 2006). Severing by cofilin increases the number of actin filament barbed ends available for polymerization and enhances filament turnover in vitro and in vivo (Chan et al., 2000; Ichetovkin et al., 2002). In several disease conditions that rely on actin dynamics, cofilin is emerging as a key player (Maloney et al., 2008; Yoder et al., 2008). A prime example is tumor cell invasion and metastasis (Wang et al., 2006; Yan et al., 2008; van Rheenen et al., 2009). In mouse and rat models of mammary cancer, expression levels of cofilin and its regulators are altered and the resulting high cofilin activity is correlated with invasion, intravasation, and metastasis (Wang et al., 2004, 2006, 2007).
The role of cofilin during cell migration and invasion is closely linked to its role in the formation of membrane protrusions, such as lamellipodia. Protrusive activity is an essential early requirement for different forms of cell motility. In invasive mammary carcinoma cells (MTLn3), cofilin is demonstrated to be an early effector of membrane protrusion, because its local activation sets the site where new lamellipodia occur as well as the direction that a migrating cell takes (Ghosh et al., 2004; Mouneimne et al., 2006; Sidani et al., 2007). We reported previously that cofilin depletion in amoeboid MTLn3 cells using siRNA has a major impact on cell morphology and results in directional cell migration, defects in cell turning and impaired chemotactic response toward an epidermal growth factor (EGF)-gradient (Sidani et al., 2007). Cofilin is shown associated with the F-actin network throughout an extending lamellipodium (Lai et al., 2008), but it is also demonstrated to be activated at sites of new lamellipodia initiation, before actin nucleating factors such as the Arp2/3 complex that are subsequently involved in lamellipodial extension (DesMarais et al., 2004, 2005). Initiating lamellipodia at new locations is one mechanism that migrating cells use to change direction, and several cofilin knockout studies (Dawe et al., 2003; Sidani et al., 2007) indicate that cofilin contributes to the cells' directional sensing machinery via its role in lamellipodia initiation.
Clues on the underlying mechanism of this activity lie in the strict cellular regulation of cofilin activity. Multiple mechanisms of cofilin regulation that are activated upon cell stimulation are known (Huang et al., 2006; Scott and Olson, 2007; Van Troys et al., 2008). Cofilin kinases (LIM kinases and testicular protein kinases) phosphorylate cofilin residue Ser3 and inactivate its ability to bind actin. Specific cofilin reactivating phosphatases have been characterized, including the Slingshot family and chronophin. In addition, cofilins are regulated by interaction with the polyphosphoinositide PI(4,5)P2 (van Rheenen et al., 2007, Van Troys et al., 2008), and by intracellular pH (Bernstein et al., 2000). pH changes have recently been shown to also directly modulate cofilin-PI(4,5)P2 interaction based on the protonation state of cofilin His133(Frantz et al., 2008). Spatio-temporal differences in these regulatory mechanisms of cofilin and the coincidence of several of these regulatory events in cells seem important for cofilin action in directional cell protrusion (Mouneimne et al., 2006; van Rheenen et al., 2007; Frantz et al., 2008; van Rheenen et al., 2009).
In this study we focus on the regulation mechanism that is based on cofilin binding to the membrane polyphosphoinositide (PI) phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2]. Many proteins that are crucial to the assembly of the actin-based migration machinery are regulated by PI(4,5)P2, and the level of this lipid is controlled by lipid kinases (PI-3K and PI-5K), phosphatases (phosphatase and tensin homologue and synaptojanins), and hydrolases (phospholipase C [PLC] isozymes). These enzymes can be activated downstream of several growth factor signaling pathways (Hilpela et al., 2004; Niggli, 2005; Ling et al., 2006; Kolsch et al., 2008). In vitro, the interaction of cofilin with PI(4,5)P2 inhibits F-actin binding (Yonezawa et al., 1990; Van Troys et al., 2000; Gorbatyuk et al., 2006). In cells, the role of the cofilin–PI(4,5)P2 interaction in actin dynamics and cell motility processes has been more difficult to address directly. Previously described cofilin mutants with decreased in vitro PI(4,5)P2 binding in most cases also display defects in actin interaction (Van Troys et al., 2000; Ojala et al., 2001), complicating their use in cells.
Indirect evidence for a role for cofilin-PI(4,5)P2 regulation has come from the demonstration that in metastatic mammary carcinoma cells (MTLn3) the early burst in actin polymerization, induced by EGF stimulation, and the resulting chemotactic response of the cells depend on both cofilin severing and PLC activation (Mouneimne et al., 2004, 2006; Sidani et al., 2007). Using different approaches including colocalization, fluorescence resonance energy transfer (FRET), and membrane fractionation, van Rheenen et al., 2007 have provided important evidence that a pool of cellular cofilin associates with the plasma membrane.
On PI(4,5)P2-hydrolysis, either induced by EGF stimulation and PLC activation or by PLC-independent means, cofilin is locally released from the membrane in an active form. Peripheral cofilin is shown to distribute into two different compartments (plasma membrane [PM] and peripheral F-actin network) that cannot be optically resolved (van Rheenen et al., 2007). In resting cells, cofilin is predominately localized in the PM compartment, where the PM binding is very transient (seconds) with a fast on/off rate. PI(4,5)P2-hydrolysis lowers the PI(4,5)P2 level and therefore slows down the on rate. Because the PM-off-rate remains unaltered, the change in on/off equilibrium causes a net translocation of cofilin from the PM (van Rheenen et al., 2007). Therefore, PLC does not need access to the cofilin-bound PI(4,5)P2 molecules to cause a translocation of cofilin from the PM (van Rheenen et al., 2009).
Although the PM and actin compartment cannot be optically resolved, the cofilin molecules located in these two compartments can be distinguished using a fluorescence loss in photobleaching (FLIP)-based assay (van Rheenen et al., 2007). In contrast to the fast PM on/off rate (<4 s), the on/off-rate of cofilin to F-actin is much slower (>25 s) (van Rheenen et al., 2007). Consequently, the exchange rate of cofilin molecules between periphery (i.e., sum of PM and peripheral F-actin compartment) and the cytosol depends on the ratio of peripheral cofilin molecules that are localized either at the PM or in the F-actin compartment (van Rheenen et al., 2007). Because cofilin translocates from the PM to the F-actin compartment upon EGF stimulation, the FLIP-based assays display different exchange kinetics with the cytosol before and after stimulation. Collectively, this approach has indicated that PI(4,5)P2 hydrolysis induces a translocation of membrane associated cofilin to the underlying F-actin and has provided the first indirect evidence that cellular cofilin activity is regulated at the membrane in a precise spatial manner after PI(4,5)P2 hydrolysis (van Rheenen et al., 2007, 2009).
Our present work directly explores how the cofilin-PI(4,5)P2 interaction affects cell motility processes, by using a cofilin mutant with enhanced PI(4,5)P2 affinity. We described previously that in actophorin, the cofilin homologue from Acanthamoeba castellanii, a point mutation in an actin-binding helix resulted in increased affinity for PI(4,5)P2 in vitro (Van Troys et al., 2000). Here, we characterize the corresponding mutant of human cofilin-1 and exploit it to directly demonstrate how an altered cofilin–PI(4,5)P2 interaction affects cell migration properties of both carcinoma cells and fibroblasts.
MATERIALS AND METHODS
Cofilin Expression Constructs
Human cofilin-1 cDNA (wild type [WT]) was cloned in the bacterial expression plasmid pKM263 (obtained via EUROSCARF, Frankfurt, Germany) to produce glutathione transferase (GST)-fusions for purification. WT cofilin in pcDNA3.1/V5-His-TOPO was described previously (Sidani et al., 2007). The D122K mutation was introduced using QuikChange mutagenesis (Stratagene, La Jolla, CA). WT and/or D122K cofilin cDNA were cloned in the eukaryotic vectors pcDNA3.1/V5-His-TOPO, pEGFP.N1, and pEGFP.C1 and in pIRES2-GFP (Invitrogen, Paisley, United Kingdom) to produce V5-, enhanced green fluorescent protein (eGFP)-tagged, and untagged cofilin, respectively. As in Sidani et al. (2007), five silent mutations were additionally present in the WT and D122K cofilin cDNA (except for constructs in pEGFP.N1), making the resulting WT and D122K cofilin-RNA resistant to down-regulation by the cofilin small interfering RNA (siRNA) used.
Proteins
Rabbit skeletal muscle actin was prepared and labeled as described in Van Troys et al. (2000). Recombinant GST-cofilin (WT or D122K mutant) was purified over glutathione-Sepharose (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) following the manufacturer's protocol but with omission of Triton X-100 in incubation and washing solutions. GST-fusion proteins were eluted with 10 mM glutathione in phosphate-buffered saline (PBS), pH 8.0, and stored in 0.02 mM EDTA and 25 mM Tris-HCl, pH 8.0, at 4°C. The stability of WT and mutant cofilin was compared by recording Trp fluorescence spectra in 1–5 M urea in 20 mM Tris-HCl, pH 8.0 (F-4500 fluorimeter; Hitachi, Tokyo, Japan) as described in Van Troys et al. (2000) (Supplemental Figure S1). Untagged cofilins were obtained by tobacco etch virus (TEV)-digest (Invitrogen) of GST-cofilin fusions (digestion during 1 h at 4°C and 0.25 U/μg protein).
PI(4,5)P2-Cofilin Binding Studies
GST-fusions of WT or D122K cofilin were incubated at 15 μM with a range of concentrations (0–200 μM) of PI(4,5)P2 (Sigma-Aldrich, St. Louis, MO) in micelles in 50 mM KCl, 0.2 mM EGTA, 0.2 mM dithiothreitol (DTT), and 25 mM Tris-HCl, pH 7.5, for 30 min on ice. We used homogeneous PI(4,5)P2 micelles, generated by brief sonication of an aqueous PI(4,5)P2 stock solution. The incubated mixtures were analyzed as in Van Troys et al. (2000). In short, the incubated mixture was analyzed by gel filtration, allowing quantification of cofilin-PI(4,5)P2 binding based on the different elution times of free and micelle-bound GST-cofilin. GST has been shown not to interact with PI(4,5)P2 in a similar assay (Zimmermann et al., 2002). The pH dependency of the PI(4,5)P2 interaction was evaluated using different incubation conditions: 50 mM KCl, 0.2 mM EGTA, and 0.2 mM DTT in either 25 mM Tris-HCl, pH 7.5, or 43 mM PIPES, pH 6.5.
PI(4,5)P2-cofilin binding at pH 7.5 was also tested using a microfiltration assay (Lambrechts et al., 1997) by using untagged cofilins (5 μM) that were prepared by TEV digest from fusion proteins; the flow-through of microfiltration through a membrane with a molecular weight cut-off of 30,000 Da (i.e., lower than the micelle size and higher than the free protein) was analyzed using SDS-polyacrylamide gel electrophoresis (PAGE) followed by Coomassie staining and quantified using ImageJ (National Institutes of Health, Bethesda, MD).
Actin Assays
Effects of cofilin (WT or D122K) on F-actin depolymerization/disassembly were monitored using light scattering at 350 nm at an angle of 90° at room temperature in an Aminco Bowman series luminescence spectrometer (Sim-Aminco, Rochester, NY). G-actin (4.5 μM) in G buffer (0.1 mM CaCl2, 0.2 mM DTT, 0.2 mM ATP, and 5 mM Tris-HCl, pH 7.7) was polymerized by adding KCl and MgCl2 to 0.1 M and 1 mM, respectively. When polymerization reached steady state, cofilin (WT or D122K mutant) or buffer (control) was added (constant volumes added that are <5% of the total sample volume), and the signal decrease was followed in time (Carlier et al., 1997).
Kinetic measurements of severing activity were carried out using an elongation assay as in (Yeoh et al., 2002). Unlabeled F-actin was added as nuclei (1 μM) to 4 μM G-actin (10% pyrene labeled) in F buffer (G buffer with 0.1 M KCl and 1 mM MgCl2), and the rate of polymerization was determined. The unlabeled seeds were added as such (control) or treated with various concentrations of WT or mutant cofilin before addition. Pyrene fluorescence was recorded using 365 nm as excitation wavelength and 388 nm as emission wavelength. Elongation rates (in Δfluorescence/s) were calculated from the increase in fluorescence in the time span between 50 and 150 s and normalized to the calculated rate in the sample containing the untreated seeds.
Cell Culture and Treatments
NIH3T3 cells were cultured in DMEM, 10% fetal bovine serum (FBS), 20 mM l-GlutaMAX I, 1% penicillin-streptomycin (Invitrogen) at 37°C and 5% CO2 and transfected using Lipofectamine 2000 (Invitrogen). MTLn3 cells were maintained, starved, and stimulated as described in DesMarais et al. (2004) and transfected using FuGENE 6 or 6HD (Roche Diagnostics, Mannheim, Germany). The cofilin siRNA used has been validated previously (Mouneimne et al., 2004; Sidani et al., 2007), and the siRNA treatment and rescue attempts with cofilin-constructs (WT or D122K) were performed exactly as in Sidani et al. (2007). The cells were transfected with the cofilin expressing constructs 8–10 h after siRNA addition.
Immunoassays
Antibodies used were anti-cofilin (Cytoskeleton, Denver, CO), anti-green fluorescent protein (GFP) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-p34 antibody (Millipore, Billerica, MA), anti-V5 antibody (Invitrogen), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Abcam, Cambridge, United Kingdom), Alexa Fluor-594 anti-chicken (Invitrogen), and IRDye 800 and 680 CW goat anti-mouse and anti-rabbit immunoglobulin G (LI-COR Biosciences, Lincoln, NE).
For immunofluorescence, MTLn3 cells were grown on glass coverslips. Cells were fixed and permeabilized as described previously (Eddy et al., 2000). In brief, cells were simultaneously fixed and permeabilized using 3.7% paraformaldehyde, 0.1% glutaraldehyde, and 0.075 mg/ml saponin (Sigma-Aldrich) in PBS. Images were recorded using an inverted microscope (model IX71; Olympus, Tokyo, Japan) or a motorized inverted microscope (model IX81L; Olympus), equipped with a computer-driven cooled charge-coupled device (CCD) camera, using an UPLanFLN(oil) 60× objective and the CellM imaging software (Olympus) and further processed using ImageJ. Relative signals in front and back of MTLn3 cells (e.g., for the Arp2/3 signal) were obtained as described in Sidani et al. (2007).
Whole cell lysates were prepared in a urea/thiourea lysis buffer containing 0.5% Triton X-100 and 0.1% DTT in the presence of a cocktail of protease and phosphatase inhibitors. For the isoelectrofocusing step of two-dimensional (2D)-SDS PAGE, Immobiline Drystrips (GE Healthcare) were used with a pH range from 3 to 10 for GFP-cofilins and (for optimal spot resolution) from 6 to 11 for endogenous cofilin, resulting in differences in separation between nonphosphorylated and phosphorylated spots for exogenous and endogenous cofilin. Assignment of phosphorylated spots was based on a shift to a more acidic pI and on the differences in 2D spot patterns before and after treatment of cell lysates with lambda phosphatase (New England Biolabs, Ipswich, MA) (data not shown). Western blotting signals were detected and quantified based on IRDye infrared dye technology (LI-COR Biosciences) on a Odyssey infrared imaging system.
Cell Morphology Scoring
Cell morphology changes of individual MTLn3 cells after siRNA treatment and/or rescue were addressed based on length to width ratio as described in Sidani et al. (2007).
Quantitative Analysis of Random Cell Migration and Membrane Protrusion
NIH3T3 cells were plated on fibronectin (5.7 μg/cm2) 24 h after transfection and allowed to spread for 16 h. Their migration at 37°C under appropriate CO2 conditions was followed by recording phase-contrast images every 5 min for 4 h by using an IX71 inverted microscope (Olympus) with a 10× numerical aperture (NA) 0.3 infinity-corrected objective and equipped with a monochrome SPOT-RT CCD camera. Image processing, and derived migration parameters are described in Supplemental Data.
MTLn3 cells were plated onto glass-bottomed dishes (MatTek, Ashland, MA) 12 h after transfection (20–22 h after siRNA) and allowed to spread for 24 h. Analysis of constitutive motility in serum was done for 45 min at a rate of two frames/min in L15 (Invitrogen), 5% FBS as described in Sidani et al. (2007) by using a 20× NA 1.4 infinity-corrected optics microscope (model IX71; Olympus), supplemented with a computer-driven cooled CCD camera and operated by IPLab Spectrum software (VayTek, Fairfield, IA). Images were processed using ImageJ.
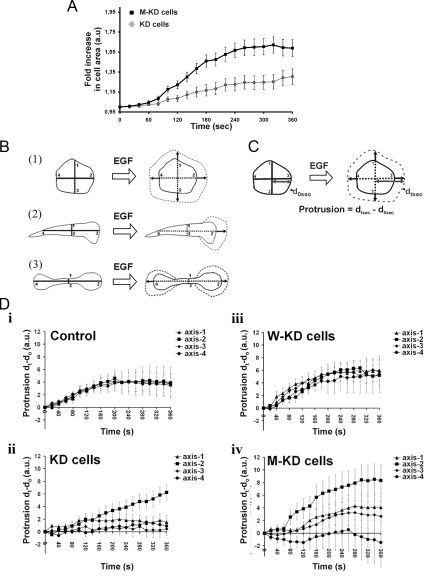
To study EGF-induced membrane protrusion, MTLn3 cells, starved in L15/0.35% bovine serum albumin for 3 h, were stimulated with 5 nM EGF (Invitrogen) at 37°C. Time-lapse series were recorded for 10 min with a time interval of 10 s. Fold changes in cell area were measured using ImageJ, standardized over the area of the cell at time 0 (before EGF stimulation), and averaged over the number of measured cells. The axial protrusion measurement is explained in Figure 7. These measurements were performed at 1-min intervals after EGF stimulation and standardized over the measurements on the corresponding cell at time 0.
Figure 7.
M-KD cells retain polarized protrusive activity upon EGF stimulation. The expression constructs are coding for WT and D122K cofilin-V5 (also used in Figures 5 and 6). (A) Mean-fold increase in cell area (as measure for protrusive activity induced by EGF) for the morphologically similar KD and M-KD cell population in function of time. The fold increase is expressed in relation to the cell area at time 0. (B) Cartoon clarifying the mode of protrusion, based upon measurements along axes. Control MTLn3 and W-KD cells follow scheme 1, whereas KD and M-KD cells follow either schemes 2 or 3. Vertical and horizontal axes pass through the geometrical center of the cells. (C) Measurement method of axial increase, t seconds after EGF stimulation (only illustrated along axis 2). Time 0 is time of EGF (5 nM) addition via bath stimulation. (D) Mean increases in distance along cell axes 1–4 in response to EGF for cells of the four MTLn3 cell populations. In total, 20 cells were used for each group: i) control cells, ii) KD cells, iii) W-KD cells, and iv) M-KD cells.
FLIP
MTLn3 cells were transiently transfected with eGFP-WT or eGFP-D122K cofilin constructs 8 h after plating on glass-bottomed dishes (MatTek). FLIP was performed 24 h after transfection using an AOBS SP5 confocal microscope (Leica, Wetzlar, Germany) at 25°C. Before the experiment, cells were starved as described above. The experiment was performed as in van Rheenen et al. (2007). In brief, at time 0, cytosol was photobleached for 10 s and fluorescence at the cell periphery (Per) and in the cytosol (Cyt) were followed for 20 s after bleaching. On bleaching, the Per/Cyt ratio increases and, because of exchange of molecules between the cytosol and periphery (sum of PM and peripheral F-actin compartment), this ratio subsequently returns to baseline levels. To normalize for bleach efficiencies, the initial ratio was set to 0, and the first point after bleaching was set to 1. The decay of the ratio follows a two-component exponential decay, with a PM component and a F-actin component. Based on biochemical properties showing that the effects on F-actin are not affected by the D122K mutation, the amplitude and the τ for the F-actin component of the decay are assumed unchanged and values are taken from van Rheenen et al. (2007): τ, 26 s; and amplitude, 0.15. Amplitude of the PM component (0.85) was taken from van Rheenen et al., (2007), whereas τPM of the membrane pool (for both the WT and D122K data sets) was varied in the fitting.
PI(4,5)P2 Hydrolysis Assay
Cells were transfected with the PI(4,5)P2 biosensor mRFP-PH(PLCδ1) (van Rheenen et al., 2005) and with either WT cofilin-GFP or D122K cofilin-GFP. The expression level of the cofilin constructs was determined by confocal acquisition of a 12-bit GFP-image that was analyzed in ImageJ. PI(4,5)P2 hydrolysis was assayed by acquiring an mRFP-confocal image every 10 s. The time series were imported into a custom-made visual studio (Professional edition 2008; Microsoft, Redmond, WA) program (JvR), in which the PI(4,5)P2 levels were analyzed by plotting the PM/cytoplasmic fluorescence over time as described in Stauffer et al. (1998), van der Wal et al. (2001), and Varnai and Balla (2008). Regions of interest (ROI) at the PM, cytoplasm, and background were determined automatically for every image in a time series. In brief, a binary mask was generated by using a thresholding step on a smoothed image. From this mask, the PM-ROI was determined by dilating the binary image for 6 pixels. The cytosol ROI was determined by dilating the binary image for another 6 pixels. The background ROI was assigned manually. These ROIs were updated for each image in the time series, and the mean fluorescence was measured. The translocation of monomeric red fluorescent protein (mRFP)-PH(PLCδ1) was expressed as the ratio of the fluorescence values for membrane and cytosol area, to correct for bleaching and cell movement.
RESULTS
D122K Cofilin-1: Properties In Vitro and in Cells
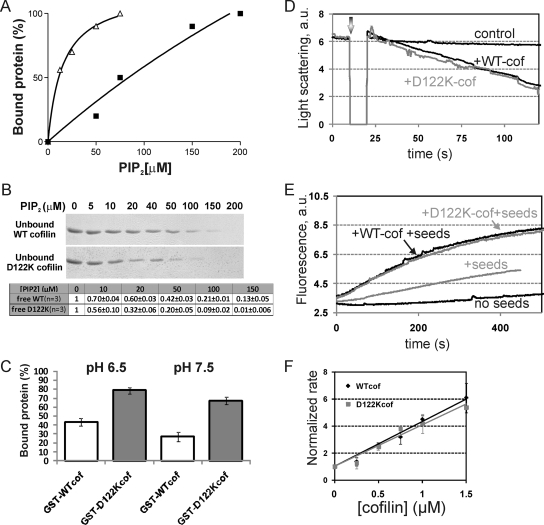
We biochemically compared the human nonmuscle cofilin-1 mutant carrying an aspartic acid-to-lysine mutation at position 122 (D122K cofilin) with the WT cofilin protein. The introduced mutation does not affect protein stability (Supplemental Figure S1). Using a gel filtration-based assay and homogeneous PI(4,5)P2 micelles as described previously in a study of actophorin mutants (Van Troys et al., 2000), we compared the PI(4,5)P2-binding capacity of GST-WT and GST-D122K cofilin. The proteins (15 μM) were incubated at pH 7.5 with micellular PI(4,5)P2, and the bound protein fraction at different total PI(4,5)P2 concentrations was quantified. Under these conditions, GST-D122K cofilin binds 7- to 10-fold more strongly to micellular PI(4,5)P2 than GST-WT cofilin (based on C50 values; binding data from 2 independent experiments; representative experiment in Figure 1A). The gain-of-function for GST-D122K cofilin was confirmed by protein–lipid overlay, by using a membrane on which PI(4,5)P2 was spotted in a range of concentrations (PIP-array; Echelon Biosciences, Slat Lake City, UT), followed by Western blotting (data not shown). Importantly, the difference in affinity for complex formation with PI(4,5)P2 was also observed, albeit less extensive, for untagged D122K versus WT cofilin by using a semiquantitative microfiltration technique (Figure 1B). The difference in PI(4,5)P2 binding between WT and D122K cofilin is apparent for ratios of PI(4,5)P2/cofilin of 10 or lower [e.g., ≤150 μM PI(4,5)P2 vs. 15 μM cofilin in Figure 1A; ≤50 μM PI(4,5)P2 versus 5 μM cofilin in Figure 1B]. The protein concentrations used here (5–15 μM) are in the range of reported cellular cofilin concentrations (e.g., 20 μM in baby hamster kidney cells; Koffer et al., 1988). Cofilin isoforms are relatively abundant actin-binding proteins (Cooper et al., 1986, Bamburg and Bray, 1987), and, although unreported, its concentration upon recruitment to the PM may thus be relatively high. Overall PI(4,5)P2 concentrations of 10 μM have been reported previously (McLaughlin et al., 2002), and PI(4,5)P2 is suggested to be homogeneously present in the plasma membrane based on cellular assays (van Rheenen et al., 2005), rather than being highly concentrated in lipid raft (as suggested by biochemical assays). The range of PI(4,5)P2 to cofilin ratios of 10 or lower, in which WT and D122K cofilin display different binding properties in vitro, may consequently be similar to local physiological ratios at the PM.
Figure 1.
D122K cofilin has an increased affinity for micellar PI(4,5)P2 but displays a WT effect on actin. (A) Percentage of GST-WT cofilin (squares) and GST-D122K cofilin (triangles) bound to homogenous PI(4,5)P2 micelles at pH 7.5 as a function of total PI(4,5)P2 concentration, based on gel filtration analysis as in Van Troys et al. (2000). C50 values ([PI(4,5)P2] for 50% cofilin binding) for GST-D122K cofilin are substantially lower than for GST-WT cofilin: 11 versus 84 μM (experiment shown), 10–12 μM versus 100–125 μM in a second independent experiment. (B) SDS-PAGE analysis and Coomassie staining of the flow-through of microfiltration of mixtures of 5 μM cofilin (WT or D122K) and PI(4,5)P2 (at the indicated concentrations and in homogenous micelles) incubated at pH 7.5. The analyzed flow-through contains unbound, free cofilin. Bottom, quantification for three independent experiments (including the one shown); levels are normalized to the sample without PI(4,5)P2. (C) pH dependency of PI(4,5)P2 binding of GST-WT and D122K cofilin (15 μM) by using the same experimental setup as in A except for different buffer conditions (either pH 6.5 or 7.5). The PI(4,5)P2-concentration used is 25 μM. Average values with standard deviations for two experiments are shown. (D) Depolymerizing effect of adding WT or D122K cofilin (3 μM) to 4.5 μM steady state F-actin at the time indicated by the arrow. Depolymerization was recorded in time by measuring 90° light scattering at 350 nm. (E) Effect of cofilin on elongation kinetics of 1 μM unlabeled F-actin nuclei (seeds) when added to 4 μM G-actin (10% pyrene labeled). The unlabeled seeds were added as such (control) or treated with 0.5 μM of WT or D122K cofilin before addition. (F) Comparison analysis of normalized elongation rates calculated from experiments as described in E performed for a range of WT (diamonds) or D122K (squares) cofilin concentrations. Mean of three data sets with SD is shown. Normalization is done versus the elongation rate of non-cofilin-treated seeds.
Cofilin-PI(4,5)P2 binding has recently been shown to display pH dependency via cofilin residue His133, with protonated His133 (at pH 6.5) promoting maximal binding to PI(4,5)P2 (Frantz et al., 2008). The increased PI(4,5)P2 binding induced by the D122K mutation, observed here, is maintained both at pH 6.5 and pH 7.5 as illustrated using 15 μM GST-fusions and 25 μM PI(4,5)P2 in Figure 1C. This suggests that WT and D122K cofilin still display the same pH dependency in their PI(4,5)P2 interaction.
Effects of cofilin (WT or D122K) on actin were next compared (Figure 1, D–F). At a ratio of 1.5 of actin over cofilin, both WT and mutant cofilin induce a similar depolymerizing effect, monitored using light scattering, upon addition to F-actin at steady state (Figure 1D). The observed level of depolymerization at this cofilin/actin ratio is in accordance with data from Carlier et al. (1997). Cofilin severing is known to generate new filament ends (Ichetovkin et al., 2002). In Figure 1, E and F, we assayed this activity by measuring the elongation rate (in the presence of actin monomers) of unlabeled F-actin seeds that have been left untreated or premixed with different concentrations of either WT or D122K cofilin, as in described in Yeoh et al. (2002). Figure 1F shows that D122K cofilin and WT cofilin have similar dose-dependent promoting effects on the actin elongation rate of the seeds. This is indicative of similar increases in elongation competent filament ends generated by the severing activity of WT or mutant cofilin. Collectively, these two experiments show that introducing the positive charge at position 122 in human cofilin-1 does not influence the actin severing or depolymerization capacity of cofilin.
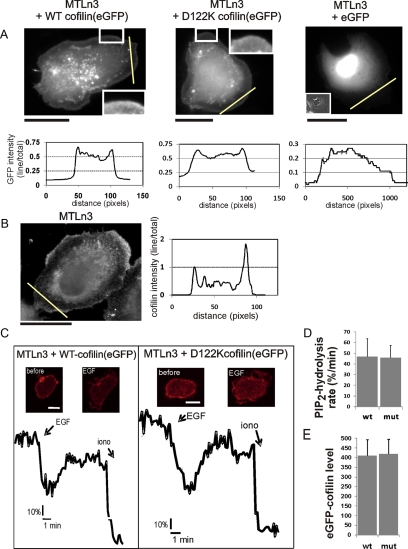
In addition, our data illustrate that overexpressed eGFP-fusions of WT and D122K cofilin display similar localization patterns in cells (Figure 2A and Supplemental Figure S2A). The overexpressed proteins are present throughout the cytoplasm of MTLn3 cells with some (peri)nuclear enrichment (Figure 2A). Notably, both WT and D122K cofilin are enriched in the extreme peripheral edge of the large flat lamellipodia of these cells. We document this by showing the intensity pattern along a line crossing a lamellipodium (Figure 2A, eGFP-expressing cells used as control). This localization pattern is overall similar to the pattern described previously for endogenous human cofilin-1 in different cell lines (Yonezawa et al., 1987). We show the localization for endogenous cofilin in MTLn3 cells for comparison (Figure 2B and Supplemental Figure S2A). Also, here peripheral enrichment is observed (Figure 2B).
Figure 2.
Localization of WT and D122K cofilin-eGFP in MTLn3 cells and effect on PI(4,5)P2 hydrolysis rate. (A) Localization of eGFP-cofilin (WT (left), D122K (middle), and eGFP (right) in MTLn3 cells. The insets show enlarged peripheral regions of the lamellipodia (left, middle) and a phase contrast image (right). The graphs show the eGFP intensity along the indicated lines (yellow) through the flat lamellipodia. Intensities are normalized to the intensity of the total cell area. eGFP-WT and D122K mutant are both targeted to the extreme leading edge; eGFP is not enriched in this region (note different y-axis for eGFP). Bars, 20 μm. (B) Immunofluorescence signal of endogenous cofilin in MTln3 showing peripheral enrichment. In the graph, the cofilin intensity along the indicated line normalized to the intensity of the total cell area is shown. Bar, 20 μm. (C) EGF-induced PI(4,5)P2 hydrolysis was measured using the PI(4,5)P2 probe mRFP-PH(PLCδ1) in MTLn3 cells transfected with eGFP-WT or eGFP-D122K cofilin and mRFP-PH(PLCδ1). Insets, confocal mRFP images before and after 5 nM EGF stimulation. The plots represent the relative PI(4,5)P2 levels over time expressed as the fluorescence ratio of plasma membrane to cytosol. To calibrate, 5 μM ionomycin was added to induce a full translocation. (D) The mean rate of PI(4,5)P2 hydrolysis (n = 5) as measured by linearly fitting the EGF-induced PI(4,5)P2 decrease between the time of EGF addition and the lowest level of PI(4,5)P2; the rate is expressed as the change in fluorescence ratio at plasma membrane to cytosol (expressed in percentage of the ratio at start of measurement) per minute. (E) The expression levels of eGFP-WT or D122K cofilin in the cells analyzed in D and E. Note that there is no significant difference.
Previous work by van Rheenen et al. (2007) demonstrated that, in MTLn3 cells, cofilin colocalizes with PI(4,5)P2 and that the membrane-bound cofilin fraction is released upon EGF-induced PI(4,5)P2-hydrolysis. The data presented in Figure 2, A and B, and Supplemental Figure S2A clearly indicate that both eGFP-WT and D122K cofilin are, similar to endogenous cofilin, targeted to the membrane compartment where PI(4,5)P2 is residing. Thus, to test whether the increased PI(4,5)P2 affinity of D122K cofilin would affect PLC-mediated PI(4,5)P2 hydrolysis in vivo, we tested EGF-induced PI(4,5)P2 hydrolysis by using the PI(4,5)P2 probe mRFP-PH(PLCδ1) in MTLn3 cells that express eGFP-D122K or WT cofilin (Figure 2, C–E). In resting cells, mRFP-PH binds to PI(4,5)P2 in the PM (Figure 2C, insets). On PI(4,5)P2 hydrolysis, mRFP-PH(PLCδ1) translocates from the PM to the cytoplasm (Figure 2C). In cells that express equal levels of eGFP-WT or D122K cofilin (Figure 2E), the membrane/cytosol fluorescence ratio of mRFP-PH was measured over time (Figure 2C). As can been seen in Figure 2, C and D, PI(4,5)P2 hydrolysis kinetics and the rate of PI(4,5)P2 hydrolysis upon EGF stimulation is not different in cells expressing either eGFP-WT or D122K cofilin. This indicates that overexpression of D122K cofilin does not influence EGF-mediated phosphatidylinositol bisphosphate hydrolysis.
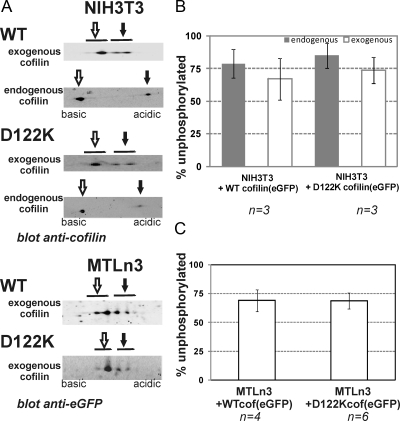
Because regulation by phosphorylation is another major regulatory mechanism of cofilin, we analyzed whether WT and D122K cofilin are phosphorylated at a similar level in cells. Using 2D-SDS-PAGE and Western blotting, we determined the ratio of exogenous and/or endogenous unphosphorylated versus phosphorylated cofilin in cell lysates. Figure 3 shows representative 2D blots (Figure 3A) and quantitative analyses (Figure 3, B and C). In NIH3T3 and MTLn3, the levels of active, unphosphorylated cofilin (Figure 3A; B and C, white bars) are not significantly different for exogenously expressed WT or mutant cofilin. Moreover, these levels are very similar to the level of unphosphorylated endogenous cofilin in the same cell lysate (determined in NIH3T3 cells; Figure 3B, gray bars) and to levels reported in resting MTLn3 (Song et al., 2006). This approach renders a static view on phosphorylation levels but clearly indicates that the introduced D122K mutation does not alter the propensity for phospho-inactivation of cofilin.
Figure 3.
D122K cofilin and WT cofilin are phosphorylated to a similar extent. (A) Representative Western blot signals for endogenous and/or exogenous (WT or D122K) cofilin in NIH3T3 and MTLn3 cells overexpressing WT or D122K cofilin(eGFP) after 2D-SDS PAGE analysis on whole cell lysates. For resolving spots of endogenous cofilin and exogenous cofilin, cell lysates were separated using pI ranges of 6–11 and 3–10, respectively. The spots corresponding to unphosphorylated and phosphorylated cofilin are indicated with open and closed arrows, respectively. (B and C) Graphs showing the percentages of unphosphorylated active cofilin for n independent experiments for NIH3T3 (B) and MTLn3 (C). Significant differences are not observed between WT and D122K cofilin(eGFP) nor between exogenous and endogenous cofilin.
In summary, the stronger PI(4,5)P2-binding D122K mutant of human cofilin-1 has unchanged properties with regard to its effects on actin dynamics and pH dependency of PI(4,5)P2 binding. In addition, it displays similar localization as well as phosphorylation levels as WT cofilin in cells and does not have a different effect on cellular PI(4,5)P2 hydrolysis rate. Together, these results indicate that the D122K-cofilin forms a unique tool for specifically elucidating the role of cofilin-PI(4,5)P2 regulation in vivo.
D122K Cofilin-overexpressing Fibroblasts Display a Faster and More Directional Migration than WT Cofilin-overexpressing Cells
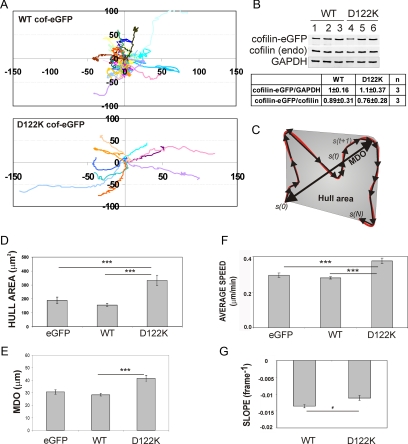
Cofilin is a regulator of actin dynamics in the leading edge of migrating cells and is important for initiating cellular protrusions and directing cell migration (DesMarais et al., 2005; Mouneimne et al., 2006; Sidani et al., 2007). Using the cofilin D122K mutant, we aimed at determining, more directly, whether these effects of cofilin during cell migration are regulated by its interaction with PI(4,5)P2. Toward this end, we followed random migration on fibronectin of fibroblast NIH3T3 cells overexpressing WT or D122K cofilin. Trajectories of fluorescent cells overexpressing cofilin-eGFP (WT or D122K) or expressing eGFP were obtained for large cell populations by automated tracking (Figure 4A). In the analyzed cell populations, the expression levels of WT and D122K cofilin-eGFP are not significantly different and also the level of overexpression over endogenous cofilin is similar. This was evaluated by Western blotting (Figure 4B) and by analytical fluorescence-activated cell sorting analysis, which revealed very similar distributions of eGFP intensity (data not shown). From the derived trajectories, several migration parameters were derived as in Debeir et al. (2005, 2008)). The maximum distance from origin (MDO) and the hull area are measurements of distance and area covered by a cell during its migratory path (Figure 4C). Overexpression of WT cofilin-eGFP has no significant effect on the mean hull area and MDO compared with mock-transfected cells (Figure 4, D and E). By contrast, overexpression of D122K cofilin-eGFP significantly increases both mean hull area and mean MDO in comparison with WT cofilin-eGFP overexpression or eGFP expression (Figure 4, D and E). These increases in distance and area covered by cells expressing the mutant cofilin are a possible reflection of changes in average cell speed, in cell turning or in both. Mean average speed of D122K cofilin-eGFP cells is indeed higher than that of control or WT cofilin-eGFP cells (Figure 4F). This not only provides evidence that D122K cofilin is active in actin dynamics in NIH3T3 cells but also that it induces faster migration on fibronectin. The mean SLOPE (Figure 4G), a parameter that is a measure for directional persistence of cell migration and calculated independently of cell speed (see Supplemental Data) is however also significantly less negative for cells with D122K cofilin compared with those with WT cofilin (p = 0.02). This indicates that D122K cofilin–eGFP-expressing cells have a tendency to migrate more on a straight path compared with cells with increased intracellular WT cofilin levels. This property is also readily detectable by visual inspection of cell trajectories, as demonstrated in Figure 4A. PI(4,5)P2 regulation of cofilin activity consequently seems to be an important factor in controlling cofilin-dependent directionality of cell migration and thus cell turning. The change in speed and in directionality may not be independent from a functional view point because it has been proposed that turning (implying repolarization) pauses moving cells.
Figure 4.
D122K cofilin overexpression results in faster and more directional fibroblast migration. (A) Cell trajectories (a selection is shown of the cell populations considered) of NIH3T3, migrating on fibronectin and either overexpressing WT (top) or D122K cofilin-eGFP (bottom). The initial position of each track is superimposed on (0,0). (B) Quantification of expression levels of WT or D122K cofilin-eGFP and ratio of cofilin-eGFP to endogenous cofilin in transfected NIH3T3 cells. Western blot with anti-cofilin signal is shown for WT (lanes 1–3) and D122K (lanes 4–6) cofilin–eGFP-expressing cells (3 independent samples). The GAPDH signal is used to correct for equal loading on the gel. In comparing WT and D122K cofilin expression levels, the cofilin-eGFP/GADPH ratio was set to 1 for WT (C) Cartoon illustrating migration parameters: cell trajectory in red, separate migration steps in black. s(0), origin, s(N), end position of trajectory. MDO and Hull area (gray) are measures of distance and area covered by a cell, respectively (see Supplemental Data). Mean value for Hull area (D), mean MDO (E), and average speed (F) for a population of NIH3T3 cells expressing eGFP (n = 155 cells), overexpressing WT (n = 390) or D122K cofilin-eGFP (n = 140). (G) Mean SLOPE, a parameter for persistence (see Supplemental Data), for NIH3T3 cell populations overexpressing WT (n = 204) or D122K cofilin-eGFP (n = 71). A less negative SLOPE value reflects a higher directionality of cell movement. (D–G) Mean or average values ± SEM; statistical significance of differences between mean values are calculated using Mann–Whitney tests and indicated by *, p < 0.05 and ***, p < 0.001.
D122K Cofilin Is Unable to Rescue Effects on Cell Polarization and Directional Migration Induced by Cofilin Knockdown (KD) in MTLn3 Cells
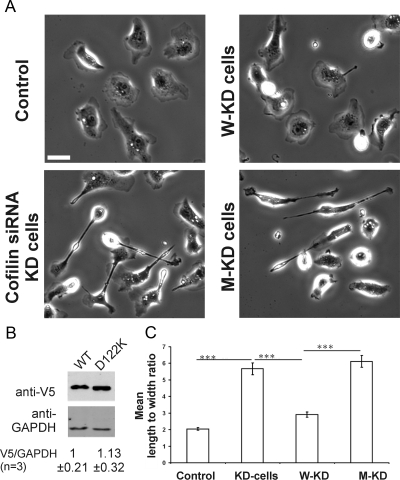
Cofilin siRNA-based KD induces in MTLn3 cells marked changes in cell morphology and migration properties (Sidani et al., 2007). To gain further insight in the cellular role of cofilin PI(4,5)P2-regulation, we chose this cell model to investigate whether D122K cofilin is able to restore the effects induced by cofilin knockdown. Cofilin knockdown was nearly complete (∼95%) in our experiments. This is in line with what is documented previously for the use of this same cofilin siRNA under identical experimental conditions [see figure 1 in Sidani et al. (2007) and Mouneimne et al. (2004)]. We here used a construct expressing D122K cofilin-V5 designed from the WT-rescue construct used in Sidani et al. (2007). The WT and mutant constructs display similar transfection efficiencies (∼60%) and showed similar expression levels determined by Western blotting (Figure 5B). MTLn3 cells treated with cofilin siRNA (KD cells) have a polarized elongated phenotype and KD cells expressing WT cofilin (W-KD) regained the rounded morphology characteristic of parental MTLn3 cells (Sidani et al., 2007) (Figure 5A). The morphology of KD cells expressing D122K cofilin (M-KD) (Figure 5A, bottom right), however, resembles that of KD cells. Measurements of the length to width ratio of control, KD, W-KD, and M-KD cells clearly demonstrate that, unlike WT cofilin, D122K cofilin expression does not rescue the elongated KD phenotype (Figure 5C). Analysis of cell perimeter, roundedness, and maximum length also showed that D122K cofilin was not able to rescue the distinct morphology of KD cells (data not shown).
Figure 5.
D122K cofilin expression does not rescue the cofilin KD morphological phenotype. (A) Morphological phenotype of control MTLn3, KD (cofilin siRNA treated), W-KD (cofilin siRNA + WT cofilin expression), and M-KD (cofilin siRNA + D122K cofilin expression) cells. Bar, 10 μm. The constructs code for V5-tagged cofilin (WT or D122K), as also used in Sidani et al. (2007). (B) Western Blot analysis demonstrating equal expression levels for WT and D122K cofilin-V5 used in A and C. Level of GAPDH is used to correct for equal loading on the gel. Below the blot, the average ratio ± SD of V5 over GAPDH signal is shown for three independent experiments; ratio for WT set to one. (C) Mean length to width ratios for the four cell populations in A; control (n = 70 cells); KD (n = 93 cells); W-KD (n = 125 cells); M-KD (n = 156 cells). Significantly different mean values, calculated between indicated pairs using t test, are shown by ***, p <0.001.
We qualitatively repeated this experiment using an internal ribosome entry site (IRES) construct that allows easy detection of cells expressing exogenous WT or D122K cofilin in the population by a coexpression of eGFP (Supplemental Figure S3). This confirmed the lack of rescue by D122K cofilin expression because, based on the eGFP signal, D122K cofilin-expressing cells are indeed still elongated (Supplemental Figure S3, right), whereas in a parallel experiment using the WT IRES construct most fluorescent cells are rounded (Supplemental Figure S3, middle).
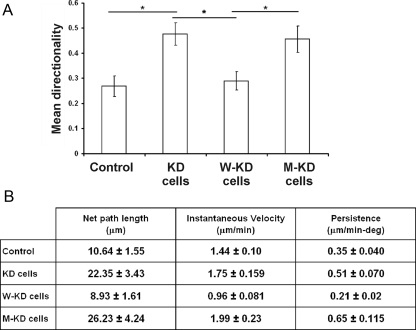
Cofilin KD cells have been described to move in a highly directional manner compared with parental and W-KD cells (Sidani et al., 2007). We thus compared the migration behavior of W-KD and M-KD MTLn3 cells. Similar to what we observed in fibroblasts (Figure 4), M-KD cells display a significantly higher directionality and higher persistence of cell migration compared with W-KD cells (Figure 6, A and B, and Supplemental Figure S5, videos 1–4). The increase in directionality is accompanied with an increase in cell speed (Figure 6B) as was also observed in NIH3T3 (Figure 4). MTLn3 cells expressing D122K cofilin consequently mimic the migration behavior of KD cells, indicating that the altered PI(4,5)P2 binding property of this mutant prevents restoring the cofilin rescue effect of inducing turning in migrating cells.
Figure 6.
D122K cofilin expression in KD cells is incapable of restoring the normal migration mode of MTLn3. (A) Mean directionality of MTLn3 control cell population (n = 19 cells) and of populations of KD (n = 17), W-KD (n = 15), and M-KD (n = 15) cells. Directionality is here the ratio of net to total path length. Unlike WT cofilin, D122K cofilin expression does not rescue the migration defects characteristic for KD cells. *p < 0.05 calculated using t test. Experimental setup and cofilin expression constructs used are as in Figure 5. Supplemental Figure S5 (videos 1–4) shows cell migration of cells from the four populations. (B) Migration parameters (mean net path length, mean cell velocity, and mean persistence) of M-KD cells compared with control, KD, and W-KD cells for same cell populations as in A. Persistence is here cell speed divided by direction change, as in Sidani et al. (2007).
Sidani et al. (2007) showed that in cofilin KD cells the Arp2/3 complex is asymmetrically distributed with a stronger presence in the front of the elongated KD cells. We therefore analyzed the distribution of the Arp2/3 complex in cofilin KD MTLn3 cells expressing either eGFP-tagged WT or mutant cofilin (W-KD vs. M-KD) (Supplemental Figure S4A). We verified the elongated morphology of eGFP-D122K cofilin-expressing cells (M-KD) (Supplemental Figure S4B) and the comparable expression levels of eGFP-WT and mutant cofilin (Supplemental Figure S4, C and D). Supplemental Figure S4E shows that the front to back ratio for the Arp2/3 signal in M-KD cells is not larger than 1 and not significantly different from that in W-KD cells. Consequently, the front enrichment for the Arp2/3 complex reported for cofilin KD (Sidani et al., 2007) seems no longer present in both W-KD and M-KD cells. Supplemental Figure S4F in addition demonstrates that also the front to back signals for eGFP in W-KD and M-KD cells are not significantly different and suggests that both WT and D122K cofilin are comparably distributed throughout the cells.
Our data show that M-KD cells differ from cofilin KD cells in Arp2/3 complex enrichment to the front but that they strongly display the same elongated morphologies and migration parameters as cofilin KD cells.
D122K Cofilin Is Incapable of Rescuing the KD Cell Defect in Initiating Multiple Lamellipodial Protrusions upon EGF Stimulation
Previous reports have demonstrated that EGF stimulation induces lamellipodial protrusive activity in MTLn3 cells. This activity follows two peaks: an early transient of actin polymerization, at 1 min after stimulation, which depends on both cofilin and PLC activity; and a late peak, at 3 min, which is PI-3K dependent (Mouneimne et al., 2004; Mouneimne et al., 2006). It was recently demonstrated, in the same cells, that EGF stimulation [inducing a PLC-dependent PI(4,5)P2-decrease] releases membrane-associated cofilin which translocates to peripheral F-actin (van Rheenen et al., 2007). To demonstrate the role of the direct PI(4,5)P2–cofilin interaction in this process, we compared EGF-induced lamellipodial protrusive activity of M-KD cells to that of control, KD, and W-KD cells. As described in Sidani et al. (2007), the increase in cell area upon EGF-induced protrusive activity is considerably lower in KD cells compared with control cells and can be restored upon rescue with WT cofilin (Supplemental Figure S5, videos 5–7). Figure 7A compares the fold increase in cell area induced by EGF stimulus for the morphologically similar KD cells and M-KD cells. The mean cell area for the M-KD cell population increases faster and the fold change is more extensive, indicating that the mutant cofilin positively contributes to actin polymerization and protrusive activity. However, EGF induced protrusive activity displayed by M-KD cells is different from that of W-KD cells (Supplemental Figure S6, videos 7 and 8). EGF-stimulated W-KD cells protrude in multiple directions and initiate protrusions with equal frequency around the entire cell perimeter (following scheme 1 in Figure 7B) (Sidani et al., 2007). In contrast and similar to KD cells, M-KD cells mainly protrude at the cell pole(s) after EGF stimulation (Figure 7B, schemes 2 or 3) and the elongated, KD-like morphology of M-KD cells consequently remains during cell stimulation. To quantitatively compare lamellipodial protrusion dynamics of round cells (control and W-KD) and elongated cells (KD and M-KD), we measured the EGF-induced increase in length of the vertical and horizontal cell axes (spanning from the geometrical center to the periphery of the cell) (Figure 7, C and D). M-KD cells display a larger increase along axis 2 versus 1 and 3 (Figure 7D, iv), which is also characteristic of elongated KD cells (Figure 7D, ii). This indicates that protrusive activity mainly occurs in a unipolar manner in M-KD, i.e., in a spatially restricted manner at one pole of the elongated cells. Protrusion along axis 2 is more efficient in M-KD cells versus KD cells (Figure 7D, ii and iv), and this underlies the higher fold increase in cell area depicted in Figure 7A for cells rescued with D122K cofilin. The protrusion phenotype in D122K–cofilin-expressing cells is thus strikingly different from WT–cofilin-expressing cells, and this strongly suggests that WT regulation of cofilin activity by PI(4,5)P2 is required to obtain apolar protrusion in vivo. Protrusions along the side of the elongated M-KD cells are only rarely observed, albeit at a slightly higher frequency compared with KD cells (28% vs. 16%).
D122K Cofilin Translocates Slower from the Plasma Membrane than WT Cofilin
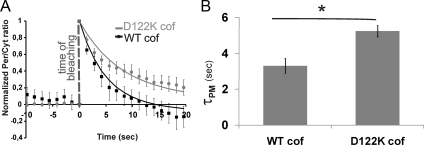
We analyzed the exchange kinetics of eGFP-D122K and eGFP-WT cofilin between the periphery (Per) and cytoplasm (Cyt) in MTLn3 cells by using a FLIP approach established in van Rheenen et al. (2007) (Figure 8). Using this approach, the localization of cofilin at the PM- and F-actin peripheral compartments can be distinguished (van Rheenen et al., 2007). In this way, we aimed at obtaining a cellular mechanistic basis for the inability of the D122K cofilin mutant to restore the phenotype induced by cofilin siRNA in MTLn3 cells. MTLn3 cells expressing either eGFP-WT or eGFP-D122K cofilin were photobleached in a cytoplasmic region. This results in an instantaneous increase of normalized Per/Cyt fluorescence (Figure 8A, time 0). Subsequently, the fluorescence in cytoplasm and in the peripheral region was followed for 20 s. As detailed in van Rheenen et al. (2007), the overall residence time of (eGFP) cofilin in the periphery is determined by residence times of two cofilin pools: cofilin on actin filaments in this peripheral region and cofilin associated with the PM. This together determines how fast fluorescent cofilin molecules translocate from periphery to cytoplasm and thus how fast the Per/Cyt ratio returns to baseline after bleaching. Figure 8A shows that the recovery of the cytoplasmic fluorescence, and thus the decrease of Per/Cyt, is slower in cells expressing eGFP-D122K cofilin compared with cells overexpressing eGFP-WT cofilin. This indicates that D122K cofilin is retained longer in the periphery.
Figure 8.
D122K cofilin translocates slower from the plasma membrane than WT cofilin. (A) FLIP was measured in MTLn3 cells overexpressing eGFP-WT cofilin (black) or eGFP-D122K cofilin (gray) after bleaching in the cytosol at time 0 as indicated. The FLIP signal is expressed as Per/Cyt (ratio of peripheral vs. cytosolic fluorescence) in time (the initial ratio is set to 0, the first measuring point after bleaching is set to 1). Data shown represent the average of four independent experiments with a total number of 31 and 33 cells for WT and D122K, respectively. The solid lines represent the two-phase exponential decay fitting based on the first 20s. (B) τPM value ± SEM (τ values or mean time constants associated with fast component of 2-phase exponential decay) derived from data for cells with eGFP-WT or eGFP-D122K cofilin shown in A; *p < 0.05 calculated using t test. Based on van Rheenen et al. (2007), τPM is representative of the exchange kinetics of plasma membrane-bound cofilin; a higher τ represents slower translocation kinetics.
van Rheenen et al. (2007) revealed that the Per/Cyt fluorescence decrease can be fitted with a two phase exponential decay consisting of a fast and a slow component, each with a mean time constant τ. These authors also provided parallel FRET-based data, assigning the fast component to the translocation of the PM-bound cofilin, whereas the slow decay component is representative for F-actin–bound cofilin. Under the assumption that amplitude and τ of the slowly decaying actin pool [0.15 and 26 s, respectively; values from van Rheenen et al. (2007)] are valid for both WT- and mutant-expressing cells (given the WT behavior of mutant cofilin in vitro; Figure 1, D–F), the best values for τ for the fast PM component (τPM) in the decay fittings were derived. Figure 8B shows τPM is significantly larger for cells expressing D122K-cofilin than for those expressing WT (5.24 ± 0.31 and 3.3 ± 0.41 s, respectively). The WT τPM-value is in good agreement with that derived for WT–cofilin-expressing cells in (van Rheenen et al., 2007) (3.7 s). Collectively, the FLIP data strongly suggest that D122K cofilin translocates slower from the PM than WT-cofilin, which is indicative for a stronger binding to PM PI(4,5)P2.
DISCUSSION
PI(4,5)P2 binding negatively regulates actin binding by cofilin in vitro (Yonezawa et al., 1990; Van Troys et al., 2000). In several cellular systems, cofilin acts downstream of PLC (Matsui et al., 2001; Mouneimne et al., 2004, 2006; Song et al., 2006; Hosoda et al., 2007; Zhou et al., 2007) and of PI(4,5)P2 hydrolysis (van Rheenen et al., 2007). However, tools demonstrating that this is based on a direct cofilin-PI(4,5)P2 interaction were lacking. The D122K cofilin mutant allowed gaining insight into the role of this interaction in vivo. D122K cofilin displays significant gain in PI(4,5)P2 affinity compared with WT in vitro. D122K cofilin, however, resembles WT in other properties: the pH dependency of its PI(4,5)P2 binding in vitro and thus its putative pH sensor role, its level of phosphorylation and localization in cells, its similar effect on cellular PI(4,5)P2 hydrolysis, and its in vitro actin severing/depolymerization activity.
Recent structural data support several of these properties of D122K cofilin. The PI(4,5)P2 binding site, delineated in chicken cofilin (Gorbatyuk et al., 2006), shows that D122 is proximal to the lipid–protein interface and that plasticity in the docking site of the inositol group allows exploiting neighboring positive residues 123–125 (Gorbatyuk et al., 2006). Frantz et al. (2008) recently showed that charge switching of His133 modulates the PI(4,5)P2 headgroup position on cofilin. The positive residues 123–125 of cofilin are used as docking site only when His133 is neutral (i.e., at pH 7.5 or higher). When protonated (pH 6.0), His133 is modeled to dock the headgroups and also interact via its side chain with the terminal phosphates of PI(4,5)P2, thus strengthening the interaction. The D122K mutation introduces a positive charge proximal to the Lys123–125-PI(4,5)P2 docking site. Based on current structural models (Gorbatyuk et al., 2006, Frantz et al., 2008), this forms a putative structural explanation for the increased affinity of D122K cofilin for PI(4,5)P2 under conditions where this site is proposed to be used in docking (i.e., at pH 7.5). Notwithstanding, our data show that the positive effect of D122K on the cofilin–PI(4,5)P2 interaction is also present at pH 6.0 in vitro and thus seems additive to the positive effect sorted by His133 protonation. The structural basis of this requires further study.
The absence of an effect of the D122K mutation on actin interaction is supported by the structure of a complex between G-actin and the C-terminal cofilin-like domain of twinfilin (Paavilainen et al., 2008). In this complex, the residue corresponding to human cofilin D122 is in the long, kinked actin-binding α-helix of cofilin but not in the actin–cofilin binding interface. Moreover, yeast cofilins in which the Asp, corresponding to D122, is mutated to Ala or Lys, behave like WT in actin filament disassembly in vitro, also in the presence of the cofilin partner Aip1 (Clark et al., 2006; Clark and Amberg, 2007).
Collectively, this implies that differences between cells expressing either D122K or WT cofilin are thus attributable to a different PI(4,5)P2–cofilin interaction. Here, by using FLIP, we demonstrated that, in cells, the mutant is retained longer at the PM than WT, underscoring it also displays stronger PI(4,5)P2 binding in vivo.
Based on the above-mentioned information, a different balance in cofilin-PI(4,5)P2 complex formation is present in D122K cofilin-expressing cells in comparison with parental or WT-expressing cells. The PI(4,5)P2 hydrolysis rate itself is not affected (Figure 2E). Our data demonstrate that this imbalance in cofilin-PI(4,5)P2 regulation induces major differences in cell migration patterns and growth factor-induced membrane protrusion. This establishes a crucial contribution for the direct cofilin-PI(4,5)P2 interaction in the regulation of cellular steering and protrusion properties, as is outlined below.
In NIH3T3 fibroblasts as well as in MTLn3 with reduced endogenous cofilin levels, D122K cofilin results in a stronger directionality of migration coupled to a higher cell speed. Cofilin activity has previously been implicated in setting the direction of migration in MTLn3 (DesMarais et al., 2004; Ghosh et al., 2004; Mouneimne et al., 2006; Sidani et al., 2007) and other cell types (Hotulainen et al., 2005). Our data demonstrate that the regulation of cofilin activity by recruitment and binding to and releasing from PI(4,5)P2 crucially contributes to this steering process. First, D122K cofilin is unable to revert the highly directional migration of MTLn3 KD cells to the random migration displayed by parental or W-KD cells. Second, the higher directionality of D122K cofilin-overexpressing fibroblasts shows that the mutant can in part compete with endogenous cofilin via its advantageous PI(4,5)P2 binding.
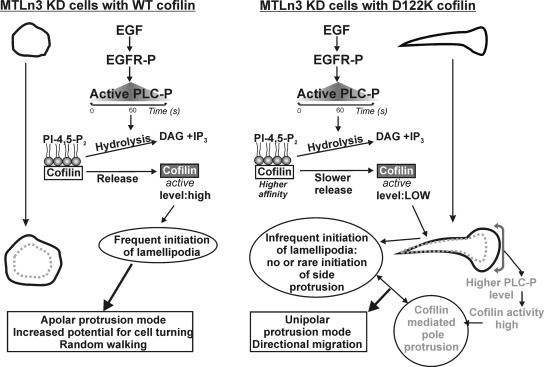
In MTLn3 cells, we provide evidence for a mechanism by which cofilin–PI(4,5)P2 interaction kinetics determines cell steering via an impact on lamellipodia initiation (Figure 9). It has been shown previously that cofilin KD MTLn3 cells are elongated and protrude in a uni- or bipolar manner (Sidani et al., 2007). This is shown to induce Arp2/3 complex enrichment in the front half of the cell and in the protruding pole. This actin nucleator is apparently unable to redistribute to other sites in absence of cofilin (Sidani et al., 2007). Cofilin activation at new sites near the membrane is proposed to generate new actin filament barbed ends that upon elongation provide sites where dendritic nucleation and actin filament extension can occur, thus leading to membrane protrusion (Ichetovkin et al., 2002; DesMarais et al., 2004; Mouneimne et al., 2004, 2006). Here, we show that D122K cofilin-expressing MTLn3 cells (M-KD) more closely resemble cofilin KD cells. Unlike what is observed upon WT cofilin rescue or in parental MTLn3, M-KD cells still show an elongated shape and do not display protrusive activity along their entire periphery. This suggests that the cofilin–PI(4,5)P2 interaction forms a crucial control point in the regulatory mechanism driving initiation of new lamellipodia in MTLn3 cells. For WT cofilin, this interaction will allow initiating protrusions in an apolar manner resulting in frequent cell turning and random migration behavior, which are needed for efficient chemotaxis in these metastatic tumor cells (Mouneimne et al., 2006; Sidani et al., 2007).
Figure 9.
PI(4,5)P2–cofilin interaction controls the mode of EGF-induced protrusion and migration in MTLn3 cells: model of underlying mechanisms in cofilin KD cells expressing either WT cofilin or D122K cofilin. The extent of release of active cofilin from PI(4,5)P2 within the timeframe of PLC activation and resulting PI(4,5)P2 hydrolysis is the key control point in lamellipodia initiation. Released cofilin is modeled to sever actin filaments in the periphery, an event followed by Arp2/3-mediated actin nucleation and lamellipodia extension. Uncompromised cofilin activation and lamellipodia initiation in WT cells results in protrusion along the entire periphery (apolar protrusion) and random walking. Due to its higher affinity for PI(4,5)P2, D122K is released slower resulting in insufficient release of cofilin and no new side protrusion formation. In the established lamellipodia in the pole, activated PLC is enriched (Chou et al., 2003; Mouneimne et al., 2006). This will locally result in stronger PI(4,5)P2 decrease, which we suggest compensates the slower D122K cofilin release and results in normal cofilin-mediated protrusion at the pole. In combination, this results in directional migration of cells expressing the mutant. EGFR-P, phosphorylated EGF receptor; PLC-P, phosphorylated (active) phospholipase C; DAG, diacylglycerol; and IP3, inositol-1,4,5-trisphosphate.
Despite these apparent similarities between M-KD cells and cofilin KD cells, our data also show that unstimulated M-KD cells do no longer display the enrichment of the Arp2/3 complex in the front protruding region that was reported for cofilin KD cells under similar conditions (Sidani et al., 2007) (Supplemental Figure S4). This indicates that whereas redistribution of the Arp2/3 is inhibited in the absence of cofilin (Sidani et al., 2007), the presence of D122K cofilin with normal severing activity still generates sufficient actin dynamics for allowing redistribution of the Arp2/3 complex within the entire cell. The absence of new lamellipodia and the retained elongated shape in M-KD cells, however, suggests this redistribution by itself is not sufficient for lamellipodia initiation along the entire periphery as observed in W-KD cells.
For D122K cofilin, the release from the PM in unstimulated cells is slower in comparison to WT (Figure 8). This is not based on slower rates of PI(4,5)P2 hydrolysis induced by the expression of D122K cofilin (Figure 2C) and thus a consequence of the higher affinity of this mutant for PI(4,5)P2 observed in vitro (Figure 1). We explain the inability of the D122K mutant to rescue defects in shape and migration of cofilin KD-cells as follows. Because protrusive activity is only restored locally in the pole and not all along the cell periphery in M-KD cells, the cofilin release event from PI(4,5)P2 is evidently a crucial local activation control switch downstream of PLC activation and subsequent PLC-induced PI(4,5)P2 hydrolysis (Figure 9). The increased affinity for PI(4,5)P2, D122K cofilin will, in M-KD cells, lead to a PM compartment filled with slower releasable cofilin molecules. PI(4,5)P2-hydrolysis will therefore release cofilin less efficiently, because the off-rate of D122K cofilin from PI(4,5)P2 is slower. On EGF stimulation, PLC is only activated during a limited time frame at the membrane, coincident with the time of cofilin activity and the first transient of actin polymerization (Mouneimne et al., 2004). PLC activation thus sets the timing and extent of cofilin release, thereby generating a critical threshold level of active cofilin at the membrane to initiate the events underlying subsequent productive membrane extension. We hypothesize that D122K cofilin expression prevents generation of new protrusions along the periphery because the threshold of local active cofilin is less frequently reached for this mutant within the limited time frame of PLC activity due to the slower release of D122K cofilin from PI(4,5)P2 (Figure 9). Together, this explains why D122K cofilin displays such a dramatic effect on cell morphology, lamellipodium initiation, and cell directionality (Figure 9).
Whereas new lamellipodial protrusion is strongly compromised in M-KD cells, protrusion at the pole is surprisingly increased. In this pole, an Arp2/3-containing lamellipodial actin network is present (Sidani et al., 2007) (Supplemental Figure S4A) in which cofilin is present throughout (Lai et al., 2008). The positive effect of D122K cofilin only in this established lamellipodium may result from its unaltered actin-binding and -remodeling activity. Activated PLC is however also shown to be enriched in existing lamellipodia in MTLn3 and other cells (Chou et al., 2003; Mouneimne et al., 2006). We hypothesize that this locally higher activation of PI(4,5)P2-hydrolyzing PLC [resulting in local lower PI(4,5)P2-levels] compensates the negative effects of the mutant (observed at other sites along the periphery) and results in sufficient mutant cofilin released in the pole to locally obtain a WT effect (Figure 9). Another possible and nonmutually exclusive mechanism underlying the locally higher release of D122K cofilin in the protruding pole may be the pH-dependent interaction with PI(4,5)P2. Proton exchange channels (such as Na+/H+ exchanger isoform 1 [NHE1]) are in several cell lines reported to be enriched in established lamellipodia, activated downstream of EGF stimulation, and important in directed motility (Denker and Barber, 2002). In line with the reported synergy between regulation by pH and by PI(4,5)P2 (Frantz et al., 2008) and assuming an enrichment of NHE1 in the poles of M-KD cells, a resulting local increase in pH might increase D122K cofilin release and result in sufficient mutant cofilin to locally obtain a WT effect.
PI(4,5)P2-regulation of cofilin forms part of the different cofilin regulatory mechanisms that will act in conjunction to result in tightly controlled, temporal and spatial cell effects such as polarization and directional movement. A role for cofilin phospho-regulation in directional migration of MTLn3 cells has already previously been demonstrated; the constitutively active and unphosphorylatable S3A cofilin has indeed been shown to inhibit directional sensing (Mouneimne et al., 2006). However, because our data suggest that D122K cofilin is similarly phosphorylated as WT cofilin (Figure 3), the defects of M-KD cells clearly establish a role for the direct cofilin–PI(4,5)P2 interaction in lamellipodial initiation and subsequent cell steering (Figure 9).
In conclusion, by unbalancing the cofilin–PI(4,5)P2 interaction in cells, we establish that this direct interaction determines local and temporal activation of cofilin downstream of EGF-induced PLC activation and is thus an important type of regulation, next to cofilin de/phosphorylation and pH regulation. Release of a crucial level of cofilin from PI(4,5)P2 locally triggers initiation of a new lamellipodium and empowers an inherently apolar and randomly migrating cell to change direction. Our data suggest that affecting the release of cofilin from PI(4,5)P2 in a negative manner forces directional migration in inherently apolar cells (MTLn3) and reinforces it in inherently polarized cells (NIH3T3). Future studies will need to show how this PI(4,5)P2-dependent spatial and temporal regulation of cofilin activity determines cell properties in other cell contexts.
Supplementary Material
ACKNOWLEDGMENTS
We thank V. Jonckheere for technical assistance and the Analytical Imaging Facility of the Gruss Lipper Biophotonics Center and the Hubrecht Imaging Center for imaging support. S. L. was a recipient of a fellowship of the Institute for Promotion of Scientific-Technological Research in Industry. C. D. is a senior research associate at the Fonds National de la Recherche Scientifique (Brussels, Belgium). This research was supported by Geconcerteerde Onderzoeksactie grant 12051401 (to C. A. and J. V.) and Fonds voor Wetenschappelijk Onderzoek G.0157.05 and Bijzonder Onderzoeksfonds 01J04806 grants (to M.V.T.) J.v.R. was supported by the Cell Migration Consortium grant U54GM064346 and National Institutes of Health grant GM-38511 (to J. C.).
Abbreviations used:
- ADF
actin depolymerizing factor
- EGF
epidermal growth factor
- FLIP
fluorescence loss in photobleaching
- FRET
fluorescence resonance energy transfer
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- KD
knockdown
- MDO
maximum distance from origin
- M-KD
MTLn3 cells with cofilin knockdown and expressing D122K cofilin
- PI(4,5)P2
phosphatidylinositol-4,5-bisphosphate
- PLC
phospholipase C
- PM
plasma membrane
- W-KD
MTLn3 cells with cofilin knockdown expressing wild-type cofilin
- WT
wild type.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0121) on September 9, 2009.
REFERENCES
- Andrianantoandro E., Pollard T. D. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Bamburg J. R. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Bamburg J. R., Bray D. Distribution and cellular localization of actin depolymerizing factor. J. Cell Biol. 1987;105:2817–2825. doi: 10.1083/jcb.105.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. W., Painter W. B., Chen H., Minamide L. S., Abe H., Bamburg J. R. Intracellular pH modulation of ADF/cofilin proteins. Cell Motil. Cytoskeleton. 2000;47:319–336. doi: 10.1002/1097-0169(200012)47:4<319::AID-CM6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Carlier M. F., Laurent V., Santolini J., Melki R., Didry D., Xia G. X., Hong Y., Chua N. H., Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. Y., Bailly M., Zebda N., Segall J. E., Condeelis J. S. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 2000;148:531–542. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Burke N. A., Iwabu A., Watkins S. C., Wells A. Directional motility induced by epidermal growth factor requires Cdc42. Exp. Cell Res. 2003;287:47–56. doi: 10.1016/s0014-4827(03)00119-8. [DOI] [PubMed] [Google Scholar]
- Clark M. G., Amberg D. C. Biochemical and genetic analyses provide insight into the structural and mechanistic properties of actin filament disassembly by the Aip1p cofilin complex in Saccharomyces cerevisiae. Genetics. 2007;176:1527–1539. doi: 10.1534/genetics.107.072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. G., Teply J., Haarer B. K., Viggiano S. C., Sept D., Amberg D. C. A genetic dissection of Aip1p's interactions leads to a model for Aip1p-cofilin cooperative activities. Mol. Biol. Cell. 2006;17:1971–1984. doi: 10.1091/mbc.E05-10-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. A., Blum J. D., Williams R. C., Jr, Pollard T. D. Purification and characterization of actophorin, a new 15,000-dalton actin-binding protein from Acanthamoeba castellanii. J. Biol. Chem. 1986;261:477–485. [PubMed] [Google Scholar]
- Dawe H. R., Minamide L. S., Bamburg J. R., Cramer L. P. ADF/cofilin controls cell polarity during fibroblast migration. Curr. Biol. 2003;13:252–257. doi: 10.1016/s0960-9822(03)00040-x. [DOI] [PubMed] [Google Scholar]
- Debeir O., Van Ham P., Kiss R., Decaestecker C. Tracking of migrating cells under phase-contrast video microscopy with combined mean-shift processes. IEEE Trans. Med. Imaging. 2005;24:697–711. doi: 10.1109/TMI.2005.846851. [DOI] [PubMed] [Google Scholar]
- Debeir O., Adanja I., Kiss R., Decaestecker C. Models of cancer cell migration and cellular imaging and analysis. In: Ampe C., Lambrechts A., editors. The Motile Actin System in Health and Disease. Kerala, India: Transworld Research Signpost; 2008. pp. 123–156. [Google Scholar]
- Denker S. P., Barber D. L. Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J. Cell Biol. 2002;159:1087–1096. doi: 10.1083/jcb.200208050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais V., Ghosh M., Eddy R., Condeelis J. Cofilin takes the lead. J. Cell Sci. 2005;118:19–26. doi: 10.1242/jcs.01631. [DOI] [PubMed] [Google Scholar]
- DesMarais V., Macaluso F., Condeelis J., Bailly M. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J. Cell Sci. 2004;117:3499–3510. doi: 10.1242/jcs.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy R. J., Pierini L. M., Matsumura F., Maxfield F. R. Ca2+-dependent myosin II activation is required for uropod retraction during neutrophil migration. J. Cell Sci. 2000;113:1287–1298. doi: 10.1242/jcs.113.7.1287. [DOI] [PubMed] [Google Scholar]
- Frantz C., Barreiro G., Dominguez L., Chen X., Eddy R., Condeelis J., Kelly M. J., Jacobson M. P., Barber D. L. Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J. Cell Biol. 2008;183:865–879. doi: 10.1083/jcb.200804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D. S., Condeelis J. S. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk V. Y., Nosworthy N. J., Robson S. A., Bains N. P., Maciejewski M. W., Dos Remedios C. G., King G. F. Mapping the phosphoinositide-binding site on chick cofilin explains how PIP2 regulates the cofilin-actin interaction. Mol. Cell. 2006;24:511–522. doi: 10.1016/j.molcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Hilpela P., Vartiainen M. K., Lappalainen P. Regulation of the actin cytoskeleton by PI(4,5)P2 and PI(3,4,5)P3. Curr. Top. Microbiol. Immunol. 2004;282:117–163. doi: 10.1007/978-3-642-18805-3_5. [DOI] [PubMed] [Google Scholar]
- Hosoda A., Sato N., Nagaoka R., Abe H., Obinata T. Activity of cofilin can be regulated by a mechanism other than phosphorylation/dephosphorylation in muscle cells in culture. J. Muscle Res. Cell Motil. 2007;28:183–194. doi: 10.1007/s10974-007-9117-6. [DOI] [PubMed] [Google Scholar]
- Hotulainen P., Paunola E., Vartiainen M. K., Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. Y., DerMardirossian C., Bokoch G. M. Cofilin phosphatases and regulation of actin dynamics. Curr. Opin. Cell Biol. 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Ichetovkin I., Grant W., Condeelis J. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 2002;12:79–84. doi: 10.1016/s0960-9822(01)00629-7. [DOI] [PubMed] [Google Scholar]
- Koffer A., Edgar A. J., Bamburg J. R. Identification of two species of actin depolymerizing factor in cultures of BHK cells. J. Muscle Res. Cell Motil. 1988;9:320–328. doi: 10.1007/BF01773875. [DOI] [PubMed] [Google Scholar]
- Kolsch V., Charest P. G., Firtel R. A. The regulation of cell motility and chemotaxis by phospholipid signaling. J. Cell Sci. 2008;121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F. P., Szczodrak M., Block J., Faix J., Breitsprecher D., Mannherz H. G., Stradal T. E., Dunn G. A., Small J. V., Rottner K. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 2008;27:982–992. doi: 10.1038/emboj.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts A., Verschelde J. L., Jonckheere V., Goethals M., Vandekerckhove J., Ampe C. The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J. 1997;16:484–494. doi: 10.1093/emboj/16.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K., Schill N. J., Wagoner M. P., Sun Y., Anderson R. A. Movin' on up: the role of PtdIns(4,5)P(2) in cell migration. Trends Cell Biol. 2006;16:276–284. doi: 10.1016/j.tcb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Maloney M. T., Kinley A. W., Pak C. W., Bamburg J. R. ADF/Cofilin, Actin Dynamics, and Disease. In: dos Remedios C. G., Chhabra D., editors. Actin-Binding Proteins and Disease. Vol. 8. New York: Springer; 2008. pp. 83–187. [Google Scholar]
- Matsui S., Adachi R., Kusui K., Yamaguchi T., Kasahara T., Hayakawa T., Suzuki K. U73122 inhibits the dephosphorylation and translocation of cofilin in activated macrophage-like U937 cells. Cell. Signal. 2001;13:17–22. doi: 10.1016/s0898-6568(00)00124-8. [DOI] [PubMed] [Google Scholar]
- McLaughlin S., Wang J., Gambhir A., Murray D. PIP(2) and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- Mouneimne G., DesMarais V., Sidani M., Scemes E., Wang W., Song X., Eddy R., Condeelis J. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 2006;16:2193–2205. doi: 10.1016/j.cub.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Mouneimne G., Soon L., DesMarais V., Sidani M., Song X., Yip S. C., Ghosh M., Eddy R., Backer J. M., Condeelis J. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 2004;166:697–708. doi: 10.1083/jcb.200405156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggli V. Regulation of protein activities by phosphoinositide phosphates. Annu. Rev. Cell Dev. Biol. 2005;21:57–79. doi: 10.1146/annurev.cellbio.21.021704.102317. [DOI] [PubMed] [Google Scholar]
- Ojala P. J., Paavilainen V., Lappalainen P. Identification of yeast cofilin residues specific for actin monomer and PIP2 binding. Biochemistry. 2001;40:15562–15569. doi: 10.1021/bi0117697. [DOI] [PubMed] [Google Scholar]
- Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int. Rev. Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- Paavilainen V. O., Oksanen E., Goldman A., Lappalainen P. Structure of the actin-depolymerizing factor homology domain in complex with actin. J. Cell Biol. 2008;182:51–59. doi: 10.1083/jcb.200803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. W., Olson M. F. LIM kinases: function, regulation and association with human disease. J. Mol. Med. 2007;85:555–568. doi: 10.1007/s00109-007-0165-6. [DOI] [PubMed] [Google Scholar]
- Sidani M., et al. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J. Cell Biol. 2007;179:777–791. doi: 10.1083/jcb.200707009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Chen X., Yamaguchi H., Mouneimne G., Condeelis J. S., Eddy R. J. Initiation of cofilin activity in response to EGF is uncoupled from cofilin phosphorylation and dephosphorylation in carcinoma cells. J. Cell Sci. 2006;119:2871–2881. doi: 10.1242/jcs.03017. [DOI] [PubMed] [Google Scholar]
- Stauffer T. P., Ahn S., Meyer T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- van der Wal J., Habets R., Varnai P., Balla T., Jalink K. Monitoring agonist-induced phospholipase C activation in live cells by fluorescence resonance energy transfer. J. Biol. Chem. 2001;276:15337–15344. doi: 10.1074/jbc.M007194200. [DOI] [PubMed] [Google Scholar]
- van Rheenen J., Achame E. M., Janssen H., Calafat J., Jalink K. PIP2 signaling in lipid domains: a critical re-evaluation. EMBO J. 2005;24:1664–1673. doi: 10.1038/sj.emboj.7600655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheenen J., Song X., van Roosmalen W., Cammer M., Chen X., Desmarais V., Yip S. C., Backer J. M., Eddy R. J., Condeelis J. S. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 2007;179:1247–1259. doi: 10.1083/jcb.200706206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheenen J., Condeelis J., Glogauer M. A common cofilin activity cycle in invasive tumor cells and inflammatory cells. J. Cell Sci. 2009;122:305–311. doi: 10.1242/jcs.031146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys M., Dewitte D., Verschelde J. L., Goethals M., Vandekerckhove J., Ampe C. The competitive interaction of actin and PIP2 with actophorin is based on overlapping target sites: design of a gain-of-function mutant. Biochemistry. 2000;39:12181–12189. doi: 10.1021/bi000816c. [DOI] [PubMed] [Google Scholar]
- Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 2008;87:649–667. doi: 10.1016/j.ejcb.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Varnai P., Balla T. Live cell imaging of phosphoinositides with expressed inositide binding protein domains. Methods. 2008;46:167–176. doi: 10.1016/j.ymeth.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Goswami S., Lapidus K., Wells A. L., Wyckoff J. B., Sahai E., Singer R. H., Segall J. E., Condeelis J. S. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- Wang W., Mouneimne G., Sidani M., Wyckoff J., Chen X., Makris A., Goswami S., Bresnick A. R., Condeelis J. S. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell Biol. 2006;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wyckoff J. B., Goswami S., Wang Y., Sidani M., Segall J. E., Condeelis J. S. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 2007;67:3505–3511. doi: 10.1158/0008-5472.CAN-06-3714. [DOI] [PubMed] [Google Scholar]
- Yan B., Yap C. T., Wang S., Lee C. K., Koh S., Omar M. F., Salto-Tellez M., Kumarasinghe M. P. Cofilin immunolabelling correlates with depth of invasion in gastrointestinal endocrine cell tumors. Acta Histochem. 2008 doi: 10.1016/j.acthis.2008.07.007. (in press) (doi: 10.1016/j.acthis.2008.07.007) [DOI] [PubMed] [Google Scholar]
- Yeoh S., Pope B., Mannherz H. G., Weeds A. Determining the differences in actin binding by human ADF and cofilin. J. Mol. Biol. 2002;315:911–925. doi: 10.1006/jmbi.2001.5280. [DOI] [PubMed] [Google Scholar]
- Yoder A., et al. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell. 2008;134:782–792. doi: 10.1016/j.cell.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa N., Nishida E., Iida K., Yahara I., Sakai H. Inhibition of the interactions of cofilin, destrin, and deoxyribonuclease I with actin by phosphoinositides. J. Biol. Chem. 1990;265:8382–8386. [PubMed] [Google Scholar]
- Yonezawa N., Nishida E., Koyasu S., Maekawa S., Ohta Y., Yahara I., Sakai H. Distribution among tissues and intracellular localization of cofilin, a 21 kDa actin-binding protein. Cell Struct. Funct. 1987;12:443–452. doi: 10.1247/csf.12.443. [DOI] [PubMed] [Google Scholar]
- Zhou L., Martinez S. J., Haber M., Jones E. V., Bouvier D., Doucet G., Corera A. T., Fon E. A., Zisch A. H., Murai K. K. EphA4 signaling regulates phospholipase Cgamma1 activation, cofilin membrane association, and dendritic spine morphology. J. Neurosci. 2007;27:5127–5138. doi: 10.1523/JNEUROSCI.1170-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Meerschaert K., Reekmans G., Leenaerts I., Small J. V., Vandekerckhove J., David G., Gettemans J. PIP(2)-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol. Cell. 2002;9:1215–1225. doi: 10.1016/s1097-2765(02)00549-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.