Abstract
We have previously identified a novel mitochondrial ubiquitin ligase, MITOL, which is localized in the mitochondrial outer membrane and is involved in the control of mitochondrial dynamics. In this study, we examined whether MITOL eliminates misfolded proteins localized to mitochondria. Mutant superoxide dismutase1 (mSOD1), one of misfolded proteins, has been shown to localize in mitochondria and induce mitochondrial dysfunction, possibly involving in the onset and progression of amyotrophic lateral sclerosis. We found that in the mitochondria, MITOL interacted with and ubiquitinated mSOD1 but not wild-type SOD1. In vitro ubiquitination assay revealed that MITOL directly ubiquitinates mSOD1. Cycloheximide-chase assay in the Neuro2a cells indicated that MITOL overexpression promoted mSOD1 degradation and suppressed both the mitochondrial accumulation of mSOD1 and mSOD1-induced reactive oxygen species (ROS) generation. Conversely, the overexpression of MITOL CS mutant and MITOL knockdown by specific siRNAs resulted in increased accumulation of mSOD1 in mitochondria, which enhanced mSOD1-induced ROS generation and cell death. Thus, our findings indicate that MITOL plays a protective role against mitochondrial dysfunction caused by the mitochondrial accumulation of mSOD1 via the ubiquitin–proteasome pathway.
INTRODUCTION
We have previously identified mitochondrial ubiquitin ligase MITOL (also known as MARCH-V) specifically localized in the mitochondria (Yonashiro et al., 2006). MITOL is a ubiquitin ligase characterized by its four transmembrane domains within the mitochondrial outer membrane, and it ubiqitinates mitochondrial fission proteins hFis1 and Drp1. Subsequently, it was found that not only the fission proteins but also the fusion proteins interact with MITOL, suggesting that MITOL regulates both mitochondrial fission and fusion (Nakamura et al., 2006; Karbowski et al., 2007). Thus, MITOL plays a critical role in regulating mitochondrial dynamics via the control of the balance between mitochondrial fission and fusion proteins. However, an essential role of MITOL in the mitochondria has not been adequately elucidated.
Intracellular quality control is a mechanism by which aberrant proteins are immediately degraded in order to protect cells from the direct toxicity of misfolded proteins, as well as from various stresses caused by the accumulation and aggregation of misfolded proteins. Endoplasmic reticulum-associated degradation (ERAD) is the most common quality control mechanism via which misfolded proteins and incompletely assembled proteins are transported from the endoplasmic reticulum (ER) to cytosol, followed by their polyubiquitination, and subsequent proteasomal degradation (Tsai et al., 2002; Meusser et al., 2005). In addition to ERAD system, recent studies have shown that there is a quality control mechanism inside the mitochondria; a mitochondrial AAA protease selectively degrades the misfolded and damaged proteins (Tatsuta and Langer, 2008). However, the AAA protease is unable to recognize the outer membrane proteins because the catalytic domain of the AAA protease is exposed to the inner membrane space or matrix. Thus, a quality control mechanism for the mitochondrial outer membrane is not thoroughly explored. Interestingly, Saccharomyces cerevisiae DOA10 (known as MARCH-VI or TEB4 in humans; Kreft et al., 2006) belonging to the MARCH family, similar to MITOL, is thought to be one of the major mediators for ubiquitinating ERAD substrates. This led us to hypothesize that MITOL is involved in the mitochondrial quality control of outer membrane proteins.
Amyotrophic lateral sclerosis (ALS) is a serious neurodegenerative disease for which therapy has not been established. Muscular atrophy, a major pathological feature of ALS, is caused by upper and lower motor neuron damage. Recently, mutant superoxide dismutase 1 (mSOD1) has been demonstrated as one of the gene products responsible for the occurrence of familial ALS (Rosen et al., 1993). The misfolded mSOD1 is accumulated in the neuronal cells of human ALS patients and ALS mice model expressing mSOD1, and it causes various cellular toxic effects such as aggregation of mSOD1, inhibition of protein degradation, axonal transport disturbance, and reduced reactive oxygen species (ROS) scavenging activity (Bendotti and Carri, 2004; Barber et al., 2006; Boillee et al., 2006). It has been believed that a malfunction in the ERAD quality control system is a main cause for neuronal death observed in ALS because ERAD-related ubiquitin ligase such as C-terminus of Hsp70 (heat-shock protein 70)-interacting protein (CHIP) attenuates the toxicity of mSOD1 by ubiquitinating it and because then it promotes its proteasomal degradation (Niwa et al., 2002; Miyazaki et al., 2004; Urushitani et al., 2004). On the other hand, a recent significant study suggested that an accumulation of mSOD1 in mitochondria is the true cause for neuronal cell death and onset of ALS (Dupuis et al., 2004; Manfredi and Xu, 2005; Lin and Beal, 2006). At present, a quality control system for this mSOD1 accumulation in mitochondria has not been reported. Therefore, mSOD1 is a good model for investigating the mechanism of mitochondrial quality control regulated by MITOL.
In this study, we report a possible role of MITOL in mitochondrial quality control. Our results indicate that MITOL attenuates the cytotoxicity of mSOD1 by selectively ubiqutinating mitochondrial mSOD1 and promoting its degradation. Taking this finding into consideration, we discuss whether it is possible to treat ALS via the activation of MITOL.
MATERIALS AND METHODS
Plasmid Constructions
SOD1 cDNA with C-terminal FLAG and myc epitope tag were obtained by RT-PCR using total RNA of HeLa cells and subcloned into pCMV5 and pcDNA3.1 myc-His vector, respectively. MULAN cDNA was obtained by RT-PCR using total RNA of HEK 293 cells and sucloned into pcDNA3.1 V5-His vector. Point mutations of SOD1 G93A, G85R, and SCAD W153R were generated by the site-directed mutagenesis kit (Stratagene, La Jolla, CA). To generate mitochondria targeting construct, a mitochondria targeting signal derived from pDs-Red-Mito vector (Clontech, Palo Alto, CA) was fused to C-terminus in SOD1 G93A. To generate ER-localized SOD1 G93A, an ER signal (KDEL) was fused to C-terminus in SOD1G93A-FLAG. To construct mouse MITOL short hairpin (shRNA) vector, two complementary oligonucleotides (sense added BamHI liker, 5′- gatcGGTGCAGAGGATCTACTAAGTCGAAACTTAGTAGATCCTCTGCACCtttttttttttt-3′; antisense added HindIII linker, 5′-agctaaaaaaGGTGCAGAGGATCTACTAAGTCGAAACTTAGTAGATCCTCTGCACC-3′) were designed and inserted into pSilencer 3.1-H1 neo vector according to the manufacturer's instructions (Ambion, Austin, TX). Human MITOL wild type (WT) and CS (CS mutant; C65S, C68S) were used as resistant vectors for mouse MITOL shRNA. Constructions of MITOL WT and CS mutant expression vectors were described previously (Yonashiro et al., 2006).
Cell Culture, Transfection, and Viability Assay
COS-7, HeLa and Neuro2a cells were cultured in DMEM supplemented with 10% fetal bovine serum. Transfection of vectors in COS-7 and Neuro2a cells were performed using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. To generate stable expression of shRNA, Neuro2a cells were transfected with shMITOL or shGFP vector and selected with 400 μg/ml G418. To induce neuronal differentiation of Neuro2a cells, cells were incubated in DMEM supplemented with 1% fetal bovine serum and 20 μM retinoic acid. For siRNA assay, sense and antisense oligonucleotides corresponding to the following target sequence were purchased from QIAGEN (Chatsworth, CA): 5′-TCTTGGGTGGAATTGCGTT-3′ and 5′-GTCCAGTGGTTTACGTCTT-3′. QIAGEN′s thoroughly tested and validated AllStars Negative Control siRNA was used as a negative control. ATP production and MTT assays were performed using CellTiter-Glo Luminescent Cell Viability Assay kit (Promega, Madison, WI) and Cell Counting Kit-8 (Dojindo Molecular Technologies, Gaithersburg, MD), respectively.
Subcellular Fractionation
Isolation of mitochondria was performed using a mitochondrial fractionation kit (Active Motif, Carlsbad, CA). Mitochondrial fractions were lysed in 0.5% Triton lysis buffer or RIPA buffer (1% Triton X-100, 0.1% SDS, 0.5% deoxycholate, 50 mM Tris, pH 7.4, 150 mM NaCl, 10 mM EDTA, 1 mM PMSF, and 10 μg/ml aprotinin) followed by SDS-PAGE or immunoprecipitations.
Immunofluorescence Microscopy
Transfected cells were fixed with 4% parafolmaldehyde in PBS(−) for 15 min at room temperature, then washed twice with 0.2% Tween20 in PBS(−), permeabilized with 0.2% Triton X-100 in PBS(−) for 10 min, washed four times with PBS(−), and blocked with 3% bovine serum albumin in PBS(−). For double staining, the cells were incubated with appropriate primary antibody for 1 h at room temperature, washed three times with 0.5% Triton X-100 in PBS(−) and then with the appropriate secondary antibody for 30 min. Mitochondria were visualized by cotransfection with pDsRed-Mito vector (Clontech). The samples were analyzed using Olympus FV1000D confocal microscope (Melville, NY). Scatter plot analysis was performed using ImageJ software (http://rsb.info.nih.gov/ij/).
Immunoprecipitation and Immunoblotting
Preparation of cell lysates, immunoprecipitation, and immunoblotting were performed as described previously (Yonashiro et al., 2006). To investigate the ubiquitination of proteins, cells were solubilized in RIPA buffer to dissociate protein complexes. Immunoprecipitates and total cell lysates were separated by SDS-PAGE and transferred to the PVDF membrane (Millipore, Bedford, MA). The blots were probed with the indicated antibody, and protein bands on the blot were visualized by the chemiluminescence reagent (Millipore).
In Vitro Ubiquitination Assay
Immunoprecipitated MITOL-HA and mSOD1-FLAG were prepared from lysates of COS-7 cells transfected with MITOL-HA or mSOD1-FLAG, individually. Immunoprecipitates were incubated with the reaction buffer containing 50 mM Tris, pH 7.4, 2 mM MgCl2, 4 mM ATP, 100 ng of E1 (Biomol, Plymouth Meeting, PA), 400 ng of E2s (Biomol), and 2 μg of His-Ub (Biomol) for 2 h at 30°C and then terminated with 3× SDS sample buffer.
Measurement of ROS by Flow Cytometry
Neuro2a cells were cotransfected with mSOD1, siRNA (scramble or MITOL specific siRNA) and pDsRed-Mito. After 24 h, cells were incubated in differentiation medium for 24 h. Cells were treated with 10 μM ROS indicator CM-H2DCFDA (Invitrogen) for 30 min at 37°C, and cells were washed with PBS(−). After gating on DsRed-Mito–positive cells, the fluorescence of H2DCFDA was analyzed by flow cytometer (Becton Dickinson, San Jose, CA).
Antibodies
Anti-MITOL rabbit polyclonal antibody was described previously (Yonashiro et al., 2006). Anti-FLAG (M2) and anti-α-tubulin were from Sigma (St. Louis, MO). Anti-c-myc antibody was from Roche (Indianapolis, IN). Anti-HA antibody was from BabCO (Richmond, CA). Anti-V5 antibody was from MBL (Nagoya, Japan). Anti-Tim23 antibody was purchased from BD Biosciences. Anti-Ubiquitin antibody (P4D1) was from Santa Cruz Biotechnology (Santa Cruz, CA).
RESULTS
MITOL Interacts with and Ubiquitinates mSOD1
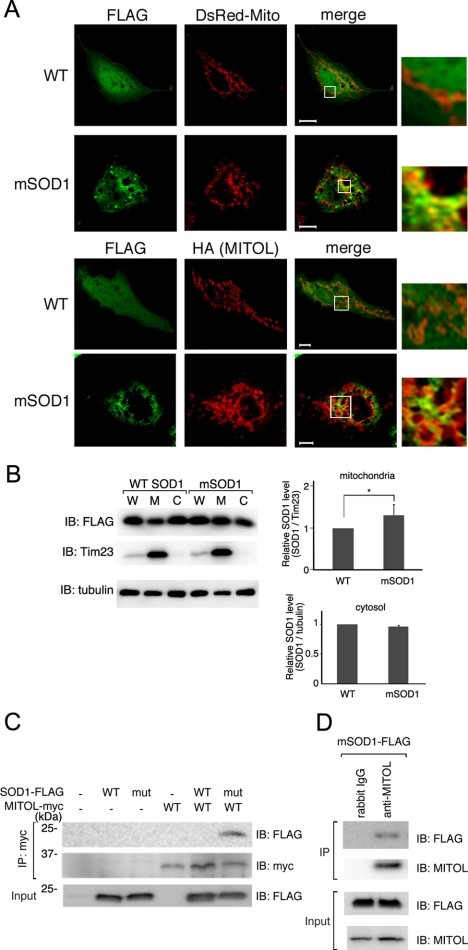
Previous studies have shown that mutant SOD1 has a greater tendency to accumulate in mitochondria than WT SOD1, which is closely associated with the onset of ALS. Therefore, we first examined whether mutant SOD1 G93A (mSOD1) accumulated in the mitochondria of our cell culture model. As shown in Figure 1A, immunofluorescence microscopy analysis indicated that mSOD1 aggregated and partially colocalized with mitochondria markers, pDsRed-Mito and MITOL. In contrast, WT SOD1 was diffusely distributed in the cytoplasm. Magnified images showed a colocalization of mSOD1 with pDsRed-Mito and MITOL. Statistical analysis indicated a more extensive mitochondrial localization of mSOD1 than that of WT SOD1 (Supplementary Figure S1A). To further confirm the mitochondrial localization of mSOD1 subcellular fractionation assay was performed. As shown in Figure 1B, the amount of mSOD1 localized in the mitochondria was greater than that of WT SOD1, whereas no significant difference was observed between the amounts of mSOD1 and WT SOD1 in the cytosolic fraction. We could not detect an increase in mSOD1 compared with WT SOD1 in total lysates in spite of accumulation mSOD1 in mitochondria. This may be due to the reflection of almost same level of SOD1 in cytosolic fraction containing a large part of proteins in total lysates. It is difficult to distinguish the distribution pattern clearly between WT SOD1 and mSOD1 in cultured cells, because mSOD1 gradually aggregates and accumulates in mitochondria in vivo. However, our results support previous observation that mSOD1 has a tendency to accumulate in mitochondria.
Figure 1.
MITOL interacts with mSOD1 but not WT SOD1. (A) Partial colocalization of mSOD1 (G93A) with mitochondria. COS-7 cells coexpressing WT SOD1-FLAG or mSOD1-FLAG with a mitochondrial marker, pDsRed-Mito (top) or MITOL-HA (bottom), were immunostained with indicated antibodies. Right panels, insets show a magnification of the merge. Bars, 10 μm. (B) Accumulation of mSOD1 in mitochondria. Whole (W), cytosolic (C), and mitochondrial (M) fractions isolated from COS-7 cells expressing WT SOD1-FLAG or mSOD1-FLAG were immunoblotted with anti-FLAG, anti-tubulin, or anti-Tim23 mitochondrial marker antibodies. The relative protein levels of SOD1 in mitochondria and cytosolic fractions were quantified by densitometry (right panels). Errors bar, SD (n = 3). * p < 0.05; Student's t test. (C) Association of MITOL with mSOD1. Mitochondrial fractions isolated from COS-7 cells transfected with indicated vectors were immunoprecipitated with anti-myc antibody, and immunoprecipitates were immunoblotted with anti-FLAG or anti-myc antibody. Mitochondrial fractions (Input) were immunoblotted with anti-FLAG antibody to confirm the expression of SOD1-FLAG. (D) Association of endogenous MITOL with mSOD1. Neuro2a (1 × 109) cells transfected with mSOD1-FLAG were treated with MG132 for 3 h. Mitochondrial fractions isolated from these cells were immunoprecipitated with control IgG or anti-MITOL antibody. Immunoprecipitates and mitochondrial fractions (Input) were immunoblotted with indicated antibodies.
Because MITOL is a mitochondrial ubiquitin ligase, we hypothesized that MITOL is involved in mitochondrial quality control and that mSOD1 may be a suitable substrate for MITOL. To test this possibility, we examined whether MITOL interacts with mSOD1. Immunoprecipitation assay revealed that MITOL specifically interacted with mSOD1 but not WT SOD1 (Figure 1C). We also confirmed the interaction of endogenous MITOL with mSOD1 (Figure 1D).
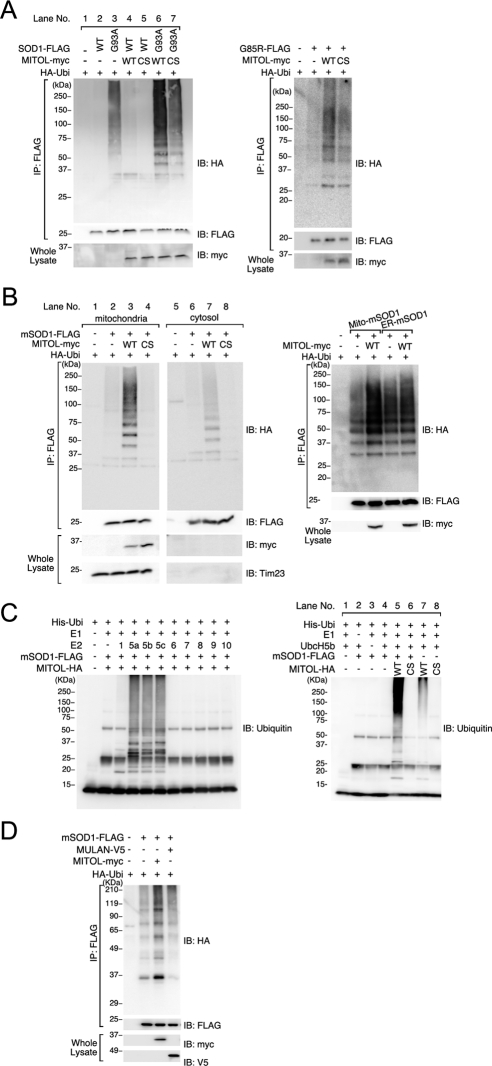
Specific interaction between MITOL and mSOD1 prompted us to examine whether MITOL ubiquitinates mSOD1. As shown in Figure 2A, WT SOD1 was not ubiquitinated in the presence or absence of MITOL coexpression (lanes 2 and 4). In contrast, mSOD1 was ubiquitinated without MITOL coexpression indicating that mSOD1 was ubiquitinated by endogenous ubiquitin ligases (lane 3). Furthermore, MITOL coexpression enhanced the ubiquitination of mSOD1 (lane 6), suggesting that MITOL ubiquitinated mSOD1. Similar to this result, MITOL CS mutant (C65S, C68S), which lacks ubiquitin ligase activity, could not enhance the ubiquitination of mSOD1. Because the MITOL CS mutant functions in a dominant-negative manner, the ubiquitination of mSOD1 coexpressed with the MITOL CS mutant (lane 7) may be catalyzed by other ubiquitin ligase(s) except endogenous MITOL. In addition to G93A, another mSOD1 (G85R) was ubiquitinated by MITOL but not by the MITOL CS mutant (Figure 2A, right). To further investigate the MITOL- dependent ubiquitination of mSOD1, a ubiquitination assay was performed after separating mitochondrial and cytosolic fractions. As shown in the left panel of Figure 2B, a higher ubiquitination of mSOD1 was observed in the mitochondrial fraction than the cytosolic fraction (Figure 2B, lanes 3 and 7). To confirm this result, we generated SOD1 mutants that are specifically directed toward either mitochondria or ER and determined the effect of MITOL on the ubiquitination of mSOD1. As shown in the right panel of Figure 2B, MITOL efficiently ubiquitinated mitochondria-targeted mSOD1 but not ER-targeted mSOD1. Consistently, MITOL promoted the degradation of mitochondria-targeted mSOD1 but not ER-targeted mSOD1 (Supplementary Figure S2A). In addition, the MITOL-dependent ubiquitination of mitochondria-targeted mSOD1 was more enhanced than that of nontagged mSOD1 (Supplementary Figure S2B). These results indicated that MITOL ubiquitinates mSOD1 but not WT SOD1, and this ubiquitination mainly occurs in the mitochondria.
Figure 2.
MITOL ubiquitinates mSOD1 in mitochondria. (A) Ubiquitinations of G93A and G85R by MITOL WT but not MITOL CS mutant. Lysates of COS-7 (left) or Neuro2a cells (right) transfected with indicated vectors were immunoprecipitated with anti-FLAG antibody, and immunoprecipitates were immunoblotted with anti-HA or anti-FLAG antibody. Whole lysates were immunoblotted with anti-myc antibody. (B) Ubiquitination of mSOD1 by MITOL mainly occurs in mitochondria. Cytosolic and mitochondrial fractions were isolated from COS-7 cells transfected with indicated vectors, each fraction was immunoprecipitated with anti-FLAG antibody, and immunoprecipitates were immunoblotted with anti-HA or anti-FLAG antibody. Mitochondrial fractions and cytosolic fractions were immunoblotted with anti-myc or anti-Tim23 antibody (left). Neuro2a cells were transfected with mitochondria targeted-mSOD1 or ER-targeted mSOD1 with MITOL WT or control vector. Lysates were immunoprecipitated with anti-FLAG antibody, and immunoprecipitates were immunoblotted with anti-HA or anti-FLAG antibody (right). (C) MITOL directly ubiquitinates mSOD1 in vitro. In vitro ubiquitination assay was performed as described in Materials and Methods. Immunopurified MITOL-HA and mSOD1-FLAG were prepared from COS-7 cells transfected with MITOL- HA and mSOD1-FLAG, independently. Identification of E2s for MITOL-dependent ubiquitination of mSOD1 (left). MITOL WT-dependent ubiquitination of mSOD1 (right). (D) MULAN fails to ubiquitinate mSOD1. Mitochondrial fractions of Neuro2a cells transfected with indicated vectors were immunoprecipitated with anti-FLAG antibody, and immunoprecipitates were immunoblotted with anti-HA antibody. Inputs were immunoblotted with anti-myc or anti-V5 antibody.
To examine whether MITOL directly ubiquitinates mSOD1, we performed in vitro ubiquitination assay. For this purpose, we first determined which ubiquitin-conjugating enzymes (E2s) are required for MITOL-dependent ubiquitination. As shown in the left panel of Figure 2C, we found that the enzymes UbcH5a, H5b, and H5c are available, but UbcH6 and UbcH9, which were previously reported to be associated with MITOL (Nakamura et al., 2006), are unavailable for ubiquitination. In vitro ubiquitination assay demonstrated that MITOL WT, but not the CS mutant, directly ubiquitinates mSOD1 (Figure 2C, right, lanes 5 and 6) compared with the autoubiquitination of MITOL WT (lane 7). A similar result was obtained by using mSOD1 G85R (data not shown). To understand the specificity of MITOL-dependent ubiquitination of mSOD1 in mitochondria, we compared MITOL with another mitochondrial ubiquitin ligase, MULAN (also known as MAPL or GIDE; Li et al., 2008; Neuspiel et al., 2008; Zhang et al., 2008). As shown in Figure 2D, MULAN failed to ubiquitinate mSOD1, suggesting that mitochondrial mSOD1 was specifically ubiquitinated by MITOL.
MITOL Overexpression Promotes mSOD1 Degradation
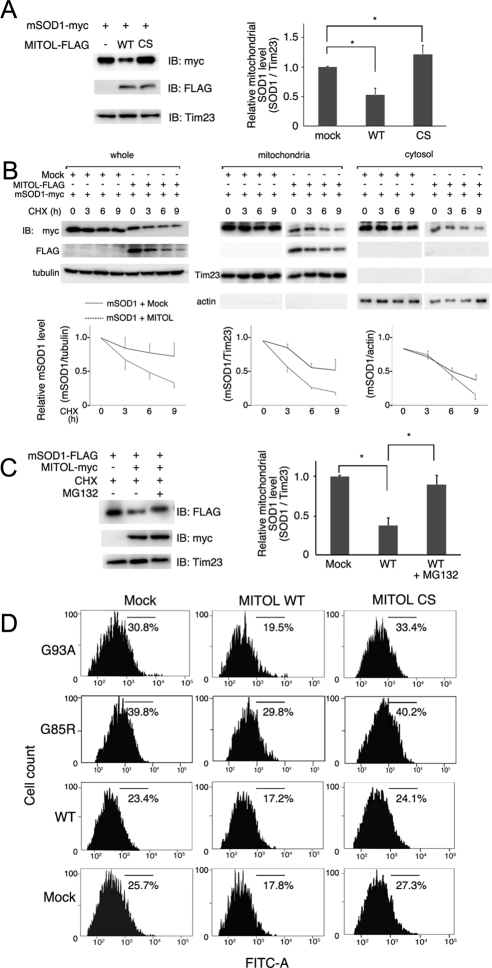
To understand the regulation of mSOD1 by MITOL, we examined the effects of overexpression of MITOL WT or CS mutant on the amount of mSOD1 in mitochondria. The amount of mSOD1 in the mitochondrial fractions isolated from Neuro2a cells transfected with mSOD1 plus control vector, mSOD1 plus MITOL WT, or mSOD1 plus MITOL CS mutant were compared. As shown in Figure 3A, the amount of mSOD1 in mitochondria was significantly decreased by MITOL WT, whereas increased by MITOL CS mutant. Statistical analysis performed on three independent experiments indicates that the amount of mSOD1 in mitochondria was reduced by half by MITOL WT. Further, we examined whether MITOL overexpression promotes the degradation of mSOD1 in the presence of the protein synthesis inhibitor cycloheximide (CHX). As shown in Figure 3B, left, CHX chase assay indicated that the degradation of mSOD1 was accelerated by overexpression of MITOL than the control vector. Statistical analysis indicates that MITOL overexpression reduced the mSOD1 level by ∼50% of the control at 9 h after CHX treatment. To further examine whether MITOL specifically reduces mitochondrial mSOD1, we compared the reduced levels of mSOD1 in mitochondrial and cytosolic fractions. As shown in Figure 3B, middle and right, the degradation of mitochondrial mSOD1 by MITOL overexpression was more remarkable than that of the cytosolic mSOD1. The degradation of mSOD1 by MITOL was inhibited in the presence of proteasome inhibitor, MG132 (Figure 3C), indicating that mSOD1 ubiquitinated by MITOL was degraded through the ubiquitin–proteasome system. Furthermore, mSOD1-dependent ROS generation was significantly reduced by MITOL overexpression (19.5%) than mSOD1 (30.8%; Figure 3D). The reason for reduction in ROS caused by MITOL overexpression is currently unknown. MITOL may be involved in the regulation of ROS production in mitochondria through an unidentified mechanism. We used another mSOD1 (G85R), which lacks the SOD activity (Rakhit and Chakrabartty, 2006). The production of ROS by G85R was higher than G93A, which possesses SOD activity, suggesting that G93A-dependent ROS generation was due to mitochondrial stress by the accumulation of unfolded mSOD1. Similarly, G85R-dependent ROS generation was reduced by the overexpression of MITOL WT (29.8%) than G85R (39.8%). In contrast, G85R-dependent ROS generation was slightly enhanced by the MITOL CS mutant. No obvious ROS generation was observed in the cells transfected with mock or WT SOD1. These results demonstrated that MITOL promoted the degradation of mSOD1 in mitochondria and inhibited mSOD1-mediated cytotoxicity.
Figure 3.
MITOL overexpression promotes mSOD1 degradation. (A) MITOL overexpression decreases the amount of mSOD1 in mitochondria. Mitochondrial fractions isolated from Neuro2a cells transfected with indicated vectors were immunoblotted with anti-myc, anti-FLAG, or anti-Tim23 antibody. The relative protein level of mSOD1 with or without MITOL was quantified by densitometry (right). Error bars, SD (n = 3). * p <0.05; Student's t test. (B) Cycloheximide (CHX)-chase assay indicates the rapid degradation of mSOD1 at mitochondria by MITOL overexpression. Neuro2a cells transfected with indicated vectors were treated with CHX (10 μg/ml) for indicated times, and each lysate of whole, mitochondrial, or cytosolic fractions was immunoblotted with indicated antibodies. The relative protein levels of mSOD1 were quantified by densitometry (bottom panels). Solid and broken lines indicate the cotransfection with mock or MITOL, respectively. Error bars SD (n = 3). (C) Degradation of mSOD1 through the ubiquitin-proteasome system. Neuro2a cells transfected with indicated vectors were incubated with CHX (10 μg/ml) with or without MG132 (50 μM) for 3 h. The relative protein levels of mSOD1 were quantified by densitometry. Error bars, SD (n = 3). * p < 0.05; Student's t test. (D) mSOD1-dependent ROS generation was reduced by MITOL WT, but was enhanced by CS mutant. Twenty-four hours after cotransfection of Neuro2a cells with indicated vectors plus pDsRed-Mito, cells were incubated in differentiation medium for 24 h. Intracellular ROS generation in DsRed-Mito–positive Neuro2a cells was measured by flowcytometric analysis using H2DCFDA staining. These represented results were from one of three experiments that produced similar results.
MITOL Knockdown Enhances the Toxicity of mSOD1
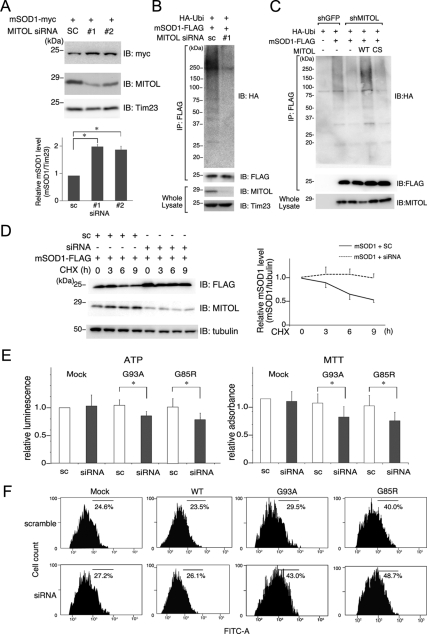
Further, we examined the effect of MITOL knockdown on mSOD1 degradation in the Neuro2a cells. As shown in Figure 4A, the amount of mSOD1 in mitochondria increased twice by MITOL depletions with two different MITOL small interfering RNAs (siRNAs). Similarly, MITOL depletion by MITOL siRNA reduced the ubiquitination of mSOD1 in the mitochondria fraction (Figure 4B). A similar result was obtained in the Neuro2a cells stably knockdown of MITOL by shMITOL, but not in the control cells by shGFP (sh green fluorescent protein; Figure 4C). A rescue of mSOD1 ubiquitination by MITOL WT resistant to shMITOL, but not CS mutant, confirmed the MITOL-dependent mSOD1 ubiquitination. Consistently, CHX-chase assay demonstrated that mSOD1 degradation was blocked in MITOL-knockdown Neuro2a cells (Figure 4D). These results indicated that endogenous MITOL is responsible for mSOD1 degradation in mitochondria.
Figure 4.
MITOL knockdown induces mSOD1 accumulation and enhances mSOD1-dependent cell toxicity. (A) Accumulation of mSOD1 in mitochondria by siRNA-mediated MITOL knockdown. Neuro2a cells were cotransfected with mSOD1 and siRNAs (scramble or two different MITOL siRNAs). Mitochondrial fractions isolated from these cells were immunoblotted with anti-myc, anti-MITOL or anti-Tim23 antibody. The relative protein level of SOD1 shown in A was quantified by densitometry. Error bars, SD (n = 3). * p < 0.05; Student's t test. (B) Inhibition of mSOD1 ubiquitination by MITOL knockdown. Mitochondrial fractions of Neuro2a cells transfected with G93A-FLAG and siRNAs (scramble or MITOL siRNA) were immunoprecipitated with anti-FLAG antibody. Immunoprecipitates and inputs were immunoblotted with indicated antibodies. (C) Inhibition of mSOD1 ubiquitination in MITOL-knockdown Neuro2a cells. Neuro2a cells stably expressing shMITOL or shGFP were transfected with indicated vectors. Human MITOL WT and CS were used as resistant vectors for mouse MITOL shRNA. Mitochondria fractions were immunoprecipitated with anti-FLAG antibody and immunoprecipitates were immunoblotted with anti-HA or anti-FLAG antibody. Inputs were immunoblotted with anti-MITOL antibody. (D) Delayed degradation of mSOD1 by MITOL depletion. Neuro2a cells were cotransfected with mSOD1 and siRNA (scramble or MITOL siRNA). Forty-eight hours after transfection, cells were treated with CHX (10 μg/ml) for indicated times, and lysates were analyzed by immunoblotting with anti-FLAG, anti-MITOL, or anti-tubulin antibody. The relative protein levels of mSOD1 were quantified by densitometry. Error bars, SD (n = 3). (E) Effects of MITOL depletion on ATP production and cell viability. Neuro2a cells were cotransfected with mSOD1 (G93A or G85R) and siRNA (scramble or MITOL siRNA). Twenty-four hours after transfection, cells were incubated in differentiation medium for 24 h. MTT assay and ATP production assay were performed. Error bars, SD (n = 3). * p < 0.05; Student's t test. (F) MITOL depletion enhances mSOD1-dependent ROS production. Twenty-four hours after cotransfection of Neuro2a cells with mSOD1 (G93A or G85R) or/and siRNAs (sc, scramble; siRNA, MITOL-specific siRNA) plus pDsRed-Mito, cells were incubated in differentiation medium for 24 h. Intracellular ROS generation in DsRed-Mito–positive Neuro2a cells was measured by flowcytometric analysis using H2DCFDA staining. These represented results were from one of three experiments that produced similar results.
To examine whether MITOL knockdown enhances mSOD1 toxicity, MTT assay and ATP production assay were used to measure the cell viability of Neuro2a cells cotransfected with mSOD1 and siRNA (scramble or MITOL siRNA). As shown in Figure 4E, MTT assay indicated that MITOL siRNA promoted mSOD1-induced cell death. Similarly, ATP production assay showed that ATP production was decreased in the Neuro2a cells cotransfected with mSOD1 and MITOL siRNA as than in the control cells (Figure 4E). To further confirm this phenomenon, we examined the effect of MITOL knockdown on ROS production caused by mSOD1 in Neuro2a cells. As shown in Figure 4F ROS production in Neuro2a cells cotransfected with G93A or G85R and MITOL siRNA (43.0 and 48.7%, respectively) was significantly increased compared with the control cells cotransfected with G93A or G85R and scramble siRNA (29.5 and 40.0%, respectively). No obvious ROS generation was observed in the cells transfected with scramble siRNA or MITOL siRNA. Our preliminary experimental results indicated that ROS production was increased in MITOL-deficient DT40 cells generated by a gene targeting method. Therefore, it is possible that a partially remaining expression of MITOL against siRNA may block the excess ROS production. Taken together, we concluded that MITOL is involved in the regulation of mSOD1-dependent cytotoxicity.
DISCUSSION
It has been previously reported that cytosolic ubiquitin ligases such as Dorfin, CHIP, and NEDL1 (Niwa et al., 2002; Miyazaki et al., 2004; Urushitani et al., 2004) ubiquitinate mSOD1 and promote its degradation. In this study, we found that MITOL interacted with and ubiquitinated mSOD1 but not WT SOD1 in Neuro2a cells. In addition, the coexpression of MITOL with mSOD1 reduced the mSOD1 level in mitochondria to approximately half of its basal level than did the expression of mSOD1 alone. In contrast, the knockdown of endogenous MITOL using MITOL-specific siRNA increased the mSOD1 level by approximately two times in mitochondria. Supporting these results, the overexpression of MITOL reduced the half-life of mSOD1, whereas the knockdown of endogenous MITOL extended the half-life of mSOD1. Taken together, we discovered a novel mechanism of eliminating the mitochondrial mSOD1, which is recognized and ubiquitinated by MITOL.
It has been believed that the mutations in SOD1 cause various forms of cell damage, including its aggregation by ER stress, inhibition of protein degradation, axonal transport disturbance, and reduced ROS scavenging activity (Bendotti and Carri, 2004; Barber et al., 2006; Boillee et al., 2006; Kabashi and Durham, 2006; Turner and Atkin, 2006); however, recent studies have shown that the accumulation of mSOD1 in mitochondria causes mitochondrial dysfunction, which is closely associated with the onset of ALS (Dupuis et al., 2004; Manfredi and Xu, 2005; Lin and Beal, 2006). Mitochondrial dysfunction caused by the accumulation of mSOD1 in mitochondria includes the following: mitochondrial vacuolization, a decrease of ATP production, an increase of ROS generation, and an impairment of calcium buffering, which ultimately lead to an apoptotic cell death. Experimental results from MTT assay and ATP production assay using MITOL-specific siRNA indicated that MITOL inhibits the loss of cell viability induced by mSOD1. In addition, flowcytometric analysis indicated that MITOL suppresses the ROS generation induced by mSOD1. These observations show that MITOL has an ability to protect mitochondria from the toxicity of mSOD1. Therefore, it is probable that the targeted expression of MITOL may be a candidate for gene therapy of familial ALS, because it selectively removes mSOD1. We are currently in the process of examining the possibility that overexpression of MITOL attenuates mSOD1-associated toxicity in vivo, by performing experiments on mice obtained by crossing MITOL-transgenic mice with mSOD1-transgenic mice, which is an animal model for studying familial ALS.
Results from recent studies have demonstrated that misfolded proteins are recognized and degraded by a quality control mechanism called ERAD. Through the ERAD system, aberrant proteins are transported from the ER to the ectoplasm, and immediately are degraded by the ubiquitin–proteasome pathway (Tsai et al., 2002; Meusser et al., 2005). We demonstrated that MITOL ubiquitinates mSOD1 aggregated around the mitochondria and promotes its degradation. This observation implies that there is a possibility of an existence of a novel mitochondrial quality control mechanism distinct from the ERAD system. Thus, MITOL may act as a “keeper-on-mitochondrion” by preventing the accumulation of those proteins at mitochondrial outer membrane. We found that not only mSOD1 but other misfolded proteins accumulated at mitochondria, such as mutant short-chain acyl CoA dehydrogenase (Pedersen et al., 2003), are also ubiquitinated by MITOL and are degraded by proteasome (Supplementary Figure S3, A and B). However, the mechanism by which MITOL recognizes these aberrant proteins is yet unknown. Because CHIP uses a molecular chaperone such as Hsp70 to recognize mSOD1 in ERAD system (Urushitani et al., 2004), it is likely that MITOL also utilizes a recognition mechanism similar to that of CHIP. To support this possibility, in our preliminary results, we observed that MITOL was associated with Hsp70 (Supplementary Figure S3C). However, unlike CHIP, MITOL does not contain Hsp-interacting domain, and therefore, the relationship between MITOL and chaperones is still unclear. Further analysis is needed to elucidate whether molecular chaperones such as Hsp70 or cochaperones are involved in the recognition of mSOD1 by MITOL.
In conclusion, MITOL promotes the degradation of aberrant proteins and protects mitochondria against their toxicity. Our findings might provide insights on definitive therapy for neurodegenerative diseases such as ALS. Further studies are needed to address the molecular mechanism underlying MITOL-dependent quality control system in mitochondria.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology and Japan Society for the Promotion of Science and a grant from the Naito Foundation.
Abbreviations used:
- ALS
amyotrophic lateral sclerosis
- CHX
cycloheximide
- ERAD
endoplasmic reticulum–associated degradation.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0112) on September 9, 2009.
REFERENCES
- Barber S. C., Mead R. J., Shaw P. J. Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. Biochim. Biophys. Acta. 2006;1762:1051–1067. doi: 10.1016/j.bbadis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Bendotti C., Carri M. T. Lessons from models of SOD1-linked familial ALS. Trends Mol. Med. 2004;10:393–400. doi: 10.1016/j.molmed.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Boillee S., Vande Velde C., Cleveland D. W. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Dupuis L., Gonzalez de Aguilar J. L., Oudart H., de Tapia M., Barbeito L., Loeffler J. P. Mitochondria in amyotrophic lateral sclerosis: a trigger and a target. Neurodegener. Dis. 2004;1:245–254. doi: 10.1159/000085063. [DOI] [PubMed] [Google Scholar]
- Kabashi E., Durham H. D. Failure of protein quality control in amyotrophic lateral sclerosis. Biochim. Biophys. Acta. 2006;1762:1038–1050. doi: 10.1016/j.bbadis.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Karbowski M., Neutzner A., Youle R. J. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft S. G., Wang L., Hochstrasser M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI) J. Biol. Chem. 2006;281:4646–4653. doi: 10.1074/jbc.M512215200. [DOI] [PubMed] [Google Scholar]
- Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., Joazeiro C. A. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. T., Beal M. F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Manfredi G., Xu Z. Mitochondrial dysfunction and its role in motor neuron degeneration in ALS. Mitochondrion. 2005;5:77–87. doi: 10.1016/j.mito.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Miyazaki K. NEDL1, a novel ubiquitin-protein isopeptide ligase for dishevelled-1, targets mutant superoxide dismutase-1. J. Biol. Chem. 2004;279:11327–11335. doi: 10.1074/jbc.M312389200. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Kimura Y., Tokuda M., Honda S., Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuspiel M., Schauss A. C., Braschi E., Zunino R., Rippstein P., Rachubinski R. A., Andrade-Navarro M. A., McBride H. M. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr. Biol. 2008;18:102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- Niwa J., Ishigaki S., Hishikawa N., Yamamoto M., Doyu M., Murata S., Tanaka K., Taniguchi N., Sobue G. Dorfin ubiquitylates mutant SOD1 and prevents mutant SOD1-mediated neurotoxicity. J. Biol. Chem. 2002;277:36793–36798. doi: 10.1074/jbc.M206559200. [DOI] [PubMed] [Google Scholar]
- Pedersen C. B., Bross P., Winter V. S., Corydon T. J., Bolund L., Bartlett K., Vockley J., Gregersen N. Misfolding, degradation, and aggregation of variant proteins. The molecular pathogenesis of short chain acyl-CoA dehydrogenase (SCAD) deficiency. J. Biol. Chem. 2003;278:47449–47458. doi: 10.1074/jbc.M309514200. [DOI] [PubMed] [Google Scholar]
- Rakhit R., Chakrabartty A. Structure, folding, and misfolding of Cu, Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim. Biophys. Acta. 2006;1762:1025–1037. doi: 10.1016/j.bbadis.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Rosen D. R. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Tatsuta T., Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;27:306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B., Ye Y., Rapoport T. A. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- Turner B. J., Atkin J. D. ER stress and UPR in familial amyotrophic lateral sclerosis. Curr. Mol. Med. 2006;6:79–86. doi: 10.2174/156652406775574550. [DOI] [PubMed] [Google Scholar]
- Urushitani M., Kurisu J., Tateno M., Hatakeyama S., Nakayama K., Kato S., Takahashi R. CHIP promotes proteasomal degradation of familial ALS-linked mutant SOD1 by ubiquitinating Hsp/Hsc70. J. Neurochem. 2004;90:231–244. doi: 10.1111/j.1471-4159.2004.02486.x. [DOI] [PubMed] [Google Scholar]
- Yonashiro R. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. GIDE is a mitochondrial E3 ubiquitin ligase that induces apoptosis and slows growth. Cell Res. 2008;18:900–910. doi: 10.1038/cr.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.