Abstract
Poly(ADP-ribose) (pADPr), made by PARP-5a/tankyrase-1, localizes to the poles of mitotic spindles and is required for bipolar spindle assembly, but its molecular function in the spindle is poorly understood. To investigate this, we localized pADPr at spindle poles by immuno-EM. We then developed a concentrated mitotic lysate system from HeLa cells to probe spindle pole assembly in vitro. Microtubule asters assembled in response to centrosomes and Ran-GTP in this system. Magnetic beads coated with pADPr, extended from PARP-5a, also triggered aster assembly, suggesting a functional role of the pADPr in spindle pole assembly. We found that PARP-5a is much more active in mitosis than interphase. We used mitotic PARP-5a, self-modified with pADPr chains, to capture mitosis-specific pADPr-binding proteins. Candidate binding proteins included the spindle pole protein NuMA previously shown to bind to PARP-5a directly. The rod domain of NuMA, expressed in bacteria, bound directly to pADPr. We propose that pADPr provides a dynamic cross-linking function at spindle poles by extending from covalent modification sites on PARP-5a and NuMA and binding noncovalently to NuMA and that this function helps promote assembly of exactly two poles.
INTRODUCTION
Mitotic spindles are normally bipolar; they are composed of two interdigitating, antiparallel microtubule arrays whose minus ends cluster at the poles and whose plus ends overlap in the center of the spindle. Chromosomes move toward spindle poles at anaphase, so accurate segregation requires bipolarity. Abnormalities, such as mono- and tripolar spindles, cause chromosome mis-segregation that generates aneuploid or inviable daughter cells. How spindles achieve bipolarity is an interesting unsolved problem. Animal cells normally contain two centrosomes at the end of G2 that are thought to catalyze assembly of two spindle poles. However, spindles often achieve bipolarity even in cells that contain too few, or too many, centrosomes, so additional mechanisms must exist to ensure bipolarity. If we understood these mechanisms, we might be able to selectively inhibit assembly of bipolar spindles in cancer cells that have too many centrosomes and thus design a cancer-selective antimitotic drug (Godinho et al., 2009). Thus, both the mechanisms that promote clustering of minus ends and the assembly of exactly two spindle poles are of basic and practical interest.
A number of proteins have been identified that localize to spindle poles, function in pole assembly, and might help determine the number of poles. These include the microtubule cross-linking protein NuMA (Compton et al., 1992;Gaglio et al., 1995; Merdes et al., 1996) and minus-end–directed motor proteins that are proposed to actively cluster minus ends by binding to one microtubule and moving toward the minus end of another (Walczak et al., 1998). Together, these proteins are thought to cluster minus ends into local foci, but it is unclear how exactly two dominant foci emerge to form the poles of a bipolar spindle. The role of nonprotein macromolecules in bipolar spindle assembly has been less studied, but at least two are known to contribute: 1) RNA is required in anastral (centrosome-lacking) meiotic spindles, in part as a component of the Rae1 complex (Blower et al., 2005; Wong et al., 2006), and 2) poly(ADP-ribose) (pADPr), the subject of this article, is also required for bipolarity in both anastral and astral spindles (Chang et al., 2004, 2005a).
pADPr, is a large, negatively charged polymer that is synthesized by poly(ADP-ribose) polymerase (PARP) enzymes using NAD+ as a substrate (Schreiber et al., 2006). Polymerization is typically initiated by covalent modification of a protein primer, which can be either the PARP enzyme itself or a nearby protein. The polymer can grow to hundreds of subunits, with occasional branches. pADPr is a highly dynamic macromolecule that can be synthesized and broken down within minutes. Breakdown is catalyzed by specific glycohydrolases, of which poly(ADP-ribose) glycohydrolase (PARG) is the most active (Ueda et al., 1972; Hatakeyama et al., 1986). Another glycohydrolase, ARH3, can hydrolyze pADPr, though it is more active at promoting hydrolysis of the small molecule acetyl-ADP-ribose (Oka et al., 2006; Ono et al., 2006). We found that pADPr is an abundant component of anastral meiotic spindles assembled in Xenopus egg extract, where it is present throughout the spindle, and also is a component of human tissue culture cell spindles, where it is enriched at the poles (Chang et al., 2004). Treatment of egg extract spindles with PARG to hydrolyze pADPr or with high concentrations of anti-pADPr IgG to coat it caused rapid breakdown of bipolar spindles, with dramatic ejection of microtubule asters as well as pole–pole elongation (Chang et al., 2004). This effect suggests pADPr functions to help keep foci of minus ends together in the spindle.
PARP-5a, also known as tankyrase, is thought to be the main enzyme that synthesizes pADPr in mitotic spindles. Various functions have been proposed for PARP-5a, including spindle pole organization, telomere maintenance, and Golgi organization (Chi and Lodish, 2000; Smith and de Lange, 2000; Chang et al., 2005a). Several lines of evidence support a key role in spindle pole organization: PARP-5a forms a complex with NuMA and localizes to spindle poles (Chang et al., 2005a,b). RNA interference (RNAi) depletion of PARP-5a caused HeLa cells (and other cell types; P. Chang, unpublished data) to arrest in mitosis with multipolar spindles. These spindles contained two poles that were associated with centrosomes as well as extra acentrosomal poles. Both types of pole were enriched in NuMA and appeared to contain locally well-focused minus ends. Mutant PARP-5a lacking catalytic activity localized to the poles of these multipolar spindles, but failed to rescue the phenotype, showing that pADPr, and not just PARP-5a polypeptide, is required for bipolarity. Recently, PARP-5a and a second PARP were identified in an RNAi screen as a protein required to form bipolar spindles in Drosophila S3 cells containing multiple centrosomes (Kwon et al., 2008). If the primary function of PARP-5a is to ensure that spindle assemble only two poles, one might expect its activity to be up-regulated during mitosis. Cell-cycle regulation of PARP-5a has not been directly measured before, but we provide biochemical evidence that it is indeed strongly up-regulated during mitosis.
How might pADPr function in spindle pole organization at the molecular level? One clue came from finding that PARP-5a covalently modifies itself, and its binding partner NuMA, with pADPr chains during mitosis (Chang et al., 2005a,b). We hypothesize that these chains extend some distance away from individual NuMA–PARP-5a complexes and somehow help condense local clusters of minus ends into two well-formed poles. This proposed function would require that a pADPr chain interact noncovalently with microtubules or with microtubule binding proteins at some distance from its covalent attachment site. Microtubules are not known to bind pADPr directly, and we consider this unlikely because both polymers are negatively charged. More likely, pADPr binds noncovalently to one or more spindle pole proteins that also bind to microtubules. pADPr-binding proteins have recently been identified or hypothesized based on various criteria (Karras et al., 2005; Ahel et al., 2008; Gagne et al., 2008), but none are known to function specifically in mitotic spindle pole organization. To test if pADPr chains growing from PARP-5a can function in pole organization, we developed a functional assay in mitotic lysate. On obtaining a positive result, we went on to identify noncovalent pADPr-binding proteins in the lysate that might mediate spindle pole organization. One of these was NuMA, which thus appears to interact both covalently and noncovalently with pADPr to promote spindle pole organization.
MATERIALS AND METHODS
Buffers
The following buffers were used: BRB80: 80 mM KPIPES, 1 mM MgCl2, and 1 mM EGTA, pH 6.8; PEM: 100 mM KPIPES, 1 mM MgCl2, and 10 mM EGTA, pH 6.8; cacodylate: 5 mM cacodylate, pH 7.0 (EMS, Hatfield, PA); AbDil: 120 mM NaCl, 10 mM Tris·Cl, pH 7.4, 0.1% Triton X-100 (TX-100), 0.1% NaN3, and 5% BSA; HBS: 150 mM NaCl, 50 mM HEPES, pH 7.4, 1 mM MgCl2, 1 mM DTT, and 1 mM EGTA; and cell lysis buffer: 150 mM NaCl, 50 mM HEPES, pH 7.4, 1 mM MgCl2, 0.5% TX-100, 1 mM DTT, and 1 mM EGTA.
Immuno-Electron Microscopy
HeLa cells were grown to 70–85% confluence on glow-discharged, poly-l-lysine–coated Aclar coverslips. Cells were permeabilized with 0.1% TX-100 in BRB80 buffer for 45 s. Samples were fixed with 4% paraformaldehyde and 0.05% gluteraldehyde in BRB80 buffer for 20 min and then washed three times in BRB80 + 0.1% TX-100. Coverslips were treated with 100 μM NaIO4 in 1× PEM + 0.1% TX-100 for 5 min and then washed with 1× PEM + 0.1% TX-100. Samples were then incubated in 100 mM succinic dihydrazide in 1× PEM + 0.1% TX-100 for 10 min and washed with 1× PEM + 0.1% TX-100. Cells were blocked with 4% donkey serum in Abdil for 30 min at room temperature (RT) and then incubated in rabbit anti-poly(ADP-ribose) LP-9610 (BD Biosciences, San Jose, CA) or chicken anti-poly(ADP-ribose) IgY (Tulip BioLabs, Lansdale, PA), or Abdil alone (for IgG only control) diluted in Abdil for 1 h at RT. Samples were then rinsed three times for 5 min in BRB80 and incubated in secondary antibody (conjugated to colloidal gold) diluted in Abdil at the concentration recommended by manufacturer for 1 h at RT. Cells were rinsed three times in BRB80 and fixed in 50 mM lysine (Sigma, St. Louis, MO; L-5501), then in 3% gluteraldehyde in BRB80 for 6 min at RT, and then in 3% glutaraldehyde in BRB80 for 12 min. Coverslips were rinsed three times in BRB80 and then three times in 0.05 M cacodylate buffer, pH 7.0, and postfixed 1% osmium in 0.8% K3Fe (CN)6 in cacodylate buffer for 15 min on ice, in a hood. Samples were rinsed three times in cacodylate buffer and then three times in distilled water. The samples then stained overnight with 1% aqueous uranyl acetate and filtered before use, at 4°C in the dark and rinsed in distilled water. Coverslips were then dehydrated using progressive lowering of temperature: 35% EtOH, 50% EtOH at 4°C for 5 min each; 50% EtOH, 80% EtOH at −20°C for 5 min each; and 95% EtOH, 100% EtOH at −40°C for 5 min each and then were brought to RT and dipped in fresh 100% EtOH and 100% propylene oxide. Samples were then infiltrated with 2:1 propylene oxide:Epon araldite, 1:2 propylene oxide:epon araldite for 30 min each and then with 100% Epon araldite for 1 h at RT, in a hood because propylene oxide fumes are toxic. Coverslips were mounted in an Aclar “sandwich” over an Aclar “washer” and polymerized at 65°C for 48 h. Samples were disassembled and affixed to microscope slides so that individual cells of the desired phenotype could be identified, excised, and remounted for sectioning. A Reichert Ultracut S ultramicrotome (Vienna, Austria) was used to make serial sections at 85 nm through the cell that were collected on 1 × 2-mm slot grids. Samples were viewed, and images were taken on a JEOL 1200 electron microscope (Peabody, MA).
Green Fluorescent Protein–tagged Proteins Expression and Purification
Green fluorescent protein (GFP)-PARP-5a or GFP-PARP-5a PARP DEAD (enzyme with a point mutant and the PARP-active site, rendering it catalytically inactive; Chang et al., 2005a) were transfected into HeLa S3 cells with 294fectin (Invitrogen, Carlsbad, CA) using an adaptation of the 293 Free Style protocol; HeLa S3 cells grown in DMEM + 10% FCS were pelleted at 400 × g for 5 min the night before transfection and resuspended to 1E6/ml in DMEM + 10% FCS. The day of transfection, 10 μl 293fectin and 10 μg DNA was used to transfect each 10 ml of HeLa S3 suspensions according to the 293 FreeStyle protocol. After 24 h cells were treated with 10 μg/ml S-trityl-l-cysteine for 11 h and prepared for GFP-PARP-5a purification. GFP-PARP-5a was purified fresh for each assay. For in vitro activity assays 250 ml of HeLa S3 cells were transfected, whereas for biochemical purification 1 l of HeLa S3 cells was used for GFP-PARP-5a expression and purification. GFP-NuMA was expressed similarly.
Preparation of Mitotic Lysates 1: Dilute Lysates for Isolation of pADPr-binding Proteins.
HeLa S3 cells were treated with 10 μM S-trityl-l-cysteine or 500 nM nocodazole for 11 h and then 1 μg/ml latrunculin B for 1 h at 37°C before generation of lysates. Cells were pelleted at 400 × g for 5 min and washed three times in ice-cold PBS. For washes, cells were pelleted at 400 × g for 5 min at 4°C. Cell pellets were then incubated in cell lysis buffer for 10 min on ice and pelleted at 14,000 × g for 5 min at 4°C. The supernatant was obtained, and 1 μg/ml nocodazole and 1 μg/ml cytochalasin B were added. The PARG inhibitor ADP-HPD at 1 μM (Calbiochem) was typically added. Extracts were prepared fresh for each experiment.
Preparation of Mitotic Lysates 2: Concentrated Lysates for Assembly of Spindle Poles.
HeLa S3 cells were treated with 10 μM S-trityl-L-cysteine for 11 h and then 1 μg/ml latrunculin A for 1 h at 37°C before generation of lysates. Cells were pelleted at 400 × g for 5 min and washed three times in ice-cold HBS. For washes, cells were pelleted at 400 × g for 5 min at 4°C. Pellets were flash-frozen in liquid nitrogen and thawed at 30°C twice and then centrifuged at 5000 × g at 4°C for 30 min. Supernatants were obtained and stored on ice, and 1 μg/ml cytochalasin B was added. The PARG inhibitor ADP-HPD 1 μM at was typically added. Extracts were prepared fresh for each experiment.
Preparation of Other Reaction Components
Mitotic centrosomes were purified from HeLa S3 cells arrested in mitosis using 10 μM S-trityl-l-cysteine using the Bornens protocol (Gosti-Testu et al., 1986). Mitotic centrosomes fractionate differently than the centrosomes normally purified from asynchronous populations of cells so care must be taken to assay each fraction for centrosomes enrichment. Mitotic chromosomes from HeLa cells were a gift from Ryoma Ohi (now at Vanderbilt Medical School).
RanQ69L fused to glutathione-S-transferase (GST) was expressed and purified as described (Wilde et al., 2001) and stored in aliquots at −80°C.
Beads Coated with GFP-PARP5-pADPr.
Anti-GFP 3E6 antibody was attached to protein A Dynabeads (Invitrogen). These beads were added to dilute mitotic lysate from cells that had been transfected with GFP-PARP5-pADPr as described above. NAD+ (100–500 μM) and ADP-HPD (1 μM) were typically added to promote pADPr polymerization. The mixture was incubated at 20°C for 30 min. Beads were recovered with a magnet and washed with cell lysis buffer containing 500 mM NaCl three times for 10 min. These beads were either added to concentrated lysate for functional assays (see Figure 3) or to more dilute lysate for identification of pADPr-binding proteins. In some cases, we substituted protein A Affiprep beads (Bio-Rad, Richmond, CA) for the biochemical experiments. ARH3 fused to GST was expressed and purified as described by Oka et al. (2006).
Figure 3.
Aster assembly trigged by pADPr on beads. Magnetic beads coated with PARP-5a lacking catalytic activity (PD-PARP-5a) or PARP-5a decorated with pADPr (PARP-5a-pADPr) were added to concentrated mitotic lysate containing Alexa 594 tubulin and 1 μM ADP-ADHP to inhibit PARG activity. Note that pADPr-coated beads induced assembly of microtubule asters. Addition of extra NAD+ (+250 μM NAD+ and + 500 μM NAD+) to the lysate to promote additional pADPr polymerization increased the density of asters and the density of microtubules within them. Microtubule appeared not to emanate directly from the pADPr-coated beads; rather they assembled into asters that associated laterally with beads.
RNA-coated Beads.
Biotinylated RNA (Ambion, Austin, TX) was attached to streptavidin Dynabeads (Invitrogen).
Assembly Reactions in Mitotic Lysates
Concentrated mitotic lysate was supplemented with Alexa 594–labeled tubulin (Invitrogen) at an empirical dilution that was optimized for each extract. Centrosomes, chromosomes, RanQ69L (1 mg/ml final), or magnetic beads coated with GFP-PARP-5a-pADPr were added, and the reaction was incubated at 20°C for 30 min. Aliquots of the reaction were spotted onto slides using wide-bore pipette tips and were imaged immediately.
Isolation of pADPr-binding Proteins
Dynabeads (or in some experiments Affiprep beads) coated with GFP-PARP-5a-pADPr were added to dilute mitotic lysates. In most experiments 1 mM NAD+ and 1 μM ADP-HPD were also added to protect and grow pADPr chains. Reactions were incubated for 1 h at 4°C and then washed three time for 10 min with cell lysis buffer containing 150 or 300 mM NaCl and one time for 10 min cell lysis buffer and then either eluted with SDS-PAGE sample buffer 95°C for 5 min or incubated with 100 μg/ml ARH3 for 30 min at 4°C followed by 20 min at 20°C. If incubated with ARH3, samples were subsequently washed three times 10 min with cell lysis buffer containing 300 mM NaCl followed by one time for 10 min with cell lysis buffer and then eluted with SDS-PAGE sample buffer.
Preparation of NuMA fragments
Three fragments of human NuMA were cloned into pET28a via PCR; the N-terminal (aa 1–216), C-terminal (aa 1701–2102), and central Rod domain (aa 217-1701). NuMA fragment bacterial expression vectors were then transformed into BL21-codon plus DE3 RIPL cells, and individual colonies were tested for expression of soluble protein. Colonies expressing maximal soluble proteins were inoculated into overnight cultures. Overnight cultures were then used to inoculate 3 l of ½ YT containing chloramphenicol and grown at 25C until OD 0.7. Cultures were induced using 100 μM IPTG overnight at 16°C and then pelleted. NuMA fragments were purified using two steps. First, fragments were bound to NiNTA (Qiagen, Chatsworth, CA) according to the Qiagen NiNTA protocol and washed using Qiagen NiNTA wash buffer. Fragments were eluted using imidazole elution according to the Qiagen protocol. Purified fragments were fractionated by gel filtration using a Bio-Rad SEC 250–5 column in cell lysis buffer and stored as frozen aliquots.
NuMA Fragment pADPr–binding Interactions
Magnetic beads coated with GFP-PARP-5a-pADPr were prepared and washed with 0.5M NaCl in cell lysis buffer as described above. About 5 μg purified NuMA fragments was incubated with the beads for 1 h at 4°C in 100 μl cell lysis buffer and then washed three times in cell lysis buffer containing 300 mM NaCl for 10 min and then one time in cell lysis buffer for 10 min. ARH3 treatment to check that binding of the rod fragment depended on pADPr was performed as described for lysate reactions above.
RESULTS
Localization of pADPr at Spindle Poles
To visualize pADPr filaments in mitotic spindles at the ultrastructural level, we used immunogold staining and thin section electron microscopy (EM). We developed a method for chemical fixation of pADPr that allowed it to pick up heavy metal stains, based on previous EM histochemistry of polysaccharides (Bradbury and Stoward, 1967). Cells were lysed in a microtubule-stabilizing buffer and briefly oxidized with periodate (IO4−) to generate reactive aldehyde groups in the ribose units in pADPr and any other sugars. Next, a bis-amine was added to convert aldehyde to amino groups and finally a formaldehyde/glutaraldehyde mixture to cross-link the now partially amine-modified pADPr to nearby proteins. pADPr was then immunolocalized using two different antibodies generated against two distinct pADPr antigens in a standard two-step preimbedding immunogold procedure. These antibodies are the most routinely used pADPr antibodies and have been well characterized. Both antibodies yielded similar immunogold localization patterns. This procedure also works well for light-level labeling, using formaldehyde alone as the fixative and fluorescent secondary antibodies (not shown). At the EM level, we observed immunogold particles specifically recognizing pADPr close to microtubules at the spindle poles (Figure 1, right and inset), confirming and extending the localization previously observed by immunofluorescence, while no staining was shown using secondary-only controls (left) (Kanai et al., 2000; Chang et al., 2004, 2005a).
Figure 1.
Ultrastructural localization of pADPr in the mitotic spindle. HeLa cells were permeabilized and fixed using a periodate/bis-amine/aldehdye protocol and then stained with two different antibodies to pADPr raised against distinct antigens (LP9610, BD Biosciences or anti-pADPr chicken, Tulip BioLabs) and control IgG or control IgY, followed by 5-nm gold conjugated to secondary antibody. Cells were then postfixed, embedded, and sectioned. A few gold particles were observed in sections from control IgG- or IgY-stained cells (left). Anti-pADPr staining (LP96–10 or pADPr) resulted in many gold particles near the spindle pole, particularly in the region between the centrosome and the spindle proper, where minus ends of spindle microtubule converge. Scale bar, 1 μm.
Spindle Pole Assembly in Mitotic Lysate
To test if local aggregates of pADPr can trigger spindle pole assembly, we developed a mitotic lysate system from HeLa cells. We were inspired by a previous lysate system that used taxol-promoted microtubule assembly to analyze the role of motor proteins in pole assembly (Mack and Compton, 2001). Our goal was to develop a lysate system from cells that we could genetically manipulate, where pole-like microtubule asters assembled in response to physiologically relevant cues, rather than taxol. For these studies we developed two kinds of mitotic lysate, a concentrated freeze-thaw lysate for functional pole assembly assays (see Figures 2 and 3) and a more conventional dilute detergent lysate for protein-binding studies (see Figures 4 and 5). To prepare lysates, suspension-cultured HeLa S3 cells were arrested in mitosis by overnight incubation with the kinesin-5 inhibitor S-trityl-l-cysteine (DeBonis et al., 2004). This arrest method has the advantage of avoiding drugs (e.g., nocodazole and taxol) that would artificially inhibit or promote microtubule assembly in the lysate. Cells were harvested and washed by centrifugation. For concentrated lysate, they were lysed by freeze-thaw of a packed pellet and clarified by centrifugation to generate a lysate containing ∼20–30 mg/ml protein. For dilute lysate, cells were lysed in detergent containing buffer, and the lysate was clarified by centrifugation. In all cases (except where noted) the PARG inhibitor ADP-HPD (Slama et al., 1995) was added at 1 μM to minimize pADPr hydrolysis. To allow visualization of PARP-5a or NuMA, we transiently transfected cells with GFP-tagged proteins before preparing lysates. GFP-PARP-5a expressed in this manner was also used as a tagged form of the protein for immunoisolation experiments, using anti-GFP IgG on magnetic beads. We showed previously that GFP-NuMA and GFP-PARP-5a localize normally to spindle poles in living cells (Chang et al., 2005a). For functional assembly reactions, we added Alexa 594-tubulin to visualize microtubule assembly in concentrated lysate, incubated reactions in tubes, and spotted aliquots onto slides for visualization.
Figure 2.
Reconstitution of aspects of spindle pole assembly in concentrated mitotic lysate. To study pADPr contribution to spindle pole assembly, we developed a human cell extract system to recapitulate spindle pole assembly. In all cases examined, the human cell extracts behaved similar to Xenopus laevis egg extract spindle pole assembly reactions. (a) Centrosome induced asters. Concentrated lysates were prepared from mitotic HeLa cells expressing GFP-PARP-5a (top row) or GFP-NuMA (bottom row) and supplemented with Alexa 594 tubulin and purified mitotic centrosomes. Extracts were imaged after 30 min. Microtubules (MT) are shown in left panels and Merge, and PARP-5a-GFP or NuMA-GFP in the middle panels, and Merge in the right panels). Note assembly of asters that recruit the known spindle pole proteins PARP-5a and NuMA to their foci. In merge MTs are shown in green, and red represents the GFP-fusion. (b) Ran-GTP induced asters. The experiment was performed as in panel a, except that 1 mg/ml RanQ69L was added in place of centrosomes. Note the abundant asters. The Ran asters are less focused than centrosome-induced asters, as judged by PARP-5a and NuMA fluorescence. In merge MTs are shown in green, and red represents the GFP-fusion. (c) Examples of structures that assembled when mitotic chromosomes alone (left) or mitotic chromosomes and centrosomes (right) were incubated in lysate. Chromosomes alone induced some asters. When chromosomes and centrosomes were mixed, spindle-like assemblies containing dense microtubule bundles were observed. Microtubules are shown in green and DNA in blue.
Figure 4.
Composition of PARP-5a-pADPr beads and identification of mitotic pADPr-binding proteins. (a) PARP-5a activity is up-regulated during mitosis. GFP-PARP-5a was isolated from lysates of asynchronous (I) and mitosis-arrested (M) HeLa S3 cells using magnetic beads (see Materials and Methods). Beads were washed with high-salt buffer and analyzed by SDS-PAGE and Coomassie blue staining (left) or anti-pADPr Western blot (right). Note much greater pADPr labeling of the M sample. This includes a strong band at the position corresponding to the well of the gel, suggesting that heavily pADPr-labeled proteins failed to enter the gel. The beads shown in the M lane are typical of those used in Figure 3. The numbers to the left show the molecular weights (kDa) of the markers used in all the gels. (b) Identification of pADPr-binding proteins. Beads like those shown in panel a (M) were incubated with mitotic lysate at 4°C and washed with 0.3 M NaCl buffer. These beads were split in two; one-half was analyzed without further treatment (5a + M-lys), the other was treated with a pADPr glycohydrolase (ARH3) and washed (5a + M-lys + ARH3). Note a number of bands that are strongly labeled with Coomassie or anti-pADPr in the lane not treated with glycohydrolase. Most of these were removed by glycohydrolase, indicating they bind to pADPr, and not the beads themselves, or by unmodified PARP-5a. (c) Cell cycle specificity of pADPr-binding proteins. PARP-5a-pADPr beads were made in mitotic lysate and washed with 500 mM NaCl in lysate buffer and then incubated with buffer alone, lysate from asynchronous (I), or mitosis-arrested (M) cells. Note that some of the heaviest bands are similar between the two cell cycle states, but in general the profiles are very different. (d) Drug specificity and requirement for NAD+. pADPr-binding proteins were isolated from cells arrested in mitosis using drugs that inhibit kinesin-5, S-tritl-l-cysteine (STC), or microtubules, nocodazole (NOC). As another test of pADPr requirement, NAD+ was omitted from the both the initial and second extract incubations (left lanes) or added at 500 μM (right lanes). Note that omission of NAD+ caused loss of most of the candidate pADPr-binding proteins. The profile of these proteins was nearly identical for the two drug arrests. (e) Comparison of pADPr and RNA-binding proteins in mitotic lysate. GFP-PARP-5a-pADPr beads were prepared as in panel a. The gel lane labeled 5a shows these beads alone. RNA beads were prepared by incubating biotinylated RNA (Ambion) with streptavidin beads. Both beads were incubated with mitotic lysate, washed, and analyzed by SDS-PAGE. Note that pADPr-coated beads recruited a similar band pattern to b, c (M), and d. RNA beads recruited what appeared to be some similar proteins by band mobility, but that had an overall very different protein profile. (f) Example of band cutting for LC/MS analysis. The boxes indicate bands from the rightmost lane in the gel shown in panel e that were excised for LC/MS analysis. This type of analysis was performed twice, in addition to some LC/MS runs on total pADPr-binding proteins. See Table 1 for the identity of the most abundant protein in each excised band.
Figure 5.
Recruitment of NuMA and CH-TOG to pADPr beads at different polymer levels. Molecular weights shown at left. GFP-PARP-5a-pADPr beads were prepared as in panel a and added to aliquots of mitotic lysate for recruitment of binding proteins. The lysates were supplemented with the indicated concentration of NAD+ and the PARG inhibitor ADP-ADHP (1 μM) except for the zero NAD+, where it was omitted. Beads were washed and blotted for pADPr, NuMA, and CH-TOG. Increasing NAD+ concentrations resulted in increased levels of pADPr on the beads, as revealed by the pADPr immunoblot. NuMA was recruited equally to the beads at all nonzero concentrations of NAD+, whereas CH-TOG recruitment increased with increasing concentrations of NAD+ and increasing amounts of pADPr on the beads.
To evaluate the ability of concentrated mitotic HeLa lysate to assemble pole-like structures in response to physiological cues, we first characterized the lysate by adapting standard pole-assembly assays used in Xenopus laevis, to our system. We tested the effects of adding mitotic centrosomes, Ran-GTP, or mitotic chromosomes. Lysate incubated without one of these cues did not assemble microtubule aggregates that were visible by fluorescence microscopy (not shown). Purified mitotic centrosomes nucleated microtubule asters that recruited both PARP-5a and NuMA to well-defined foci at their centers (Figure 2a). RanQ69L, a mutant of the small GTPase Ran that binds but does not hydrolyze GTP (Klebe et al., 1995; Palacios et al., 1996) also induced formation of asters that recruited PARP-5a and NuMA to their foci (Figure 2b). Minus ends, NuMA, and PARP-5a were not as precisely focused in these “Ran asters” as in mitotic centrosome-nucleated asters (cf. Figure 2, a and b), presumably because the Ran asters self-organize using motors and cross-linkers. RanQ69L is known to induce asters and spindles in mitotic Xenopus egg extract (Carazo-Salas et al., 1999; Wilde and Zheng, 1999), but to our knowledge this is the first demonstration that it can induce assembly of asters in a somatic cell lysate. Mitotic chromosomes also induced assembly of microtubule asters, and coincubation of mitotic chromosomes and centrosomes triggered assembly of structures that bore some morphological resemblance to mitotic spindles (Figure 2c). We conclude that this concentrated mitotic lysate system can assemble pole-like structures in vitro in response to known physiological cues and may be useful for investigating the mechanisms by which Ran-GTP and chromosomes trigger microtubule assembly during mitosis.
We next tested if aggregates of pADPr were capable of triggering spindle pole assembly in this system. As a physiological source of spindle pole pADPr, we used magnetic beads coated with mitotic PARP-5a self-modified with pADPr. To make these beads, we prepared dilute mitotic lysate from cells expressing GFP-PARP-5a and supplemented it with 1 mM NAD+ and 1 μM ADP-HPD (a PARG inhibitor) to promote pADPr synthesis. The GFP-PARP-5a, self-modified with pADPr, was recruited onto magnetic beads coated with anti-GFP IgG during the polymerization reaction, and the beads were collected and washed. Biochemical characterization of these beads is described below. As a control, we expressed a GFP fusion of catalytically inactive PARP-5a (GFP-PARP-5a PD) and purified it from lysate on magnetic beads using the same procedure. These beads are coated with PARP-5a polypeptide, but not with pADPr chains. Beads coated with catalytically inactive PARP-5a were unable to assemble microtubule asters in concentrated mitotic lysate (Figure 3, top left). Beads coated with mitotic PARP-5a from which pADPr chains were extended induced robust formation of microtubule asters (Figure 3, top right), suggesting that the presence of pADPr on PARP-5a is required for microtubule aster assembly around the beads. Asters assembled around the PARP-5a-pADPr beads were always located close to beads or bead aggregates, but the microtubules did not seem to emanate directly from the beads. Rather, the asters had distinct foci and appeared to associate laterally with the beads near these foci. To determine if the amount of pADPr on the beads affects microtubule aster assembly, we added increasing concentrations of NAD+ to the reaction (which already contained PARG inhibitor). Addition of 250 μM and 500 μM NAD+ resulted in a concentration-dependent increase in the number of microtubule asters found associated with the beads, and the microtubule density in each aster (Figure 3, bottom left and right, respectively). These data show that pADPr elongated from PARP-5a is capable of inducing assembly of pole-like microtubule asters in mitotic cytoplasm, arguing that it has a functional role in pole assembly in cells. We hypothesized that induction of microtubule asters by pADPr-coated beads depended on recruiting specific proteins from the mitotic lysate onto the pADPr polymers and set out to identify these proteins, as describe below.
PARP-5a Is More Active in Mitosis than Interphase
We first tested whether PARP-5a is cell cycle regulated. Dilute lysates were prepared from cells that were transfected with GFP-PARP-5a and either not arrested (I), or arrested in mitosis with S-trityl-l-cysteine (M). Both lysates were supplemented with NAD+ and PARG inhibitor. Magnetic beads coated with anti-GFP IgG were added, and the lysate was incubated at 25°C to promote simultaneous polymerization of pADPr and immunocapture of modified PARP-5a. Similar amounts of GFP-PARP-5a were recovered from I and M lysates, though the mitotic protein was smeared upward on the gel and thus appeared less abundant (Figure 4a, Coomassie-stained lanes). Immunoblotting for pADPr revealed that mitotic PARP-5a, and proteins that coimmunoprecipitate with it, were much more heavily modified with pADPr than interphase PARP-5a (Figure 4a, immunoblot). We suspect that a significant fraction of the pADPr-modified protein from the mitotic lysate is decorated with so much polymer that it does not enter the gel, because we noted a strong signal in the position corresponding to the well in the immunoblot. This experiment shows that PARP-5a is much more active in mitosis than interphase and that the mitotic form is suitable for isolation of pADPr-binding proteins. To our knowledge, this is the first demonstration that PARP-5a activity is cell cycle regulated, and our system will be useful for elucidating the mechanism of this regulation.
Identification of Mitotic pADPr-binding Proteins
To identify pADPr-binding proteins, beads coated with PARP-5a decorated with pADPr (similar to those in the M lanes in Figure 4a) were transferred to a second batch of lysate containing PARG inhibitor (mitotic or interphase), incubated at 4°C to recruit binding proteins, and washed with buffer of approximately physiological ionic strength. Bound proteins were eluted with hot sample buffer and resolved by SDS-PAGE. We tested the effect of varying factors such ionic strength of lysis and wash buffers, incubation time/temperature, extent of pADPr modification, and g-force during lysate clarification, so as to maximize the efficiency and specificity of the capture reaction. In Figure 4 we report data from our current protocol, acknowledging that further experimentation is required to define the affinity and specificity of candidate pADPr-binding proteins (see also Figure 5).
Mitotic lysate proteins that bound to pADPr are shown in Figure 4b, lane 3. To test which proteins bound specifically to the pADPr modification, as opposed to beads alone or PARP-5a alone, we treated the beads with a pADPr glycohydrolase and a wash, before SDS-PAGE (Figure 4b, lane 2). We used ARH3 for hydrolysis because it is easer to express in bacteria than PARG. Immunoblotting for pADPr confirmed that most of the polymer was removed by ARH3 treatment (Figure 4b, immunoblot). Hydrolysis of pADPr caused most of the binding proteins to be lost (Figure 4b, lane 2), indicating that pADPr is the component on the beads that recruits most of the binding proteins from mitotic lysate.
To evaluate which of the pADPr-binding proteins were mitosis specific, we prepared PARP-5a-pADPr beads in mitotic lysate and then added them to mitotic or interphase lysate supplemented with PARG inhibitor, followed by incubation at 4°C and washing (Figure 4c). Binding proteins were recovered in both cases, and the band pattern was fairly different, indicating that many of the candidate pADPr-binding proteins recovered in mitotic lysate are mitosis-specific, especially at molecular weights > 70 kDa. Some bands appeared to be common to both cell cycle states. These included three bands between 40 and 60 kDa that are the most abundant binding proteins in either extract. These bands contain abundant heat-shock protein (HSP) 70, tubulin, and actin by LC/MS and immunoblot (Table 1 and data not shown). A number of low-molecular-weight bands corresponding to ribosomal proteins by LC/MS (Table 1) are also common to both cell cycle states. We are suspicious that binding of abundant chaperone, cytoskeleton, and ribosomal proteins to pADPr is an artifact, for example, because of aggregation of the proteins in the lysate. These common bands were recovered in lower amounts from the interphase lysate, perhaps because the pADPr on the beads was slowly degraded in this lysate, whereas it continues to be synthesized in mitotic lysate.
Table 1.
Proteins that bind to pADPr in mitotic lysate from Figure 4, lane 3, identified by LC-MS
| Band number | Protein | Bandnumber | Protein | Bandnumber | Protein |
|---|---|---|---|---|---|
| 1 | PLEC1r | 6 | PPLr | 12 | DNAJA3, Actinr |
| 2 | AHNK | 7 | DXH9r, TKNS1BP1, LARP1, PHLDB2r, LMO7r | 13 | Ribosomal subunitsr |
| Heavy banda | Spectrinr, filaminr | 8 | HERC5, SND1r, HRPUr | 14 | Ribosomal subunitsr |
| 3 | DSPr | 9 | CLTC, IF3Ar | 15 | Ribosomal subunitsr |
| 4 | NuMA1r | 10 | HSP70r (several isoforms), PABPC3, DDX3Xr,PABPC4, XRCC6r, HNRNPMr | 16 | ADPRHL2 (ARH3) |
| 5 | Myosin II heavy chainsr | 11 | RL3r, DNAJA3 | 17 | ADPRHL2 (ARH3) |
Proteins are listed by gene symbol except in cases where multiple isoforms of the same protein or multiple proteins of the same type were identified. Proteins in one band are listed in decreasing order of number of peptides recovered. This experiment was performed three times¤ cutting out different numbers of gel bands, and proteins that were seen more than once in LC/MS runs are noted with a superscript r (r).
a The heavy band, which is marked with an asterisk (*) in Figure 4f and contains mostly spectrin and filamin from two other experiments, is included in this table.
In Figure 4d we show an experiment where we varied the drug used to promote mitotic arrest (STLC vs. nocodazole) and also tested the effect of omitting NAD+ and PARG inhibitor from the initial pADPr synthesis + immunocapture step in the protocol, which greatly reduces pADPr accumulation. Essentially identical proteins were recovered in lysate from cells arrested with either drug, indicating that the precise method of mitotic arrest does not matter and also indicating the reproducibility of the pADPr-binding protein profile when the incubation and wash protocols were kept constant. Omission of NAD+ and PARG inhibitor resulted in loss of most binding proteins, confirming that most of the binding proteins are recruited to pADPr on the beads, not to the PARP-5a polypeptide or the bead matrix.
As a specificity control, we compared proteins that bind to RNA and pADPr in mitotic lysate; RNA was chosen as a control polymer because it has similar chemical functionality. Figure 4e shows proteins that bound to biotinylated RNA on magnetic beads (lane 2) with pADPr-binding proteins (lane 4). Lane 3 in this gel shows the GFP-PARP-5a beads before incubation in the second batch of lysate for reference. RNA appeared to recruit some of the same proteins as pADPr, for example, the low-molecular-weight ribosomal proteins, but overall the profile of binding proteins was fairly different between the two polymers, at least as judged by SDS-PAGE bands. This suggests many of the mitotic pADPr-binding proteins are specific for that polymer and do not bind to any negatively charged polymer. However, subsequent LC/MS analysis (Table 1) showed than a significant fraction of pADPr-binding proteins are annotated as RNA-binding proteins, so the specificity issue remains to be addressed.
To identify proteins that bind to pADPr in mitotic lysate, we performed LC/MS on gel bands excised from the experiment shown in Figure 4e, lane 4, as indicated by the boxes on the gel lane (shown in Figure 4F). We avoided bands that seemed to be common to the RNA and pADPr lanes and also with an abundant high-molecular-weight band (indicated with an asterisk in Figure 4f) that we already knew to be mostly filamin and spectrin. The identity of abundant proteins in each gel band (three or more peptides identified) is indicated in Table 1. We performed several LC/MS experiments, and most of the proteins in Table 1 were observed in more than one experiment, as indicated with a superscript r in Table 1. Although all the proteins in Table 1 require pADPr to bind to the beads, several of them are of questionable specificity. In this category we include large cytoskeletal proteins (plectin, filamin, spectrin, desmoplakin, and myosin-II), ribosomes, HSP70, and HSP40. All of these are abundant and are common contaminants in immunopurification assays. Several RNA-binding proteins were recovered. This might reflect nonspecific binding or possibly an RNA-mimic function of pADPr.
On the basis of known biology of pADPr, we suspect that several of the bands in Table 1 are specific pADPr-binding proteins. These include: NuMA, which is implicated in spindle pole assembly; TNKS1BP1, which is a known PARP-5a–binding protein of unknown function (Seimiya and Smith, 2002); XRCC6 (aka KU70), which is part of the DNA damage detection/repair pathway (Boulton and Jackson, 1996) and may bind to pADPr generated by PARP-1 near DNA strand breaks; and ADPRHL2 (aka ARH3), an enzyme with pADPr hydrolytic activity—the same one we used for hydrolysis of the polymer in Figure 4a. Of these proteins, the most obvious candidate for a role in spindle pole assembly was NuMA. This protein was found in all LC/MS analyses of pADPr-binding proteins and was confirmed in multiple Western blot experiments (e.g., see Figure 5). NuMA was not found in the RNA-binding proteins from mitotic lysate as assayed by Western blot (not shown), so it binds pADPr specifically. The amount of NuMA recovered in the binding fraction was variable; in most experiments it was a Coomassie-stained band, though in the particular experiment shown in Figure 4e, it was less abundant for unknown reasons.
Dependence of NuMA Binding on the pADPr Modification Level
NuMA has been previously shown to bind to PARP-5a independent of pADPr (Sbodio and Chi, 2002); therefore, its recovery in the pADPr-binding proteins might be due to a direct interaction with PARP-5a polypeptide. To test this and also the effect of varying the amount of pADPr on the beads, we titrated NAD+ into mitotic lysate and used Western blotting to measure the amount NuMA that bound. Magnetic beads coated with anti-GFP were added to lysate containing GFP-PARP-5a, and varying amounts of NAD+. PARP inhibitor was added to all batches except the zero NAD+. Beads were washed, eluted with sample buffer, and immunoblotted for pADPr, NuMA, and CH-TOG (Figure 5). CH-TOG, also known as XMAP215 and CKAP5, is enriched at spindle poles and has an important role in microtubule nucleation and elongation (Becker et al., 2003). It was indentified by LC/MS as a potential pADPr-binding protein in early experiments, but after refining our protocol it was less evident. Increasing concentrations of NAD+ resulted in increased levels of pADPr on the beads, as expected (Figure 5, pADPr immunoblot). It is important to note that all known pADPr antibodies fail to detect polymers shorter than ∼10 ADPr residues. Given that the PARP-5a used in our assay is purified from mitotic extract, it is probable that a relevant amount if pADPr is associated with PARP-5a in the 0 μM NAD+, but is not visualized on the pADPr immunoblot. A small amount of NuMA was recruited to the beads without added NAD+, probably by direct protein–protein interaction with PARP-5a or perhaps due to binding to undetected pAPDr. When NAD+ and PARG inhibitor were added, the amount of NuMA recovered was greatly increased, suggesting that this protein is mostly recruited by interaction with pADPr. An alternative interpretation is that the NuMA must be covalently modified with pADPr in solution to bind, but we consider this less likely because the NuMA band was not smeared upward on the gel and also from the pure protein-binding data below. NuMA recruitment plateaued at the lowest NAD+ concentration suggesting a high affinity for pADPr. CH-TOG was also recruited to the beads; however, recruitment increased with increasing concentrations of NAD+, suggesting its binding affinity is lower than NuMA.
A Recombinant NuMA Fragment Binds Directly to pADPr
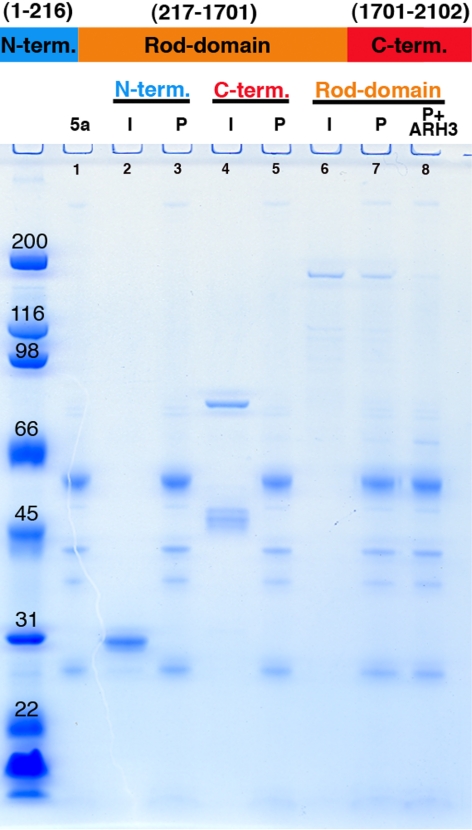
The foregoing experiment did not reveal whether NuMA recruitment to pADPr-coated beads involved direct or indirect binding interactions or whether NuMA itself must be covalently modified by pADPr to bind to PARP-5a beads in mitotic lysate. To address these questions and to begin to narrow down the pADPr-binding site, we turned to bacterially expressed NuMA. Prokaryotes do not contain PARPs, so recombinant proteins expressed in them are free of pADPr modifications. Full-length NuMA is highly insoluble as a pure protein (Harborth et al., 1999), so we split it into three fragments for expression and purification (Figure 6, top). These approximately correspond to the domain organization of NuMA, representing the C- and N-terminal globular domains, and to a central, coiled-coil rod domain. Each segment was expressed in Escherichia coli as a His6 fusion, and purified. All three fragments were soluble. PARP-5a beads modified with pADPr were prepared in mitotic lysate as in Figures 4 and 5 and washed with 0.5 M NaCl to remove the majority of binding proteins from the lysate, including endogenous NuMA. Removal of NuMA and other binding proteins from the beads was confirmed by SDS-PAGE and immunoblots (not shown). The washed PARP-5a-pADPr beads were then incubated with the recombinant NuMA fragments in buffer, collected, washed, and analyzed by SDS-PAGE. The rod domain bound to the beads in this assay, whereas the N- and C-terminal domains did not (Figure 6, gel). To determine if rod domain binding to GFP-PARP-5a-pADPr was specific for the pADPr component, we added ARH3 after incubating the rod domain with beads. pADPr hydrolysis dramatically reduced the amount of rod domain bound to the beads (Figure 6, gel, lane 8). This experiment suggests that NuMA binds directly to pADPr, by an interaction involving its coiled-coiled rod domain. Thus, a protein known to play a central role in spindle pole assembly binds noncovalently to pADPr, both in mitotic lysate and as a pure, expressed fragment.
Figure 6.
NuMA rod domain binds directly to p(ADPr). Molecular weights are shown at left. NuMA was expressed in bacteria as three nonoverlapping fragments tagged with His6. Top, these corresponded approximately to the main structural (terminal) domains in NuMA: N-term., aa 1–216; C-term., aa1701–2102; and Rod domain, aa. 217-1701. Bottom, fragments were incubated with PARP-5a-pADPr (5a) bound to magnetic beads and then were washed and analyzed by SDS-PAGE. In all cases I refers to input and p to the washed PARP-5a-input samples. Only the Rod domain bound to the beads. For lane 8, beads from a Rod domain incubation were treated with the glycohydrolase ARH3 and washed. Note Rod domain binding to PARP-5a-pARPr was lost, indicating it binds to the pADPr on the beads and not to the beads themselves or to PARP-5a polypeptide.
Previously published reports identified a direct binding interaction between NuMA and PARP-5a. This binding was shown to be mediated through the RXXPDG1748 motif on the C-terminal domain of NuMA (Sbodio and Chi, 2002). Our results did not identify a similar binding interaction between the C-terminal domain of NuMA and PARP-5a. We believe the discordant results are due to differences in pADPr modification on the C-terminal NuMA fragments used in the two assays. Our binding assays utilized unmodified recombinant fragments, whereas previously published reports used a cotransfection/copurification assay in BOSC cells; FLAG-tagged PARP-5a and GST-tagged NuMA fragments (aa 1605–2016) were transiently transfected, and then binding between the two polypeptides was analyzed using GST or FLAG tag purification followed by immunoblotting (Sbodio and Chi, 2002). Both PARP-5a and the C-terminal NuMA fragment were transiently expressed in their assays. Because NuMA is a known PARP-5a substrate, it is possible that their C-terminal NuMA fragment was modified by pADPr. This could have resulted in binding to PARP-5a directly or via cross-linking through other pADPr-binding proteins.
DISCUSSION
Here, we showed that beads coated with PARP-5a modified with pADPr induce assembly of microtubule asters in a concentrated mitotic lysate system, we show PARP-5a is much more active in mitosis than interphase and that p(ADPR) elongated from PARP-5a recruits multiple binding proteins in mitotic lysate, almost entirely through the pADPr polymer. One of these binding proteins is NuMA, which appears to bind directly to the polymer through its rod domain, as assayed with bacteria-expressed NuMA fragments.
Our finding that PARP-5a is much more active as a PARP in mitosis than interphase (Figure 4a) has important implication for its regulation and function of this protein. Several functions have been proposed for this protein, but both its localization and RNAi knockdown phenotype point to a role in spindle pole assembly. Finding that its PARP activity is strongly activated during mitosis supports a primary function in spindle assembly, without ruling out other functions such as telomere and Golgi maintenance. An obvious candidate regulatory mechanism is cell cycle–specific phosphorylation. PARP5 is phosphorylated in mitosis, in part by GSK3 (Yeh et al., 2006). Whether its PARP activity is positively regulated by GSK3 or Cdk1 phosphorylation has yet to be tested.
Much of our effort went into developing methods for identifying proteins that bind noncovalently and specifically to pADPr in mitotic cells and might function together with pADPr in spindle pole assembly. We took the approach of incubating PARP-5a modified with pADPr in mitotic lysate and identifying proteins that bound to it. This approach has the advantage that the pADPr is generated by the pole-relevant PARP in mitotic lysate and thus presumably has a physiological pADPr structure. We do not know how the amount of pADPr attached to the PARP-5a in lysate compares to that in spindle poles in living cells. This is difficult to control in lysates, because PARG tends to hydrolyze pADPr, even when a PARG inhibitor was added, so we needed to actively promote PARP activity to observe PARP-5a modification and binding protein recruitment. NuMA was recruited efficiently even at relatively low levels of pADPr modification, unlike another candidate binding protein CH-TOG (Figure 5). This suggests a relatively high-affinity interaction between NuMA and pADPr that is independent of the exact chain length or local density of pADPr.
We identified a large number of proteins that bind to pADPr in mitotic lysates (Figures 4 and 5 and Table 1). If a significant fraction of these are physiologically relevant, the implication is that the polymer interacts with multiple cellular systems. An important issue is the specificity of the binding interaction. All of our candidate binding proteins were specific for pADPr by the criterion that they were absent when the polymer was deliberately hydrolyzed (Figures 4b and 6) or lost by incubation in lysate without NAD+ (Figures 4d and 5, no NAD+ lanes). However, the pADPr-coated beads recruited large amounts of several proteins that are abundant and potentially sticky, such as large cytoskeletal proteins, ribosomes, and HSC70/40. The specificity of these interactions are suspect. We systematically varied experimental conditions in an attempt to prevent their recruitment, but were never fully successful. Thus we cannot rule out the possibility that these interactions are in fact specific, and pADPr physiologically interacts with several abundant cellular components, though we dis-favor this interpretation. We chose RNA as a comparison polymer for addressing specificity, because its chemical functionality resembles pADPr. Although the overall binding profile was quite different for the two polymers (Figure 4E), some proteins in lysates did appear to bind to both by SDS-PAGE mobility, and some of the candidate pADPr-binding bands identified by LC-MS are known RNA-binding proteins (Table 1). It is possible that proteins that bind to both pADPr and RNA are binding to pADPr nonspecifically. However, it is also possible that pADPr can acts as a RNA mimetic under some circumstances, and this dual-binding specificity is physiologically relevant. RNA is required for spindle assembly (Blower et al., 2005), and it is possible that some proteins in the spindle interact with both RNA and pADPr. Our list of candidate pADPr-binding proteins in mitotic cytoplasm has little overlap with recently published lists of candidate pADPr-binding proteins from other groups (Karras et al., 2005; Ahel et al., 2008; Gagne et al., 2008). This might reflect that fact that mitotic pADPr-binding proteins appear mostly distinct from interphase binding proteins (Figure 4c) and also the different techniques used. Biochemical characterization of pADPr-binding proteins is still in its infancy, and elucidating the biological function of the polymer in different contexts will require considerable effort on this front. Elucidating molecular recognition of pADPr by binding proteins will be easiest in cases where binding can be mapped to a specific motif or domain, such as the recently identified pADPr-binding zinc finger (PDZ domain) in CHFR (Ahel et al., 2008). In the case of NuMA, where pADPr binding was preliminarily mapped to a long coiled-coil rod domain (Figure 6), it will be more difficult.
NuMA is an excellent candidate for mediating aster assembly in extracts, and spindle pole focusing in cells, by binding noncovalently to pADPr. It is known to have aster-promoting activity in mitotic HeLa lysate (Gaglio et al., 1995) and meiotic Xenopus egg extract (Nachury et al., 2001). In cells, it is known to have minus-end focusing and spindle pole-assembly activity (Fant et al., 2004). NuMA is multimeric. It may have as many as six microtubule-binding sites per assembled unit, and these units tend to polymerize (Harborth et al., 1999). Previous models for its function in aster and pole assembly proposed that it uses these microtubule binding sites to cross-link microtubules at the poles, perhaps acting in concert with cytoplasmic dynein (Merdes et al., 1996; Fant et al., 2004). Finding that it binds pADPr noncovalently and is also covalently modified by pADPr during mitosis (Chang et al., 2005a; Chang et al., 2005b) suggests the model in Figure 7. In this model, NuMA and minus-end–directed motors are together sufficient to assembly local clusters of minus ends. PARP-5a is recruited to these clusters by interaction with NuMA, where it modifies itself and NuMA with pADPr chains. These chains grow outward and are captured noncovalently by NuMA on nearby clusters. This helps stitch clusters together over longer distances and thus helps to define two discrete poles (Figure 7). Why might evolution select pADPr for this proposed long-distance cross-linking function? One possibility is structural; pADPr may make longer, and more multivalent, cross-links than would be possible using a protein. Another is that rapid turnover of pADPr by synthesis and hydrolysis is important. This feature allows the cell to synthesize pADPr rapidly at the onset of mitosis, when PARP-5a is turned on (Figure 4a) and to degrade it again rapidly at anaphase. Spindle poles fragment in late anaphase (Rusan and Wadsworth, 2005), perhaps because they loose the cross-liking activity of pADPr. Although hydrolysis of pADPr is usually mediated by PARG, our finding ARH3 as a pADPr-binding protein in mitotic lysate (Table 1) suggest this enzyme may also participate. We speculate that rapid pADPr turnover at spindle poles by synthesis-degradation might allow for some kind of adaptive control of minus-end cross-linking that receives information from spindle structure, and in response controls pADPr levels, to ensure robust assembly of exactly two poles.
Figure 7.
Model for pADPr function at spindle poles. Microtubules are shown as green lines with their polarity indicated by arrowheads. NuMA (yellow pentagon) and minus-directed motors (not shown) are sufficient to locally cluster minus ends. PARP-5a (blue rectangle) is recruited to minus-end clusters by protein–protein interaction with NuMA. Polymerization of pADPr (red lines) from priming sites on PARP-5a and NuMA recruits more NuMA, which promotes further condensation of minus-end clusters into two poles. Note that this diagram is drawn to illustrate a conceptual hierarchy of interactions, not a temporal sequence of events. The blow-up illustrates that the pADPr chains attach covalently to PARP-5a and NuMA (red dots at end of polymer) and interact noncovalently with NuMA.
ACKNOWLEDGMENTS
We thank Puck Ohi (Vanderbilt University) for mitotic chromosomes, Aaron Groen (Yale University) for RanGTP-Q69L, Duane Compton (Dartmouth Medical School) for the full-length NuMA clone and NuMA antibodies, and Micha Rape and the members of the Mitchison lab for experiment discussions. P.C. is a Rita Allen and Kimmel Foundation Scholar, and a Howard S. and Linda B. Stern Career Development Assistant Professor. This work was supported National Institutes of Health (NIH) Grants GM39565 (T.J.M.) and NIH 5F32GM070090-02 (P.C.).
Abbreviations used:
- pADPr
poly(ADP-ribose)
- PARP
poly(ADP-ribose) polymerase
- PARG
poly(ADP-ribose) glycohydrolase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-06-0477) on September 16, 2009.
REFERENCES
- Ahel I., Ahel D., Matsusaka T., Clark A. J., Pines J., Boulton S. J., West S. C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Becker B. E., Romney S. J., Gard D. L. XMAP215, XKCM1, NuMA, and cytoplasmic dynein are required for the assembly and organization of the transient microtubule array during the maturation of Xenopus oocytes. Dev. Biol. 2003;261:488–505. doi: 10.1016/s0012-1606(03)00330-0. [DOI] [PubMed] [Google Scholar]
- Blower M. D., Nachury M., Heald R., Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Boulton S. J., Jackson S. P. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Bradbury S., Stoward P. J. The specific cytochemical demonstration in the electron microscope of periodate-reactive mucosubstances and polysaccharides containing vic-glycol groups. Histochemie. 1967;11:71–80. doi: 10.1007/BF00326613. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R. E., Guarguaglini G., Gruss O. J., Segref A., Karsenti E., Mattaj I. W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Chang P., Coughlin M., Mitchison T. J. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 2005a;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- Chang P., Jacobson M. K., Mitchison T. J. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- Chang W., Dynek J. N., Smith S. NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem. J. 2005b;391:177–184. doi: 10.1042/BJ20050885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi N. W., Lodish H. F. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- Compton D. A., Szilak I., Cleveland D. W. Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J. Cell Biol. 1992;116:1395–1408. doi: 10.1083/jcb.116.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBonis S., Skoufias D. A., Lebeau L., Lopez R., Robin G., Margolis R. L., Wade R. H., Kozielski F. In vitro screening for inhibitors of the human mitotic kinesin Eg5 with antimitotic and antitumor activities. Mol. Cancer Ther. 2004;3:1079–1090. [PubMed] [Google Scholar]
- Fant X., Merdes A., Haren L. Cell and molecular biology of spindle poles and NuMA. Int. Rev. Cytol. 2004;238:1–57. doi: 10.1016/S0074-7696(04)38001-0. [DOI] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Compton D. A. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J. P., Isabelle M., Lo K. S., Bourassa S., Hendzel M. J., Dawson V. L., Dawson T. M., Poirier G. G. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho S. A., Kwon M., Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- Gosti-Testu F., Marty M. C., Berges J., Maunoury R., Bornens M. Identification of centrosomal proteins in a human lymphoblastic cell line. EMBO J. 1986;5:2545–2550. doi: 10.1002/j.1460-2075.1986.tb04533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J., Wang J., Gueth-Hallonet C., Weber K., Osborn M. Self assembly of NuMA: multiarm oligomers as structural units of a nuclear lattice. EMBO J. 1999;18:1689–1700. doi: 10.1093/emboj/18.6.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama K., Nemoto Y., Ueda K., Hayaishi O. Purification and characterization of poly(ADP-ribose) glycohydrolase. Different modes of action on large and small poly(ADP-ribose) J. Biol. Chem. 1986;261:14902–14911. [PubMed] [Google Scholar]
- Kanai M., Uchida M., Hanai S., Uematsu N., Uchida K., Miwa M. Poly(ADP-ribose) polymerase localizes to the centrosomes and chromosomes. Biochem. Biophys. Res. Commun. 2000;278:385–389. doi: 10.1006/bbrc.2000.3801. [DOI] [PubMed] [Google Scholar]
- Karras G. I., Kustatscher G., Buhecha H. R., Allen M. D., Pugieux C., Sait F., Bycroft M., Ladurner A. G. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe C., Bischoff F. R., Ponstingl H., Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 1995;34:639–647. doi: 10.1021/bi00002a031. [DOI] [PubMed] [Google Scholar]
- Kwon M., Godinho S. A., Chandhok N. S., Ganem N. J., Azioune A., Thery M., Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack G. J., Compton D. A. Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. Proc. Natl. Acad. Sci. USA. 2001;98:14434–14439. doi: 10.1073/pnas.261371298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J. D., Cleveland D. W. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Nachury M. V., Maresca T. J., Salmon W. C., Waterman-Storer C. M., Heald R., Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Oka S., Kato J., Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- Ono T., Kasamatsu A., Oka S., Moss J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc. Natl. Acad. Sci. USA. 2006;103:16687–16691. doi: 10.1073/pnas.0607911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I., Weis K., Klebe C., Mattaj I. W., Dingwall C. RAN/TC4 mutants identify a common requirement for snRNP and protein import into the nucleus. J. Cell Biol. 1996;133:485–494. doi: 10.1083/jcb.133.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan N. M., Wadsworth P. Centrosome fragments and microtubules are transported asymmetrically away from division plane in anaphase. J. Cell Biol. 2005;168:21–28. doi: 10.1083/jcb.200409153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbodio J. I., Chi N. W. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J. Biol. Chem. 2002;277:31887–31892. doi: 10.1074/jbc.M203916200. [DOI] [PubMed] [Google Scholar]
- Schreiber V., Dantzer F., Ame J. C., de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Seimiya H., Smith S. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182) J. Biol. Chem. 2002;277:14116–14126. doi: 10.1074/jbc.M112266200. [DOI] [PubMed] [Google Scholar]
- Slama J. T., Aboul-Ela N., Goli D. M., Cheesman B. V., Simmons A. M., Jacobson M. K. Specific inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J. Med. Chem. 1995;38:389–393. doi: 10.1021/jm00002a021. [DOI] [PubMed] [Google Scholar]
- Smith S., de Lange T. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 2000;10:1299–1302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- Ueda K., Oka J., Naruniya S., Miyakawa N., Hayaishi O. Poly ADP-ribose glycohydrolase from rat liver nuclei, a novel enzyme degrading the polymer. Biochem. Biophys. Res. Commun. 1972;46:516–523. doi: 10.1016/s0006-291x(72)80169-4. [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Vernos I., Mitchison T. J., Karsenti E., Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- Wilde A., Lizarraga S. B., Zhang L., Wiese C., Gliksman N. R., Walczak C. E., Zheng Y. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat. Cell Biol. 2001;3:221–227. doi: 10.1038/35060000. [DOI] [PubMed] [Google Scholar]
- Wilde A., Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Wong R. W., Blobel G., Coutavas E. Rae1 interaction with NuMA is required for bipolar spindle formation. Proc. Natl. Acad. Sci. USA. 2006;103:19783–19787. doi: 10.1073/pnas.0609582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh T. Y., Sbodio J. I., Chi N. W. Mitotic phosphorylation of tankyrase, a PARP that promotes spindle assembly, by GSK3. Biochem. Biophys. Res. Commun. 2006;350:574–579. doi: 10.1016/j.bbrc.2006.09.080. [DOI] [PubMed] [Google Scholar]