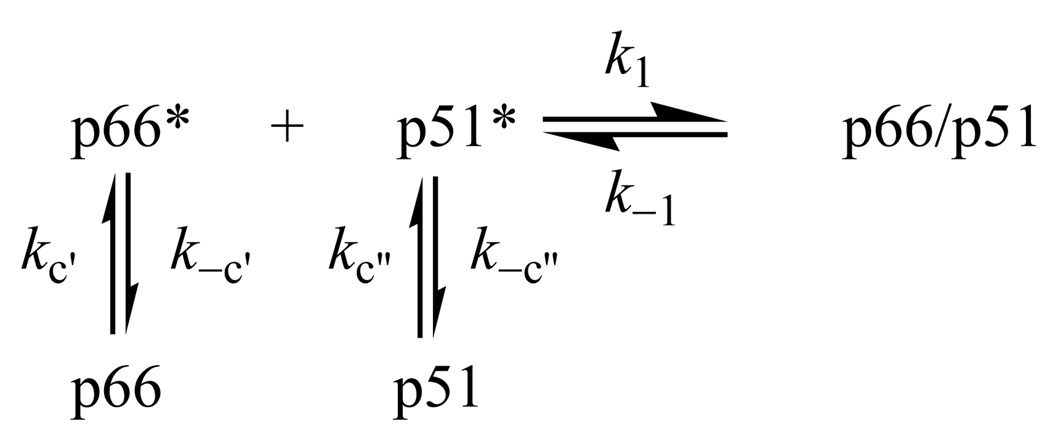Abstract
The biologically active form of HIV-1 reverse transcriptase is the p66/p51 heterodimer. The process of maturation of the heterodimer from precursor proteins is poorly understood. Previous studies indicated that association of p66 and p51 is very slow. Three techniques, a pre-steady-state activity assay, intrinsic tryptophan fluorescence, and a FRET assay, were used to monitor the dimerization kinetics of RT. Kinetic experiments were carried out with purified p66 and p51 proteins in aqueous buffer. All three techniques gave essentially the same results. The dissociation kinetics of p66/p51 were first-order with rate constants kdiss of ~ 4 × 10−6 s−1 (t1/2 = 48 h). The association kinetics of p66 and p51 were concentration dependent with second-order rate constants kass of ~ 1.7 M−1s−1 for the simple bimolecular association reaction. The implications of slow dimerization of p66/p51 for the maturation process are discussed. A reaction-controlled model invoking conformational selection is proposed to explain the slow protein-protein association kinetics.
HIV-11 reverse transcriptase is a critical player in the life cycle of HIV, the virus that causes AIDS. This multifunctional enzyme catalyzes three reactions: RNA- and DNA-dependent DNA polymerization and RNA hydrolysis. RT is an asymmetric heterodimer composed of two subunits, p66 and p51 (2). The structural asymmetry is quite astonishing considering that the two subunits are products of the same gene and exhibit identical N-terminal amino acid sequences (3). The p66 subunit contains both a polymerase and an RNase H domain. The polymerase domain comprises four subdomains: fingers, palm, thumb, and connection. The p51 subunit contains the same four N-terminal subdomains as p66, but lacks the C-terminal RNase H domain (4). The four subdomains common to the two subunits have different orientations relative to one another in the RT structure (5). The p66 subunit is described as having a flexible “open” conformation which contains the polymerase active site (6), while the p51 subunit has a compact “closed” conformation that conceals the active site residues, making it catalytically inactive (7). The subunit interactions are also asymmetric; the interfaces of p66 and p51 use completely different amino acid residues. Even contacts made between the connection subdomains of each subunit use entirely separate surfaces (5). Two models have been proposed for heterodimer formation: a concerted model where p66 and p51 are cleaved by HIV protease from separate gag-pol precursors and a sequential model where the RNase H domain is cleaved from one subunit of a p66/p66 homodimer intermediate (8). Further investigation of these subunit interactions is vital to understanding the mechanism of RT heterodimer maturation.
Previous studies of the dimerization kinetics of RT, using a combination of polymerase activity and fluorescence, proposed that subunit association is a two-step process (Scheme 1). The initial phase involves a rapid bimolecular association of p66 and p51 (104 M−1s−1) to form an inactive heterodimeric intermediate p66*p51, followed by slow isomerization (10−5 s−1) to the catalytically active heterodimer p66/p51 (9–11). Subunit dissociation was monophasic with a dissociation rate constant on the order of 10−3 s−1 (11, 12). The above studies achieved subunit dissociation of RT by addition of acetonitrile and subsequent subunit association by dilution of the resulting monomeric subunits into aqueous buffer. However, RT dissociation in the absence of acetonitrile was considerably slower (13), suggesting that acetonitrile may have an effect on the dimerization kinetics.
Scheme 1.
Two-Step Dimerization Mechanism
This paper presents the dimerization kinetics of RT in aqueous solution using separately prepared p66/p51, p66, and p51 proteins. Our prior thermodynamic study of RT subunit equilibria allows us to design kinetic experiments without resorting to organic solvent (1). With knowledge of homo- and heterodimer dissociation constants, reactions are set up at appropriate protein concentrations to monitor RT heterodimer dissociation or association. Three complementary techniques, including a pre-steady-state activity assay, intrinsic tryptophan fluorescence, and FRET, are used to monitor the dimerization kinetics of RT. These three techniques report protein structure in different parts of the enzyme. The implications of these findings for the maturation of RT heterodimer are discussed.
EXPERIMENTAL PROCEDURES
Materials
A488 C5-maleimide and QSY9 C5-maleimide were purchased from Invitrogen (Eugene, OR). Oligodeoxynucleotides were purchased from Operon Technologies (Alameda, CA). dCTP (>99% purity) was from Pharmacia (Peapack, NJ). [γ-33P]ATP was from PerkinElmer Life Sciences (Boston, MA). Biochemical reagents were purchased from Roche Applied Science (Indianapolis, IN). Other chemicals were purchased from Sigma Chemicals (St. Louis, MO) and Fisher Scientific (Pittsburgh, PA). RT buffer D is 0.05 M Tris (RNase, DNase free; pH 7.0), 25 mM NaCl, 1 mM EDTA, and 10% (v/v) glycerol (molecular biology grade redistilled). RT buffer D used for kinetic experiments monitored by enzymatic activity and tryptophan fluorescence also contained 1 mM TCEP; the final pH was 7.0.2 RT reaction buffer is 0.05 M Tris (pH 7.8), 0.01 M MgCl2, 0.08 M KCl, and 5 mM DTT. PBS buffer is 137 mM NaCl, 11.9 mM phosphates (pH 7.4), and 2.7 mM KCl. TBE buffer is 0.089 M Tris, 0.089 M boric acid (pH 8.0), and 2 mM EDTA. TE buffer is 0.01 M Tris (pH 8.0) and 1 mM EDTA. Loading buffer is 90% formamide, 10% 10X TBE, 0.025% bromophenol blue, and 0.025% xylene cyanol. The acrylamide solution in TBE contains 20% acrylamide:bis-acrylamide (19:1) and 7.8 M urea.
Synthesis of N-Acetylcysteine-A488
N-acetylcysteine (0.5 mg, 81.1 mmol) was placed in a 1.5 mL microcentrifuge tube and dissolved in 0.5 mL methanol. A488 C5-maleimide (1.5 equiv, 123.0 mmol) was added and the reaction was mixed for 1.5 h at room temperature. Upon completion of the reaction, the volume was reduced under vacuum and the product was purified via silica gel flash chromatography. The crude reaction mixture was loaded onto an 8 cm × 0.5 cm column equilibrated with DCM. The column was washed with four column volumes of 4:1 DCM:methanol to elute any residual starting materials and then with 1:2 DCM:methanol to obtain the clean product.
Protein Preparation
RT proteins were purified as described previously (1, 14). Protein concentration is determined from absorbance at 280 nm (1, 15). All RT protein concentrations are expressed as monomer.
RT proteins with a single cysteine at position 3 were prepared from two plasmids containing the S3C/C38V/C280S triple mutation. The S3C mutation was introduced into a plasmid containing the C38V/C280S double mutation using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA): p6H RTC38V/C280S for p66 and p6H RT51C38V/C280S for p51 (1). Mutagenic oligonucleotide primer sequences for the S3C mutation were: forward, 5'–GTACAGTCTCAATAGGGCAAATGGGAAGCTGGGATC–3'; reverse, 5'–GATCCCAGCTTCCCATTTGCCCTATTGAGACTGTAC–3'. All mutations were confirmed by DNA sequencing at Cleveland Genomics (Cleveland, OH).
Mutated proteins p66S3C/C38V/C280S and p51S3C/C38V/C280S were labeled with either A488 or QSY9 maleimide using the following procedure. The protein was treated with a 4-fold molar excess of DTT overnight at 5 °C followed by dialysis into PBS for 3 h. A fresh stock solution of maleimide probe was prepared in dimethylformamide. The protein was reacted for 1 h at room temperature three times with a 10-fold excess of A488 maleimide or a 5-fold excess of QSY9 maleimide. Labeled protein was then affixed to 0.5 mL Ni-NTA Superflow resin (Qiagen, Valencia, CA) in a 3 mL disposable column (Pierce, Rockford, IL) and washed with PBS containing 25 mM imidazole until the absorbance at 495 nm < 0.005. The column was capped on the bottom and 0.5 mL of PBS containing 0.3 M imidazole was added; after gentle agitation, the column was allowed to sit for 20 min. Labeled protein was eluted with PBS containing 0.3 M imidazole, concentrated against sucrose, and dialyzed into RT buffer D. Labeling stoichiometry was estimated from absorption spectra using extinction coefficients ε(495) = 7.1 × 104 M−1cm−1 for A488 and ε(562) = 8.8 × 104 M−1cm−1 for QSY9 (16, 17). Molar ratios of protein to probe were 1.0–1.08. Labeling specificity was confirmed by tandem mass spectrometry using a LCQ Deca XP Max ion trap mass spectrometer (Thermo Finnigan, San Jose, CA).
Pre-Steady-State Activity Assay
RT dimer concentration was determined by active-site titration at saturating DNA substrate concentration. The 25/36-mer random sequence DNA described previously was used as substrate (18). The oligonucleotides were purified; the duplex was annealed, purified, and 5’-end labeled with [γ-33P]ATP (19). DNA concentration was determined by absorbance at 260 nm (15). 10 µL of RT solution was mixed with 90 µL of RT reaction buffer containing 1 µM DNA substrate and 0.1 mM dCTP at 37 °C. 5 µL aliquots were removed and mixed with 5 µL of 0.1 M EDTA to quench the reaction. Time points were taken at 20 s intervals for the first 4 min and at 1 min intervals for an additional 4 min. The remaining reaction mixture was left overnight at 37 °C to go to completion and a final time point was taken to determine 3’-OH concentration. 10 µL of loading buffer was added to the quenched reaction mixtures and products were separated on a 20% denaturing polyacrylamide gel. The gel was exposed on a phosphorimaging screen (Bio-Rad Laboratories, Hercules, CA); the resulting image was quantified using a Storm Phosphorimager (Molecular Dynamics, Piscataway, NJ) and OptiQuant software (Packard Instrument Co., Meriden, CT); and the reaction was quantified as described (18). The time course of dCMP addition y was fit by linear regression to
| (1) |
where B is the burst amplitude at t = 0 and kss is the steady-state rate. The burst amplitude is the active enzyme concentration in dimer units, because one heterodimer binds per labeled DNA; [p66/p51] = 2B, where [p66/p51] is in monomer units.
Dissociation and association kinetics of p66/p51 were measured in RT buffer D at 5 °C. Dissociation reactions were started by 200-fold dilution of a 10 µM solution of p66/p51 to 50 nM, so that reassociation can be ignored in the data analysis. After dilution, 10 µL aliquots were assayed at 0.17, 0.5, 1, 2, 3, and 6 h; every 6 h to 36 h; every 12 h to 96 h; and then every 24 h to 10 days. The burst amplitudes B(t) at various times t after dilution of p66/p51 were fit to
| (2) |
where kdiss is the dissociation rate constant and the Ci are constants. Association reactions were started by mixing equal volumes of 1 µM p66 with either 1 or 5 µM p51; 10 µL aliquots of the reaction mixture were assayed at 1 or more hour intervals up to 10 days. At early times, the bimolecular association p66 + p51 → p66/p51 can be assumed to be an irreversible reaction and an association rate constant kass can be calculated from the slope of the integrated rate equations. For equal initial concentrations of [p66]0 and [p51]0,
| (3) |
and for unequal initial concentrations,
| (4) |
An observed rate constant kobs was obtained by fitting B(t) at time t after mixing p66 and p51 to
| (5) |
where C is a constant. The association kinetics are exponential only under pseudo first-order conditions.
Steady-State Fluorescence
Absorbance was measured on a Cary 3E UV–Vis spectrophotometer at 5 °C. Fluorescence was measured on a PC1 Photon Counting spectrofluorometer (ISS, Champaign, IL) in ratio mode under magic angle conditions using 4-nm excitation and 16-nm emission band-pass. Temperature was maintained at 5 °C by a TC 425 Peltier unit (Quantum Northwest, Liberty Lake, WA) and the sample compartment was flushed with nitrogen to prevent condensation. Samples were placed in 45 µL quartz cells with 3 mm path length (Starna Cells, Inc., Atascadero, CA). Absorbance at the long wavelength maximum was < 0.3 to avoid inner filter effects. Fluorescence quantum yields Φ were measured relative to fluorescein in 0.1 M NaOH at 450-nm excitation wavelength, 5 °C. A value of 0.95 at 22 °C was used for the quantum yield of fluorescein (20).
Dissociation and association kinetics of p66/p51 in RT buffer D were monitored by fluorescence using Vinci 1.6.SP7 software (ISS, Champaign, IL). Fast kinetic intensity data were collected from samples and a reference fluorophore every 5 s (signal averaged over 1 s) for the first 2 h. This was followed by one 16-h period of slow data acquisition every 5 min (signal averaged over 10 s) and subsequent 24-h periods for 6–10 days. Reactions of samples containing two labeled subunits were run simultaneously with a control reaction containing one A488-labeled subunit and one unlabeled subunit, and the background fluorescence from the control reaction was subtracted. Fluorescence intensity F = Is / Ir was calculated from the ratio of sample intensity Is to reference intensity Ir to correct for instrumental drift.
Intrinsic tryptophan fluorescence was measured at 295-nm excitation and 340-nm emission wavelength using NATA in water as reference. A488 fluorescence was measured at 450-nm excitation and 540-nm emission wavelength using fluorescein in 0.1 M NaOH as reference. Dissociation reactions were started by 200-fold dilution of a 10 µM solution of unlabeled or labeled p66/p51. Association reactions were started by mixing equal volumes of 1 µM solutions of unlabeled or labeled p66 with 1–5 µM solutions of unlabeled or labeled p51. Samples were mixed in the cell and placed in the fluorometer within 5 s. The fractional fluorescence increase or decrease due to dissociation or association of RT was fit to an exponential function.
| (6a) |
| (6b) |
where F0 is intensity at t = 0, F(t) is intensity at time t, and F∞ is intensity of the last time point. Here too, the association kinetics are exponential only under pseudo first-order conditions.
Time-Resolved Fluorescence
Fluorescence lifetimes were measured by time-correlated single photon counting as described (21). Decay data were collected at 450-nm excitation and 540-nm emission wavelength in 1024 channels of 18 and 24 ps/channel. Intensity decays I(t) with amplitudes αi and lifetimes τi were fit to sums of exponentials.
| (7) |
Decay curves on different time scales were deconvolved by global analyses with the lifetimes linked. Amplitude-weighted lifetimes τ̄ are calculated from
| (8) |
Fractional intensities fi of individual decay components were calculated from lifetime data and the relative steady-state intensity F(540) at 540 nm.
| (9) |
RESULTS
RT subunits undergo three coupled dimerization reactions in solution:
| (10a) |
| (10b) |
| (10c) |
with Kd(p66/p51) = 310 nM, Kd(p66/p66) = 4.2 µM, and Kd(p51/p51) = 230 µM (1).2 The relative concentrations of the three dimeric species must be taken into consideration when designing kinetic experiments. The RT concentrations can be calculated from eqs 10a–10c using Mathematica as described previously (1).
Kinetics Monitored by Enzymatic Activity
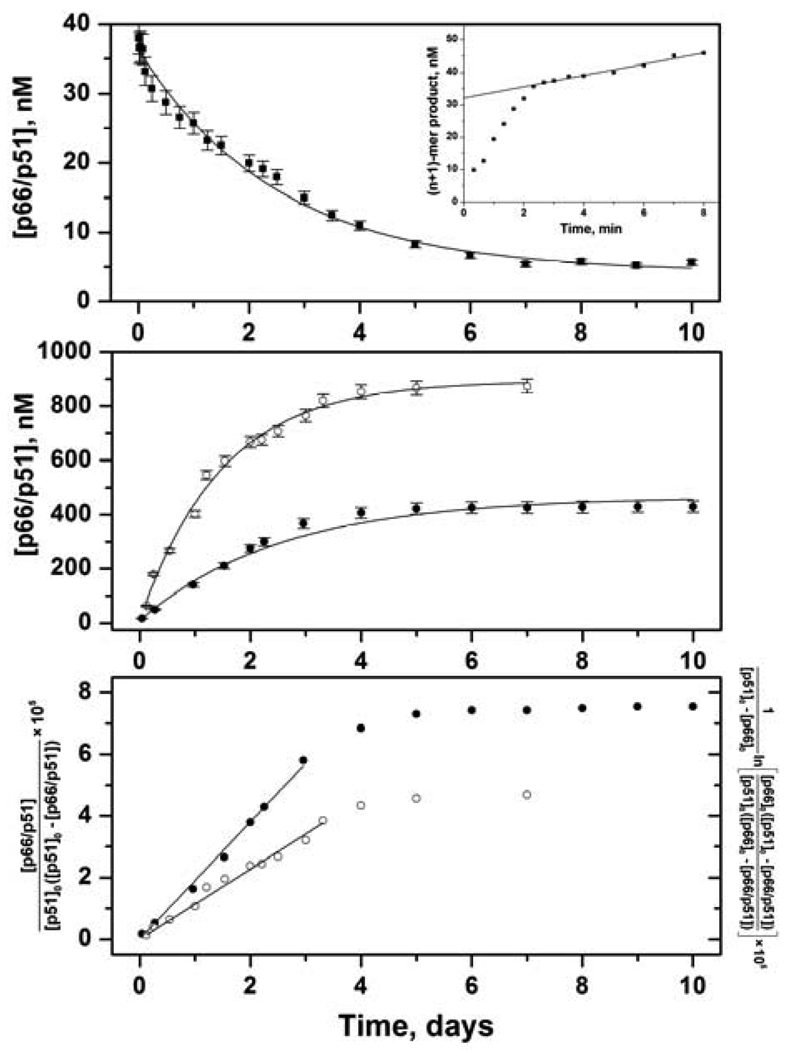
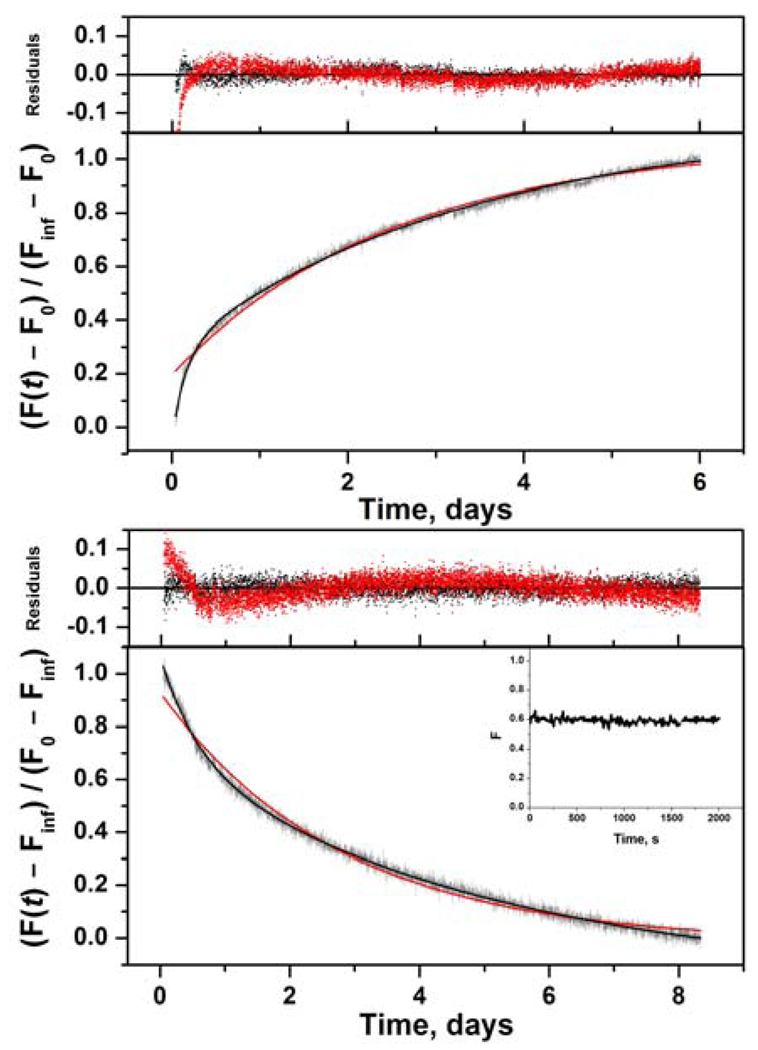
Dimerization is absolutely required for both the DNA polymerase and RNase H activities of RT (10, 22). Monomeric forms of RT are not catalytically active and do not bind primer/template substrate (15). In the pre-steady-state assay, the time course of a single nucleotide addition experiment has a rapid initial burst followed by a slow linear steady-state phase (Figure 1, upper panel inset). The slope of the steady-state phase is the steady-state rate kss; the intercept is the burst amplitude B, which gives the amount of active enzyme at saturating concentrations of DNA substrate. For all experiments, the burst phase occurred within the first 2 min of each 8-min reaction. Thus, a pre-steady-state assay at each time point in the dimerization reaction measures dimer concentration directly, unlike the conventional steady-state DNA polymerase assay.
FIGURE 1.
Kinetics of RT subunit dissociation and association monitored by pre-steady-state activity assay. Error bars are standard deviations of at least 3 experiments. (upper panel) Dissociation of 50 nM p66/p51 at various times (■) fit to eq 2 (solid line). Inset shows a time course of single nucleotide addition to DNA substrate. Slope of line is steady-state rate kss; intercept is burst amplitude B = [p66/p51] / 2. (middle panel) Association of 0.5 µM p66, 0.5 µM p51 (●) and 0.5 µM p66, 2.5 µM p51 (○) fit to eq 5 (solid lines). (lower panel) Association of 0.5 µM p66, 0.5 µM p51 (●) fit to eq 3 and 0.5 µM p66, 2.5 µM p51 (○) fit to eq 4.
Dissociation kinetics for RT were determined under irreversible conditions using a concentration jump to a value < Kd to induce dissociation. The lower limit for detection in the pre-steady-state assay was established to be less than 5 nM of an equilibrated solution of RT heterodimer. In an equilibrated solution of p66/p51 at [monomer]tot = 10 µM, 74% of total monomer is heterodimer: [p66/p51] = 7.4 µM, [p66/p66] = 0.38 µM, [p51/p51] = 0.022 µM, [p66] = 0.9 µM, and [p51] = 1.3 µM. This compares to an equilibrated solution at [monomer]tot = 50 nM, in which only 6.8% of total monomer is heterodimer: [p66/p51] = 3.4 nM, [p66/p66] = 0.24 nM, [p51/p51] = 0.0048 nM, [p66] = 23 nM, and [p51] = 23 nM. Each dissociation experiment was performed three times, and burst amplitudes for the same time point were averaged and plotted versus time (Figure 1, upper panel). Active p66/p51 concentrations approach, but never reach, zero because 50 nM RT is 6.8% heterodimer at equilibrium. The dissociation data were analyzed with eq 2 assuming a first-order reaction
| (11) |
with a dissociation rate constant kdiss = 4.4 ± 0.2 × 10−6 s−1 (Table 1).
Table 1.
Dissociation and Association Rate Constants of Wild-Type RTa
| method | protein solnsb | kdiss, 10−6 s−1 | kobs, 10−6 s−1 | kass, M−1s−1 |
|---|---|---|---|---|
| dissociation reaction | ||||
| pre-steady-state assay | 50 nM p66/p51 | 4.4 ± 0.2 | ||
| Trp fluorescencec | 50 nM p66/p51 | 3.9 ± 0.3 | ||
| association reaction | ||||
| pre-steady-state assay | 0.5 µM p66, 0.5 µM p51 | 5.8 ± 0.2 | 2.2 ± 0.2 d | |
| 0.5 µM p66, 2.5 µM p51 | 7.8 ± 0.3 | 1.3 ± 0.2 e | ||
| 1.4 ± 0.2 f | ||||
| Trp fluorescencec | 0.5 μM p66, 0.5 μM p51 | 5.4 ± 0.4 | ||
To study association kinetics, the initial solutions of p66 and p51 should be dilute enough to ensure predominantly monomers prior to mixing, but concentrated enough to form heterodimer after mixing. We compromised with a 1 µM solution of p66, in which [p66/p66] = 0.13 µM, and a 5 µM solution of p51, in which [p51/p51] = 37 nM. After mixing equal volumes of these solutions, the [monomer]tot = 3 µM; at equilibrium, 90% of the p66 will be heterodimer: [p66/p51] = 900 nM, [p66/p66] = 2 nM, [p51/p51] = 51 nM, [p66] = 47 nM, and [p51] = 2 µM. Burst phases typically occurred within the first 3 min of each reaction. Burst amplitudes for the same time points from three association reactions were averaged and plotted versus time (Figure 1, middle panel, open symbols). In the upper curve, active p66/p51 concentrations never go above 900 nM, because only 90% of p66 is heterodimer at equilibrium. Analogous experiments with p66 in excess of p51 were not performed because an equilibrated solution of 5 µM p66 is 53% homodimer. However, experiments were performed using 1 µM solutions of p66 and p51 to determine the molecularity of the association reaction and facilitate comparison with tryptophan fluorescence experiments (Figure 1, middle panel, filled symbols). At these monomer concentrations, [p66/p51] = 440 nM at equilibrium. For a simple reversible association reaction (Scheme 2),
| (12) |
The data at early times in the association reactions were fit to integrated rate equations for a second-order reaction (eq 3 and eq 4; Figure 1, lower panel). The kass = 2.2 ± 0.2 M−1s−1 for equal initial concentrations and kass = 1.3 ± 0.2 M−1s−1 for unequal initial concentrations. The entire time courses were fit to eq 5 to obtain observed rate constants kobs of 5.8 ± 0.2 × 10−6 s−1 and 7.8 ± 0.3 × 10−6 s−1. For a simple association equilibrium,
| (13) |
The concentration dependence of kobs indicates that the rate-limiting step for association of RT subunits is bimolecular. The association rate constant kass was calculated from kobs = 7.8 ± 0.3 × 10−6 s−1 determined under pseudo first-order conditions using eq 13 with [p51] = [p51]0 = 5 µM, giving kass = 1.4 ± 0.2 M−1s−1 in agreement with the value calculated using eq 4 (Table 1).
Scheme 2.
Bimolecular Dimerization Mechanism
Kinetics Monitored by Tryptophan Fluorescence
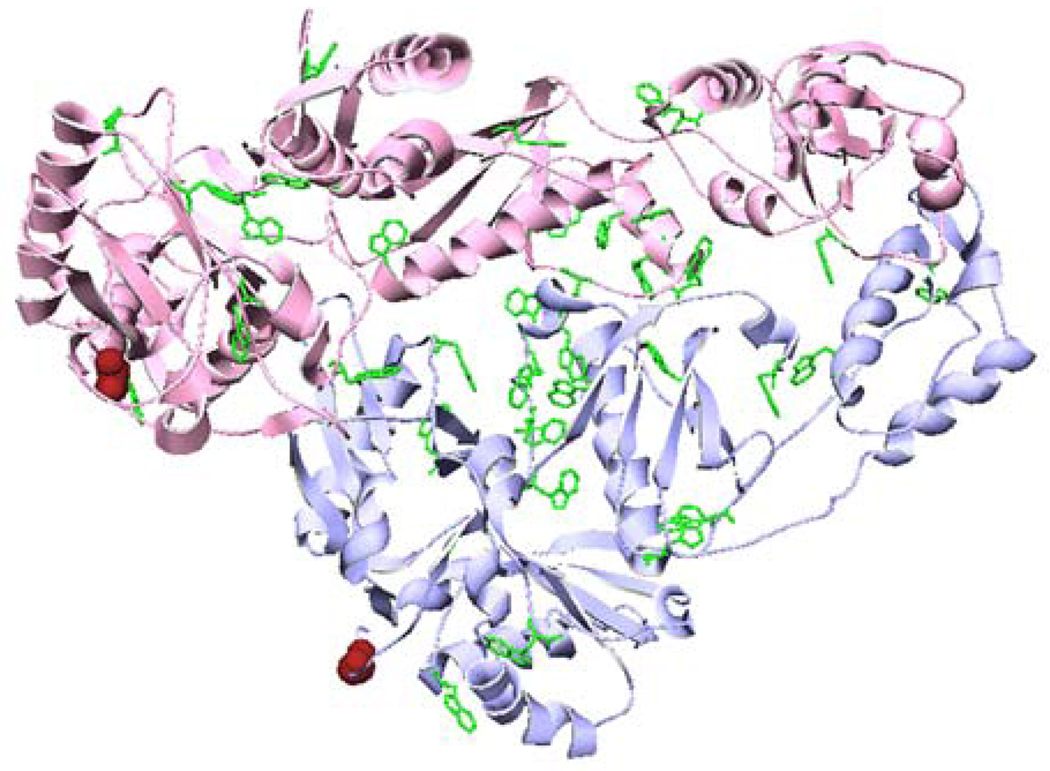
RT heterodimer contains 37 Trp residues distributed throughout the structure, any of which may report a conformational change due to dimerization (Figure 2). A cluster of six Trps in the connection subdomain has been shown to play a major role in dimerization (23, 24). As reported previously, the dissociation of RT heterodimer by 20% acetonitrile is accompanied by a 25% increase of tryptophan fluorescence intensity and a shift in the emission spectrum from 338 to 345 nm (11). In the absence of acetonitrile the fluorescence intensity changes due to dimerization are about 25% with no shift in the emission maximum of 337 nm. We exploited this change in tryptophan fluorescence intensity to determine the dissociation and association kinetics of RT under identical conditions as the pre-steady-state assay. The fractional increase or decrease in fluorescence intensity is plotted versus time in Figure 3. The dissociation data were fit to eq 6a to obtain kdiss = 3.9 ± 0.3 × 10−6 s−1 (Figure 3, upper panel), within error of the value measured in the pre-steady-state assay (Table 1). However, inspection of Figure 3 (upper panel) shows that a single exponential does not adequately fit the data for the first 8 h. A better fit is obtained by including a second exponential term in eq 6a, which gives kdiss(1) = 3.6 ± 0.4 × 10−6 s−1 and kdiss(2) = 7.6 ± 0.5 × 10−5 s−1 with relative amplitudes of 0.72 ± 0.1 and 0.28 ± 0.1. The slower rate constant kdiss(1), which is the major component, is about the same as the value obtained from the single exponential fit; the minor component kdiss(2) is 20-fold higher.
FIGURE 2.
Ribbon diagram of unliganded HIV-1 p66/p51 RT (1DLO; pink for p66, blue for p51). Tryptophan sidechains are shown in green; labeling sites are indicated in red, using the van der Waals radius of S3.
FIGURE 3.
Kinetics of RT subunit dissociation and association monitored by intrinsic tryptophan fluorescence. (upper panel) Dissociation of 50 nM p66/p51 fit to eq 6a (red line) and eq 6a with a second exponential term (black line). Residuals for both fits shown above. (lower panel) Association of 0.5 µM p66, 0.5 µM p51 fit to eq 6b (red line) and eq 6b with a second exponential term (black line). Residuals for both fits shown above. Inset shows the initial 2000 s.
For the association reaction, fluorescence intensity was measured at 5 s intervals in an attempt to resolve a fast bimolecular step. However, as seen in Figure 3 (inset), the fluorescence did not change detectably for the first 2 h after mixing. The association data measured at 5 min intervals were fit to eq 6b to obtain kobs = 5.4 ± 0.4 × 10−6 s−1, in good agreement with the value obtained from enzymatic activity (Table 1). However, because p66 has 19 Trps and p51 has 18 Trps, it proved impossible to use excess p51 to achieve pseudo first-order conditions. In an experiment using 1 µM p66 and 3 µM p51, the excess p51 contributed sufficient background fluorescence to obscure the decrease in fluorescence due to dimerization. Figure 3 (lower panel) shows that a single exponential does not adequately fit the data for the first 12 h. A better fit is obtained here as well by including a second exponential term in eq 6b, which gives kobs(1) = 2.3 ± 0.1 × 10−6 s−1 and kobs(2) = 2.1 ± 0.2 × 10−5 s−1 with relative amplitudes of 0.70 ± 0.1 and 0.30 ± 0.1. The slower rate constant kobs(1), which is the major component, is a factor of 2.3 lower then the value obtained from the single exponential fit; the minor component kobs(2) is 4-fold higher.
Kinetics Monitored by FRET
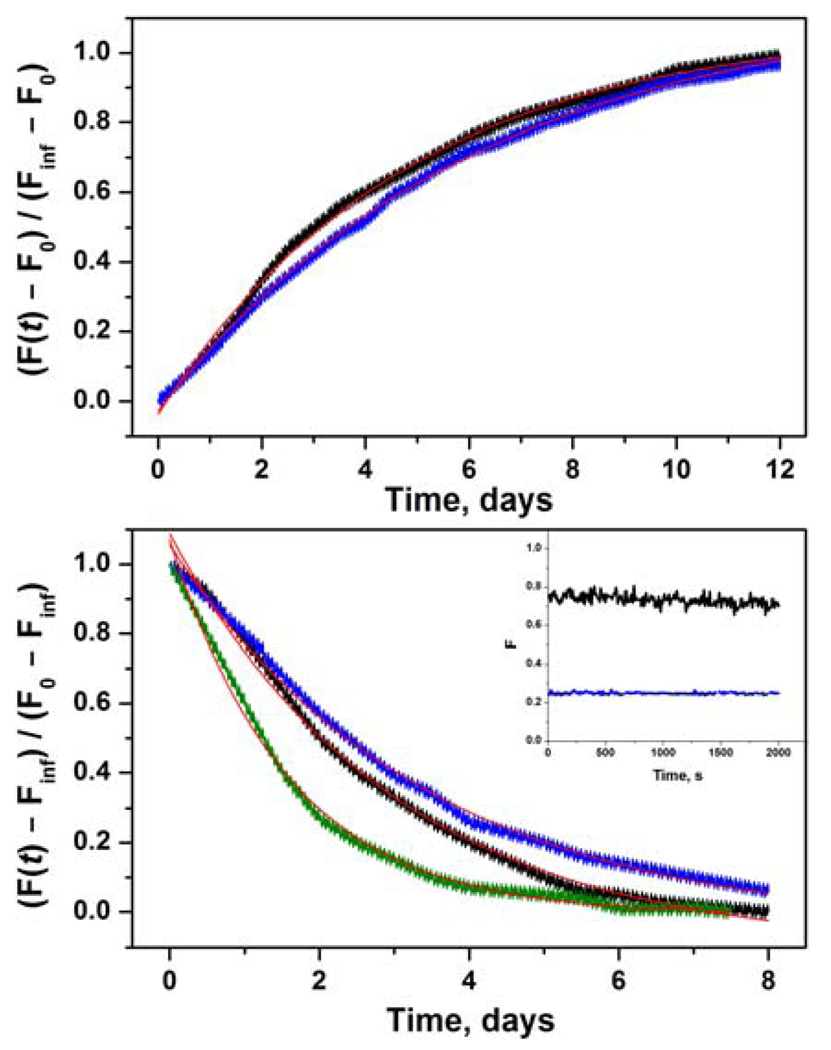
The 10 N-terminal residues of both subunits are loops in the crystal structure of the heterodimer (Figure 2). We attached fluorescence probes to a single cysteine at position 3 of the RT sequence. These labeled proteins were used in a FRET assay with A488/QSY9 as the donor/acceptor pair. The Förster distances R0 are 60 Å for p66-A488/p51-QSY9 and 59 Å for p66-QSY9/p51A-488, assuming random orientation of the transition dipoles (25). This compares to a distance of ~40 Å between the S3 residues of p66 and p51 in the crystal structure (26). The A488 donor has high photostability and quantum yield; the QSY9 acceptor is nonfluorescent, which eliminates background fluorescence due to direct excitation of the acceptor. Therefore, the fluorescence intensity of A488 should be highly quenched in the heterodimer relative to the monomers. The upper panel of Figure 4 displays the dissociation kinetics of heterodimer after dilution of p66-A488/p51-QSY9 and of p66-QSY9/p51-A488. A control reaction of heterodimer with donor alone was run in parallel to correct for any change in donor fluorescence due to a protein conformational change. The values of the dissociation rate constant obtained by fitting these data to eq 6a were independent within error of the position of the labels: kdiss = 2.5 ± 0.5 × 10−6 s−1 for p66-A488/p51-QSY9 and kdiss = 1.9 ± 0.4 × 10−6 s−1 for p66-QSY9/p51-A488 (Table 2). The dissociation rate constants of the labeled RT are about a factor of two lower than for wild-type RT.
FIGURE 4.
Kinetics of RT subunit dissociation and association monitored by FRET. (upper panel) Dissociation of 50 nM p66-A488/p51-QSY9 (black) and 50 nM p66-QSY9/p51-A488 (blue) fit to eq 6a (red lines). (lower panel) Association of 0.5 µM p66-A488, 0.5 µM p51-QSY9 (black); 0.5 µM p66-QSY9, 0.5 µM p51-A488 (blue); and 0.5 µM p66-A488, 2.5 µM p51-QSY9 (olive) fit to eq 6b (red lines). Inset shows the initial 2000 s of 0.5 µM p66-A488, 0.5 µM p51-QSY9 (black) and 0.5 µM p66-QSY9, 0.5 µM p51-A488 (blue).
Table 2.
Dissociation and Association Rate Constants of Labeled RTa
| protein solnsb | kdiss, 10−6 s−1 | kobs, 10−6 s−1 | kassc, M−1s−1 |
|---|---|---|---|
| dissociation reaction | |||
| 50 nM p66-A488/p51-QSY9 | 2.5 ± 0.5 | ||
| 50 nM p66-QSY9/p51-A488 | 1.9 ± 0.4 | ||
| association reaction | |||
| 0.5 µM p66-A488, 0.5 µM p51-QSY9 | 4.1 ± 0.4 | ||
| 0.5 µM p66-QSY9, 0.5 µM p51-A488 | 3.4 ± 0.4 | ||
| 0.5 µM p66-A488, 1.5 µM p51-QSY9 | 4.8 ± 0.3 | 1.6 ± 0.2 | |
| 0.5 µM p66-A488, 2.0 µM p51-QSY9 | 5.7 ± 0.4 | 1.6 ± 0.2 | |
| 0.5 µM p66-A488, 2.5 µM p51-QSY9 | 7.1 ± 0.3 | 1.8 ± 0.4 | |
RT buffer D, 5 °C. Errors are standard deviations of 3 experiments. λex = 450 nm, λem = 520 nm
Final concentrations
Calculated from eq 13.
The lower panel in Figure 4 shows the association data from experiments mixing p66-A488 with p51-QSY9 and p66-QSY9 with p51-A488. The fluorescence intensity did not change for the first 2 h after mixing 1 µM solutions of the two subunits (inset), and the observed rate constant kobs obtained from the fit to eq 6b was independent of labeling position (Table 2). Additional experiments were performed under pseudo first-order conditions, using 1 µM p66-A488 and 3–5 µM p51-QSY9. As expected for a bimolecular reaction, the value of the observed rate constant kobs is concentration dependent (Table 2). The association rate constant kass = 1.7 M−1s−1 calculated from eq 13 for labeled RT agrees well with the value for wild-type RT (Table 1).
DISCUSSION
RT heterodimer maturation is a complex process that, despite its importance, is not understood. Central to this process are the interactions of its subunits. The concentration of RT in the virion, estimated from the number of RT molecules and the size of the viral particle, is roughly 250 µM. At these concentrations the dimerization reaction would be ~105-fold faster and occur on a time scale of minutes rather than days. A complete replication cycle of HIV takes ~48 h (27). RT is translated as part of the p160 gag-pol polyprotein and subsequently cleaved by the pol-encoded protease. The processing of a 90-kDa pol polyprotein to yield p66/p51 has been examined using an inducible bacterial expression vector (8). The full-length polyprotein was the first product formed, followed by p66; p51 only appeared after a significant amount of p66 had accumulated. Both the concerted and sequential models proposed for heterodimer formation posit a subunit dimerization step. In the concerted model, p66 and p51 associate to form mature heterodimer. In the sequential model, p66 self-associates to form homodimer, followed by removal of the RNase H domain from one subunit by HIV protease. These results support the sequential model. The previously reported association rate constant kass = 1.4 × 10−2 M−1s−1 for p66/p66 homodimer formation is two orders of magnitude lower than the value of ~1.7 M−1s−1 obtained here for p66/p51 heterodimer formation (10). A faster association rate constant for formation of p66/p51 heterodimer compared to p66/p66 homodimer suggests that RT formation may proceed via the concerted model. However, the homodimer association kinetics were measured using organic solvent (10). Whatever the model, these results suggest that RT heterodimer maturation appears to be a slow process. A chaperone protein in the cell may facilitate RT dimerization, but so far no such protein has been identified. The nucleocapsid protein cleaved from the gag-pol polyprotein was a plausible candidate, though in preliminary experiments it had no effect on dimerization.
In this work we used three techniques to monitor dimerization: enzymatic activity, tryptophan fluorescence, and FRET. These techniques report different aspects of RT structure: presence of primer/template binding cleft and polymerase active site; local environment of tryptophan residues, presumably the tryptophan cluster in the connection subdomains; and proximity of the N-termini of the two subunits. Our results support a simple reversible bimolecular association reaction (Scheme 2). We obtained similar values from each method for both the association and dissociation rate constants. Heterodimer dissociation is a first-order reaction with kdiss = 4 × 10−6 s−1 for wild-type heterodimer and 2 × 10−6 s−1 for labeled heterodimer. Association of the two monomers is a second-order reaction dependent on monomer concentrations with kass ≅ 1.7 M−1s−1. There is no evidence of a fast bimolecular association followed by slow isomerization to active RT. If this were the case, we would have been able to observe a biphasic reaction in both the tryptophan fluorescence and FRET experiments. According to the law of mass action the equilibrium dissociation constant Kd(p66/p51) = kdiss / kass. The Kd values calculated from the ratio of the rate constants are 4–10-fold larger than the value determined by sedimentation equilibrium (1). Possible contributions to this difference are: (1) buffer pH was 7.0 in kinetic experiments compared to 6.5 in ultracentrifugation experiments;2 and (2) p66 solutions prior to mixing contained about 13% p66/p66 homodimer, which is accounted for in the Kd(p66/p51) but not in the rate constants. Protein degradation does not occur during these slow reactions, as RT is stable for at least a month in RT buffer D at 5 °C (1).
Protein-protein association kinetics with rate constants ranging from < 103 to > 109 M−1s−1 have been examined experimentally and theoretically (28). A kinetic mechanism similar to Scheme 1, which proceeds through a transient complex or kinetic intermediate, is often used to describe these systems. Diffusion-controlled rate constants, which involve local conformational changes between unbound proteins and the native complex, fall on the upper end of the above time scale, while reaction-controlled rate constants, which involve sizeable domain movement-type changes, fall on the lower end. Studies of slow protein-protein interactions on the time scale of RT subunit dimerization are not unprecedented. Homodimerization of BirA, a biotin regulatory protein in E. coli, is governed by very slow forward (kon = 20 M−1s−1) and reverse (koff = 3 × 10−4 s−1) rate constants (29).
The extremely slow association of RT subunits is quite an intriguing result. RT faces the dilemma of forming asymmetric dimers from two subunits with identical amino acid sequence. HIV-1 RT is an asymmetric heterodimer in the 127 crystal structures currently available (30). Although there are no crystal structures of p66/p66 and p51/p51 homodimers, both of which have polymerase activity (10), the homodimers would also have to be asymmetric to create the primer/template binding cleft and polymerase active site. There are also no crystal structures of p66 and p51 monomers. However, hydrogen exchange mass spectrometry shows that p51 monomer and the polymerase domain of p66 monomer have similar solution structures (unpublished data), which are comparable to the solution structure of the p51 subunit in p66/p51 (31). In order to adopt the asymmetric structure of the heterodimer, the conformations of p66 monomer and, to a lesser extent, p51 monomer must change. Three models for the slow association reaction are: (1) weakly bound dimer intermediate, (2) unfavorable orientation effects, and (3) conformational selection. Model 1 postulates a two-step mechanism involving a transient complex (Scheme 1). If k−1 » k2 the association reaction will exhibit second-order kinetics with a rate constant kass ∝ Kak2, where Ka = k1 / k−1 is the equilibrium constant for formation of the complex. The slow association rate constant kass implies that Ka is very small (mM−1). At the µM concentrations of the kinetic experiments, the intermediate never comprises a detectable fraction of RT and occurs at such low concentrations that accumulation of active heterodimer proceeds very slowly. Model 1 is analogous to the previously proposed mechanism based on Scheme 1, except that isomerization k2 from inactive to active heterodimer has to be faster than isomerization k−2 from active to inactive heterodimer in order to observe active p66/p51. Model 2 invokes direct binding of p66 and p51 with unfavorable orientation effects (Scheme 2). The slow bimolecular step implies that productive collisions of p66 and p51 are extremely rare. Model 3 proposes conformational selection from a population of dynamic monomer structures (32). Only two monomers in the right conformations can combine to form the asymmetric dimer. The individual subunits undergo separate conformational changes prior to the bimolecular reaction to form active heterodimer (Scheme 3). The unimolecular rate constants kc and k−c in this scheme would be undetectable by enzymatic activity or FRET, because these techniques only monitor dimerization. The conformational equilibrium constant Kc = kc / k−c is unfavorable, and the overall rate constant kass ∝ Kck1 is thereby slow. Because our data provide scant evidence for complexity of either the association or dissociation kinetics, we cannot distinguish between the three models based on the results in this paper. However, we prefer Model 1 because of the flexibility of RT together with the different structures of the subunits in the asymmetric heterodimer (31). Future studies of the effects of temperature, pH, osmolytes, and mutational analysis on the dimerization kinetics will test these models.
Scheme 3.
Reaction-Controlled Dimerization Mechanism
The biphasic association and dissociation kinetics resolved by tryptophan fluorescence introduce a wrinkle into these models, although the rate constants of the second minor component are still very slow. Attempts to fit the enzymatic activity and FRET data to two exponential functions failed to resolve two components with different rate constants. The deviation of the tryptophan fluorescence data from a single exponential function is most apparent in the first 8 h of the dissociation reaction and the first 12 h of the association reaction (Figure 3). In the case of the enzymatic activity data, there are sufficient points at early times in the dissociation experiment, but not in the association experiment, to have resolved the biphasic kinetics. The data acquisition protocols for the tryptophan fluorescence and FRET experiments were identical, yet no evidence of biphasic kinetics is apparent in the FRET data. If there are two forms of heterodimer that undergo simultaneous association and dissociation reactions, then the biphasic kinetics should be manifest by all three techniques. The minor component may represent either the conformational equilibria of the monomers in Scheme 3 or inactive heterodimer p66*p51 in Scheme 1, which associates and dissociates slowly. An inactive intermediate would not be seen by enzymatic activity; it would be seen by FRET unless the close positioning of the N-termini is one of the last conformational changes to occur.
Previously RT dimerization kinetics were monitored by fluorescence and polymerase activity, and the results were interpreted according to the two-step mechanism in Scheme 1 (9–12). Rapid association and dissociation kinetics yielding a second-order rate constant for the first step in the association reaction, k1 = 2.8 × 104 M−1s−1, and a first-order rate constant for the overall dissociation reaction, kd = k−1k−2 / (k−1 + k2) = 4.5 × 10−3 s−1 were determined by tryptophan fluorescence. Slow association kinetics yielding a first-order rate constant for the second step in the association reaction, k2 = 3–6 × 10−5 s−1, were observed by polymerase activity. Biphasic association kinetics monitored by ANS fluorescence were decomposed into fast and slow phases with a second-order rate constant k1 = 3.9 × 104 M−1s−1 and a first-order rate constant k2 = 2.8 × 10−5 s−1. The dimerization mechanism proposed in Scheme 1 with the above rate constants is implausible. If k−1 > k2, then kd ~ k−2, and k2 has to be larger then k−2 in order to observe active heterodimer. However, the overall dissociation rate constant kd ~ k−2 is two orders of magnitude faster than the rate-limiting step k2 in the association reaction. A possible reason for the fast kinetics is the use of organic solvent to produce p66 and p51 monomers. All previous experiments achieved dissociation of p66/p51 by addition of 20% acetonitrile and subsequent association by a 12-fold dilution of the monomeric subunits into aqueous buffer (9–12). The acetonitrile-induced dissociation rate constant kd = 4.5 × 10−3 s−1 (11, 12) monitored by fluorescence is three orders of magnitude higher than the dilution-induced dissociation rate constants kdiss = 2–4 × 10−6 s−1 obtained here, while the fast association rate constant k1 = 2.8 × 104 M−1s−1 is four orders of magnitude higher than our association rate constants kass ≅ 1.7 M−1s−1. A likely explanation of the fast changes (1000 s) in fluorescence is that addition of acetonitrile produces protein unfolding as well as subunit dissociation. Conversely, dilution out of acetonitrile induces fast refolding and slow subunit association. The presence of acetonitrile causes increases in tryptophan and ANS fluorescence intensities (33, 34), which would augment the magnitude of intensity changes; it also causes shifts in the emission spectra. On the other hand, the slow recovery of polymerase activity after dilution into buffer, interpreted as an isomerization rate constant k2 = 3–6 × 10−5 s−1 (9), is only 10-fold higher than the kobs = 3.4–5.8 × 10−6 s−1 values obtained for the equimolar association reactions monitored by enzymatic activity, tryptophan fluorescence, and FRET. At least part of this difference can be attributed to different experimental conditions: pH 8.0, 25 °C for the published value versus pH 7.0, 5 °C in this paper.
ACKNOWLEDGEMENT
The authors are grateful to Dr. Michael E. Ignatov for assistance with the pre-steady-state assay and to Ms. Kathryn J. Howard for assistance with the QSY9 labeling reaction. We also thank Dr. Vernon E. Anderson for advice about analysis of the kinetics data and helpful discussions about the reaction mechanisms, Dr. Angela Gronenborn for suggesting Model 1, and Dr. Jonathan Karn for critical reading of the manuscript.
APPENDIX
A488 Fluorescence
The fluorescence of A488 appears to be environmentally sensitive; the intensity of labeled p66 is greater than labeled p51 (Figure 4, lower panel inset). Johnson and coworkers observed a 60–70% decrease in the fluorescence of A488-labeled calmodulin upon binding of a small peptide, which they attributed to proximity of the fluorescence probe and a tryptophan residue in the peptide (35). We pursued these environmental effects by measuring the fluorescence quantum yield and lifetime of A488-labeled RT monomers and heterodimers as well as N-acetylcysteine-A488. Table A1 gives the quantum yield data. The absorption and emission maxima are 488 and 520 nm for all A488 conjugates. The p66-A488 monomer has the same quantum yield as labeled N-acetylcysteine, which is 3-fold greater than the quantum yield of p51-A488 monomer. The values of the quantum yield decrease slightly if at all upon dimerization. As expected, the quantum yields of heterodimers containing the donor/acceptor pair are lower than those of heterodimers with donor alone by factors of eight for p66-A488/p51-QSY9 and three for p66-QSY9/p51-A488.
Table A1.
Quantum Yields and Amplitude-Weighted Lifetimesa
| protein solnsb | Φ | τ̄, ns |
|---|---|---|
| 0.5 µM p66-A488, | 0.80 ± 0.02 | 1.05 |
| 2.5 µM p51 | 0.75 ± 0.05 | 1.55 |
| 2.5 µM p51-QSY9 | 0.10 ± 0.03 | 1.37 |
| 0.5 µM p51-A488, | 0.26 ± 0.01 | 1.23 |
| 2.5 µM p66 | 0.25 ± 0.02 | 1.41 |
| 2.5 µM p66-QSY9 | 0.079 ± 0.004 | 1.41 |
| N-acetylcysteine-A488 | 0.82 ± 0.02 | |
| N-acetylcysteine-A488c | 0.80 ± 0.02 |
RT buffer D, 5 °C. λex = 450 nm. Errors are range of average values of 2–4 experiments
Final concentrations
RT buffer D-D2O, 5 °C.
Table A2 gives the fluorescence lifetime data for A488 conjugates. N-acetylcysteine-A488 is a control with no protein interaction. The fluorescence decay is best fit by a biexponential function with a major 4.1-ns component and a minor 0.54-ns component, which has a negative amplitude. A negative term in a fluorescence decay signifies an excited-state reaction with a fluorescent product. The absence of a deuterium isotope effect on the quantum yield suggests that the excited-state reaction does not involve proton transfer (36). The fluorescence decays of all RT conjugates gave good fits to three exponential functions with lifetimes of about 3.6-, 1-, and 0.05-ns. The long lifetime of the RT conjugates is shorter than the 4.1-ns lifetime of N-acetylcysteine-A488. In all the conjugates, the short lifetime component has the largest amplitude, but contributes only about 2% of total intensity (Figure A1, upper panel). Amplitude-weighted lifetimes τ̄ of about 1 ns are calculated from eq 8 (Table A1). The quantum yield drop of A488 in the environment of p66 compared to p51 is accompanied by about 1.2-fold increase in amplitude-weighted lifetime. Likewise, the quantum yields of A488-labeled heterodimers decrease in the presence of QSY9 acceptor, whereas the amplitude-weighted lifetimes increase 1.2–1.3-fold. A quantum yield drop without a parallel lifetime drop is emblematic of static quenching. Static quenching occurs when a species becomes effectively nonfluorescent with intensity or lifetime too small to measure.
Table A2.
Fluorescence Decay Parametersa
| protein solnsb | α1c | τ1d, ns | α2c | τ2d, ns | τ3d, ns | χr2 |
|---|---|---|---|---|---|---|
| 0.5 µM p66-A488, | 0.24 ± 0.02 | 3.59 | 0.17 ± 0.01 | 0.99 | 0.035 | 1.25 ± 0.03 |
| 2.5 µM p51 | 0.35 ± 0.02 | 3.68 | 0.22 ± 0.02 | 1.08 | 0.049 | 1.31 ± 0.04 |
| 2.5 µM p51-QSY9 | 0.31 ± 0.02 | 3.66 | 0.20 ± 0.02 | 1.05 | 0.046 | 1.36 ± 0.03 |
| 0.5 µM p51-A488, | 0.28 ± 0.01 | 3.63 | 0.19 ± 0.02 | 0.98 | 0.057 | 1.21 ± 0.05 |
| 2.5 µM p66 | 0.33 ± 0.01 | 3.51 | 0.24 ± 0.01 | 0.93 | 0.066 | 1.40 ± 0.01 |
| 2.5 µM p66-QSY9 | 0.33 ± 0.04 | 3.44 | 0.27 ± 0.01 | 0.90 | 0.075 | 1.29 ± 0.02 |
| N-acetylcysteine-A488 | 0.84 ± 0.01 | 4.10 | −0.16 ± 0.01 | 0.54 | 1.30 ± 0.02 | |
| N-acetylcysteine-A488e | 0.85 ± 0.03 | 4.05 | −0.15 ± 0.01 | 0.50 | 1.35 ± 0.03 |
RT buffer D, 5 °C. λex = 450 nm, λem = 540 nm
Final concentrations
Σαi = 1. Errors in αi are range of average values from global analysis of two decay curves
Errors in τi ~ 5%
RT buffer D-D2O, 5 °C.
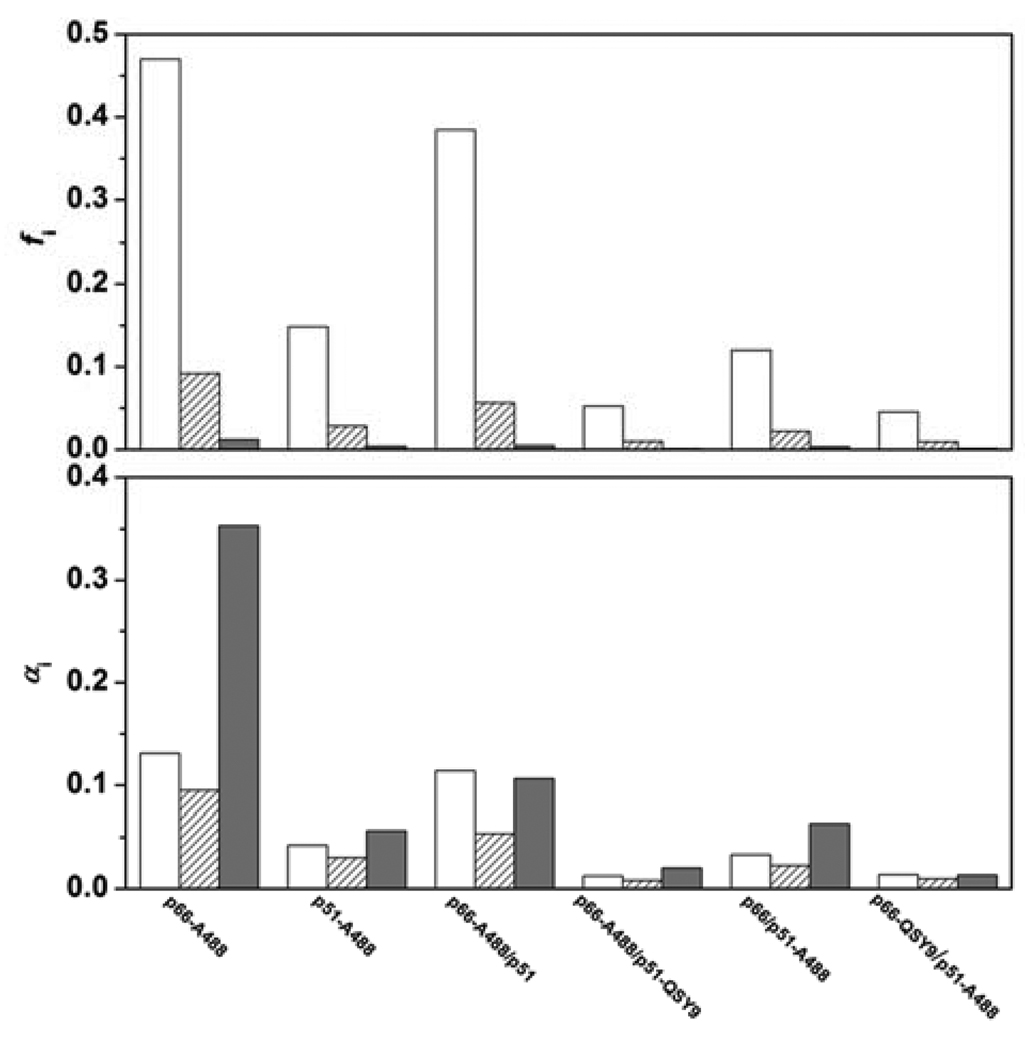
FIGURE A1.
Fractional intensities and amplitudes of 3.6-, 1-, and 0.05-ns decay components of A488-labeled RT monomers and heterodimers. 3.6-ns component (open bars), 1.0-ns component (hatched bars), and 0.05-ns component (filled bars). (upper panel) Fractional intensities calculated from data in Table A2 using eq 9. (lower panel) Amplitudes normalized to the steady-state intensity of N-acetylcysteine-A488 at 540 nm.
The lower panel of Figure A1 shows the amplitudes of the three lifetime components of the RT conjugates normalized to the steady-state intensity of N-acetylcysteine-A488 at 540 nm. We assume that the 3.6-, 1-, and 0.05-ns lifetimes of the RT conjugates report three ground-state species of A488 in distinct environments and that the same three species occur in all the RT conjugates. Then the quenching of A488 fluorescence in p51 relative to p66 is due to conversion of the three fluorescent species to nonfluorescent species, as seen by the amplitude losses. Tryptophan and to a much lesser extent tyrosine have been shown to quench bimane fluorescence by both dynamic and static quenching mechanisms (37, 38). If bimane and tryptophan are close (10–15 Å), the quenching is dynamic with parallel drops in quantum yield and lifetime. For very close distances (5–10 Å), the quenching is static and only the quantum yield decreases with no change in lifetime. We propose that the 3.6-, 1-, and 0.05-ns lifetimes of A488 on p66 arise from dynamic quenching by a nearby aromatic residue, possibly W212, which is 5–13 Å away from S3 in the crystal structure (26). The three lifetimes may arise from different positions of A488 on a 10 Å pentamethylene tether. The static quenching of A488 on p51 would invoke an additional position in which A488 is very close to W212.
The FRET quenching of A488 fluorescence in the two heterodimers with donor/acceptor pairs reflects static quenching due to loss of amplitude, not lifetime quenching. FRET quenching is normally dynamic quenching, which shortens the fluorescence lifetime. For FRET quenching of the A488/QSY9 pair to manifest static quenching, the distance between donor and acceptor has to be < 30 Å, certainly feasible for two probes on 10 Å tethers.
Footnotes
This work was supported by NIH grant GM071267. C.F.V. was supported by NIH Training Grant HL07653.
Abbreviations: A488, Alexa Fluor 488; ANS, 1-anilino-8-naphthalenesulfonate; DCM, dichloromethane; DTT, dithiothreitol; EDTA, ethylaminediaminetetraacetic acid; FRET, Förster resonance energy transfer; HIV-1, human immunodeficiency virus type 1; NATA, N-acetyltryptophanamide; NTA, nitrilotriacetic acid; p66-A488, p66S3C/C38V/C280S labeled with A488; p66-QSY9, p66S3C/C38V/C280S labeled with QSY9; p51-A488, p51S3C/C38V/C280S labeled with A488; p51-QSY9, p51S3C/C38V/C280S labeled with QSY9; RT, reverse transcriptase; TCEP, tris(2-carboxyethyl)phosphine; Tris, tris(hydroxymethyl)aminomethane.
These values were determined in RT buffer D containing 1 mM TCEP, which was added after adjusting pH (1). The addition of TCEP lowered the pH to 6.5.
REFERENCES
- 1.Venezia CF, Howard KJ, Ignatov ME, Holladay LA, Barkley MD. Effects of efavirenz binding on the subunit equilibria of HIV-1 reverse transcriptase. Biochemistry. 2006;45:2779–2789. doi: 10.1021/bi051915z. [DOI] [PubMed] [Google Scholar]
- 2.di Marzo Veronese F, Copeland TD, DeVico AL, Rahman R, Oroszlan S, Gallo RC, Sarngadharan MG. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986;231:1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]
- 3.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 4.Hizi A, McGill C, Hughes SH. Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc. Natl. Acad. Sci. U.S.A. 1988;85:1218–1222. doi: 10.1073/pnas.85.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Smerdon SJ, Jager J, Kohlstaedt LA, Rice PA, Friedman JM, Steitz TA. Structural basis of asymmetry in the human immunodeficiency virus type 1 reverse transcriptase heterodimer. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7242–7246. doi: 10.1073/pnas.91.15.7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobo-Molina A, Ding J, Nanni RG, Clark AD, Jr, Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, Hizi A, Hughes SH, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Grice SFJ, Naas T, Wohlgensinger B, Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sluis-Cremer N, Arion D, Abram ME, Parniak MA. Proteolytic processing of an HIV-1 pol polyprotein precursor: insights into the mechanism of reverse transcriptase p66/p51 heterodimer formation. Int. J. Biochem. Cell Biol. 2004;36:1836–1847. doi: 10.1016/j.biocel.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 9.Divita G, Rittinger K, Geourjon C, Deleage G, Goody RS. Dimerization kinetics of HIV-1 and HIV-2 reverse transcriptase: a two step process. J. Mol. Biol. 1995;245:508–521. doi: 10.1006/jmbi.1994.0042. [DOI] [PubMed] [Google Scholar]
- 10.Restle T, Muller B, Goody RS. Dimerization of human immunodeficiency virus type 1 reverse transcriptase. A target for chemotherapeutic intervention. J. Biol. Chem. 1990;265:8986–8988. [PubMed] [Google Scholar]
- 11.Divita G, Restle T, Goody RS. Characterization of the dimerization process of HIV-1 reverse transcriptase heterodimer using intrinsic protein fluorescence. FEBS Lett. 1993;324:153–158. doi: 10.1016/0014-5793(93)81383-b. [DOI] [PubMed] [Google Scholar]
- 12.Divita G, Rittinger K, Restle T, Immendorfer U, Goody RS. Conformational stability of dimeric HIV-1 and HIV-2 reverse transcriptases. Biochemistry. 1995;34:16337–16346. doi: 10.1021/bi00050a014. [DOI] [PubMed] [Google Scholar]
- 13.Wohrl BM, Krebs R, Thrall SH, Le Grice SFJ, Scheidig AJ, Goody RS. Kinetic analysis of four HIV-1 reverse transcriptase enzymes mutated in the primer grip region of p66. Implications for DNA synthesis and dimerization. J. Biol. Chem. 1997;272:17581–17587. doi: 10.1074/jbc.272.28.17581. [DOI] [PubMed] [Google Scholar]
- 14.Le Grice SFJ, Cameron CE, Benkovic SJ. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 1995;262:130–144. doi: 10.1016/0076-6879(95)62015-x. [DOI] [PubMed] [Google Scholar]
- 15.Ignatov ME, Berdis AJ, Le Grice SFJ, Barkley MD. Attenuation of DNA replication by HIV-1 reverse transcriptase near the central termination sequence. Biochemistry. 2005;44:5346–5356. doi: 10.1021/bi048000p. [DOI] [PubMed] [Google Scholar]
- 16.Haugland RP. Coupling of monoclonal antibodies with fluorophores. Methods Mol. Biol. 1995;45:205–221. doi: 10.1385/0-89603-308-2:205. [DOI] [PubMed] [Google Scholar]
- 17.Molecular Probes. http://probes.invitrogen.com/media/pis/mp00143.pdf.
- 18.Berdis AJ, Stetor SR, Le Grice SFJ, Barkley MD. Molecular mechanism of sequence-specific termination of lentiviral replication. Biochemistry. 2001;40:12140–12149. doi: 10.1021/bi010354i. [DOI] [PubMed] [Google Scholar]
- 19.Wong I, Patel SS, Johnson KA. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 20.Brannon JM, Magde D. Absolute quantum yield determination by thermal blooming. Fluorescein. J. Phys. Chem. 1978;82:705–709. [Google Scholar]
- 21.Watrob HM, Pan C-P, Barkley MD. Two-step FRET as a structural tool. J. Am. Chem. Soc. 2003;125:7336–7343. doi: 10.1021/ja034564p. [DOI] [PubMed] [Google Scholar]
- 22.Restle T, Muller B, Goody RS. RNase H activity of HIV reverse transcriptases is confined exclusively to the dimeric forms. FEBS Lett. 1992;300:97–100. doi: 10.1016/0014-5793(92)80172-d. [DOI] [PubMed] [Google Scholar]
- 23.Baillon JG, Nashed NT, Kumar A, Wilson SH, Jerina DM. A leucine zipper-like motif may mediate HIV reverse transcriptase subunit binding. New Biol. 1991;3:1015–1019. [PubMed] [Google Scholar]
- 24.Tachedjian G, Aronson HE, de los Santos M, Seehra J, McCoy JM, Goff SP. Role of residues in the tryptophan repeat motif for HIV-1 reverse transcriptase dimerization. J. Mol. Biol. 2003;326:381–396. doi: 10.1016/s0022-2836(02)01433-x. [DOI] [PubMed] [Google Scholar]
- 25.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd ed. New York: Springer; 2006. [Google Scholar]
- 26.Hsiou Y, Ding J, Das K, Clark AD, Jr, Hughes SH, Arnold E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Åresolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 27.Kim SY, Byrn R, Groopman J, Baltimore D. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 1989;63:3708–3713. doi: 10.1128/jvi.63.9.3708-3713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber G, Haran G, Zhou HX. Fundamental aspects of protein-protein association kinetics. Chem. Rev. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao H, Beckett D. Kinetic partitioning between alternative protein-protein interactions controls a transcriptional switch. J. Mol. Biol. 2008;380:223–236. doi: 10.1016/j.jmb.2008.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.RCSB Protein Data Bank. http://www.rcsb.org/pdb/home/home.do.
- 31.Seckler JM, Howard KJ, Barkley MD, Wintrode PL. Solution structural dynamics of HIV-1 reverse transcriptase heterodimer. Biochemistry. 2009 doi: 10.1021/bi900790x. DOI: 10.1021/bi900790x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weikl TR, von Deuster C. Selected-fit versus induced-fit protein binding: kinetic differences and mutational analysis. Proteins. 2009;75:104–110. doi: 10.1002/prot.22223. [DOI] [PubMed] [Google Scholar]
- 33.Slavik J. Anilinonaphthalene sulfonate as a probe of membrane composition and function. Biochim. Biophys. Acta. 1982;694:1–25. doi: 10.1016/0304-4157(82)90012-0. [DOI] [PubMed] [Google Scholar]
- 34.Yu H-T, Colucci WJ, McLaughlin ML, Barkley MD. Fluorescence quenching in indoles by excited-state proton transfer. J. Am. Chem. Soc. 1992;114:8449–8454. [Google Scholar]
- 35.Slaughter BD, Urbauer RJ, Urbauer JL, Johnson CK. Mechanism of calmodulin recognition of the binding domain of isoform 1b of the plasma membrane Ca2+-ATPase: kinetic pathway and effects of methionine oxidation. Biochemistry. 2007;46:4045–4054. doi: 10.1021/bi602481u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laws WR, Brand L. Analysis of two-state excited-state reactions. The fluorescence decay of 2-naphthol. J. Phys. Chem. 1979;83:795–802. [Google Scholar]
- 37.Sato E, Sakashita M, Kanaoka Y, Kosower EM. Organic fluorescent reagents. XIV. Novel fluorogenic substrates for microdetermination of chymotrypsin and aminopeptidase: bimane fluorescence appears after hydrolysis. Bioorg. Chem. 1988;16:298–306. [Google Scholar]
- 38.Mansoor SE, McHaourab HS, Farrens DL. Mapping proximity within roteins using fluorescence spectroscopy. A study of T4 lysozyme showing that ryptophan residues quench bimane fluorescence. Biochemistry. 2002;41:2475–2484. doi: 10.1021/bi011198i. [DOI] [PubMed] [Google Scholar]