Abstract
Background
We advocate unifying classical and genomic classification of bacteriophages by integration of proteomic data and physicochemical parameters. Our previous application of this approach to the entirely sequenced members of the Podoviridae fully supported the current phage classification of the International Committee on Taxonomy of Viruses (ICTV). It appears that horizontal gene transfer generally does not totally obliterate evolutionary relationships between phages.
Results
CoreGenes/CoreExtractor proteome comparison techniques applied to 102 Myoviridae suggest the establishment of three subfamilies (Peduovirinae, Teequatrovirinae, the Spounavirinae) and eight new independent genera (Bcep781, BcepMu, FelixO1, HAP1, Bzx1, PB1, phiCD119, and phiKZ-like viruses). The Peduovirinae subfamily, derived from the P2-related phages, is composed of two distinct genera: the "P2-like viruses", and the "HP1-like viruses". At present, the more complex Teequatrovirinae subfamily has two genera, the "T4-like" and "KVP40-like viruses". In the genus "T4-like viruses" proper, four groups sharing >70% proteins are distinguished: T4-type, 44RR-type, RB43-type, and RB49-type viruses. The Spounavirinae contain the "SPO1-"and "Twort-like viruses."
Conclusion
The hierarchical clustering of these groupings provide biologically significant subdivisions, which are consistent with our previous analysis of the Podoviridae.
Background
We recently described methods aimed at unifying classical and genomic classification of bacteriophages by integration of protein sequence data and physicochemical parameters. We developed two protein sequence similarity-based tools, CoreExtractor and CoreGenes [1], to parse-out and quantify relationships between pairs of phages resulting in a single correlation score [2]. This analysis is followed by a deconstruction and literature analysis of the known morphological and physicochemical characteristics of these phages. The biological interpretation of molecular correlations between 55 fully sequenced Podoviridae show that this approach agrees with the current phage classification of the International Committee on Taxonomy of Viruses (ICTV) and suggests that, generally, horizontal gene transfer only partially masks evolutionary relationships between phages. Using a cut-off value of 40% homologous proteins, we verified relationships between phages known to be similar and identified several new bacteriophage genera. At the 20-30% homology level, we identified relationships of a higher order justifying the introduction of the subfamily taxonomical category.
The Myoviridae in the VIIIth ICTV Report comprise five genera of bacteriophages (Mu, P1, P2, SPO1, and T4-like viruses) and one genus of archeal viruses, phiH. I3 and phiKZ-like phages have been recently proposed as additional genera http://www.ncbi.nlm.nih.gov/ICTVdb/Ictv/fs_myovi.htm. These genera include only a small fraction of presently known myoviruses with fully sequenced genomes [3]. We analyze and interpret here the correlations between 102 Myoviridae genomes found in the National Center for Biotechnology Information (NCBI) and the Tulane University T4 Genome databases.
Results and discussion
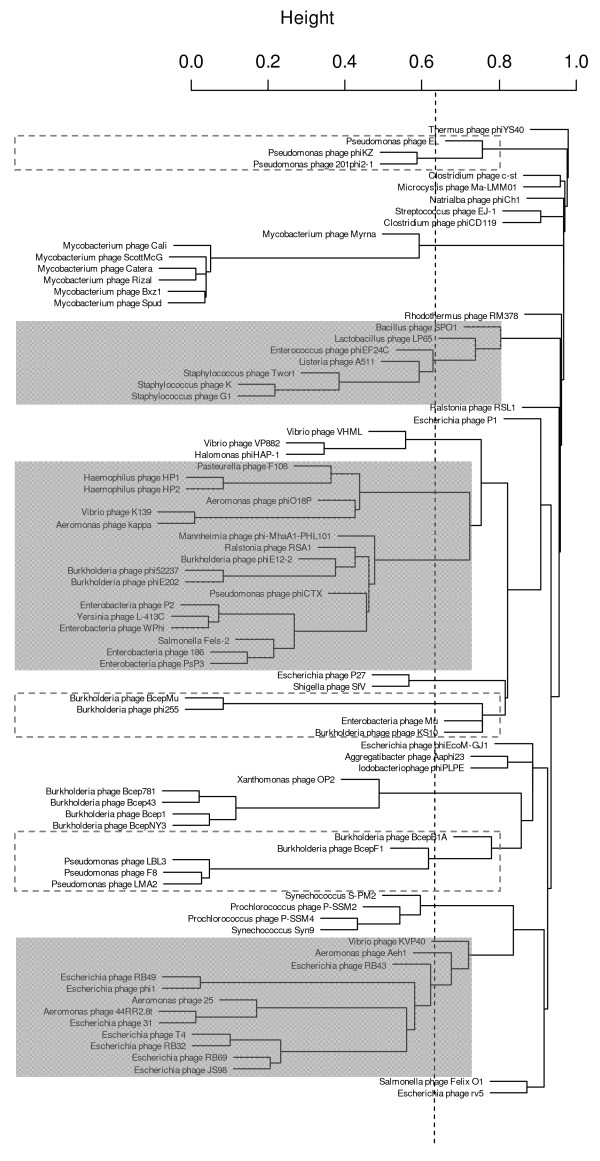
Figure 1 shows the correlation, based on the CoreExtractor distance measure, among all available Myoviridae genomes in the NCBI databases. To verify and more subtly compare individual correlations, the CoreGenes approach was applied to subsets of related phages, including several genomes not currently available in public databases (Table 1). As in previous analyses of the Podoviridae [2], threshold values of 40% and 20% (and 0.6 and 0.8 relative dissimilarity, respectively) of homologous proteins strongly suggest genus and subfamily boundaries, respectively (Additional file 1). They are corroborated by morphological, molecular or physiological data and discussed in the paragraphs below.
Figure 1.
Hierarchical cluster dendrogram of the analyzed Myoviridae. The relative dissimilarity between the phage proteomes (between 0.0 and 1.0) forms the basis for the proposed groupings. The dotted lines reflects the cut-off value used for the establishment of genera, used consistently for all Myoviridae and the previously defined Podoviridae [107]. Subfamily and tentative subfamily groupings are indicated in the grey and dotted boxes, respectively.
Table 1.
Comparison of CoreExtractor and CoreGenes and the classification of fully sequenced members of the Myoviridae
| I. TEEQUATROVIRINAE | ||||
|---|---|---|---|---|
| Percent identity | ||||
| 1. The T4-like viruses | Accession No. | CoreExtractor | CoreGenes | |
| T4-type phages | ||||
| Escherichia phage T4 | NC_000866 | 100 | 100.0 | |
| Escherichia phage JS10 | NC_012741 | Not determined | 72.7 | |
| Escherichia phage JS98 | NC_010105 | 77 | 74.1 | |
| Escherichia phage RB14 | NC_012638 | Not determined | 83.5 | |
| Escherichia phage RB32 | NC_008515 | 88 | 84.2 | |
| Escherichia phage RB51 | NC_012635 | Not determined | 85.6 | |
| Escherichia phage RB69 | NC_004928 | 73 | 73.4 | |
| 44RR2.8-type phages | ||||
| Aeromonas phage 44RR2.8t | NC_005135 | 100 | 100.0 | |
| Escherichia phage 31 | NC_007022 | 98 | 97.6 | |
| Aeromonas phage 25 | NC_008208 | 82 | 82.5 | |
| RB43-type phages | ||||
| Escherichia phage RB43 | NC_007023 | 100 | 100.0 | |
| Escherichia phage RB16 | Tulane | Not determined | 84.2 | |
| RB49-type phages | ||||
| Escherichia phage RB49 | NC_005066 | 100 | 100.0 | |
| Escherichia phage JSE | NC_012740 | Not determined | 93.6 | |
| Escherichia phage φ1 | NC_009821 | 97 | 97.1 | |
| 2. The KVP40-like viruses | ||||
| Vibrio phage KVP40 | NC_005083 | 100 | 100.0 | |
| Vibrio phage nt-1 | Tulane | Not determined | 80.8 | |
| Acinetobacter phage 133 | Tulane | Not determined | 39.9 | |
| Aeromonas phage Aeh1 | NC_005260 | 28 | 35.6 | |
| Aeromonas phage 65 | Tulane | Not determined | 34.9 | |
| II PEDUOVIRINAE | ||||
| 1. The P2-like viruses | ||||
| Enterobacteria phage P2 | NC_001895 | 100 | 100.0 | |
| Enterobacteria phage Wφ | NC_005056 | 89 | 90.7 | |
| Yersinia phage L-413C | NC_004745 | 95 | 88.4 | |
| Enterobacteria phage 186 | NC_001317 | 72 | 74.4 | |
| Enterobacteria phage PsP3 | NC_005340 | 70 | 72.1 | |
| Salmonella Fels-2 | NC_010463 | 65 | 67.4 | |
| Salmonella SopEφ | AY319521 | Not determined | 62.8 | |
| Burkholderia phage φE202 | NC_009234 | 51 | 55.8 | |
| Mannheimia phage φ-MhaA1-PHL101 | NC_008201 | 51 | 55.8 | |
| Pseudomonas phage φCTX | NC_003278 | 53 | 53.5 | |
| Burkholderia phage φ52237 | NC_007145 | 49 | 51.2 | |
| Ralstonia phage RSA1 | NC_009382 | 49 | 51.2 | |
| Burkholderia phage φE12-2 | NC_009236 | 49 | 48.8 | |
| 2. The HP1-like viruses | ||||
| Haemophilus phage HP1 | NC_001697 | 100 | 100.0 | |
| Haemophilus phage HP2 | NC_003315 | 97 | 85.7 | |
| Pasteurella phage F108 | NC_008193 | 57 | 59.5 | |
| Vibrio phage K139 | NC_003313 | 51 | 54.8 | |
| Vibrio phage κ | NC_010275 | 49 | 54.8 | |
| Aeromonas phage ΦO18P | NC_009542 | 44 | 50.0 | |
| III. SPOUNAVIRINAE | ||||
| 1. The SPO1-like viruses | ||||
| Bacillus phage SPO1 | NC_011421 | 100 | 100.0 | |
| 2. The Twort-like viruses | ||||
| Staphylococcus phage Twort | NC_007021 | 100 | 100.0 | |
| Staphylococcus phage K | NC_005880 | 74 | 43.5 | |
| Staphylococcus phage G1 | NC_007066 | 97 | 56.9 | |
| Listeria phage P100 | NC_007610 | 51 | 34.8 | |
| Listeria phage A511 | NC_009811 | 51 | 35.4 | |
| Peripherally related: | ||||
| Enterococcus phage φEC24C | NC_009904 | 32 | 31.8 | |
| Lactobacillus phage LP65 | NC_006565 | 25 | 26.2 | |
| OTHER ICTV-RECOGNIZED GENERA | ||||
| 1. The Mu-like viruses | ||||
| Enterobacteria phage Mu | NC_000929 | 100 | 100.0 | |
| 2. The P1-like viruses | ||||
| Escherichia phage P1 | NC_005856 | 100 | 100.0 | |
| Escherichia phage P7 | AF503408 | Not determined | 87.3 | |
| PROPOSED GENERA WITHIN THE MYOVIRIDAE | ||||
| 1. The Bcep781-like viruses | ||||
| Burkholderia phage Bcep781 | NC_004333 | 100 | 100.0 | |
| Burkholderia phage Bcep43 | NC_005342 | 98 | 95.5 | |
| Burkholderia phage Bcep1 | NC_005263 | 85 | 90.9 | |
| Burkholderia phage BcepNY3 | NC_009604 | 87 | 92.4 | |
| Xanthomonas phage OP2 | NC_007710 | 52 | 50.0 | |
| 2. The BcepMu-like viruses | ||||
| Burkholderia phage BcepMu | NC_005882 | 100 | 100.0 | |
| Burkholderia phage φE255 | NC_009237 | 89 | 86.8 | |
| 3. The FelixO1-like viruses | ||||
| Salmonella phage Felix O1 | NC_005282 | 100 | 100.0 | |
| Escherichia phage wV8 | EU877232 | Not determined | 92.4 | |
| Erwinia phage φEa21-4 | NC_011811 | Not determined | 52.7 | |
| 4. The HAP1-like viruses | ||||
| Halomonas phage φHAP-1 | NC_010342 | 100 | 100.0 | |
| Vibrio phage VP882 | NC_009016 | Not determined | 73.9 | |
| 5. The Bzx1-like viruses | ||||
| Mycobacterium phage Bzx1 | NC_004687 | 100 | 100.0 | |
| Mycobacterium phage Catera | NC_008207 | 95 | 95.4 | |
| Mycobacterium phage Cali | NC_011271 | 92 | 93.6 | |
| Mycobacterium phage ScottMcG | NC_011269 | 93 | 94.5 | |
| Mycobacterium phage Rizal | NC_011272 | 95 | 95.9 | |
| Mycobacterium phage Spud | NC_011270 | 97 | 98.2 | |
| Mycobacterium phage Myrna | NC_011273 | 39 | 46.3 | |
| 6. The phiCD119-like viruses | ||||
| Clostridium phage ΦCD119 | NC_007917 | Not determined | 100.0 | |
| Clostridium phage ΦCD2 | NC_009231 | Not determined | 50.6 | |
| Clostridium phage ΦCD27 | NC_011398 | Not determined | 36.7 | |
| 7. The phiKZ-like viruses | ||||
| Pseudomonas phage φKZ | NC_004629 | 100 | 100.0 | |
| Pseudomonas phage 201φ2-1 | NC_010821 | 50 | 51.0 | |
| Peripherally related: | ||||
| Pseudomonas phage EL | NC_007623 | 30 | 21.9 | |
| 8. The PB1-like viruses | ||||
| Pseudomonas phage PB1 | NC_011810 | Not determined | 100.0 | |
| Pseudomonas phage F8 | NC_007810 | Not determined | 95.7 | |
| Pseudomonas phage LBL3 | NC_011165 | 97 | 89.2 | |
| Pseudomonas phage LMA2 | NC_011166 | 97 | 95.7 | |
| Pseudomonas phage SN | NC_011756 | Not determined | 92.5 | |
| Pseudomonas phage 14-1 | NC_011703 | Not determined | 92.5 | |
| Burkholderia phage BcepF1 | NC_009015 | 44 | 43.0 | |
| Peripherally related: | ||||
| Burkholderia phage BcepB1A | NC_005886 | 22 | 24.7 | |
| PRELIMINARY GROUPINGS AND UNRELATED PHAGES | ||||
| (cyanomyoviridae) | ||||
| Synechococcus S-PM2 | NC_006820 | 100 | 100.0 | |
| Synechococcus Syn9 | NC_008296 | 41 | 41.5 | |
| Prochlorococcus phage P-SSM2 | NC_006883 | 35 | 40.3 | |
| Prochlorococcus phage P-SSM4 | NC_006884 | 35 | 39.8 | |
| (phage SfV and relatives) | ||||
| Shigella phage SfV | NC_003444 | 100 | 100.0 | |
| Escherichia phage P27 | NC_003356 | 42 | 43.1 | |
| Aggregatibacter phage Aaφ23 | NC_004827 | 100 | 100.0 | |
| Clostridium phage c-st | NC_007581 | 100 | 100.0 | |
| Escherichia phage rV5 | NC_011041 | 100 | 100.0 | |
| Escherichia phage P4 | NC_001609 | 100 | 100.0 | |
| Escherichia phage φEcoM-GJ1 | NC_010106 | 100 | 100.0 | |
| Iodobacteriophage phiPLPE | NC_011142 | 100 | 100.0 | |
| Lactobacillus phage Lb338-1 | NC_012530 | 100 | 100 | |
| Microcystis phage Ma-LMM01 | NC_008562 | 100 | 100.0 | |
| Natrialba phage φCh1 | NC_004084 | 100 | 100.0 | |
| Ralstonia phage RSL1 | NC_010811 | 100 | 100.0 | |
| Rhodothermus phage RM378 | NC_004735 | 100 | 100.0 | |
| Streptococcus phage EJ-1 | NC_005294 | 100 | 100.0 | |
| Thermus phage φYS40 | NC_008584 | 100 | 100.0 | |
A. Myoviridae Subfamilies
I. Teequatrovirinae
1. T4-like viruses nova comb
The ICTV currently lists only six sequenced viruses as members of the T4 phage genus, namely enterobacterial phage T4, Acinetobacter phage 133, Aeromonas phages Aeh1, 65 and 44RR2.8t, and Vibrio phage nt-1. However, the scientific literature and public databases abound with descriptions of "T4-like" phages and the analysis of complete genome sequences indicates that the T4-related phages constitute one of the largest groups of bacterial viruses. This corroborates ecogenomic studies on the diversity of these viruses as apparent in the heterogeneity of capsid (gp23) genes in isolates from Japanese rice fields [4], marine systems [5,6], and from Lithuania [7], Bangladesh and Switzerland [8]. These studies suggest that the fully sequenced T4 phages are but a small fraction of the T4-related genomes in nature. Nevertheless, there are clear commonalities among all sequenced "T4-like" genomes from different host groups, including the cyanophages, namely a set of 33-35 genes that have persisted during the evolution of genomes with sizes from 160 to 250 kb [9]. This core of genes seems to have resisted divergence throughout evolution. Nevertheless, these horizontal substitutions do not erase the evidence of the global relationship between phages and clear hybrid phages within this group have not been identified to date [10,11]. Work done at Tulane University [10,11], led to the tentative conclusion that it takes about 33 T4 genes to determine a genetic program that controls lytic phage development in the host cell.
Based on the Myoviridae cluster dendrogram (Figure 1), the current ICTV genus "T4-like viruses" can be subdivided into two genera and several subgroups. By analogy to the T7-related podoviruses, now named the Autographivirinae, the former ICTV genus was raised to the rank of a subfamily, the Teequatrovirinae, named after the best-studied of these phages, coliphage T4. The first genus, the "T4-like viruses", includes what were previously termed the T-even and "pseudo-T-even" phages [12,13]. Our name perpetuates the old ICTV nomenclature, but is now limited to enterobacterial and Aeromonas phages. The KVP40 phages, consisting of two former members of the "schizo-T-evens" [14] form the other genus.
The "T4-like viruses" are morphologically indistinguishable and have moderately elongated heads of about 110 nm in length, 114 nm long tails with a collar, base plates with short spikes, and six long kinked tail fibers. Within this assemblage, we identified four distinct subtypes with >70% protein similarity. These are the T4-type phages (phages T4, JS10, JS98, RB14, RB32, RB51, RB69), 44RR-type (phages 44RR2.8t, 31, 25), RB43-type (RB43, RB16), and the RB49-type viruses (RB49, JSE, φ1). They can be subdivided by the presence of specific encoded proteins as outlined in Table 2. In the subtype T4 phages, three specific proteins with defined functions (Pin, MotB, ModA) were found. Pin is an inhibitor of the host's Lon protease [15,16], while the other two proteins function to modulate transcription [17,18].
Table 2.
Type-specific proteins in T4 phages
| Type (host) | Genome size (in kb) | Type-specific proteins |
|---|---|---|
| T4 (E. coli) | 165.9-170.5 | NP_049650, 049704, 049747, 049694 (Pin), 049626 (MotB), 049635 (ModA) |
| 44RR2.8t (Aeromonas) | 161.5-173.6 | NP_932430, 932451, 932460, 932567, 932569, 932577 |
| RB49 (E. coli) | 164.1 | NP_891619, 891621, 891622, 891626, 891736, 891753, 891760, 891800, 891816 |
| RB43 (E. coli) | 178.7 | YP_239033, 239034, 239054, 239086, 239094, 239097, 239130, 239215, 239216, 239241 |
Heteroduplex analyses indicate that coliphages T2, T4 and T6 share >85% sequence similarity [19], warranting their inclusion, in spite of lack of detailed sequence data for T2 and T6, into the T4-type subgroup. The DNA of the T-even phages contains 5-hydroxymethylcytosine (5-HMC). While this modified nucleotide is common in T4-related phages [20], its presence has not been ascertained biochemically in the other phages (JS98, RB14, RB32, RB69) included in this subgroup. T4 gp42 dCMP hydroxymethylase and Alc that blocks transcription from cytosine containing DNA are required for the incorporation of 5-HMC rather than cytosine into T-even DNA. Genes specifying homologs of the T4 gp42 and Alc proteins are also present in the 44RR2.8t-type phages.
2. KVP40-like viruses
The KVP40 viruses comprise two marine vibriophages, KVP40 and nt-1, with genomes of approximately 246 kb. KVP40 infects Vibrio parahaemolytius and was isolated from seawater. Phage nt-1 infects Vibrio natriegens and originates from a coastal marsh. The phages differ from T4 in head length (137 nm vs. 111 nm), but are identical to phage T4 in tail morphology. KVP40 has a feather of decoration proteins on its head [21,22].
Three other T4 phages do not fit into these groups: Acinetobacter phage 133, Aeromonas hydrophila phage Aeh1 and Aeromonas salmonicida phage 65. Morphologically, phage 133 is identical to T4, whereas Aeh1 and 65 have the same heads of 133 nm in length as Vibrio phages KVP40 and nt-1. They were considered to be part of the "schizo-T-even" group [23] and have a T4-type tail structure [20]. CoreGenes and our supplementary phylogenetical analyses indicate that these phages are too dissimilar, by our criteria, to be included into one of the genera listed above.
The four marine cyanophages (P-SSM2, P-SSM4, S-PM2 and Syn9) infect Synechococcus or Prochlorococcus strains and harbor T4 genes causing this group to be named the "exo-T-evens" [24,25]. These phages have isometric heads and much longer tails than T4. CoreGenes analysis indicates that they form a group sharing >40% proteins in common. While P-SSM2, P-SSM4 and Syn9 share 93 proteins, they show considerable dissimilarity in appearance, size, and DNA content (Table 3). Phylogenetic analysis based upon sequence alignments of gp20 (portal vertex protein [26]) and photosystem II protein D1 [27,28] indicate considerable diversity exist among cultured and environmental cyanophages. This is also confirmed by an analysis of data from the marine virome from the Sorcerer II Global Ocean Sampling expedition [29]. Based upon these observations, we feel that the creation of genera within cyanophage myoviruses is premature at the present time.
Table 3.
T4 cyanophages
| Phage | Head, nm | Tail length, nm | DNA size, kb | ORFs | References |
|---|---|---|---|---|---|
| P-SSM2 | 110* | 100* | 252 | 327 | [103] |
| P-SSM4 | 70* | 200* | 178 | 198 | [103] |
| S-PM2 | 67 | 200 | 187 | 239 | [104,105] |
| Syn9 | 87 | 150 | 173 | 226 | [106] |
*From published micrographs.
Rhodothermus marinus phage RM378 (NC_004735) is a virus said to have a head of 95 × 85 nm and a tail of 150 nm in length [30]. It was called a "ThermoT-even phage" by Filée et al. [6], but our CoreGenes analysis reveals that its proteins shows minimal sequence similarity to any T4-related virus.
II. Peduovirinae
This subfamily is a large phage group derived from the ICTV genus "P2-like phages" and is named the Peduovirinae. Virions have heads of 60 nm in diameter and tails of 135 × 18 nm. Phages are easily identified because contracted sheaths tend to slide off the tail core. The subfamily falls into three different groups. As shown by CoreExtractor and CoreGenes analyses, and using the 40% similarity criterion for inclusion into the same genus, phage HP1 has only 9 genes in common P2. Even if other P2 phages are considered, HP1 shares only 17 genes with any phage of the "P2-like" genus. Using the 40% similarity criterion for inclusion into the same genus, it is therefore justified to consider P2 and HP1 as members of different genera and to upgrade the present genus "P2 phages" to a subfamily.
1. P2-like viruses nova comb
This genus includes P2 itself and its extensively studied relative, coliphage 186. Both originate from the Pasteur Institute in Paris, France. Phage P2 is one of three phages (P1, P2, P3) isolated by G. Bertani in the beginning of the 1950's from the "Li" (Lisbonne and Carrère) strain of E. coli [31]. Later on, F. Jacob and E. Wollman isolated phage 186 and many other viruses from enterobacteria collected by L. Le Minor [32]. The reason for the early interest in these phages was that P2 and 186 are temperate. The analysis of the genetic control of these two modes was the starting point for ongoing fertile research on phage biology and molecular biology in general.
The genomes of phage P2 and 186 were the first P2 genomes to be fully sequenced and analyzed. Almost all P2 and 186 genes have been assigned a function [33-35]. Coliphages WΦ and L-413C are very similar to P2 in both gene content and gene order. They are closely related to each other, sharing all but one protein. The only genes of these phages that differ from P2 are the lysogeny-related genes, which may have been horizontally acquired and are totally different, but have been inserted at the same locations into all genomes. The only exception to this is that phage P2 has a 786 bp ORF (orf30) with unknown function inserted between the S and V genes. There is no such insertion in WΦ and L-413C, but Pseudomonas phage ΦCTX (see below) has another uncharacterized ORF located at this position. Enterobacterial phages 186, PSP3, Fels-2, and SopEΦ also share their overall gene order and many genes with P2, but the genes are more diverged. Unlike P2, these phages are UV-inducible due to the presence of the tum gene. In addition, they have a different lysis-lysogeny switch region. P2 phages seem to have either of two different proteins for repression of the lytic cycle. P2, WΦ and L-413C have the repressor gene C whereas 186, PSP3, Fels-2, SopEΦ, HP1, HP2, and K139 (below) instead have the sequence-unrelated genes CI and CII, both of which are equally needed for establishing lysogeny.
Mannheimia phage Φ-MhaA1-PHL101, Pseudomonas phageΦCTX, and Ralstonia phage RSA1 have many P2 genes and an overall order of structural genes that is P2-like, although interspersed with some uncharacterized genes. Their presumed regulatory gene regions include additional putative and uncharacterized ORFs. Phage ΦCTX has only the P2 regulatory gene ogr (transcriptional activator of the late genes) and the recombination enzyme int (integrase), Φ-MhaA1-PHL101 has repressor (CI) and antirepressor (Cro) equivalents which are most closely related to the regulatory proteins of the P22-like enterobacteria phage ST104 than to P2.
Phage RSA1 seems to have only one P2-related regulatory gene, the ogr gene, although it is more related to the Ogr/Delta-like gene in ΦCTX. The RSA1 integrase is more similar to the integrases of the P2-like Burkholderia phages (ΦE202, Φ52237, and ΦE12-2 and P22-like viruses.
2. HP1-like viruses
The genome architecture of HP1 [36] and its close relative, HP2, resembles that of P2 although their cos sites, as with Pseudomonas ΦCTX [37], are located next to attP rather than downstream of the portal protein-encoding gene as it is in P2. The P2 gene order is also conserved in Vibrio phages K139 [38] and κ and the Pasteurella phage F108 [39]. As in P2, the genomes can be divided into blocks of structural and regulatory genes. The structural genes are more similar in HP1 and HP2 than the regulatory genes. The six genes coding for capsid proteins are arranged in the same order in HP1 phages and many P2 phages. The other structural genes, coding mainly for tail components, show generally no similarity to those of P2 phages. Only some of the regulatory genes are similar in both HP1 and P2 phages, e.g., int, CI, and repA. Regulatory genes in general are more conserved within the HP1 group.
Aeromonas phage ΦO18P [40] is included into the HP1 phages. It contains slightly more genes related to HP1 than to P2, although, when looking at individual proteins, it sometimes appears to have an intermediate position. Its Rep protein is very similar to the DNA replication protein of Salmonella phage PSP3 and the A protein of phages K139, F108, WΦ, and P2 homologs. The ΦO18P major capsid protein is similar to the capsid proteins of phages K139, ΦCTX, 186, and the Burkholderia phages.
III. The Spounavirinae
This proposed subfamily contains the ICTV-recognized genus "SPO1-like viruses" and, on the basis of our results, a proposed new genus (the "Twort-like viruses") and two peripherally related viruses, Lactobacillus plantarum phage LP65 [41] and Enterococcus faecalis phage φEF24C [42,43]. All of these are virulent, broad-host range phages which infect members of the Firmicutes. They possess isometric heads of 87-94 nm in diameter and conspicuous capsomers, striated 140-219 nm long tails, a double base plate, and globular structures at the tail tip. The latter have been resolved as base plate spikes and short kinked tail fibers with six-fold symmetry [44]. Members of this group usually possess large (127-142 kb) nonpermuted genomes with 3.1-20 kb terminal redundancies [45,46]. The proposed name for this subfamily is derived from SPO plus una (latin for "one").
While the head diameter of Bacillus phage SPO1, of 87 nm [47], is consistent with membership in the group, its tail is significantly shorter than that of most members (140-150 nm) [3,48], and, the DNA contains 5-hydroxymethyluracil (HMU) rather than thymine. The outliers of this group comprise phages LP65 [41] and φEF24C [42,43]. At 193 nm, the tail of phage LP65 is similar in length to that of other members of this group, but its genome is not terminally redundant [41]. Lastly, the genome size (142 kb), proteome and morphology of Enterococcus phage φEF24C is clearly consistent with membership in this group (head diameter 93 nm; tail length 204 nm), but its genome is circularly permuted. Their close relationship was discussed in a recent paper [44].
Using a BLASTP raw threshold score of 100 and CoreGenes 3.0 http://binf.gmu.edu:8080/CoreGenes3.0/ to compare the proteomes of Twort, A511, LP65, and φEF24C against SPO1, we identified two clusters of genes which are conserved. These corresponded to packaging and morphogenesis genes (SPO1 gp2.11 to gp16.2); and the cluster of replication genes, including helicase, exonuclease, primase, and resolvase (SPO1 gp19.5 - gp24.1). The DNA polymerases (SPO1 gp31 and homologs) of these phages are related more closely to bacterial-type I DNA polymerases than other phage deoxynucleotide polymerizing enzymes. The presence of host-related proteins in viruses has been observed by Dinsdale et al. [49] and elegantly explained by Serwer [50]. Metagenomic studies by the former group indicate the presence of numerous host-related proteins, including those related to motility and chemotaxis, in the virome fractions. While the functional significance of photosynthetic protein psbA in cyanophage genomes has been conclusively shown [51,52], the presence of host-related sequences should still be considered with healthy skepticism if the only data is the presence of homologs.
1. SPO1-like viruses
The current ICTV genus "SPO1 viruses" comprises some 10 Bacillus phages and Lactobacillus phage 222a; only the genome of SPO1 has been sequenced [53]. All SPO1-like Bacillus phage genomes that have been studied contain 5-hydroxymethyluracil (HMU) instead of thymine and encode dUMP hydroxymethylase activity (SPO1 gp29). This phage also contains the unique 171-amino acid head decoration protein gp29.2. Whether this is unique to members of this genus will require the sequencing of additional genomes. Using cryo-electron microscopy, Duda and coworkers [54] confirmed the earlier observation [47] that the icosahedral head of SPO1 head has the triangulation number T = 16 rather than the more common T = 25. This feature is also shared with eukaryotic herpesviruses.
2. Twort-like viruses
The phages form a fairly homogeneous group of virulent phages infecting staphylococci (Twort, G1, K) [55] and Listeria (A511, P100) [56]. The group is named after phage "Twort," which may be a descendant of the original bacteriophage described by F.W. Twort in 1915 [57]. Apparently, this phage was deposited at the Pasteur Institute of Paris in 1947 when Twort was invited there to retell the story of his discovery (personal communication to H.-W.A. by J.-F. Vieu, curator of the phage collection of the Pasteur Institute; 1983).
B. Additional ICTV-recognized genera
1. Mu-like viruses
Phage Mu is morphologically almost identical to phage P2. Although phage Mu shares features (e.g. replicative transposition) with BcepMu [58] and two siphoviruses, Pseudomonas phages B3 and D3112 [59,60], this phage holds a unique position within the Myoviridae, since its proteome displays only limited homology to any other completely sequenced phage genome.
Mu and P2 have only 4 proteins in common (overall 9.8% similarity). P2 differs from Mu by genome size (33.6 kb vs. 36.7 kp in Mu), the number of proteins (43 proteins vs. 55 in Mu), gene order, and the presence of a single capsid protein and cohesive ends in its DNA. By contrast, Mu has two capsid proteins and two sets of tail fiber genes and replicates via transposition, which is a very rare mode of replication. Mu shares this characteristic with BcepMu, but BcepMu has no tail fiber inversion system and only a limited proteomic correlation to Mu (9 gene homologs; 16.4% similarity).
Only coliphage D108, as shown by heteroduplex analysis, shows significant similarity to Mu to warrant inclusion in the Mu genus [61]. Unfortunately, only portions of the genome of D108 have been sequenced. Putative Mu proviruses have been reported in a wide range of bacteria [62-64]. CoreGenes analysis revealed that only some of them can be reasonably described as Mu proviruses, namely, Escherichia blattae prophage MuEb [65], Haemophilus influenzae Rd prophage Hin-Mu [66], and Shewanella oneidensis prophage MuSo2 [NC_004347].
2. P1-like viruses
This small genus comprises coliphage P1, famous for its cre recombinase and the large-insert cloning vectors engineered on the basis of the phage genome [67], P7 (AF503408) and its not yet sequenced putative relative D7. Phage P1 stands out from any of the phages described here by its morphology. Phage P1 differs from the phages described here by size and morphology. It has a very large head of approximately 85 nm in diameter and a very long tail of 228 × 18 nm in the extended state. Tails have base plates and 90 nm long, kinked fibers. The tails of related, not yet sequenced phages of enterobacteria and Aeromonas vary between 170 and 240 nm in length. All phages of this group produce three types of head-size variants (small, normal, intermediate).
C. Additional genera within the Myoviridae
1. Bcep781-like viruses
"Bcep" stands for Burkholderia cepacia, and phages with this designation infect bacteria belonging to the B. cepacia genomic complex. The Bcep781 phages form a group of virulent myophages of which the genome sequence of five members, Bcep781, Bcep1, Bcep43, BcepNY3 and Xanthomonas phage OP2, is known [68,69]. The Bcep781 phages are small viruses with distinctly shorter tails than P2, Mu, and BcepMu [68].
The genomes of these phages range from 46 to 49 kb in size and encode 66 to 71 proteins. The four Bcep phages encode a single tRNA each. They form a homogeneous phage group not just in terms of sequence, but also by their distinctive genome organization compared to other groups. The genomes of the Bcep781 phages are divided into four gene clusters encoded on alternate strands such that, using Bcep781 as the example, genes 1 through 19 and 29 through 51 are present on the bottom strand while genes 20 through 28 and 52 through 66 are present on the top strand. Head genes are located in the first cluster and tail genes are located in the third cluster. The virion major capsid and decoration proteins, Bcep781 gp12 and gp13, were identified by protein sequencing and show some similarity to head proteins from the "PB1-like viruses" group. Several tail morphogenesis proteins, corresponding to Bcep781 gp29 through gp52, can be linked to P2 tail genes by PSI-BLAST. In contrast to structural genes, genes for DNA replication and lysis are scattered throughout the genome. The lysis genes of these phages are not organized into a cassette but instead overlapping Rz and Rz1 genes are separated from the endolysin and holin genes [70]. A distinctive feature of these phages is the presence of highly, maybe completely, circularly permuted genomes. The terminases of these phages are strongly related to other pac-type phages that also have highly permuted genomes [71].
2. BcepMu-like viruses
This group was named "BcepMu-like viruses" because, like Mu and unlike most other phages, its members utilize transposition for replication. The distinctive genomic feature implicating the use of replicative transposition is the presence of random host DNA sequences at either end of the packaged virion DNA [58]. These host sequences are derived from excision of prophage DNA from random sites scattered over the host genome. This requires fundamental differences in terminase function as compared to more typical terminases that utilize concatemers of phage genomic DNA as a substrate. This is reflected by the homology between BcepMu TerL and Mu TerL. Another genome feature shared by BcepMu and Mu is the presence of genomic terminal CA dinucleotide repeats, a feature common in many transposons. Furthermore, BcepMu and Mu seem to be morphologically identical.
Despite these similarities, BcepMu and its close relative φE255 have marked differences in genome organization and minimal overall protein sequence similarity to Mu, explaining why they have not been grouped together. The putative BcepMu transposase is not related to the Mu transposase, TnpA, but instead is a distant member of the Tn552-IS1604 transposase family. The BcepMu genome is organized into two clusters, with genes 1 through 13 encoded on the bottom strand and genes 17 through 52 on the top strand. The cluster of bottom strand genes includes transcription regulators, the transposase, and a number of small genes of unknown function. The lysogeny control region is likely to include genes 16 and 17, located at the interface of the bottom strand/top strand gene clusters. This is followed by a lysis cassette consisting genes encoding a holin, endolysin, Rz and Rz1. Proteins 27 through 51 encompass the head and tail morphogenesis cassette. The BcepMu tail biosynthetic cassette proteins are recognizably related both in sequence and in gene order to those of coliphage P2.
BcepMu is present as a prophage in many B. cenocepacia strains of the human pathogenic ET2 lineage [58,72]. Phage φE255 is a phage of the soil saprophyte B. thailandensis [NC_009237]. BcepMu phages, however, are not limited to Burkholderia hosts as related prophage elements have been identified in the genomic sequence of many other bacteria, for example Chromobacterium violaceum [NP_901809].
3. Felix O1-like viruses
Salmonella phage Felix O1 has a relatively large head (70 nm in diameter) and a tail of 138 × 18 nm characterized by subunits overlapping each other like roof tiles and showing a criss-cross pattern like phages PB-1 and F8. Notably, it exhibits small collars and eight straight tail fibers. Upon contraction, the base plate separates from the sheath. The type virus Felix O1 is widely known as a diagnostic Salmonella-specific phage [21]. Until recently, the genomic sequence (86.1 kb) of phage Felix O1 was unique and was considered, as such, a "genomic orphan", but two related genomes have been recently characterized, though their sequences have yet to be deposited to the public databases. They are coliphage wV8 and Erwinia amylovora phage φEa21-4 (DNA sizes 88.5 and 84.6 kb, respectively [73,74].
4. HAP1-like viruses
This genus contains two marine phages, Vibrio parahaemolyticus phage VP882 (NC_009016) and Halomonas aquamarina phage φHAP-1 [75]. Both are temperate viruses possessing 38-43 kb genomes which lack integrase genes. While our proteomic analysis and the literature suggests that Vibrio harveyi phage VHML [76,77] should be included in this genus, there is no evidence that this phage can be propagated: it is only produced after induction, does not plaque, and must be considered a defective prophage. The data presented by Mobberley et al. [78] show that φHAP-1 exists as a linear prophage in lysogens and possesses a protelomerase (ORF34, YP_001686770.1) and a partitioning protein (ParA homolog, ORF33, YP_001686769.1) which are homologous to proteins encoded by VHML and VP882. While these viruses share some homology with the coliphage P2, this is largely restricted to the genes associated with tail morphogenesis V (gpV, W, J, I, H, G) and F operons (gpFI, FII, E, T, U, D). Based upon their radically different life cycle from the other P2 phages, we have chosen not to include them in the Peduovirinae.
5. Bzx1-like or I3-like viruses
Myoviruses are exquisitely rare in the Actinobacteria (only an estimated 1% of all attempts to isolate phages from cultures was successful [79]). Phages I3, Bzx1 and Catera are characterized by heads of 80 nm in diameter and unusually short tails of 80 nm in length with a cup-shaped base plate. They do not resemble any other mycobacteriophages nor any other myovirus. We propose that this genus contains the following eight Mycobacterium smegmatis bacteriophages: I3, Bxz1, Cali, Catera, Myrna, Rizal, ScottMcG and Spud. Phage I3, which has been the first to be described, is the type virus of the newly proposed myovirus genus although it has not yet been fully sequenced. Within this assemblage, we identified a distinct subtype which show >90% protein similarity to Bxz1 (Cali, Catera, Rizal, ScottMcG and Spud) and genomes of 154-156 kb [80,81]. Mycobacteriophage Myrna, with a genome of 164 kb, shares approximately 45% of proteins with the Bxz1 subgroup phages. Interesting features include the presence of adenylosuccinate synthase homologs among the Bxz1 subgroup (gp250) and its absence in the genome of Myrna. The latter possesses several proteins not present in the Bxz1 group, including the large hypothetical proteins gp187 (YP_002225066.1) and gp243 (YP_002225120.1), a putative nicotinate phosphoribosyltransferase (gp263, YP_002225140.1) and ATP-dependent protease (gp262, YP_002225139.1).
6. phiCD119-like viruses
These are all integrative temperate phages of Clostridium difficile with genomes ranging from 51-60 kb in size and a mol%G+C of 28.7-29.4 [82-84]. The genus is named after its first fully sequenced member. In each case, the electron micrographs are of poor quality [84,85] or the measurements are very variable with large standard deviations [85]. Virus head diameters are given as 50-65 nm and tail lengths are said to range from 110 to 210 nm [82-84]. In certain cases, their annotation is also questionable, The multiple repressor/antirepressors annotated in the genomes of ΦCD27 and φC2 do not appear to contain helix-turn-helix or other DNA binding motifs [86]; nor the presence, in the latter phage, of ParA/ParB homologs. What unites these viruses, in addition to similar proteomes, is the presence in each of a cytosine-C5 specific DNA methylase (pfam00145, DNA_methylase, C-5 cytosine-specific DNA methylase; ΦCD119 protein YP_529611.1) and a DNA replication cassette composed of three proteins: a DnaD (primosome recruiting protein, presumably analogous to lambda gpO and P22 gp18; ΦCD119 protein YP_529603.1), a hypothetical protein (misidentified in ΦCD27 as a putative resolvase/integrase and missed entirely in the annotation of ΦCD119) and a single-stranded DNA binding protein.
7. phiKZ-like viruses
Phages φKZ and EL are members of a group of giant phages isolated, to date, only in Pseudomonas species. Their heads are isometric, 120 nm in diameter, and they possess 190 nm-long tails. The phage heads contain an inner body. The DNA of φKZ is over 280 kb in size and has 306 ORFs, most of which are unrelated to ORFS of any known protein [87], while EL contains 201 ORFs within its 211 kb genome [88]. These two phages and Pseudomonas phage Lin68 have recently been proposed as part of a genus "phiKZ viruses" [89]. We now consider that the differences (number of ORFs, mol%G+C, protein homologs) between φKZ and EL exclude EL from membership in the same genus. Indeed, the recent analysis of novel Pseudomonas phage 201φ2-1 [90] showed this phage to have a strong correlation to φKZ (167 similar proteins), suggesting that it is a true member of the phiKZ virus genus.
8. PB1-like viruses
This genus is named after the first isolated member of this group (PB1) [91]. Morphological and DNA-DNA hybridization studies by V. Krylov indicated that the following Pseudomonas phages were related: E79, 16, 109, 352, 1214, FS, 71, 337, φC17, SL2, B17 [92]. The sequences of a number of viruses belonging to this genus, namely F8, BcepF1, PB1, 14-1, LBL3, LMA2, and SN (P.-J. Ceyssens, personal communication) have now been completed. None of these phages encodes a recognizable integrase, suggesting that they are virulent.
Phage F8 is one of the Pseudomonas typing phages from the Lindberg set which includes six more similar phages [93,94]. It possesses a 70-nm wide head with visible capsomers and a 138 nm-long tail, four short straight tail fibers and a base plate that separates from the sheath upon contraction. The tail exhibits no transverse striations, but presents a criss-cross pattern [95]. This criss-cross pattern is a rare feature that has only been observed in phage Felix O1.
BcepF1 was isolated from soil by enrichment culture [96] using a Burkholderia ambifaria strain as its host (E.J. Summer and C.F. Gonzalez, unpublished). The BcepF1 genome is 72 kb in size and encodes 127 proteins while the genome of F8 is 66 kb and encodes 91 proteins [97]. Both genomes are organized into four alternating, unequal gene clusters on the top and bottom strands. The phages share 43 recognizable homologous proteins. The shared proteins specify virion morphogenesis, DNA metabolism and packaging and include a number of hypothetical proteins of unknown function.
A striking feature of both F8 and BcepF1 is the large number of small genes, all encoding hypothetical proteins and clustered together. In BcepF1, the first 20 kb of the genome, encoding 62 proteins, is devoted almost exclusively to these. In F8, there are two clusters of 8 kb (encoding gp1 through gp16, except gp4, TerL) and 4 kb (encoding proteins gp77 through gp91) of primarily small hypothetical novel genes. These heterogeneous regions are largely responsible for the difference in genome size and protein content between the two phages. It has generally been assumed that these small proteins are involved in host take-over (E. Kutter, personal communications) which appears to be substantiated by the results of Liu and coworkers [98].
Phages F8 and BcepF1 have some similarity to myophage BcepB1A, which is itself related in a mosaic fashion to the Bcep781 group of phages [68]; however, these similarities are essentially limited to morphogenetic proteins. As in the Bcep781 phages, several putative tail assembly proteins of F8 and BcepF1 can be linked to those of P2 by PSI-BLAST.
C. Single phages
In addition to the phage groups listed above, complete genome sequences are available for phages without apparent relatives, namely Aggregatibacter (formerly Actinobacillus) phage Aaφ23; Bacillus thuringiensis phage 0305φ8-36, Clostridium phages c-st, Escherichia phages φEcoM-GJ1 and rV5; Microcystis phage Ma-LMM01, Ralstonia phage RSL1, Rhodothermus phage RM378; Streptococcus phage EJ-1, and Thermus phage φYS40. References to these phages may be found in the NCBI RefSeq database.
General summary
The comparison of proteomes by CoreGenes/CoreExtractor BLASTP programs appears to be a decisive progress in classifying tailed bacteriophages, i.e., our results corroborate the existing ICTV classification of the Myoviridae and are generally well compatible with other informatics-based studies (Table 4), like the reticulate clustering based on gene families [99] (Lima-Mendez, personal communication). Our studies also refine certain relationships and suggest new ones. Specifically, we propose three new subfamilies (Peduovirinae, Teequatrovirinae, Spounavirinae) and eight new genera (Bcep781, BcepMu, Bzx1, Felix, HAP1, PB1, phiCD119 and phiKZ-like viruses). The individualization of genera containing two or three members as well as of genomic orphans, e.g. coliphage P1 without apparent homologs, is taxonomically as valuable and important as the confirmation of the large T4 and P2 groups and in total agreement with previous informatics-based classifications (Table 4). Our studies once again prove the utility of the dual CoreGenes/CoreExtractor approach to defining relationships between large numbers of virus genomes. These relationships carry evolutionary relevance, since our proteomic analyses, combined with the phylogenetic studies [100], suggest that the Myoviridae are mainly influenced by vertical evolution rather than by horizontal gene transfer. As observed in the Cluster dendrogram, the clusters are populated unevenly - several include only one phage while two, the largest, include dozens phages. This reflects the fact that past phage research has focused on coliphages, and suggests that we should broaden our research to include phages from a broader range of bacteria.
Table 4.
Concordance of classifications
| Classification | ICTV | Proteomic Tree 2 | ---- | Phage_Finder | This work |
|---|---|---|---|---|---|
| Reference | ICTV VIIIth Report, 2005 | Edwards and Rohwer, 2005 | Serwer et al., 2004 | Fouts, 2006 | |
| Approach | Traditional | Signature genes | Large terminase | CoreGenes | |
| Phage or phage group | T4, Aeh1, KVP40, RB43, RB49, 25, 31 44RR2.8t, 65 | T4 | T4, KVP40, RB49 | T4, Aeh1, KVP40, RB43, RB49, 25, 31 44RR2.8t, 65 | |
| P1 | P1 | P1 | |||
| P2, Fels-2, HP1, HP2, K139, φCTX, 186 | P2. HP1, HP2, φCTX | P2, Fels-2, HP1, HP2, L413-C, 186; Mu | P2, φCTX, 186 (HP1 occupies a separate position) | P2, Fels-2, HP1, HP2, K139, L-413C, φCTX, 186 | |
| Mu | Mu | Mu | |||
| SPO1 | K | P100, Twort | SPOl, K, P100, Twort | ||
| ΦH | |||||
Comparison of our results with those of the ICTV (ICTV VIIIth Report, 2005), Proteomic Tree 2 (Edwards and Rohwer, 2005), Phage_Finder (Fouts, 2006) and phylogeny of terminases (Serwer et al., 2004).
Among the 102 analyzed Myoviridae, phage Mu displayed the most significant evidence of horizontal gene exchange. This virus is related to three members of pilus-specific Siphoviridae infecting Pseudomonas aeruginosa (DMS3, D3112, B3 [59,60,101]), sharing 20 to 40% of its genes with each of them. These phages can be viewed as true hybrids, produced by recombination of different ancestors and, like the couple lambda/P22 (to be described in a future paper), cross family boundaries based on tail morphology. Nonetheless, the majority of Myoviridae, when forced to cluster, do so in a logical manner: upgrading of the ICTV genus "P2 phages" to the Pduovirinae with two genera ("P2 viruses" and "HP1 viruses") is a straightforward proposal and the same is true for the Spounavirinae (SPO1 viruses and Twort viruses).
Relationships among T4-like phages are more complicated. We reject the postulated inclusion of the cyanophages since their overall similarity to T4 is too low for consideration, at least according to our criteria. Comeau and Krisch [29] have recently recognized three groups of T4-related phages. The "Near T4" group containing the T-evens, Pseudo T-evens, and Schizo T-evens; the "Far T4" clade including Exo-T4 phage RM378; and, the "Cyano T4" assemblage. We believe that the latter are sufficiently different from the other T4 viruses to be excluded from the Teequatrovirinae at this time. This implies that this subfamily currently contains two distinct genera: T4 and KVP40 viruses. Within our restricted "T4 phages" genus, four subtypes were identified (T4-type, 44RR2.8t-type, RB43-type and the RB49-type viruses). This is confirmed by the phylogenetic studies of Filée et al. [5] and our unpublished results. Since these subtypes include different species, no equivalent taxonomic level is currently available in the official ICTV classification. Perhaps the introduction of a "subgenus" level should be considered in order to account for the complexity of T4-related phages. Alternately, a general elevation of all taxonomic levels (from the subfamily level) may be envisioned.
This study illustrates the great diversity and biological richness of tailed phages. The number of independent genera is not surprising in view of the antiquity of tailed bacteriophages, which are found in archaea and bacteria and may predate the separation of these domains. It can be expected that many more phage groups will be found or individualized in the future. For example, this study does not include giant Bacillus phage G, the largest bacterial virus with a genome of 497,513 bp and 684 genes [102] whose sequence is not yet available for comparison.
We reiterate our statement in our publication on the taxonomy of the Podoviridae, "We highly recommend that the entire genome of any newly sequenced phage be thoroughly screened (BLASTX) against the Entrez Query "Viruses [ORGN]" databases to reveal all similarities for quick identification of potential relationships. A validation step using CoreGenes is essential and more precise for individual comparisons [2]."
Conclusion
Myoviridae can be classified by their proteomes into subfamilies and genera. This classification is in close agreement with ICTV - and other informatics-based classifications.
Methods
Phages and bioinformatic tools
This study is limited to the genomes of completely sequenced, viable Myoviridae from the databases of NCBI http://www.ncbi.nlm.nih.gov/ and the Tulane University at New Orleans, LA (GT4P, "Genomes of the T4 Phages"; http://phage.bioc.tulane.edu/, excluding prophages without a virion stage. We follow here the ICTV which classifies viable viruses only. Prophages and proviruses, prophage fragments, defective viruses, phage-like "bacteriocins", virus-like or phage-likes particles from sections or the environment, viroids, satellite viruses, plasmids, or transposons, or artificial virus hybrids are not considered. CoreExtractor and CoreGenes software were used as described previously [2]. In the case of CoreExtractor, the BLASTX analysis of phage gene products was performed using the NCBI Batch BLAST server, http://greengene.uml.edu/programs/NCBI_Blast.html hosted by the University of Massachusetts at Lowell, MA. Searches were performed against the NCBI nonredundant database (BLOSUM45 matrix, with a 0.05 expectancy cut-off value) (Additional Figure 2). Several versions of CoreGenes are available, with each upgrade incorporating previous functions, at http://www.binf.gmu.edu/genometools.html. In particular, for the current study, a version, CoreGenes3.0beta, was developed specifically for tallying the total number of genes contained in the genomes. It also displays a percent value of genes in common with a specific genome. Additionally, this version finds unique genes between two genomes. The BLASTP stringency setting was set at its default value (75). Proteins containing at least 132 amino acid residues were subjected to BLASTP analysis at NCBI or Tulane University.
Hierarchical cluster dendrogram
Cluster analysis was used to visualize the structure of the proteomic data. We constructed a dissimilarity matrix from the CoreExtractor matrix. The dissimilarity between two phage genomes was taken as one (1) minus the average of the two reciprocal correlation scores in the CoreExtractor matrix (Figure S1B). Subsequently, single linkage hierarchical clustering was performed using "The R Project for Statistical Computing" software http://www.r-project.org/.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
All the authors contributed to the writing of this manuscript. RL and AMK planned and executed the comparisons. RL, PM and DS developed the software used. Cluster dendrograms were generated by PD.
Supplementary Material
CoreExtractor comparison of Myoviridae phages. A. This Excel figure shows relative correlation scores (above 10%), based on the number of homologous proteins between two phages. Colour tags are added to visualize these correlations (from green to red for increasing correlation scores). B. Corresponding dissimilarity matrix.
Contributor Information
Rob Lavigne, Email: Rob.Lavigne@biw.kuleuven.be.
Paul Darius, Email: paul.darius@biw.kuleuven.be.
Elizabeth J Summer, Email: elizsum@tamu.edu.
Donald Seto, Email: dseto@gmu.edu.
Padmanabhan Mahadevan, Email: pmahadev@gmu.edu.
Anders S Nilsson, Email: anders.nilsson@gmt.su.se.
Hans W Ackermann, Email: ackermann4@gmail.com.
Andrew M Kropinski, Email: kropinsk@queensu.ca.
Acknowledgements
The authors thank Michael Graves (Greengene, University of Massachusetts at Lowell, MA) for access to the NCBI Batch BLAST server and Erika Lingohr (Laboratory for Foodborne Zoonoses) for her computer assistance. We also thank Ian Molineux, Elizabeth Kutter, Arianne Toussaint, Gipsi Lima-Mendez, Arcady Mushegian, Martin Loessner, Viktor Krylov, Harald Brüssow, David Prangishvili and Jim Karam for helpful discussions. A.K. is supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada. RL, H-WA and AK are members of the ICTV Subcommittee for Viruses of Prokaryotes. DS wishes to congratulate his graduate advisor Professor Maurice J. Bessman of The Johns Hopkins University on the occasion of his emeritus status after 50 contiguous years of funded research and upon his 80th birthday July 2008.
References
- Zafar N, Mazumder R, Seto D. CoreGenes: a computational tool for identifying and cataloging "core" genes in a set of small genomes. BMC Bioinformatics. 2002;3:12. doi: 10.1186/1471-2105-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne R, Seto D, Mahadevan P, Ackermann H-W, Kropinski AM. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Research in Microbiology. 2008;159:406–414. doi: 10.1016/j.resmic.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball A. In: VIIIth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball A, editor. New York, NY: Elsevier Academic Press; 2005. Virus Taxonomy; pp. 35–85. [Google Scholar]
- Jia Z, Ishihara R, Nakajima Y, Asakawa S, Kimura M. Molecular characterization of T4-type bacteriophages in a rice field. Environmental Microbiology. 2007;9:1091–1096. doi: 10.1111/j.1462-2920.2006.01207.x. [DOI] [PubMed] [Google Scholar]
- Filée J, Bapteste E, Susko E, Krisch HM. A selective barrier to horizontal gene transfer in the T4-type bacteriophages that has preserved a core genome with the viral replication and structural genes. Molecular Biology & Evolution. 2006;23:1688–1696. doi: 10.1093/molbev/msl036. [DOI] [PubMed] [Google Scholar]
- Filée J, Tétart F, Suttle CA, Krisch HM. Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12471–12476. doi: 10.1073/pnas.0503404102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausa V, Piesiniene L, Staniulis J, Nivinskas R. Abundance of T4-type bacteriophages in municipal wastewater and sewage. Ekologija (Vilnius) 2003;1:47–50. [Google Scholar]
- Zuber S, Ngom-Bru C, Barretto C, Bruttin A, Brüssow H, Denou E. Genome analysis of phage JS98 defines a fourth major subgroup of T4-like phages in Escherichia coli. Journal of Bacteriology. 2007;189:8206–8214. doi: 10.1128/JB.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau AM, Bertrand C, Letarov A, Tétart F, Krisch HM. Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology. 2007;362:384–396. doi: 10.1016/j.virol.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Nolan JM, Petrov V, Bertrand C, Krisch HM, Karam JD. Genetic diversity among five T4-like bacteriophages. Virology Journal. 2006;3:30. doi: 10.1186/1743-422X-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov VM, Nolan JM, Bertrand C, Levy D, Desplats C, Krisch HM, Karam JD. Plasticity of the gene functions for DNA replication in the T4-like phages. Journal of Molecular Biology. 2006;361:46–68. doi: 10.1016/j.jmb.2006.05.071. [DOI] [PubMed] [Google Scholar]
- Desplats C, Dez C, Tétart F, Eleaume H, Krisch HM. Snapshot of the genome of the pseudo-T-even bacteriophage RB49. Journal of Bacteriology. 2002;184:2789–2804. doi: 10.1128/JB.184.10.2789-2804.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod C, Repoila F, Kutateladze M, Tétart F, Krisch HM. The genome of the pseudo T-even bacteriophages, a diverse group that resembles T4. Journal of Molecular Biology. 1997;267:237–249. doi: 10.1006/jmbi.1996.0867. [DOI] [PubMed] [Google Scholar]
- Miller ES, Heidelberg JF, Eisen JA, Nelson WC, Durkin AS, Ciecko A, Feldblyum TV, White O, Paulsen IT, Nierman WC, Lee J, Szczypinski B, Fraser CM. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. Journal of Bacteriology. 2003;185:5220–5233. doi: 10.1128/JB.185.17.5220-5233.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Takahashi H. A novel expression system for production of a labile protein in Escherichia coli by infection with cytosin-substituting T4 phage. Agricultural and Biological Chemistry. 1991;55:2507–2513. [PubMed] [Google Scholar]
- Skorupski K, Tomaschewski J, Rüger W, Simon LD. A bacteriophage T4 gene which functions to inhibit Escherichia coli Lon protease. Journal of Bacteriology. 1988;170:3016–3024. doi: 10.1128/jb.170.7.3016-3024.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann B, Depping R, Gineikiene E, Kaliniene L, Nivinskas R, Ruger W. ModA and ModB, two ADP-ribosyltransferases encoded by bacteriophage T4: catalytic properties and mutation analysis. Journal of Bacteriology. 2004;186:7262–7272. doi: 10.1128/JB.186.21.7262-7272.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulitzer JF, Colombo M, Ciaramella M. New control elements of bacteriophage T4 pre-replicative transcription. Journal of Molecular Biology. 1985;182:249–263. doi: 10.1016/0022-2836(85)90343-2. [DOI] [PubMed] [Google Scholar]
- Kim JS, Davidson N. Electron microscope heteroduplex study of sequence relations of T2, T4, and T6 bacteriophage DNAs. Virology. 1974;57:93–111. doi: 10.1016/0042-6822(74)90111-1. [DOI] [PubMed] [Google Scholar]
- Ackermann H-W, Krisch HM. A catalogue of T4-type bacteriophages. Archives of Virology. 1997;142:2329–2345. doi: 10.1007/s007050050246. [DOI] [PubMed] [Google Scholar]
- Ackermann H-W, DuBow MS. Viruses of Prokaryotes. Boca Raton, FL: CRC Press; 1987. [Google Scholar]
- Ackermann H-W, Kasatiya SS, Kawata T, Koga T, Lee JV, Mbiguino A, Newman FS, Vieu JF, Zachary A. Classification of Vibrio bacteriophages. Intervirology. 1984;22:61–71. doi: 10.1159/000149535. [DOI] [PubMed] [Google Scholar]
- Tétart F, Desplats C, Kutateladze M, Monod C, Ackermann H-W, Krisch HM. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. Journal of Bacteriology. 2001;183:358–366. doi: 10.1128/JB.183.1.358-366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats C, Krisch HM. The diversity and evolution of the T4-type bacteriophages. Research in Microbiology. 2003;154:259–267. doi: 10.1016/S0923-2508(03)00069-X. [DOI] [PubMed] [Google Scholar]
- Hambly E, Tétart F, Desplats C, Wilson WH, Krisch HM, Mann NH. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11411–11416. doi: 10.1073/pnas.191174498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short CM, Suttle CA, Short CM, Suttle CA. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Applied & Environmental Microbiology. 2005;71:480–486. doi: 10.1128/AEM.71.1.480-486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I, Tzahor S, Williamson S, Shmoish M, Man-Aharonovich D, Rusch DB, Yooseph S, Zeidner G, Golden SS, Mackey SR, Adir N, Weingart U, Horn D, Venter JC, Mandel-Gutfreund Y, Beja O. Viral photosynthetic reaction center genes and transcripts in the marine environment. ISME Journal. 2007;1:492–501. doi: 10.1038/ismej.2007.67. [DOI] [PubMed] [Google Scholar]
- Tzahor S, Man-Aharonovich D, Kirkup BC, Yogev T, Berman-Frank I, Polz MF, Beja O, Mandel-Gutfreund Y. A supervised learning approach for taxonomic classification of core-photosystem-II genes and transcripts in the marine environment. BMC Genomics. 2009;10:229. doi: 10.1186/1471-2164-10-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau AM, Krisch HM. The capsid of the T4 phage superfamily: The evolution, diversity, and structure of some of the most prevalent proteins in the biosphere. Molecular Biology & Evolution. 2008;25:1321–1332. doi: 10.1093/molbev/msn080. [DOI] [PubMed] [Google Scholar]
- Blondal T, Hjorleifsdottir SH, Fridjonsson OF, Aevarsson A, Skirnisdottir S, Hermannsdottir AG, Hreggvidsson GO, Smith AV, Kristjansson JK. Discovery and characterization of a thermostable bacteriophage RNA ligase homologous to T4 RNA ligase 1. Nucleic Acids Research. 2003;31:7247–7254. doi: 10.1093/nar/gkg914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. Journal of Bacteriology. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Wollman EL. Sur les processus de conjugaison et de recombinaison chez Escherichia coli. I. - L'induction par conjugaison ou induction zygotique. Annales de l'Institut Pasteur. 1956;91:486–510. [PubMed] [Google Scholar]
- Bertani LE, Bertani G. Genetics of P2 and related phages. Advances in Genetics. 1971;16:199–237. doi: 10.1016/s0065-2660(08)60359-4. [DOI] [PubMed] [Google Scholar]
- Portelli R, Dodd IB, Xue Q, Egan JB. The late-expressed region of the temperate coliphage 186 genome. Virology. 1998;248:117–130. doi: 10.1006/viro.1998.9263. [DOI] [PubMed] [Google Scholar]
- Nilsson AS, Haggård-Ljungquist E. In: The Bacteriophages. Second. Calendar R, editor. New York: Oxford University Press; 2006. The P2-like bacteriophages; pp. 365–390. [Google Scholar]
- Esposito D, Fitzmaurice WP, Benjamin RC, Goodman SD, Scocca JJ. The complete nucleotide sequence of bacteriophage HP1 DNA. Nucleic Acids Research. 1996;24:2360–2368. doi: 10.1093/nar/24.12.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Kanaya S, Ohnishi M, Terawaki Y, Hayashi T. The complete nucleotide sequence of fCTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: implications for phage evolution and horizontal gene transfer via bacteriophages. Molecular Microbiology. 1999;31:399–419. doi: 10.1046/j.1365-2958.1999.01158.x. [DOI] [PubMed] [Google Scholar]
- Kapfhammer D, Blass J, Evers S, Reidl J. Vibrio cholerae phage K139: complete genome sequence and comparative genomics of related phages. Journal of Bacteriology. 2002;184:6592–6601. doi: 10.1128/JB.184.23.6592-6601.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoy S, Aranda J, Alvarez G, Barbe J, Llagostera M. Isolation and sequencing of a temperate transducing phage for Pasteurella multocida. Applied & Environmental Microbiology. 2006;72:3154–3160. doi: 10.1128/AEM.72.5.3154-3160.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilstein F, Dreiseikelmann B. Temperate bacteriophage FO18P from an Aeromonas media isolate: characterization and complete genome sequence. Virology. 2008;373:25–29. doi: 10.1016/j.virol.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Chibani-Chennoufi S, Dillmann ML, Marvin-Guy L, Rami-Shojaei S, Brüssow H. Lactobacillus plantarum bacteriophage LP65: a new member of the SPO1-like genus of the family Myoviridae. Journal of Bacteriology. 2004;186:7069–7083. doi: 10.1128/JB.186.21.7069-7083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama J, Rashel M, Maeda Y, Takemura I, Sugihara S, Akechi K, Muraoka A, Wakiguchi H, Matsuzaki S. Isolation and characterization of a novel Enterococcus faecalis bacteriophage fEF24C as a therapeutic candidate. FEMS Microbiology Letters. 2008;278:200–206. doi: 10.1111/j.1574-6968.2007.00996.x. [DOI] [PubMed] [Google Scholar]
- Uchiyama J, Rashel M, Takemura I, Wakiguchi H, Matsuzaki S. In silico and in vivo evaluation of bacteriophage fEF24C, a candidate for treatment of Enterococcus faecalis infections. Applied & Environmental Microbiology. 2008;74:4149–4163. doi: 10.1128/AEM.02371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp J, Dorscht J, Lurz R, Bielmann R, Wieland M, Zimmer M, Calendar R, Loessner MJ. The terminally redundant, nonpermuted genome of Listeria bacteriophage A511: a model for the SPO1-like myoviruses of gram-positive bacteria. Journal of Bacteriology. 2008;190:5753–5765. doi: 10.1128/JB.00461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan BJ, Davies P, Carstens EB, Kropinski AM. Characterization of the genome of Pseudomonas aeruginosa bacteriophage phi PLS27 with particular reference to the ends of the DNA. Journal of Virology. 1989;63:1587–1594. doi: 10.1128/jvi.63.4.1587-1594.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkus ME, Shub DA. Mapping the genes in the terminal redundancy of bacteriophage SPO1 with restriction endonucleases. Journal of Virology. 1985;56:40–48. doi: 10.1128/jvi.56.1.40-48.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker ML, Ralston EJ, Eiserling FA. Bacteriophage SPO1 structure and morphogenesis. II. Head structure and DNA size. Journal of Virology. 1983;46:250–259. doi: 10.1128/jvi.46.1.250-259.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker ML, Eiserling FA. Bacteriophage SPO1 structure and morphogenesis. I. Tail structure and length regulation. Journal of Virology. 1983;46:239–249. doi: 10.1128/jvi.46.1.239-249.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L, McDaniel L, Moran MA, Nelson KE, Nilsson C, Olson R, Paul J, Brito BR, Ruan Y, Swan BK, Stevens R, Valentine DL, Thurber RV, Wegley L, White BA, Rohwer F. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- Serwer P. Evolution and the complexity of bacteriophages. Virol J. 2007;4:30. doi: 10.1186/1743-422X-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard A, Clokie MRJ, Shub DA, Mann NH. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:27. doi: 10.1073/pnas.0401478101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clokie MR, Shan J, Bailey S, Jia Y, Krisch HM, West S, Mann NH, Clokie MRJ, Shan J, Bailey S, Jia Y, Krisch HM, West S, Mann NH. Transcription of a 'photosynthetic' T4-type phage during infection of a marine cyanobacterium. Environmental Microbiology. 2006;8:827–835. doi: 10.1111/j.1462-2920.2005.00969.x. [DOI] [PubMed] [Google Scholar]
- Stewart CR, Houtz JE, Smith A, Ford M, Peebles C, Casjens SR. The genome of Bacillus subtilis bacteriophage SPO1. 17th Evergreen International Phage Biology Meeting, Evergreen Olympia, WA, August 12-17. 2007
- Duda RL, Hendrix RW, Huang WM, Conway JF. Shared architecture of bacteriophage SPO1 and herpesvirus capsids [erratum appears in Curr Biol. 2006 Feb 21;16(4):440] Current Biology. 2006;16:R11–R13. doi: 10.1016/j.cub.2005.12.023. [DOI] [PubMed] [Google Scholar]
- Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5174–5179. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton RM, Noordman WH, Biswas B, de Meester ED, Loessner MJ. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regulatory Toxicology & Pharmacology. 2005;43:301–312. doi: 10.1016/j.yrtph.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Twort FW. An investigation on the nature of the ultramicroscopic viruses. Lancet. 1915;189:1241–1243. [Google Scholar]
- Summer EJ, Gonzalez CF, Carlisle T, Mebane LM, Cass AM, Savva CG, LiPuma J, Young R. Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. Journal of Molecular Biology. 2004;340:49–65. doi: 10.1016/j.jmb.2004.04.053. [DOI] [PubMed] [Google Scholar]
- Braid MD, Silhavy JL, Kitts CL, Cano RJ, Howe MM. Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. Journal of Bacteriology. 2004;186:6560–6574. doi: 10.1128/JB.186.19.6560-6574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PW, Chu L, Guttman DS. Complete sequence and evolutionary genomic analysis of the Pseudomonas aeruginosa transposable bacteriophage D3112. Journal of Bacteriology. 2004;186:400–410. doi: 10.1128/JB.186.2.400-410.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill GS, Hull RC, Curtiss R IIIrd. Mutator bacteriophage D108 and its DNA: an electron microscopic characterization. Journal of Virology. 1981;37:420–430. doi: 10.1128/jvi.37.1.420-430.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canchaya C, Proux C, Fournous G, Bruttin A, Brüssow H. Prophage genomics. Microbiology & Molecular Biology Reviews. 2003;67:238–276. doi: 10.1128/MMBR.67.2.238-276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Research. 2006;34:5839–5851. doi: 10.1093/nar/gkl732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan GJ, Hatfull GF, Casjens S, Hendrix RW. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. Journal of Molecular Biology. 2002;317:337–359. doi: 10.1006/jmbi.2002.5437. [DOI] [PubMed] [Google Scholar]
- Andres S, Wiezer A, Bendfeldt H, Waschkowitz T, Toeche-Mittler C, Daniel R. Insights into the genome of the enteric bacterium Escherichia blattae: cobalamin (B12) biosynthesis, B12-dependent reactions, and inactivation of the gene region encoding B12-dependent glycerol dehydratase by a new mu-like prophage. Journal of Molecular Microbiology & Biotechnology. 2004;8:150–168. doi: 10.1159/000085788. [DOI] [PubMed] [Google Scholar]
- Saariaho AH, Lamberg A, Elo S, Savilahti H. Functional comparison of the transposition core machineries of phage Mu and Haemophilus influenzae Mu-like prophage Hin-Mu reveals interchangeable components. Virology. 2005;331:6–19. doi: 10.1016/j.virol.2004.09.041. [DOI] [PubMed] [Google Scholar]
- Lobocka MB, Rose DJ, Plunkett G III, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR. Genome of bacteriophage P1. Journal of Bacteriology. 2004;186:7032–7068. doi: 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summer EJ, Gonzalez CF, Bomer M, Carlile T, Morrison W, Embry A, Kucherka AM, Lee J, Mebane L, Morrison WC, Mark L, King MD, LiPuma MJ, Vidaver AK, Young R. Divergence and mosaicism among virulent soil phages of the Burkholderia cepacia complex. Journal of Bacteriology. 2006;188:255–268. doi: 10.1128/JB.188.1.255-268.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Matsuura T, Ohara T, Azegami K. Sequence analysis of the genome of OP2, a lytic bacteriophage of Xanthomonas oryzae pv. oryzae. Journal of General Plant Pathology. 2006;72:104–110. [Google Scholar]
- Summer EJ, Berry J, Tran TA, Niu L, Struck DK, Young R. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. Journal of Molecular Biology. 2007;373:1098–1112. doi: 10.1016/j.jmb.2007.08.045. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Gilcrease EB, Winn-Stapley DA, Schicklmaier P, Schmieger H, Pedulla ML, Ford ME, Houtz JM, Hatfull GF, Hendrix RW. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. Journal of Bacteriology. 2005;187:1091–1104. doi: 10.1128/JB.187.3.1091-1104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley R, Kenna DT, Vandamme P, Ure R, Govan JR. Lysogeny and bacteriophage host range within the Burkholderia cepacia complex. Journal of Medical Microbiology. 2003;52:483–490. doi: 10.1099/jmm.0.05099-0. [DOI] [PubMed] [Google Scholar]
- Villegas A, She YM, Kropinski AM, Lingohr EJ, Mazzocco A, Ojha S, Waddell TE, Ackermann HW, Moyles DM, Ahmed R, Johnson RP. The genome and proteome of a virulent Escherichia coli O157:H7 bacteriophage closely resembling Salmonella phage Felix O1. Virology Journal. 2009;6:41. doi: 10.1186/1743-422X-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman SM, Kropinski AM, Castle AJ, Svircev AM. Complete genome of the broad-host-range Erwinia amylovora phage fEa21-4 and its relationship to Salmonella phage felix O1. Applied & Environmental Microbiology. 2009;75:2139–2147. doi: 10.1128/AEM.02352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobberley JM, Authement RN, Segall AM, Paul JH. The temperate marine phage fHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J Virol. 2008;82:6618–6630. doi: 10.1128/JVI.00140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakey HJ, Cullen BR, Owens L, Oakey HJ, Cullen BR, Owens L. The complete nucleotide sequence of the Vibrio harveyi bacteriophage VHML. Journal of Applied Microbiology. 2002;93:1089–1098. doi: 10.1046/j.1365-2672.2002.01776.x. [DOI] [PubMed] [Google Scholar]
- Oakey HJ, Owens L, Oakey HJ, Owens L. A new bacteriophage, VHML, isolated from a toxin-producing strain of Vibrio harveyi in tropical Australia. Journal of Applied Microbiology. 2000;89:702–709. doi: 10.1046/j.1365-2672.2000.01169.x. [DOI] [PubMed] [Google Scholar]
- Mobberley JM, Authement RN, Segall AM, Paul JH. The temperate marine phage FHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. Journal of Virology. 2008;82:6618–6630. doi: 10.1128/JVI.00140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann H-W. 5500 Phages examined in the electron microscope. Archives of Virology. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- Hatfull GF, Pedulla ML, Jacobs-Sera D, Cichon PM, Foley A, Ford ME, Gonda RM, Houtz JM, Hryckowian AJ, Kelchner VA, Namburi S, Pajcini KV, Popovich MG, Schleicher DT, Simanek BZ, Smith AL, Zdanowicz GM, Kumar V, Peebles CL, Jacobs WR Jr, Lawrence JG, Hendrix RW. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genetics. 2006;2:e92. doi: 10.1371/journal.pgen.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR Jr, Hendrix RW, Hatfull GF. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- Mayer MJ, Narbad A, Gasson MJ. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. Journal of Bacteriology. 2008;190:6734–6740. doi: 10.1128/JB.00686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh S, Ong PF, Song KP, Riley TV, Chang BJ. The complete genome sequence of Clostridium difficile phage fC2 and comparisons to fCD119 and inducible prophages of CD630. Microbiology. 2007;153:676–685. doi: 10.1099/mic.0.2006/002436-0. [DOI] [PubMed] [Google Scholar]
- Govind R, Fralick JA, Rolfe RD. Genomic organization and molecular characterization of Clostridium difficile bacteriophage FCD119. Journal of Bacteriology. 2006;188:2568–2577. doi: 10.1128/JB.188.7.2568-2577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh S, Riley TV, Chang BJ. Isolation and characterization of temperate bacteriophages of Clostridium difficile. Appl Environ Microbiol. 2005;71:1079–1083. doi: 10.1128/AEM.71.2.1079-1083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan G, Bu C, Gao Y, Wang X, Xu N, Mathee K. Mining protein sequences for motifs. Journal of Computational Biology. 2002;9:707–720. doi: 10.1089/106652702761034145. [DOI] [PubMed] [Google Scholar]
- Mesyanzhinov VV, Robben J, Grymonprez B, Kostyuchenko VA, Bourkaltseva MV, Sykilinda NN, Krylov VN, Volckaert G. The genome of bacteriophage phiKZ of Pseudomonas aeruginosa. Journal of Molecular Biology. 2002;317:1–19. doi: 10.1006/jmbi.2001.5396. [DOI] [PubMed] [Google Scholar]
- Hertveldt K, Lavigne R, Pleteneva E, Sernova N, Kurochkina L, Korchevskii R, Robben J, Mesyanzhinov V, Krylov VN, Volckaert G. Genome comparison of Pseudomonas aeruginosa large phages. Journal of Molecular Biology. 2005;354:536–545. doi: 10.1016/j.jmb.2005.08.075. [DOI] [PubMed] [Google Scholar]
- Krylov VN, Dela Cruz DM, Hertveldt K, Ackermann H-W. "phiKZ-like viruses", a proposed new genus of myovirus bacteriophages. Archives of Virology. 2007;152:1955–1959. doi: 10.1007/s00705-007-1037-7. [DOI] [PubMed] [Google Scholar]
- Thomas JA, Rolando MR, Carroll CA, Shen PS, Belnap DM, Weintraub ST, Serwer P, Hardies SC. Characterization of Pseudomonas chlororaphis myovirus 201varphi2-1 via genomic sequencing, mass spectrometry, and electron microscopy. Virology. 2008;376:330–338. doi: 10.1016/j.virol.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway BW, Egan JB, Monk M. Lysogeny in Pseudomonas aeruginosa. Australian Journal of Experimental Biology. 1960;38:321–330. doi: 10.1038/icb.1960.34. [DOI] [PubMed] [Google Scholar]
- Krylov VN, Tolmachova TO, Akhverdian VZ. DNA homology in species of bacteriophages active on Pseudomonas aeruginosa. Archives of Virology. 1993;131:141–151. doi: 10.1007/BF01379086. [DOI] [PubMed] [Google Scholar]
- Bergan T. A new bacteriophage typing set for Pseudomonas aeruginosa I. Selection procedure. Acta Pathologica et Microbiologica Scandinavica B. 1972;80:117–180. doi: 10.1111/j.1699-0463.1972.tb00148.x. [DOI] [PubMed] [Google Scholar]
- Lindberg RB, Latta RL. Phage typing of Pseudomonas aeruginosa: clinical and epidemiological considerations. Journal of Infectious Diseases. 1974;130:S33–S43. doi: 10.1093/infdis/130.supplement.s33. [DOI] [PubMed] [Google Scholar]
- Ackermann H-W, Cartier C, Slopek S, Vieu J-F. Morphology of Pseudomonas aeruginosa typing phages of the Lindberg set. Annales de l'Institut Pasteur/Virologie. 1988;139:389–404. doi: 10.1016/s0769-2617(88)80075-3. [DOI] [PubMed] [Google Scholar]
- Van Twest R, Kropinski AM. Bacteriophage enrichment from water and soil. Methods in Molecular Biology. 2009;501:15–21. doi: 10.1007/978-1-60327-164-6_2. [DOI] [PubMed] [Google Scholar]
- Kwan T, Liu J, DuBow M, Gros P, Pelletier J, Kwan T, Liu J, Dubow M, Gros P, Pelletier J. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. Journal of Bacteriology. 2006;188:1184–1187. doi: 10.1128/JB.188.3.1184-1187.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T, McCarty J, Srikumar R, Williams D, Wu JJ, Gros P, Pelletier J, DuBow M. Antimicrobial drug discovery through bacteriophage genomics. Nature Biotechnology. 2004;22:185–191. doi: 10.1038/nbt932. [DOI] [PubMed] [Google Scholar]
- Lima-Mendez G, van HJ, Toussaint A, Leplae R. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol Biol Evol. 2008;25:762–777. doi: 10.1093/molbev/msn023. [DOI] [PubMed] [Google Scholar]
- Rohwer F, Edwards R. The Phage Proteomic Tree: a genome-based taxonomy for phage. Journal of Bacteriology. 2002;184:4529–4535. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzik JM, Rosche WA, Rietsch A, O'Toole GA. Isolation and characterization of a generalized transducing phage for Pseudomonas aeruginosa strains PAO1 and PA14. Journal of Bacteriology. 2004;186:3270–3273. doi: 10.1128/JB.186.10.3270-3273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedulla ML, Lewis JA, Hendrickson HL, Ford ME, Houtz JM, Peebles CL, Lawrence JG, Hatfull GF, Hendrix RW. Bacteriophage G: analysis of a bacterium-sized phage genome. Proceeding of the 103rd Annual Meeting of the American Society for Microbiology, Washington, DC. 2003.
- Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW, Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. Plos Biology. 2005;3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann NH, Clokie MR, Millard A, Cook A, Wilson WH, Wheatley PJ, Letarov A, Krisch HM. The genome of S-PM2, a "photosynthetic" T4-type bacteriophage that infects marine Synechococcus strains. Journal of Bacteriology. 2005;187:3188–3200. doi: 10.1128/JB.187.9.3188-3200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann NH. The third age of phage. Plos Biology. 2005;3:e182. doi: 10.1371/journal.pbio.0030182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigele PR, Pope WH, Pedulla ML, Houtz JM, Smith AL, Conway JF, King J, Hatfull GF, Lawrence JG, Hendrix RW. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environmental Microbiology. 2007;9:1675–1695. doi: 10.1111/j.1462-2920.2007.01285.x. [DOI] [PubMed] [Google Scholar]
- Lavigne R, Seto D, Mahadevan O, Ackermann H-W, Kropinski AM. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Research in Microbiology. 2008;159:406–414. doi: 10.1016/j.resmic.2008.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CoreExtractor comparison of Myoviridae phages. A. This Excel figure shows relative correlation scores (above 10%), based on the number of homologous proteins between two phages. Colour tags are added to visualize these correlations (from green to red for increasing correlation scores). B. Corresponding dissimilarity matrix.