Abstract
The plant viral re-initiation factor transactivator viroplasmin (TAV) activates translation of polycistronic mRNA by a re-initiation mechanism involving translation initiation factor 3 (eIF3) and the 60S ribosomal subunit (60S). QJ;Here, we report a new plant factor—re-initiation supporting protein (RISP)—that enhances TAV function in re-initiation. RISP interacts physically with TAV in vitro and in vivo. Mutants defective in interaction are less active, or inactive, in transactivation and viral amplification. RISP alone can serve as a scaffold protein, which is able to interact with eIF3 subunits a/c and 60S, apparently through the C-terminus of ribosomal protein L24. RISP pre-bound to eIF3 binds 40S, suggesting that RISP enters the translational machinery at the 43S formation step. RISP, TAV and 60S co-localize in epidermal cells of infected plants, and eIF3–TAV–RISP–L24 complex formation can be shown in vitro. These results suggest that RISP and TAV bridge interactions between eIF3-bound 40S and L24 of 60S after translation termination to ensure 60S recruitment during repetitive initiation events on polycistronic mRNA; RISP can thus be considered as a new component of the cell translation machinery.
Keywords: Cauliflower mosaic virus (CaMV), eIF3, large ribosomal subunit (60S), re-initiation supporting protein (RISP), transactivator viroplasmin (TAV)
Introduction
The translation initiation pathway operating on most eukaryotic mRNAs is cap-dependent linear scanning, in which initiation occurs exclusively at the most 5′-proximal AUG codon in a favourable initiation context (Kozak, 1999). First, translation initiation factors (eIFs) 1, 1A and 3, and a ternary complex (TC, eIF2–GTP–Met-tRNAiMet) bind to the 40S ribosomal subunit (40S) to form a 43S pre-initiation complex (PIC) that is then loaded onto the capped 5′-end of the mRNA (which is pre-bound with eIFs 4F, 4A and 4B) before scanning to the initiation start site. Translation initiation proceeds through the formation of 48S PIC at this AUG codon followed by the joining of a 60S ribosomal subunit (60S)—a process mediated by eIF5B (Pestova et al, 2007). After elongation and termination of translation, ribosomes normally dissociate from the mRNA, except if the first translated upstream ORF (uORF) is short (<∼30 codons), in such a case translation re-initiation at a downstream ORF is permitted (Morris and Geballe, 2000).
Translation re-initiation, that is, when ribosomes having terminated translation of an ORF give rise to 40S subunits capable of resuming scanning and reinitiating at a downstream AUG, is a mechanism used to down or upregulate the production of potent proteins, such as growth factors, protein kinases and transcription factors. Several disorders in humans are due to perturbation of such regulation (Kozak, 2002). The ability of the 40S subunits to reinitiate depends on the time required to translate the uORF and the intergenic distance between the two ORFs (Morris and Geballe, 2000; Kozak, 2001). It was proposed that eIFs bound to 40S during the first initiation event might remain loosely bound to the 80S ribosome and decay gradually during the first few elongation cycles. Remaining initiation factors might help 40S subunits to resume scanning and allow them to remain initiation-competent (Kozak, 2001). In order to reinitiate, the 40S must recruit the TC de novo to initiate translation (Dever et al, 1992), and recruit 60S subunits to start elongation. Several canonical eIFs have been shown to influence reinitation efficiency after sORF translation: eIF3, eIF4F and eIF4A (Kim et al, 2004; Pöyry et al, 2004; Szamecz et al, 2008).
Although eIF3 has multiple functions in translation initiation, it is also required for translation re-initiation (for a review see Hinnebusch, 2006). In yeast, resumption of scanning by post-termination ribosomes on GCN4 mRNA after uORF1 translation depends on eIF3 subunit a, which can provide a link between mRNA and 40S (Szamecz et al, 2008). In plants, eIF3 subunit h is critically required to overcome the inhibitory effect of multiple short uORFs (Kim et al, 2004).
A specialized ‘termination-re-initiation' mechanism found in mammalian viruses and LINE-1 retrotransposons has been invoked to accomplish rather exceptional cases of re-initiation after the translation of long ORFs (Horvath et al, 1990; Alisch et al, 2006; Luttermann and Meyers, 2007). This mechanism is used for translation of the downstream ORF on a bicistronic mRNA with closely spaced termination and initiation codons if the uORF bears a cis-acting element near its stop codon that renders terminating ribosomes highly re-initiation-competent (Meyers 2003, 2007; Luttermann and Meyers, 2007; Powell et al, 2008). Recently, Jackson et al (Pöyry et al, 2007) characterized such a cis-element located at the 3′-end of ORF 2 on the bicistronic subgenomic mRNA of feline calicivirus as a binding site for eIF3, and showed that eIF3 has a crucial role in translation re-initiation of ORF 3 in rabbit reticulocyte lysate.
Cauliflower mosaic virus (CaMV) and some related pararetroviruses are able to achieve translation of their polycistronic RNA in plants by a ‘virus-activated re-initiation' mechanism that is strictly dependent on the viral protein transactivator viroplasmin (TAV), which is an essential, multifunctional protein in CaMV (Bonneville et al, 1989; Ryabova et al, 2006). TAV is a viral re-initiation factor that is able to promote translation of viral or artificial polycistronic RNAs, and transactivation efficiency in plant protoplasts depends neither on the size of the uORF nor on the distance between the ORFs to be translated (Fütterer and Hohn, 1991, 1992). To accomplish the translation of polycistronic RNA, TAV interacts with eIF3 subunit g, and also with 60S subunits through the 60S ribosomal protein L24 (Park et al, 2001). TAV binding to L18 (Leh et al, 2001) or L13 (Bureau et al, 2004) has also been reported. The proposed model states that TAV prevents dissociation of eIF3 from the translating ribosome during the long elongation event, and positions this latter factor for downstream ORF translation (Park et al, 2001, 2004).
Cellular examples of re-initiation after long ORF translation are extremely rare. Nevertheless, about 12% of 5′-UTRs from human and plant mRNAs have a uORF, and about 7% harbour multiple uORFs (Suzuki et al, 2000; Kawaguchi and Bailey-Serres, 2005). Strikingly, in plants about 10% of all uORFs are longer than 35 codons (Hauden and Jorgensen, 2007; Tran et al, 2008), and their translation would be expected to strongly diminish initiation at the main ORF in any eukaryotic system. One such mRNA—an ETTIN (ETT) mRNA—is translated by a putative re-initiation mechanism that depends on the 60S ribosomal protein L24 (Nishimura et al, 2005). The cellular mechanisms used for the translation of such polycistronic mRNAs remain elusive, and specific cellular proteins that can directly regulate translation re-initiation events on cellular mRNAs remain to be identified.
Here, we used CaMV–TAV as a tool to fish for cellular proteins that could function in the activation of polycistronic mRNA translation. We report a new plant protein—‘Re-Initiation Supporting Protein' (RISP)—that interacts with TAV, eIF3 and the large 60S ribosomal subunit, and stimulates TAV-activated translation re-initiation.
Results
A new plant protein interacts with the essential transactivation domain of viral re-initiation factor TAV
The central domain of TAV has been implicated in re-initiation as an essential minimal transactivation domain (MAV) (De Tapia et al, 1993). To identify factors that interact directly with MAV, we carried out a yeast two-hybrid screening of an Arabidopsis cDNA library using an amino terminal portion of TAV (NTAV) (Figure 1A), which led to the identification of a protein (Mr=45 kDa; accession number NM_125513.1 encoded by rispa) that we call RISP. (A second gene, rispb (accession number NP_196406), was later found in plant genome, which encodes a protein with a high degree of similarity to RISP; for an alignment see Supplementary Figure 1S). RISP was found to be expressed in Arabidopsis and turnip plants, and had mobility in SDS–PAGE corresponding to its predicted size of 45 kDa and similar to that of recombinant RISP purified from Escherichia coli (Figure 2C). Immunofluorescence analysis of the sub-cellular localization of RISP in leaves of Brassica rapa plants using anti-RISP antibodies suggested a cytoplasmic distribution (Figure 3C, left panels). We screened the yeast, human and other mammalian genomes for RISP-like proteins, but no orthologs have yet been found.
Figure 1.
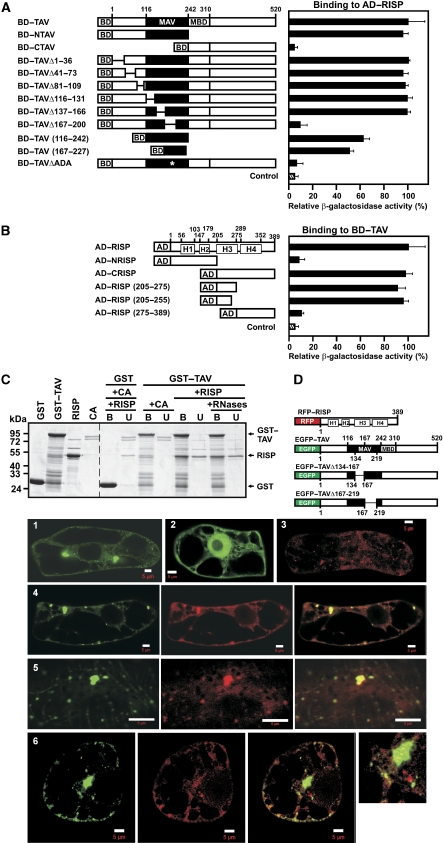
Association of re-initiation supporting protein (RISP) with transactivator viroplasmin (TAV) and mapping of their interaction domains. (A) Interaction between TAV and its deletion mutants fused to the Gal4 binding domain (BD) and RISP fused to Gal4 activation domain (AD) in the yeast two-hybrid system was quantified by measuring β-galactosidase activity. The highest value of β-galactosidase activity in diploids transformed with both full-length constructs was taken as 100% (12 Miller units). MAV, minimal segment of TAV; and MBD, multiple protein-binding domain. (B) Quantification of interactions between RISP and its deletion mutants fused to Gal4 AD and BD-TAV. H1–H4 predicted coiled-coil domains. (C) GST and GST–TAV bound to glutathione beads were incubated with either purified recombinant RISP or Conalbumin (CA, 75 kDa). Lanes (+ RNases) show the experiment carried out in the presence of an RNase cocktail. The beads were washed, and the unbound (U) and bound (B) fractions were analyzed by SDS–PAGE followed by Coomassie blue staining. Right panel, Interactions of RISP with GST or GST–TAV; and left panel, purified GST, GST–TAV and RISP. (D) Schematic representation of full-length RISP fused to the C-terminus of RFP, and full-length TAV (or truncated versions) fused to the C-terminus of enhanced green fluorescent protein (EGFP). Panels 1–6: Imaging fluorescence assays showing tobacco BY-2 cells transiently expressing EGFP–TAV (green, 1), EGFP alone (green, 2), or RFP-RISP (red, 3). 4 Left: EGFP–TAV, central: red fluorescent protein (RFP)–RISP, and right: merged. 5 Left: EGFP–TAVΔ134–167, central: RFP–RISP, right: merged. 6 Left: EGFP–TAVΔ167–219, central: RFP–RISP, right: merged; far right panel: high magnification image of part of the cell. Scale bars, 5 μm.
Figure 2.
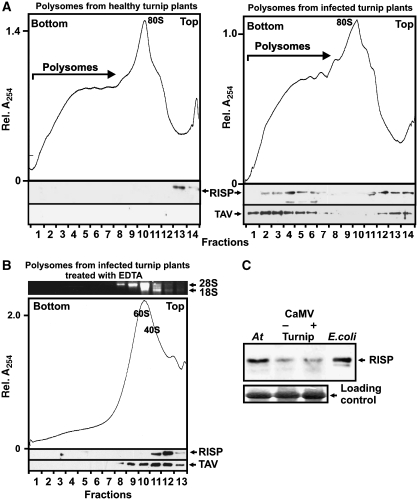
Re-initiation supporting protein (RISP) and transactivator viroplasmin (TAV) accumulate in polyribosomes during viral infection. (A, B) Ribosomal profiles of polyribosomes and ribosomal species from healthy (left) and Cauliflower mosaic virus (CaMV)-infected (right) turnip plants (A) untreated and (B) treated with 30 mM EDTA. A, B show the UV profile of the gradient with 40S and 60S, monosomes (80S) and polysomes indicated are shown. 1 ml aliquot fractions were either precipitated with 10% TCA and analyzed by SDS–PAGE and immunoblotting using polyclonal antibodies against TAV and RISP (lower panels); or analyzed by agarose gel electrophoresis (upper panel in B). Positions of 18S and 28S rRNAs are indicated. (C) Immunoblotting using polyclonal antibodies against RISP of extracts isolated from Arabidopsis (At), healthy (Turnip −) and CaMV-infected (Turnip +), turnip plants and recombinant RISP expressed in Escherichia coli. Loading controls are shown (lower panel).
Figure 3.
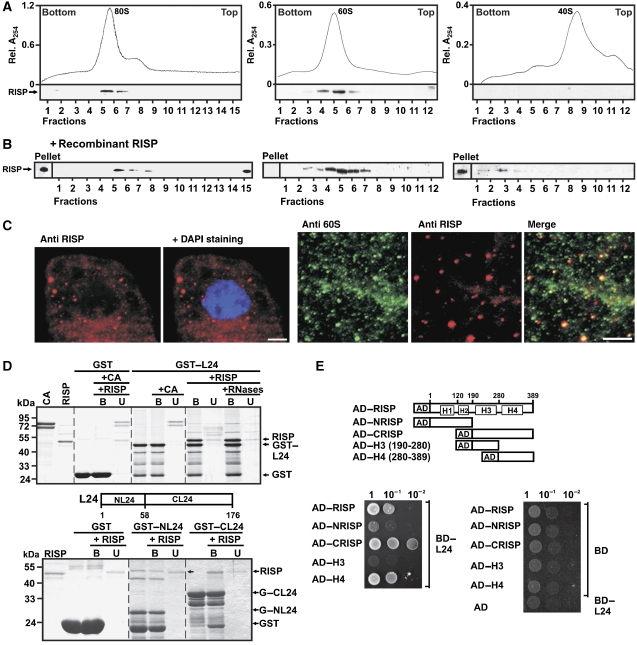
Re-initiation supporting protein (RISP) binds the 60S ribosomal protein L24 and co-sediments with 60S and 80S. (A) RISP co-sediments with 80S (left), and 60S (middle) but not with 40S (right) ribosomes. Lower panels: immunostaining of gradient fractions with antibodies against RISP. (B) 80S, 60S and 40S were incubated with recombinant RISP at approximately 1:1 molar ratio before being subjected to sucrose density gradient centrifugation as in (A) followed by western blot analysis with antibodies against RISP. (C) Left panels: Immunofluorescence assays showing localization of RISP (red) within cytoplasm of BY-2 cells. Nucleus was stained with DAPI (blue); right panels: colocalization of endogenous 60S (green) and RISP (red) in BY-2 tobacco cells; only part of the cytoplasmic compartment is shown. Anti-60S and anti-RISP images were merged in the right-most panel. Scale bar, 5 μm. (D) RISP interacts with the C-terminal region of L24 in GST pull-down assay. RISP or the control protein CA was incubated with recombinant L24 fused to GST (GST–L24, upper panel). Lanes labelled +RNases show the experiment carried out in the presence of an RNase cocktail. RISP was mixed with either the N- or C-terminus of L24 fused to GST (GST–NL24 and GST–CL24; bottom panel) bound to glutathione beads. The beads were washed, and purified bound (B) and unbound (U) proteins were resolved by SDS–PAGE and stained with Coomassie blue. (E) Yeast two-hybrid interactions between BD–L24 and RISP and its deletion mutants fused to AD. Equal OD600 units and 1:1, 1:10 and 1:100 dilutions were spotted from left to right and incubated for 2 days.
We mapped TAV and RISP interactions using the yeast two-hybrid assay (Figure 1A and B). RISP can be divided tentatively into four parts, each characterized by a coiled-coil structure predicted with high probability by computer analysis (helices H1–H4; Supplementary Figure S1). TAV and RISP truncation and deletion mutants were tested to delineate regions important for binding (Figure 1A and B). The N-terminal part of TAV (aa 1–242) interacted as strongly with RISP as complete TAV, whereas the C-terminal part (aa 243–520) was inactive. More than half of the binding activity was retained in the isolated MAV region (aa 116–242) or a sub-fragment thereof (aa 167–227). An internal deletion of TAV (aa 167–200) that had previously been shown to abolish transactivation activity (Kobayashi and Hohn, 2003) also diminished RISP binding (Figure 1A), suggesting that RISP could support TAV in its re-initiation function. According to secondary structure predictions (Evans and Bycroft, 1999), the RISP binding site (aa 167–227) consists of the C-terminal half of the putative dsRNA binding domain (aa 167–182), followed by an unstructured region starting with Ala183. Deletion of three amino acids (183AlaAspAla; ΔADA) within this unstructured region virtually abolished the interaction of TAV with RISP (Figure 1A).
Mapping of regions of RISP participating in interaction with TAV showed that a segment spanning residues 205–255 within the C-terminal part (AD—RISP (205–255)) interacted with TAV as strongly as full-length RISP (Figure 1B). This domain harbours the putative coiled-coil domain H3, which might mediate this interaction. The interaction between RISP and TAV was confirmed in a GST-pull down assay using GST–TAV and RISP (Figure 1C). RISP was present in the bound fraction after incubation with GST–TAV, but in the unbound fraction after incubation with GST alone. A purified control protein, conalbumin (CA), remained in the unbound fraction with GST–TAV, confirming the specificity of the TAV–RISP interaction (Figure 1C). To show that the interaction of TAV with RISP is not mediated by RNA, the mixture of interacting partners was treated with an RNase cocktail (Figure 1C). Again RISP was found in the bound fraction when incubated with GST–TAV. This result suggests that RNA does not mediate the TAV–RISP interaction.
We also overexpressed TAV in fusion with enhanced green fluorescent protein (EGFP–TAV) and RISP with red fluorescent protein (RFP–RISP; Figure 1D) in cultured BY-2 tobacco cells. EGFP–TAV formed cytoplasmic aggregates of different sizes and shapes (Figure 1D-1) as described earlier (Haas et al, 2005), consistent with its role as a major component of viral inclusion bodies, whereas EGFP alone was diffused between the nucleus and the cytoplasm (Figure 1D-2). RFP–RISP appeared as small dots (Figure 1D-3). Upon transient co-expression of both fluorescent fusions, RFP–RISP formed structures that exactly followed the shape and size of EGFP–TAV aggregates (Figure 1D-4), indicating that the RFP–RISP fusion protein retained its ability to interact with EGFP–TAV in the cytoplasm of BY-2 cells. However, no co-localization was observed between EGFP–TAVΔ167–219 and RFP–RISP (Figure 1D-6), in good agreement with our observation that the deleted region in TAV contains a binding site for RISP. In contrast, deletion of the N-terminal MAV domain (aa 134–167) of TAV did not perturb co-localization (Figure 1D-5).
TAV promotes RISP association with polysomes in planta
The presence of TAV on polysomes (Park et al, 2001) and its ability to bind RISP led us to investigate whether RISP associates with polysomes in planta. Polysomes from healthy or CaMV-infected plants were fractionated on sucrose gradients, and TAV and RISP were analyzed by western blots using anti-TAV and anti-RISP antisera (Figure 2A). In extracts from infected plants, a significant proportion of TAV and RISP accumulated at the position of polysomes, whereas in extracts from healthy plants, RISP was detected at the top of the gradient (Figure 2A). In contrast, there was no significant shift of TAV and RISP to polysome-containing fractions after EDTA treatment of polysomes isolated from infected plants (Figure 2B). These results indicate the relatively stable association of RISP with polysomes isolated from infected plants, whereas RISP is removed from polysomes from healthy plants and/or from ribosomal species during sedimentation through sucrose (Figure 2B). Western blot analysis of extracts isolated from healthy and CaMV-infected turnip plants did not show significant variations in endogenous RISP levels (Figure 2C). Thus, RISP is recruited or stabilized in polysomes in CaMV-infected cells, and may have a role in TAV-mediated transactivation.
RISP domain H4 interacts with ribosomal protein L24, and can mediate a RISP–60S link
We next analyzed whether RISP alone can interact with ribosomes, especially ribosomal subunits. Western blot analysis of purified, salt-washed wheat germ mono-ribosomes (80S), 60S and 40S ribosomal subunits showed that endogenous RISP associated with washed 80S and 60S subunits, but not 40S subunits (Figure 3A). To investigate whether ribosomes can be loaded with additional molecules of RISP, recombinant RISP prepared in E. coli was incubated with isolated ribosomes or 60S or 40S subunits at a molar ratio of about 1:1, and the mixtures were fractionated on sucrose gradients. The amount of RISP co-sedimenting with 60S increased; however, some RISP did not co-sediment with 80S or 40S, but was detected in the pellet and at the top of the sucrose gradient (Figure 3B). We speculate that, in the absence of 60S, RISP may interact with some heavy cytoskeleton-like fractions that co-purify with 40S and especially 80S ribosomes, which are thus pelleted together during ribosome isolation steps. We conclude that mono-ribosomes and 60S ribosomal subunits isolated from wheat germ contain endogenous RISP, and that 60S subunits but not complete ribosomes can bind to additional RISPs, suggesting that the presence of the 40S subunit may preclude loading of RISP onto the 60S subunit of the complete ribosome. Thus, RISP co-sediments with 80S monosomes, and may interact with the surface of the 60S subunit that makes a contact with 40S. A GST pull-down analysis confirmed the direct physical interaction between RISP and 60S (Supplementary Figure S2A). Likewise, we found that RISP co-precipitated with the 60S subunit fraction (see Figure 5A), confirming that RISP associates with 60S in vivo.
Figure 5.
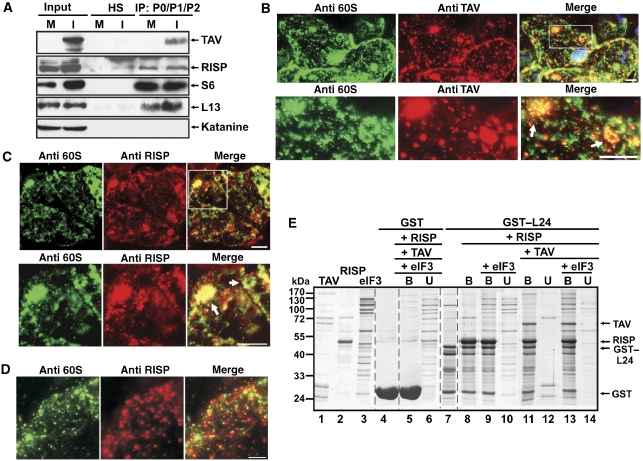
The re-initiation supporting protein (RISP)–transactivator viroplasmin (TAV) complex mediates contacts between 60S ribosomal protein L24 and eIF3. (A) Mock-inoculated (M) and cauliflower mosaic virus (CaMV)-infected (I) turnip plants were used for co-immunoprecipitation with anti-P0–P1–P2 antibodies. The panels show immunoblotting of RISP, TAV, S6 or L13 present in input, normal human serum (HS) and the entire immunoprecipitate (IP) using appropriate rabbit polyclonal antibodies. (B–D) Immunofluorescence assay showing colocalization of endogenous 60S and TAV (B) 60S and RISP in systemically CaMV-infected epidermal cells of leaves of B. rapa plants at 15 days post-inoculation (dpi; C) and 60S and RISP in mock-infected epidermal cells (D). Double-immunolabelling was carried out using anti-TAV and anti-P0–P1–P2 antibodies (anti-60S) (B) or anti-60S and anti-RISP antibodies (C, D). The nucleus was stained with DAPI (blue). The lower panels in (B) and (C) represent higher magnification images of the insets in the upper panels. In the merge, DAPI fluorescence is blue, TAV is red, 60S is green and RISP is red. Round-shaped structures are indicated by arrows. Scale bars, 5 μm. (E) GST pull-down assays. GST–L24 attached to glutathione Sepharose 4B beads (lane 7) was mixed either with RISP (lane 8), or RISP and eIF3 (lanes 9 and 10), or RISP and TAV (lanes 11–12), or RISP, TAV and eIF3 (lanes 13–14); purified TAV, RISP and eIF3 are shown in lanes 1, 2 and 3, respectively. Unbound (U) and bound (B) fractions were analyzed by SDS–PAGE followed by Coomassie blue staining.
In vivo immunofluorescence analysis showed that endogenous RISP is present mainly in the cytoplasm of BY-2 cells as large (0.5–1 μm) and small granules (anti-RISP; Figure 3C, right panels), whereas prominent anti-60S labelling was shown in the cytoplasm as numerous granules of different size (Figure 3C, anti-60S). Superimposition of these images (Figure 3C, merge) showed that around 90% of RISP particles co-localized with a sub-population of 60S ribosomal subunits, as confirmed by high values of Pearson's correlation coefficient, Rr=0.577 (see Materials and methods section).
Previously, we showed that TAV binds to the N-terminus of the 60S ribosomal protein L24 (Park et al, 2001), which corresponds to the full-length protein in archaebacteria (Hatakeyama et al, 1989). RISP interacting with 60S-bound TAV could be in close spatial vicinity to L24, and thus interact with it. GST pull-down analysis confirmed the interaction between RISP and either full-length L24 (Figure 3D, upper panel) or its C-terminus (GST–CL24; aa 58–176) (Figure 3D, bottom panel), whereas there was no interaction of RISP with GST alone, and no interaction of the control protein CA with GST–L24 (Figure 3D). Interactions of L24 with RISP were not affected by treatment of the mixture of interacting partners with an RNase cocktail, suggesting that the RNA does not mediate these interactions (Figure 3D, upper panel). We found no interaction with ribosomal proteins L18 or L13, which were also implicated in TAV binding using GST pull-down assay (data not shown). Yeast two-hybrid analysis confirmed a specific interaction between BD–L24 and AD–RISP and showed the conserved H4 domain of RISP to be the L24-binding site (Figure 3E). Thus, the C-terminal end of RISP may contact the 60S ribosomal subunit directly.
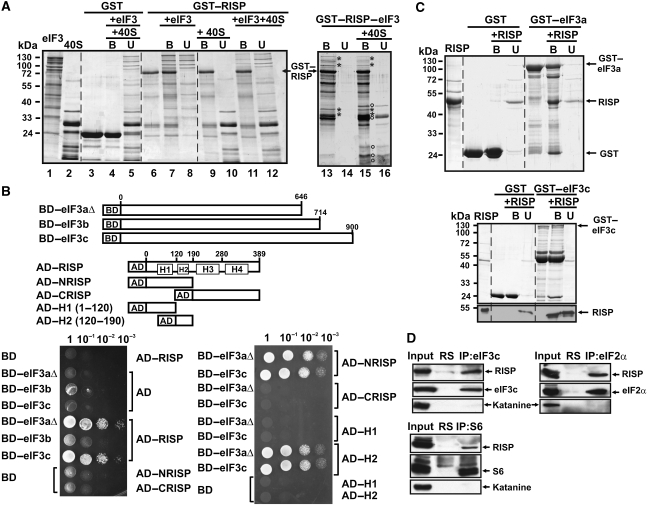
RISP pre-bound to eIF3 via subunits a/c can interact with 40S
Owing to its interaction with TAV, we hypothesized that RISP might interact with translation initiation factor eIF3 implicated in TAV binding. Interaction of RISP with eIF3 was first indicated by GST pull-down assays (see Figure 4A): GST–RISP forms a complex with purified wheat germ eIF3 (Figure 4A, lane 7), but does not show significant binding to 40S-bound eIF3 (Figure 4A, cf lanes 11 and 12). This result provided the clue that subunits of eIF3 contacting the 40S ribosome might be involved in RISP binding. Three subunits of eIF3 implicated in 40S binding in yeast (a, b and c) (Valášek et al, 2003; Nielsen et al, 2006) were tested for their ability to interact with RISP in the yeast two-hybrid system (Figure 4B). We used Arabidopsis thaliana eIF3 subunit 3aΔ—a fragment spanning the first 646 amino acid residues of 3a (total length 988 aa) that can still participate in 40S binding in yeast (Valášek et al, 2003)—and full-length subunits b and c (Figure 4B); subunits a and c were able to interact with RISP in yeast (Figure 4B). The RISP fragment containing helix 2 (H2) mediates the interaction with either eIF3aΔ or eIF3c (Figure 4B). These findings were confirmed by a GST-pull down assay (Figure 4C). Thus, RISP interaction with eIF3 can be mediated by at least two subunits. These data strongly suggest the formation of a complex between RISP and eIF3, and prompted us to analyze binding of this complex to 40S. Surprisingly, we found that a preformed RISP–eIF3 complex (Figure 4A, lane 13) can interact with 40S ribosomal subunits provided at a 2-fold molar excess (Figure 4A, lane 15). Thus, the RISP–eIF3 complex could enter the 43S PIC via its interaction with 40S. To confirm this hypothesis in vivo, three key components of the 43S-PIC—eIF3, eIF2 and 40S—were pelleted using corresponding antibodies (Figure 4D). Consistently, endogenous RISP, but not with the control endogenous plant protein, katanine co-immunoprecipitated with endogenous eIF3 (eIF3c was used as a marker), eIF2 (marker: eIF2α) and 40S (marker: RP S6) in soluble cell extracts (Figure 4D), thus suggesting that RISP is part of the 43S-PIC in plants.
Figure 4.
Re-initiation supporting protein (RISP) binding to eIF3, through direct interaction with eIF3 subunits a and c, mediates its interaction with 40S. (A) GST–RISP binds to wheat eIF3 and 40S. Interactions of GST (lane 3), and GST–RISP (lane 6) bound to glutathione Sepharose 4B beads with eIF3 (lanes 7 and 8), 40S (lanes 9–10) or eIF3 and 40S (lanes 4–5 for GST and 11–12 for GST–RISP) are shown. The left panel (lanes 1 and 2) shows eIF3 and 40S. Interactions between the GST–RISP–eIF3 complex (lane 13) and 40S are shown in lanes 15–16 (right panel). Asterisks: characteristic eIF3 subunits; open circles: characteristic 40S proteins specifically co-precipitated with GST–RISP–eIF3. (B) The H2 domain of RISP interacts with eIF3aΔ and eIF3c in vivo. Schematic representation of BD–eIF3aΔ, BD–eIF3c and RISP, and its truncated versions fused to AD. Yeast two-hybrid analysis was carried out with RISP, AD–NRISP, AD–CRISP, and AD–H1 and AD–H2 against eIF3aΔ, eIF3b or eIF3c. Four dilutions of the transformation mixture are shown. (C) The H2 domain of RISP interacts with eIF3aΔ and eIF3c in GST pull-down assays. RISP was incubated with recombinant eIF3aΔ (upper panel) and eIF3c (bottom panel) fused to GST (GST–eIF3aΔ and GST–eIF3c) bound to glutathione beads. Purified proteins, and bound (B) and unbound (U) material were resolved by SDS–PAGE and stained with Coomassie blue. RISP distribution between the B and U fractions for GST–eIF3c was analyzed by western blot with polyclonal anti-RISP antibodies (lower panel). (D) Co-immunoprecipitation of RISP with eIF3c, or S6, or eIF2α. Arabidopsis suspension cultures were used for co-immunoprecipitation with either anti-eIF3c (upper left panel, or anti-eIF2α (upper right panel) or anti-S6 (bottom panel) antibodies. Each panel shows immunoblotting of RISP, eIF3c, S6, eIF2α and the control protein katanine present in input, normal rabbit serum (RS) and the entire immunoprecipitate (IP).
Identification of a multifactor complex between 60S (L24), TAV, RISP and eIF3
Our present results showing complex formation between 60S and RISP, together with earlier results demonstrating TAV binding to 60S in vitro (Park et al, 2001), prompted us to investigate whether RISP and TAV could associate with the 60S subunits in planta. To test for association between TAV, RISP and 60S, we immunoprecipitated 60S from extracts prepared from leaves of mock- and CaMV-inoculated turnip plants using anti-P0/P1/P2 antibodies (Figure 5A). Endogenous RISP and 40S (marker: RP S6) co-immunoprecipitated with 60S subunits in extracts prepared from healthy plants; and RISP, 40S and TAV proteins associated with 60S in CaMV-infected plants (Figure 5A), suggesting association between RISP and 60S, or TAV, RISP and 60S, with or without 40S. Western blot analysis of purified 60S ribosomal subunits pre-incubated with recombinant TAV and/or RISP after sucrose-gradient centrifugation showed that RISP and TAV co-sediment with 60S subunits (Supplementary Figure S2B), again suggesting that the TAV–RISP complex can be loaded on 60S.
To address the biological relevance of the RISP–TAV–60S complex in planta, we carried out immunofluorescence analysis of epidermal cells in leaves of CaMV-infected Brassica rapa plants. Cells infected with CaMV contain typical large and small virus-induced inclusion bodies, of which TAV is the main component (see Haas et al, 2005). Prominent anti-60S labelling was revealed in both the cytoplasm and nucleus of infected cells, whereas TAV aggregates were distributed in the cytoplasm and around the nucleus (Figure 5B). Co-localization between 60S and TAV was detected mainly within large TAV viroplasms, as round-shaped structures appearing during viral infection (Figure 5B). To locate RISP in CaMV-infected epidermal cells in relation to 60S ribosomal subunits, we used another pair of antibodies (anti-RISP and anti-60S ribosomal proteins P0–P1–P2; Figure 5C). Again 60S subunits were seen as large round-shaped structures (enlarged on the bottom panel in Figure 5C). Strong RISP labelling co-localized with these large viroplasms, particularly within the round-shaped 60S structures (arrow in Figure 5C). We did not detect these structures in the epidermal cells of healthy plants (Figure 5D). These data support the in vivo formation of large complexes (possibly large polysomes) containing RISP, TAV and ribosomes in CaMV-infected plants, which could represent a pool of re-initiation-competent ribosomes loaded with RISP and TAV.
Furthermore, our earlier experiments had shown that TAV interacts with the N-terminal domain of L24 (Park et al, 2001), whereas we now know that RISP interacts with its C-terminal part (Figure 3E). Thus, we wished to confirm that L24 could serve as a scaffold for RISP and TAV, and mediate their simultaneous interaction with 60S. To test whether L24 can interact with both RISP and TAV in vitro, a GST–L24–RISP complex was reconstituted by incubating GST–L24 with a fivefold excess of RISP (Figure 5E, lane 8). Unbound RISP was then removed and the complex was further incubated with a roughly equimolar amount of TAV. The data obtained (Figure 5E) suggest that RISP does not compete with TAV for L24 binding, and that RISP, L24 and TAV form a ternary complex. Moreover, this ternary complex can expand to a quaternary complex by strongly binding eIF3 (eIF3 was found only in GST–L24-bound fraction; Figure 5E, lane 13). As predicted, L24–RISP also binds eIF3, but less strongly (Figure 5E, lane 9). Indeed, eIF3 binds GST–L24 strongly in the presence of RISP (Supplementary Figure S3). Thus, L24 mediates binding between TAV and RISP likely on the 60S ribosomal subunit, and the TAV–RISP complex may bridge interactions between 60S and eIF3.
RISP is indispensable for TAV-mediated transactivation in planta
We next tested the effect of RISP on TAV transactivation capacity using transient expression of mono- and bi-cistronic reporter constructs in plant protoplasts. pmonoCAT contains a single chloramphenicol acetyltransferase (CAT) ORF, whereas pbiGUS contains two consecutive ORFs: CaMV ORF VII and β-glucuronidase (GUS) (Figure 6A) (Bonneville et al, 1989). In Nicotiana plumbaginifolia protoplasts, efficient translation of the GUS-ORF requires the presence of TAV, whereas the expression of CAT acts as a control for transfection/translation efficiency. Although RISP alone could not replace TAV (as a control we used a plasmid containing a GST-ORF), when overexpressed above its endogenous level in the presence of TAV, it strongly enhanced GUS-ORF translation (Figure 6A). The RISP deletion mutant Δ205–255, lacking the TAV-interaction domain, was unable to increase the level of TAV-mediated transactivation (Figure 6A). No significant stimulation effect was observed when TAV was co-transfected together with the RISPΔH2 or ΔH4 deletion mutants lacking the eIF3- or L24-binding domain, respectively (Figure 6A).
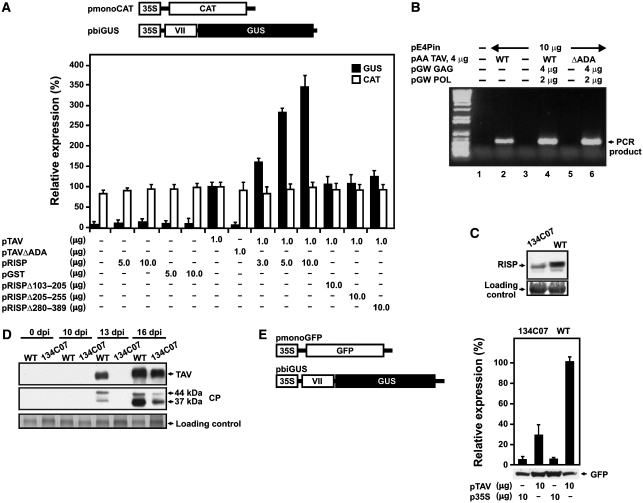
Figure 6.
Re-initiation supporting protein (RISP) participates in transactivator viroplasmin (TAV)-mediated transactivation in plant protoplasts. (A) Schematic diagram of the monocistronic chloramphenicol acetyltransferase (pmonoCAT) and dicistronic β-glucuronidase (pbiGUS) reporter constructs. Nicotiana plumbaginifolia protoplasts were co-transfected with the two reporter plasmids shown, as well as effector plasmids in the amounts indicated below the graph. All reporter and effector constructs were expressed under the control of the CaMV 35S promoter (35S). The amount of CAT (open bars) and GUS enzymatic activity (closed bars) synthesized in N. plumbaginifolia protoplasts is indicated. Results shown represent the means obtained in three independent experiments. Results are expressed as a percentage, with the amount of CAT and the enzymatic activity of GUS synthesized in protoplasts transfected with pTAV only being set as 100%. (B) Ability of TAV and TAVΔADA mutant to support Cauliflower mosaic virus (CaMV) replication. Semiquantitative reporter-targeted PCR analysis (25 cycles) of total LMW DNA from transfected protoplasts. Lane 1, mock-transfected protoplasts; lane 2, transfection with pE4Pin (10 μg) and pAATAV (wild-type; 4 μg); lane 3, pE4Pin (10 μg) alone; lane 4, pE4Pin (10 μg), pAA TAV (wild-type; 4 μg), pGW GAG (4 μg), pGW POL (2 μg); lane 5, pE4Pin (10 μg), pAA TAVΔADA (mutant, 4 μg); lane 6, pE4Pin (10 μg), pAA TAVΔADA (mutant, 4 μg); pGW GAG (4 μg), pGW POL (2 μg). LMW DNA, low molecular weight DNA, GAG, capsid protein precursor; POL, polyprotein with protease, reverse transcriptase, and RNase H activity. (C) Accumulation of RISP in wild-type and 134C07 Arabidopsis plants. Extracts from 0.1 g leaves were analyzed with rabbit polyclonal anti-RISP antibodies. (D) Kinetics of TAV and CP protein accumulation in CaMV infected wild-type and 134C07 plants. (E) Efficiency of TAV-mediated transactivation in mesophyll protoplasts prepared from wild-type (WT) and mutant (134C07) plants. GUS activity is shown as black bars; green fluorescent protein (GFP) accumulation was analyzed by western blot using anti-GFP antibodies. The data shown are the means of three independent assays, and error bars indicate s.d.
Thus, RISP lacking the TAV-binding domain lost its capacity to support TAV-mediated transactivation. To confirm the role of RISP in transactivation we tested the TAV three amino acid deletion mutant (ΔADA) that does not interact with RISP. This mutant was not active in transactivation (Figure 6A), suggesting a direct involvement of TAV in sequestration of RISP. Next, we investigated whether ΔADA affects viral fitness and the re-initiation capacity of TAV in planta. CaMV replication can be tested in single cells (in plant protoplasts) and requires TAV-activated polycistronic translation. We found that the TAVΔADA mutant supported CaMV replication in plant protoplasts only if GAG and POL were provided in trans, strongly suggesting that deletion of these three amino acids specifically affected TAV transactivation function (see Figure 6B). (Loss of transactivation function can be complemented by co-expressing CaMV capsid protein (GAG) and polyprotein (protease-reverse transcriptase/RNAseH; POL) from separate mono-cistronic plasmids (Kobayashi and Hohn, 2003)).
To study the importance of RISP for viral replication, we identified an Arabidopsis T-DNA insertion mutant for RISP, with a T-DNA insertion within the rispa (134C07; see Supplementary Figure S4A); this mutant has no particular phenotype (Supplementary Figure S4B). RT–PCR analysis failed to detect transcripts originating from rispa in the mutant, but not in wild-type Col0 Arabidopsis plants, suggesting rispa disruption. Levels of transcripts from rispb or a polyubiquitin gene in the mutant were not affected significantly (Supplementary Figure S4C). Western blot analysis with polyclonal antibodies against recombinant RISP indicated a roughly threefold decrease in the level of a 45 kDa protein—the expected size of RISP—in mutant plants as compared with wild-type plants (Figure 6C). We speculate that the observed protein in the mutant may correspond to a protein expressed from rispb. To determine whether rispa gene disruption has any impact on CaMV infection, we mechanically inoculated wild-type and RISPΔ Arabidopsis plants with CaMV and compared virus replication kinetics in the inoculated plants by monitoring accumulation of two viral proteins: coat protein (CP) and TAV (Figure 6D). Mutant plants eventually showed development of symptoms identical to those caused by CaMV in wild-type plants. However, symptom appearance and viral protein accumulation was delayed. In wild-type plants, TAV and CP accumulation was first observed at 13 days after inoculation (dpi), but at 16 dpi in mutant plants (Figure 6D). On the basis of accumulated data, we hypothesize that this delay is due to defects in virus-mediated transactivation.
The effect of rispa gene disruption on transactivation was studied more directly using Arabidopsis mesophyll protoplasts prepared from wild-type and mutant plants (Yoo et al, 2007). Protoplasts were transformed with the same plasmid constructions used for N. plumbaginifolia protoplasts, but using pmonoGFP (encoding GFP; Figure 6E, left panel), as a mono-cistronic reporter construct. We found that mesophyll protoplasts prepared from mutant plants are at least threefold less efficient in TAV-mediated re-initiation of the downstream GUS-ORF compared with protoplasts prepared from wild-type plants, whereas GFP expression from the 5′-proximal ORF was equally as efficient in both types of protoplast. Unfortunately, knockdown of both rispa and rispb genes using an amiRNA approach (Schwab et al, 2006) proved lethal (data not shown).
Discussion
Here, we report the isolation of a novel plant protein that we have named RISP for several reasons: (1) in plant protoplasts RISP stimulates the function of a re-initiation factor of viral origin, TAV, in re-initiation of translation; (2) RISP mediates interaction with the transactivation domain of TAV (MAV), and the mutant TAVΔADA, which is defective in RISP binding, failed to support re-initiation in either a transient expression or a viral amplification system in plant protoplasts; and (3) RISP targets two key components of the cellular translation machinery, eIF3 and 60S, that have been shown before to be interaction partners of TAV. The mutants TAVΔADA or RISPΔH3, which disrupt the bridge between RISP and TAV, abolished the effect of TAV or RISP, respectively, on transactivation in non-host (N. plumbaginifolia) and host (Arabidopsis protoplasts; data not shown) plant protoplasts. The RISP gene-specific knockout caused a delay in viral replication and a significant reduction of the TAV-mediated transactivation level in mesophyll protoplasts, although a residual level of transactivation could be still supported by a RISP homologue. These observations substantiate the idea that RISP acts to promote CaMV-mediated transactivation.
Our results suggest that TAV recruits this cellular protein to reinforce its interaction with eIF3 and 60S, and ensures polycistronic translation. How could RISP accomplish its effect on TAV-mediated re-initiation in plants? First, RISP interacts strongly with eIF3 both in vivo and in vitro. The N-terminal α-helical H2 domain of RISP interacts in vivo with the two largest plant core subunits of eIF3, eIF3a and eIF3c. In yeast, both these subunits have been implicated in 40S binding (Valášek et al, 2003; Nielsen et al, 2006). In our in vitro assays, addition of 40S to eIF3 strongly decreased formation of the RISP–eIF3 complex, suggesting that the 40S- and RISP-binding sites on eIF3 partially overlap, that the RISP binding site on 40S-bound eIF3 is not accessible, or that different binding states could be necessary. However, RISP can interact with 40S in vitro if already paired with eIF3. In vivo, RISP is co-immunoprecipitated not only with eIF3 and 40S but eIF2α as well, indicating that RISP can be a part of the 43S-PIC.
We show that while the N-terminal H2 domain of RISP interacts with eIF3, its C-terminal H4 domain is required to bind the C-terminal half of L24 (Figure 3D and E). Furthermore, disruption of the H4 domain almost abolished the activity of RISP in TAV-mediated transactivation (Figure 6A), indicating the significance of L24 binding for RISP function. We therefore favour a model in which, L24 forms an important intermolecular bridge between RISP and TAV on the 60S ribosomal subunit as a secondary contact, in addition to their direct contact as observed in vitro (Figure 5E). Accordingly, L24 was found to be critical for a putative re-initiation mechanism proposed for the expression of ARF encoding genes (Nishimura et al, 2005). L24 is predicted to be located at the periphery of the large subunit surface of the 60S subunit, and serves as a bridge between the large and small ribosomal subunits by interacting with helix 44 of the 16S rRNA (Ban et al, 2000; Spahn et al, 2001). In yeast, L24 (as part of the ribosome) was reported to enhance translational efficiency, and its depletion causes the appearance of so called ‘halfmers', when polysomes are deficient in active 60S subunits, although it is not essential for cell viability (Baronas-Lowell and Warner, 1990; Dresios et al, 2000). Thus RISP, with or without TAV, likely binds the internal surface of 60S very close to the main factor binding site (Spahn et al, 2001). Accordingly, it appears most likely that binding of TAV or RISP to 60S via L24 interferes with L24 function in mediating 40S and 60S contact, and thereby negatively affects the elongation efficiency of the entire ribosome. We note, for instance, that the interaction between RISP and L24 may contribute to re-initiation at either termination and/or scanning steps, especially when strengthened by TAV. However, this hypothesis requires functional testing of the corresponding complexes on purified ribosomal subunits.
Our findings presented here and in previous studies, strongly suggest that recruitment or joining of 60S might be the second limiting step in polycistronic translation. Indeed, a rate-limiting step in translation initiation in the majority of well-studied cases is formation of the 48S-PIC at the proper initiation codon (Jackson, 1996; Mathews et al, 1996). Retention of eIF3 on translationally active ribosomes increases their re-initiation capacity (Park et al, 2001; Szamecz et al, 2008). Our results suggest that the next step in re-initiation—recruitment of functionally active 60S subunits—is also a rate-limiting step that can be targeted by inhibitory mechanisms. Indeed, suppression of the 60S subunit joining step was suggested to regulate the first translation initiation event on LOX mRNA in Drosophila (Ostareck et al, 2001), and mutations identified in several 60S ribosomal proteins can cause derepression of GCN4 translation by altering the 60S subunit joining step in yeast (Foiani et al, 1991; Martín-Marcos et al, 2007). Examples of regulated 60S recruitment can involve eIF6, which interacts with 60S and precludes 80S formation (Ceci et al, 2003). Consistently, it was demonstrated recently that eIF6 is rate-limiting for translation in mammals (Gandin et al, 2008). Thus, it is not unusual for re-initiation factors to target both 40S and 60S subunits, suggesting control of eIF3 recruitment by TAV or TAV–RISP and 60S recruitment/joining 60S to the PIC by RISP or by TAV–RISP.
What is the functional role of the RISP–TAV interaction with 60S? The most striking feature that has been revealed by our work is the construction of a bridge between L24 and eIF3 in vitro (Figure 5E). In infected plants, this bridge may consist of the RISP–TAV complex. This connection would facilitate recruitment of 60S to the eIF3-containing scanning complex or allow re-use (re-cycling) of the 60S ribosomal subunit during subsequent initiation events, with RISP working in concert with TAV.
Previously, we proposed a model in which TAV can mediate binding of eIF3 to 60S, possibly through an association with L18/L13 ribosomal proteins (Park et al, 2001). This interaction allows eIF3–TAV to travel with elongating ribosomes until the termination codon is reached. At the termination step, the eIF3–TAV complex is transferred back to 40S to regenerate a PIC capable of binding TC (Park et al, 2001). Here, we speculate that the RISP–eIF3 complex binds 40S at the 43S-PIC formation step, where it can later serve as a TAV-binding site upon subunit joining. During elongation, RISP together with TAV–eIF3 can be stabilized on 80S, likely by the TAV–60S interaction; accumulation of eIF3 and RISP in polysomes is seen in the presence of TAV (Park et al, 2001; Figure 2A). At the termination step, the function of the RISP–TAV complex would be to bridge the relaxed 40S–60S interactions through contact with 40S-bound eIF3 and, at the same time, to re-establish contact with 60S, but now through L24 (Figure 7A). It was proposed that binding of eIFs 3, 1 and 1A to post-termination complexes (postTC) normally causes 60S to dissociate from the mRNA in vitro (Pisarev et al, 2007). We suggest that, when the RISP–TAV complex bridges L24–eIF3 interactions, 60S dissociation could be delayed during the termination event, supporting 60S re-joining during the re-initiation event.
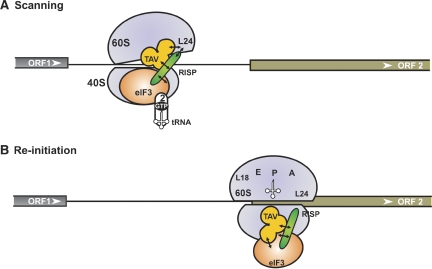
Figure 7.
Proposed model of re-initiation supporting protein (RISP) function in 60S recruitment during virus-activated re-initiation. We propose the following scenario: during ORF1 elongation, the RISP–transactivator viroplasmin (TAV)–80S complex can be stabilized by transfer of TAV–RISP–eIF3 to the solvent surface of 60S through TAV binding to L18/L13. During termination, the TAV–RISP–eIF3 complex is relocated back to 40S to reconstruct a pre-initiation complex (PIC) competent for re-initiation. (A) RISP–TAV establishes a bridge between 40S-bound eIF3 and 60S through the ribosomal protein L24, preventing, for a short time, removal of 60S. During scanning, RISP bridges the relaxed 40S–60S interactions through contact with 40S-bound eIF3, while simultaneously stabilizing TAV–L24 contacts (open conformation of 80S). This open 80S conformation allows eIF3-bound 40S to continue scanning and search for a downstream start codon. (B) Codon–anticodon recognition and positioning of Met-tRNAiMet in the ribosomal P-site would then displace TAV and RISP from L24 followed by the formation of 80S ready for elongation. eIF3, TC, RISP, TAV, L24, 40S and 60S are indicated.
A re-cycling model seems attractive; this model includes two alternative conformations of the 40S–60S complex in the presence of TAV: (A) ‘open' scanning, and (B) ‘closed' re-initiation configurations (Figure 7). According to our earlier results (Park et al, 2001) TAV binds either to eIF3 or to L24, and eIF3g outcompetes L24 for TAV binding. During scanning, RISP strengthens TAV binding to L24 and at the same time bridges the relaxed 40S–60S interaction by direct binding to the eIF3 complex (Figure 7A). TAV interacting with 60S-bound RISP would maintain an open conformation of 80S, thus allowing scanning. Somewhere around the codon–anticodon interaction, binding of RISP–TAV to L24 will be disrupted by factors to allow the switch from scanning mode to initiation followed by elongation mode (Figure 7B).
The ‘60S re-cycle' model may explain why the efficiency of TAV-activated polycistronic translation is not dependent on the distance between two ORFs (Fütterer and Hohn, 1991, 1992). Indeed, TAV can ensure re-use or immediate recruitment of 60S and eIF3, as well as keeping them travelling with 40S during scanning to a downstream ORF. In another virus-specific re-initiation strategy—‘termination-re-initiation'—eIF3 recruitment occurs through a cis-acting RNA element (Pöyry et al, 2007). This strategy requires close spacing between the stop and start codons (Meyers, 2003; Pöyry et al, 2007). This juxtaposition of stop–start codons could favour the re-use of 60S in subsequent re-initiation events.
Comparative analysis of RISP and TAV function could open up the possibility of answering important questions about the regulatory mechanisms that limit polycistronic translation in eukaryotic cells. It seems reasonable to suppose that RISP is a component of the plant translational machinery and that it functions in translation initiation, which would not be surprising as it targets factors essential for the initiation process: eIF3, which is required at least for de novo recruitment of TC (eIF2–GTP–Met-tRNAiMet); and the 60S ribosomal subunit, the second essential component of each initiation step.
Materials and methods
Plasmids
The partial RISP cDNA was isolated from an Arabidopsis yeast two-hybrid library (41–389 aa; Clontech). The 5′-part of its coding region was amplified from an Arabidopsis cDNA library using a 5′-RACE kit (NM_125513.1; Invitrogen; Supplementary Figure S1). Description of plasmids and their construction, and details of biochemical methods can be found in the Supplementary data.
Plant growth and viral infection
Arabidopsis wild-type, T-DNA mutant (134C07) plants (Col0 background) were raised from seeds under standard conditions. At 4–5 weeks post-germination, seedlings were inoculated mechanically with CaMV Cabb-JI.
Purification of proteins, ribosomes and polysomes
Wheat germ eIF3 was kindly provided by Professor K Browning (University of Texas at Austin, USA). Conalbumin was obtained from a GE Healthcare Gel Filtration calibration kit. GST-fusion proteins were expressed in E. coli BL21 DE3 RIL (Stratagene). GST was removed by on-column cleavage with PreScission protease (GE Healthcare). 80S ribosomes, 60S and 40S subunits were isolated from wheat germ according to Lax et al (1986) and Spermulli et al (1977). Polysome isolation and density centrifugation was carried out as described by Park et al (2001) (see Supplementary data). Ribosomal complexes were prepared as described in the Supplementary data.
Two-hybrid strategy
The mating strategy for two-hybrid screening and two-hybrid analysis has been described previously (Park et al, 2001). The two-hybrid interaction analysis carried out in AH109 is described in the Supplementary data.
Pull-down experiments
Glutathione-S-transferase pull-down assays were carried out as described by Park et al (2001). Co-immunoprecipitation experiments were done, as described, with modifications (Haas et al, 2008; see Supplementary data). Rabbit polyclonal anti-TAV antibodies were raised against recombinant 6His-TAV. Rabbit polyclonal antibodies against RISP, L13, eIF3c and S6 were raised against the full-length Arabidopsis recombinant proteins produced in E. coli. Human anti-ribosomal P antigen antibodies to 60S ribosomal proteins P0–P1–P2 were purchased from Immunovision. Rabbit polyclonal anti-eIF2α and anti-eIF3c antibodies were a kind gift from Professor K Browning. Rabbit polyclonal anti-katanine antibodies were a kind gift from Marylin Vantard (CEA Grenoble, France).
Transient expression
Leaf protoplasts derived from N. plumbaginifolia were prepared and samples of 6 × 105 protoplasts were used for polyethylene glycol-mediated transfection as described by Kobayashi et al (1998). The GUS-expressing di-cistronic reporter plasmid (pbiGUS; 2.5 μg) was always co-transfected with a CAT-expressing plasmid (pmonoCAT; 2 μg). For transactivation, 1 μg pTAV (or pTAVΔADA) or 5–10 μg pRISP (or pGST) was added as indicated. TAV, TAVΔADA, RISP and RISP deletion mutants were well expressed in N. plumbaginifolia protoplasts, as controlled by western blotting (data not shown). CAT and GUS activities were determined as described (Pooggin et al, 2000). The values given are the means from more than three independent experiments.
Arabidopsis mesophyll protoplasts were prepared from 3- to 4-week-old plantlets (Col-0 and 134C07) and transfected by the method of Yoo et al (2007). For transactivation, 10 μg pbiGUS, 10 μg pmonoGFP and either 10 μg pTAV (or 10 μg empty vector p35S) were transfected. pmonoGFP expression was monitored by western blot with anti-GFP antibodies kindly provided by D Gilmer (IBMP, Strasbourg).
Detection of CaMV replication by reporter-targeted PCR
Principals and details of the experiments were described in Kobayashi and Hohn (2003). Turnip protoplasts were transfected with the following plasmids in the indicated combinations pE4Pin (viral clone with PCR reporter, 10 μg); pAA TAV, Wild-type (WT) or three-aa deletion mutants, ΔADA, 4 μg; pGW GAG (mono-cistronic CaMV GAG), 4 μg; and pGW POL (mono-cistronic CaMV POL), 2 μg. The total amount of plasmid was adjusted to 20 μg with pBluescript. At 3 days post-transfection, protoplasts were harvested and low molecular weight DNA was extracted. Replication of CaMV DNA was assayed by reporter-targeted PCR using PSS and LAS primers through 25 reaction cycles.
Immunolabelling in tissue and BY2 cells
Endogenous RISP, 60S and TAV were localized in epidermal cells of infected turnip plants using appropriate primary antibodies, which were visualized by Alexa Fluor 568- or 488-conjugated anti-human or anti-rabbit secondary antibodies (Invitrogen).
BY2 cells were harvested and attached to poly-L-lysine-coated coverslips, fixed with a mixture of 1% glutaraldehyde and 1.5% paraformaldehyde and immunolabelled using combinations of appropriate primary antibodies (see Supplementary data). Nuclei were stained with 1 μg/ml 4,6′-diamidino-2-phenylindole (DAPI) solution.
Imaging analysis
Samples were analyzed with a Zeiss LSM510 confocal microscope (Jena, Germany). The quantitative colocalization analyses were performed using the NIH ImageJ software with the Colocalization Finder plugin, available at http://rsb.info.nih.gov/ij/plugins/. This software was used to determine the Rr, which describes the extent of overlap between image pairs. It is a value between −1 and +1, with −1 being no overlap and +1 being perfect overlap of two images.
Supplementary Material
Supplementary Figure 1S
Supplementary Figure 2S
Supplementary Figure 3S
Supplementary Figure 4S
Supplementary Figure Legends
Supplementary Materials and Methods
Review Process File
Acknowledgments
We thank K Browning for wheat germ eIF3 and anti-eIF2α and anti-eIF3c antibodies and M Taliansky for his expertize in immunolabeling in tissue. Thanks to H Rothnie for critical reading of the manuscript. This work was supported by grant BCMS364 from Fonds National de la Science (ACI, France), grant BLAN06-2_135889 from Agence Nationale de la Recherche (France), and grant INE 20031114123 from Fondation pour la Recherche Médicale (France) to LR and by a PhD fellowship from the Ministry of Research to OT.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV (2006) Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev 20: 210–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Baronas-Lowell DM, Warner JR (1990) Ribosomal protein L30 is dispensable in the yeast Saccharomyces cerevisiae. Mol Cell Biol 10: 5235–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneville J-M, Sanfaçon H, Fütterer J, Hohn T (1989) Posttranscriptional transactivation in cauliflower mosaic virus. Cell 59: 1135–1143 [DOI] [PubMed] [Google Scholar]
- Bureau M, Leh V, Haas M, Geldreich A, Ryabova L, Yot P, Keller M (2004) P6 protein of Cauliflower mosaic virus, a translation reinitiator, interacts with ribosomal protein L13 from Arabidopsis thaliana. J Gen Virol 85: 3765–3775 [DOI] [PubMed] [Google Scholar]
- Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhäuser N, Narchisio PC, Biffo S (2003) Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426: 579–584 [DOI] [PubMed] [Google Scholar]
- De Tapia M, Himmelbach A, Hohn T (1993) Molecular dissection of the cauliflower mosaic virus translational transactivator. EMBO J 12: 3305–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG (1992) Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68: 585–596 [DOI] [PubMed] [Google Scholar]
- Dresios J, Derkatch IL, Liebman SW, Synetos D (2000) Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 39: 7236–7244 [DOI] [PubMed] [Google Scholar]
- Evans SP, Bycroft M (1999) NMR structure of the N-terminal domain of Saccharomyces cerevisiae RNase HI reveals a fold with a strong resemblance tothe N-terminal domain of ribosomal protein L9. J Mol Biol 291: 661–669 [DOI] [PubMed] [Google Scholar]
- Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG (1991) GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol 11: 3203–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J, Hohn T (1991) Translation of a polycistronic mRNA in the presence of the cauliflower mosaic virus transactivator protein. EMBO J 10: 3887–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J, Hohn T (1992) Role of an upstream open reading frame in the translation of polycistronic mRNAs in plant cells. Nucleic Acids Res 20: 3851–3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Miluzio A, Barbieri A-M, Beugnet A, Kiyokawa H, Marchisio PC, Biffo S (2008) Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 455: 684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas G, Azevedo J, Moissiard G, Geldreich A, Himber C, Bureau M, Fukuhara T, Keller M, Voinnet O (2008) Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J 27: 2102–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M, Geldreich A, Bureau M, Dupuis L, Leh V, Vetter G, Kobayashi K, Hohn T, Ryabova L, Yot P, Keller M (2005) The open reading frame VI product of Cauliflower mosaic virus is a nucleocytoplasmic protein: its N terminus mediates its nuclear export and formation of electron-dense viroplasms. Plant Cell 17: 927–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama T, Kaufmann F, Schroeter B, Hatakeyama T (1989) Primary structures of five ribosomal proteins from the archaebacterium Halobacterium marismortui and their structural relations to eubacterial and eukaryotic ribosomal proteins. Eur J Biochem 185: 685–693 [DOI] [PubMed] [Google Scholar]
- Hauden CA, Jorgensen RA (2007) Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol 30: 5–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2006) eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 31: 553–562 [DOI] [PubMed] [Google Scholar]
- Horvath CM, Williams MA, Lamb RA (1990) Eukaryotic coupled translation of tandem cistrons: identification of the influenza B virus BM2 polypeptide. EMBO J 9: 2639–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ (1996) A comparative view of initiation site selection mechanisms. In Translational Control, Hershey JWB, Mathews MB, Sonenberg N (eds), pp 71–112. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Kawaguchi R, Bailey-Serres J (2005) mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res 33: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-H, Kim B-H, Yahalom A, Chamovitz DA, von Arnim AG (2004) Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 16: 3341–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Hohn T (2003) Dissection of cauliflower mosaic virus transactivator/viroplasmin reveals distinct essential functions in basic virus replication. J Virol 77: 8577–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Tsuge S, Nakayashiki H, Mise K, Furusawa I (1998) Requirement of cauliflower mosaic virus open reading frame VI product for viral gene expression and multiplication in turnip protoplasts. Microbiol Immunol 42: 377–386 [DOI] [PubMed] [Google Scholar]
- Kozak M (1999) Initiation of translation in prokaryotes and eukaryotes. Gene 234: 187–208 [DOI] [PubMed] [Google Scholar]
- Kozak M (2001) Constraints on reinitiation of translation in mammals. Nucleic Acids Res 29: 5226–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (2002) Emerging links between initiation of translation and human diseases. Mamm Genome 13: 401–410 [DOI] [PubMed] [Google Scholar]
- Lax SR, Lauer SJ, Browning KS, Ravel JM (1986) Purification and properties of protein synthesis initiation and elongation factors from wheat germ. Methods Enzymol 118: 109–128 [DOI] [PubMed] [Google Scholar]
- Leh V, Yot P, Keller M (2001) The cauliflower mosaic virus translational transactivator interacts with the 60S ribosomal subunit protein L18 of Arabidopsis thaliana. Virology 266: 1–7 [DOI] [PubMed] [Google Scholar]
- Luttermann C, Meyers G (2007) A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J Biol Chem 282: 7056–7065 [DOI] [PubMed] [Google Scholar]
- Martín-Marcos P, Hinnebusch AG, Tamame M (2007) Ribosomal protein L33 is required for ribosome biogenesis, subunit joining, and repression of GCN4 translation. Mol Cell Biol 27: 5968–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Sonenberg N, Hershey JWB (1996) The pathway and mechanism of eukaryotic protein synthesis. In Translational Control, Hershey JWB, Mathews MB, Sonenberg N (eds), pp 1–29. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Meyers G (2003) Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J Biol Chem 278: 34051–34060 [DOI] [PubMed] [Google Scholar]
- Meyers G (2007) Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus. J Virol 81: 9623–9632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DR, Geballe AD (2000) Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20: 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KH, Valášek L, Sykes C, Jivotovskaya A, Hinnebusch AG (2006) Interaction of the RNP1 motif in PRT1 with HCR1 promotes 40S binding of eukaryotic initiation factor 3 in yeast. Mol Cell Biol 26: 2984–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K (2005) The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17: 2940–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck D, Ostareck-Lederer A, Shatsky I, Hentze M (2001) Lipoxygenase mRNA silencing in erythroid differentiation: the 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell 104: 281–290 [DOI] [PubMed] [Google Scholar]
- Park HS, Browning KS, Hohn T, Ryabova LA (2004) Eucaryotic initiation factor 4B controls eIF3-mediated ribosomal entry of viral reinitiation factor. EMBO J 23: 1381–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Himmelbach A, Browning KS, Hohn T, Ryabova LA (2001) A plant viral ‘reinitiation' factor interacts with the host translational machinery. Cell 106: 723–733 [DOI] [PubMed] [Google Scholar]
- Pestova T, Lorsch JR, Hellen CUT (2007) The mechanism of translation initiation in eukaryotes. In Translational Control in Biology and Medicine, Mathews MB, Sonenberg N, Hershey JWB (eds), pp 87–128. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Pisarev A, Hellen T, Pestova T (2007) Recycling of eukaryotic posttermination ribosomal complexes. Cell 131: 286–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooggin MM, Hohn T, Fütterer J (2000) Role of a short open reading frame in ribosome shunt on the cauliflower mosaic virus RNA leader. J Biol Chem 275: 17288–17296 [DOI] [PubMed] [Google Scholar]
- Powell ML, Napthine S, Jackson RJ, Brierley I, David Brown TDK (2008) Characterization of the termination-reinitiation strategy employed in the expression of influenza B virus BM2 protein. RNA 14: 2394–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Connell EJ, Fraser CS, Jackson RJ (2007) The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev 21: 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Jackson RJ (2004) What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev 18: 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabova LA, Pooggin MM, Hohn T (2006) Translation reinitiation and leaky scanning in plant viruses. Virus Research 119: 52–62 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn C, Beckmann R, Eswar N, Penczek P, Sali A, Blobel G, Frank J (2001) Structure of the 80S ribosome from Saccharomyces cerevisiae–tRNA-ribosome and subunit–subunit interactions. Cell 107: 373–386 [DOI] [PubMed] [Google Scholar]
- Spermulli LL, Walthall BJ, Lax SR, Ravel JM (1977) Purification and properties of a Met-tRNA binding factors from wheat germ. Arch Biochem Biophys 178: 565–575 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ishihara D, Sasaki M, Nakagawa H, Hata H, Tsunoda T, Watanabe M, Komatsu T, Ota T, Isogai T, Suyama A, Sugano S (2000) Statistical analysis of the 5′ untranslated region of human mRNA using ‘Oligo-Capped' cDNA libraries. Genomics 64: 286–297 [DOI] [PubMed] [Google Scholar]
- Szamecz B, Rutkai E, Cuchalová L, Munzarová V, Herrmannová A, Nielsen KH, Burela L, Hinnebusch AG, Valášek L (2008) eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev 22: 2414–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran MK, Schultz CJ, Baumann U (2008) Conserved upstream open reading frames in higher plants. BMC Genomics 31: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L, Mathew AA, Shin B-S, Nielsen KH, Szamecz B, Hinnebusch AG (2003) The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev 17: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1S
Supplementary Figure 2S
Supplementary Figure 3S
Supplementary Figure 4S
Supplementary Figure Legends
Supplementary Materials and Methods
Review Process File