Abstract
The oxidative phosphorylation (OXPHOS) system consists of five multimeric complexes embedded in the mitochondrial inner membrane. They work in concert to drive the aerobic synthesis of ATP. Mitochondrial and nuclear DNA mutations affecting the accumulation and function of these enzymes are the most common cause of mitochondrial diseases and have also been associated with neurodegeneration and aging. For this reason, several approaches for the assessment of the OXPHOS system enzymes have been developed. Based on the methods described elsewhere, we present here assays for a biochemical characterization of the OXPHOS system in cells and mitochondria isolated from cultured cells or tissues.
Keywords: Electron transport chain, Mitochondria, OXPHOS, Polarography, Spectrophotometry
INTRODUCTION
The oxidative phosphorylation system consists of four multimeric complexes, coenzyme Q and cytochrome c forming the mitochondrial respiratory chain (I–IV), which transfer electrons from reducing equivalents to water, creating a proton gradient across the inner mitochondrial membrane, which is used by a fifth complex, the F1F0 ATPase, to drive the synthesis of ATP. Mitochondrial and nuclear DNA mutations affecting the accumulation and function of the electron transport and OXPHOS system enzymes are the most common cause of mitochondrial diseases and have also been associated with neurodegeneration and aging (DiMauro and Schon, 2003, Fernandez-Vizarra et al., 2009, Reeve et al., 2008).
Polarographic studies of oxygen consumption (Basic Protocols 1 and 2) and spectrophotometric analyses of the mitochondrial respiratory chain enzymes (Basic Protocol 3) are classical powerful tools for the characterization of mitochondrial respiratory capacity in mitochondria-enriched fractions prepared from tissues or cultured cells, in tissue homogenates, or in whole cells (Barrientos, 2002, Birch-Machin and Turnbull, 2001, Munnich and Rustin, 2001, Rustin et al., 1991, Rustin et al., 1994, Villani et al., 1998). Based on methods described elsewhere, we present here a set of assays for biochemical characterization of the OXPHOS system in cells and mitochondria isolated from cultured cells or tissues.
Basic Protocol 1
POLAROGRAPHIC OXYGEN CONSUMPTION STUDIES USING INTACT CELLS
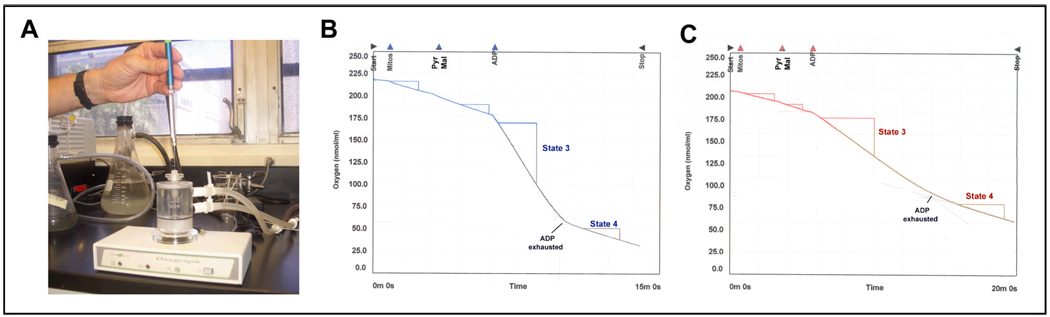
A significant amount of our knowledge of electron transport in mitochondria comes from oxygen electrode recordings. The oxygen concentration in a sealed incubation chamber is continuously monitored, and the effects of adding different compounds to the chamber can be recorded. In a typical experiment, after the polarograph (Figure 1A) has been calibrated, the respiratory buffer containing phosphate is introduced into the chamber followed by the biological material (permeabilized cells or isolated mitochondria). Subsequently, the substrates, ADP (which causes a sudden burst in oxygen uptake as the ADP is converted to ATP), and specific inhibitors that block respiration are added. Respiration in the presence of ADP is called State 3; when the ADP is exhausted, respiration turns to a resting state or State 4 (Figure 1B and C). A replicate experiment can be done in the presence of an uncoupler such as carbonyl cyanide m-chlorophenylhydrazone (CCCP), which leads to a permanently high rate of respiration in the absence of ADP.
Figure 1.
Polarographic assessment of mitochondrial respiratory chain performance. (A) Polarographic chamber (Hansatech). (B) and (C) Typical oxygen consumption recording chart using mitochondria isolated from either wild type cells or cells with partial respiratory deficiency, respectively. Pyruvate plus malate were used as site I substrates.
The rate of oxygen diffusion to the cathode of the monitoring electrode (and hence the current) depends on the oxygen concentration in the main incubation chamber. It also depends on several other factors that must be controlled: temperature, membrane thickness and permeability, sample viscosity, and stirring speed. There is no intrinsic calibration of an oxygen electrode. At regular intervals, or if the instrument is disassembled, it must be recalibrated with a reducing agent (usually sodium dithionite) against a known standard (usually air). In addition, the electrode should be clean and free of oxidized material.
Among the different models that are currently available, the use of a polarograph with an adjustable chamber volume (i.e., from 0.25 up to 1 ml) is important when working with small amounts of biological material. The amount of mitochondria and cells suggested for use in the assays described in this section is optimized for 0.3 ml of standard medium with a Clark oxygen electrode in a water-jacketed microcell, magnetically stirred, at 37 °C (Hansatech Instruments Limited, Norfolk, United Kingdom) (Figure 1A). The biological material, substrates and inhibitors are introduced into the chamber through a capillary hole in the plunger using a graduated syringe (e.g. Hamilton) as illustrated in Figure 1A.
Materials
Adenine nucleotides (Sigma): 40 mM ADP, 40 mM ATP
Bovine serum albumin (BSA)(Sigma)
Digitonine high purity (Calbiochem)
Inhibitors (Sigma): 2 mM Rotenone, 1M Malonate, 0.2 mM Antimycin A, 80 mM potassium cyanide (KCN), 1.5 mM oligomycin.
Substrates (Sigma): 50 mM Pyruvate, 175 mM Malate, 175 mM L-Glutamate, 50 mM Succinate, 50 mM Duroquinone, 50 mM Ascorbate, 500 mM L-glycerol 3-phosphate (G3P). All the substrates are dissolved in H2O and the pH adjusted to pH 7.4. 6 mM N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD)
Uncouplers (Sigma): 1mM carbonyl cyanide m-chlorophenylhydrazone (CCCP)
Measure intact cell-coupled endogenous respiration
This measurement is performed in intact, non-permeabilized, cells and provides a general assessment of the ability of the cells to respire using endogenous substrates.
Collect exponentially growing cells (in medium changed 2–4 hours before) by trypsinizing and pelleting them at 500 × g.
Resuspend the pellet at 7.5×106 cells/ml in respiration buffer (RB).
Inject 300 µl of cell suspension into the polarographic chamber and measure the intact cell-coupled endogenous cell respiration.
Add KCN (to 700 µM) to block respiration and continue measuring respiration for additional 2 min.
The respiratory rate in the presence and absence of KCN is obtained by dividing the µmols of oxygen consumed per min (rate provided by the computer program running the polarograph) by the number of cells used in the experiment. The final respiratory rate is obtained by subtracting the KCN-insensitive respiration.
Measure substrate oxidation in digitonin-permeabilized cells
To permeabilize the plasma membrane to different respiratory substrates, add 60 µg digitonin/ml to the cell suspension in RB (7.5×106 cells/ml) and incubate for 1–2 min at room temperature (Barrientos, 2002, Villani et al., 1998).
Add 5 vol RB supplemented with 1 mg/ml bovine serum albumin (BSA) to the cell suspension to stop excessive membranes solubilization.
Pellet the cells by centrifugation at 500 × g.
Resuspend the pellet at 7.5×106 cells/ml RB supplemented with 1 mg/ml BSA and 0.5 mM ADP. The RB should be maintained air-equilibrated at 37 °C. The suspension of permeabilized cells is ready to use in oxygen consumption experiments.
Immediately after permeabilization, inject 300 µl cell suspension into the polarographic chamber. Start oxygen consumption measurements with the different substrates as explained in steps 6 and 7. If necessary, add extra ADP.
-
It is possible to measure oxygen consumption with different substrates by either performing a single experiment for each substrate or by combining several sets of substrates and inhibitors as explained in this section. With one cell suspension sample in the polarographic chamber follow the following substrates oxidation in the order listed, always starting by site I substrates and following with site II and III substrates. Complexes I, II and III are successively inhibited prior to adding the next substrates.
-
6a.
Add pyruvate (8 mM) plus malate (0.2 mM), record oxygen consumption for 2 min; add glutamate (15 mM) (site I substrates) and record oxygen consumption for additional 2–5 min. Add rotenone (3 µM) to inhibit the reaction and monitor oxygen consumption for 2–5 min.
-
6b.
Subsequently, add succinate (10 mM) (site II substrate) and ATP (130 µM) and record oxygen consumption for 2–5 min. Add malonate (10 mM) to inhibit complex II and monitor oxygen consumption for 2–5 additional min.
-
6c.
Finally, add duroquinol (0.6 mM) (site III substrate) and record oxygen consumption for 2 min. Add antimycin A (1 µM) to inhibit complex III and monitor oxygen consumption for 2–5 additional min.
-
6d.
For each substrate, the respiratory rate in the presence and absence of the specific inhibitor is obtained by dividing the µmols of oxygen consumed per min (rate provided by the computer program running the polarograph) by the number of cells used in the experiment. The final respiratory rate is obtained by subtracting the inhibitor-insensitive respiration.
-
6a.
Measure complex IV activity polarographically in either intact or permeabilized cells
Inject 300 µl of intact or digitonine-permeabilized cell suspension at 7.5×106 cells/ml into the polarographic chamber.
Add ascorbate (10 mM) plus N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD, 0.2 mM) which give electrons directly to cytochrome c, thus providing with an estimation of oxidation through complex IV. Record oxygen consumption for 2–5 min.
Add KCN (to 700 µM) to block respiration and continue measuring respiration for additional 2–5 min.
The ascorbate-TMPD oxidation rate in the presence and absence of KCN is obtained by dividing the µmols of oxygen consumed per min (rate provided by the computer program running the polarograph) by the number of cells used in the experiment. The final respiratory rate is obtained by subtracting the KCN-insensitive respiration.
Basic Protocol 2
OXYGEN CONSUMPTION STUDIES IN ISOLATED MITOCHONDRIA
The accuracy and reproducibility of polarographic studies in isolated mitochondria greatly depend on the quality of the mitochondrial preparation. Methods for the isolation of mitochondria from human tissues or cultured cells can be found elsewhere (Birch-Machin and Turnbull, 2001, Hartwig et al., 2009, Pallotti and Lenaz, 2001, Rustin et al., 1994). A minimum of 30–60 µg of mitochondrial protein is used in each experiment. Some of the assays that can be performed in intact mitochondria include:
oxidation of glutamate (20 mM) plus malate (20 mM) and oxidation of pyruvate (20 mM) plus malate (1 mM) in the absence (state 4) and presence (state 3) of 0.3 mM ADP (the reaction is sensitive to 0.3 mM KCN)
oxidation of succinate (12 mM) in the presence of 3 µM rotenone and 0.2 mM ATP, inhibited with 10 mM malonate followed by the oxidation of glycerol 3-phosphate (G3P, 20 mM) and the oxidation of duroquinol (0.6 mM) which is fully inhibited with 0.3 mM KCN
oxidation of ascorbate (10 mM) plus TMPD (0.2 mM), which is inhibited with 0.3 mM KCN.
The respiratory control (which measures the coupling between electron flux and ATP synthesis) associated with the oxidation of each substrate can be determined by two means: (a) by calculating the State 3/State 4 ratio and (b) comparing, the oxygen consumption rates, following state 3, obtained in the presence of the specific F0F1-ATPase inhibitor oligomycin (10 µM), and in the presence of the uncoupler CCCP (10 µM).
It is possible to calculate a P:O ratio (the relationship between ATP synthesis and oxygen consumption) by measuring the decrease in oxygen concentration during the rapid burst of state 3 respiration after adding a known amount of ADP until it is finished (which results in an extensive reduction of the respiratory rate). The amount of oxygen consumed is then related to the quantity of ADP added. The P:O ratio varies with the substrate utilized and gives an estimation of OXPHOS efficiency.
Basic Protocol 3
ENZYMATIC ACTIVITY STUDIES
The measurement of the specific activity of the individual complexes of the respiratory chain can be performed spectrophotometrically. Some of the activities can be measured directly by using whole cells. For this purpose, cells growing exponentially in medium changed a few hours before starting the experiment are collected by trypsinization, pelleted, and resuspended in cold phosphate-buffered saline medium to a concentration of 5×106 cells/ml. The suspension is aliquoted and frozen at −80 °C until used for the different enzymatic assays. The same assays can be performed using tissue homogenates.
The activity of the five OXPHOS enzymes can also be confidently measured in isolated mitochondria. A total of 5–40 µg of mitochondrial protein must be used to determine the activity of each complex. The assays described here are performed at 37 °C (except citrate synthase activity, at 30 °C) in 1 ml of medium basically as reported (Barrientos, 2002, Rustin et al., 1991, Rustin et al., 1994).
To compare OXPHOS activities among cells, tissues or individuals, researchers commonly normalize OXPHOS enzymatic activities measured in isolated mitochondria to the activity of citrate synthase, an enzyme of the tricarboxylic acids cycle (see Support Protocol 1).
Materials
Adenine nucleotides (Sigma): 40 mM ADP, 40 mM ATP
Bovine serum albumin (BSA)
Electron donors and acceptors (Sigma): 40 mM NADH, 20 mM Decylubiquinone, 50 mM succinate, 0.2 M 2, 6-dichlorophenolindophenol (DCPIP), 400mM phosphoenolpyruvate (PEP)
Enzymes (Sigma): Lactate dehydrogenase, pyruvate kinase
Inhibitors (Sigma): 0.2 mM Antimycin A (AA), 80 mM KCN, 2mM rotenone, 100 mM thenoyltrifluoroacetone (TTFA)
Lithium borohydrate
50 mM Tris-HCl (pH 8.0)
Uncouplers (Sigma): carbonyl cyanide m-chlorophenylhydrazone (CCCP)
Detergents: 10M Lauryl maltoside.
Measure the rotenone-sensitive NADH-decylubiquinone oxidoreductase (NQR): complex I activity
This assay involves decylubiquinone as the electrons acceptor and NADH as donor. It is not possible to measure NQR in whole cells due to the absence of activation of the enzyme with decylubiquinone. Because the accuracy of the method relies on the access of NADH to its binding site in complex I, mitochondrial membranes should be disrupted first by freeze–thawing the samples two or three times in hypotonic medium (25 mM K2PO4 pH 7.2, 5 mM MgCl2) (Birch-Machin and Turnbull, 2001) followed by a hypotonic shock in H2O (Chretien et al., 1990).
-
1a.
In 800 µl of H2O add mitochondria (20–40 µg of protein, pretreated as mentioned earlier) and incubate 1–2 min at 37 °C.
The incubation of mitochondria in H2O must not be longer than 3 min, because the rotenone-sensitive activity can be affected.
-
2a.
Add 200 µl of 50 mM Tris (pH 8.0) medium supplemented with 5 mg/ml BSA, 0.8 mM NADH as donor, 240 µM KCN, and 4 µM antimycin A (AA).
-
3a.
Start the reaction by the addition of 50 µM of the acceptor decylubiquinone.
-
4a.
Measure activity by following the decrease in absorbance at 340 nm resulting from the oxidation of NADH for 3 min.
-
5a.
Add 4 µM rotenone and keep monitoring activity for 3 additional min to measure the rotenone-sensitive activity.
Measure rotenone-sensitive NADH-cytochrome c oxidoreductase activity (NCCR): complex I + III activity
This assay is performed following the increase in absorbance at 550 nm resulting from the reduction of freshly prepared cytochrome c. The assay involves oxidized cytochrome c as the electrons acceptor and NADH as donor.
-
1b.
In 800 µl of H2O add mitochondria (20–40 µg of protein) and incubate 1–2 min at 37 °C.
-
2b.
Add 200 µl of 50 mM Tris-HCl (pH 8.0) medium supplemented with 5 mg/ml BSA, 40 µM oxidized cytochrome c as the acceptor, and 240 µM KCN. Incubate for 4 min.
-
3b.
Start the reaction by addition of 0.8 mM NADH as donor. Measure activity for 3 min.
-
4b.
Add 4 µM rotenone and keep monitoring activity for 3 additional min to measure the rotenone-sensitive activity.
Measure succinate decylubiquinone DCPIP reductase (SQR): complex II activity
This assay involves DCPIP as the electrons acceptor and succinate as donor. Since the activity depends on the disruption of the inner mitochondrial membrane, biological material must be freeze–thawed before using. To fully activate complex II (which can be partially deactivated due to the tight binding of the competitive inhibitor oxaloacetate) it is absolutely necessary to preincubate the biological material with succinate.
-
1c.
In 1 ml of SQR medium add mitochondria (20–40 µg of protein) or whole cells (3–10×104 cells), 80 µM DCPIP as acceptor, 4 µM rotenone, and 0.2 mM ATP.
-
2c.
Add 10 mM succinate as the donor and incubate 10 min at 30 °C.
-
3c.
Start the reaction by addition of 80 µM decylubiquinone. Measure activity for 5 min.
-
4c.
Follow the decrease in absorbance at 600 nm resulting from the reduction of 2,6-dichlorophenolindophenol (DCPIP).
-
5c.
Add 10 mM malonate (competitive inhibitor) to inhibit the oxidation of succinate.
Measure succinate cytochrome c reductase (SCCR): complex II + III activity
The assay is performed following the increase in absorbance at 550 nm resulting from the reduction of freshly prepared cytochrome c. The assay involves oxidized cytochrome c as the electrons acceptor and succinate as donor. Also in this case, to fully activate complex II it is absolutely necessary to preincubate the biological material with succinate.
-
1d.
In 1 ml of SCCR medium add mitochondria (20–40 µg of protein) or whole cells (3–10×104 cells), 240 µM KCN, 4 µM rotenone, and 0.2 mM ATP.
-
2d.
Add 10 mM succinate as the donor and incubate 10 min at 30 °C.
-
3d.
Start the reaction by addition of 40 µM oxidized cytochrome c. Measure activity for 5 min.
-
4d.
Add 10 mM malonate to inhibit the oxidation of succinate.
SCCR activity is sensitive to malonate and TTFA. However, in whole cell experiments, the inhibition with malonate is weak (less than 50/60%) and use of TTFA recommended.
Measure ubiquinol cytochrome c reductase (QCCR) complex III activity
The assay is performed following the increase in absorbance at 550 nm resulting from the reduction of cytochrome c. The assay involves oxidized cytochrome c as the electrons acceptor and decylubiquinol as donor.
-
1e.
Decylubiquinol must be freshly prepared (Barrientos, 2002, Ragan and Cottingham, 1985). To the decylubiquinone solution, add a few crystals of lithium borohydrate and mix it by pipetting until the solution becomes transparent. Eliminate the excess of borohydrate with a few microliters of concentrated HCl, until no bubbles are produced.
The final pH should be 2–3.
-
2e
Biological material should be freeze–thawed in isotonic medium before using to gently disrupt the inner mitochondrial membrane.
It has been described that addition of final 0.6M lauryl maltoside helps to disrupt the membranes and liberate the maximum QCCR activity (Birch-Machin and Turnbull, 2001).
-
3e.
In 1 ml of QCCR medium add mitochondria (20–40 µg of protein) or whole cells (3–10×104 cells), 80 mM decylubiquinol as the donor, 240 µM KCN, 4 µM rotenone, and 200 µM ATP.
-
4e.
Start the reaction by addition of 40 µM oxidized cytochrome c. Measure activity for 5 min.
-
5e.
Add 0.4 µM antimycin A to distinguish between the reduction of cytochrome c catalyzed by complex III and the non-enzymatic reduction of cytochrome c by the reduced quinone.
-
6e.
Include background controls (without biological material and with and without AA) and substract from the rate obtained with biological material.
The components of the reaction produce an unspecific oxidation of decylubiquinol, and also the inhibitor AA affects the redox state of decylubiquinol.
The background activity depends on the length and the composition of the side chain of the quinol used. Decylubiquinol is the most suitable substrate because of its low background activity (Barrientos, 2002, Chretien et al., 2004, Krahenbuhl et al., 1994).
Measure cytochrome c oxidase: complex IV activity
The assay is performed following the decrease in absorbance at 550 nm resulting from the oxidation of reduced cytochrome c, which is the electrons donor.
-
1f.
When working with whole cells, it is recommended the preparation to be frozen–thawed two or three times in isotonic medium before using. When using mitochondrial fractions, they can be freshly prepared or aliquoted and kept frozen until use.
-
1f.
In 1 ml of isosmotic COX medium, add mitochondria (5–20 µg of protein) or whole cells (3–10×104 cells) and 10 µM reduced cytochrome c.
Cytochrome c solution should be freshly reduced by adding some crystals of sodium dithionite.
-
2f.
Permeabilize the outer mitochondrial membrane to cytochrome c by adding 2.5 mM lauryl maltoside. Follow the decrease in absorbance at 550 nm for 3 min.
When working with cells, it is suggested that the detergent be added to the COX medium with the reduced cytochrome c and start the reaction with the biological material.
-
3f.
Inhibit the reaction with 240 µM KCN and monitor absorbance at 550 nm for additional 2 min.
Measure COMPLEX V (F1F0-ATP SYNTHASE) ACTIVITY
Complex V activity (ATP hydrolysis) can be measured spectrophotometrically by a coupled assay using lactate dehydrogenase and pyruvate kinase as the coupling enzyme (Barrientos, 2002, Rustin et al., 1991). For each ATP molecule that is hydrolyzed, a molecule of NADH is oxidized.
-
1g.
In a microtube prepare a medium containing 200 µl 50 mM Tris (pH 8.0), 5 mg/ml BSA, 20 mM MgCl2, 50 mM KCl, 15 µM CCCP, 5 µM antimycin A, 10 mM PEP, 2.5 mM ATP, 4 units of lactate dehydrogenase and pyruvate kinase, and 1 mM NADH. Incubate the medium 5 min at 37 °C.
-
2g.
Incubate mitochondria (20–40 µg of protein) in distilled water for 30 s at 37 °C (use the spectrophotometer's chamber).
-
3g.
Add the medium to the chamber to start the reaction. Follow the reaction for 3 min.
-
4g.
Add 3 µM oligomycin and follow the reaction for an additional 3 min to distinguish the ATPase activity coupled to the respiratory chain. The expected inhibition with oligomycin is 60–90%.
-
5g.
Follow the increase in absorbance at 340 nm resulting from NADH oxidation.
It is not possible to follow this measurement in cells because of a strong oligomycin-insensitive ATPase activity.
Support Protocol 1
NORMALIZATION OF OXPHOS ACTIVITIES
Respiratory and enzymatic activities are usually expressed by cell or by unit of cellular or mitochondrial protein. To compare OXPHOS activities among cells, tissues or individuals, researchers commonly normalize OXPHOS enzymatic activities measured in isolated mitochondria to the activity of citrate synthase, an enzyme of the tricarboxylic acids cycle.
Materials
Electron donors and acceptors (Sigma): 50 mM oxalacetic acid (adjust to pH 7.4), 10 mM acetyl CoA, 0.1M 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB)
10 mM Tris-HCl (pH 7.5)
Detergents: 10% Triton X-100 (w/v).
Prepare 1 ml of 10 mM Tris–HCl, pH 7.5 buffer containing 0.2% Triton X-100, 0.1 mM DTNB, 0.2 mM acetyl-CoA and 20–40 µg of mitochondrial proteins.
Incubate 5 min at 30 °C. Measure the baseline at 412 nm for 5 min.
Start the reaction by adding freshly prepared 0.5 mM oxalacetic acid. Follow the reaction for 5 min, by measuring the increase in absorbance at 412 nm resulting from the reduction of 0.1 mM DTNB (5,5′-dithiobis(2-nitrobenzoic acid)).
Obtain the rate of citrate synthase activity.
Normalize OXPHOS enzyme activity by dividing the enzymatic activity rates corresponding to each enzyme by the rate of citrate synthase activity. The ratios can be used to compare enzymatic activities in different samples.
REAGENTS AND SOLUTIONS
Assay buffer for COX (isosmotic COX medium): 10 mM KH2PO4, pH 6.5, 0.25 M sucrose, and 1 mg/ml BSA. Store at 4°C. BSA should be added freshly.
Assay buffer for SQR / SCCR / QCR: 10 mM KH2PO4 (pH 7.8), 2 mM EDTA, and 1 mg/ml BSA. Store at 4°C. BSA should be added freshly.
Hypotonic medium (25 mM K2PO4 pH 7.2, 5 mM MgCl2). Store at 4°C.
Respiratory buffer (RB): 0.3 M mannitol, 10 mM KCl, 5 mM MgCl2, and 10 mM K2PO4, pH 7.4. Store at 4°C.
COMMENTARY
Background Information
Nuclear and mitochondrial genetic OXPHOS defects account for a large variety of clinical symptoms in both childhood and adulthood. Additionally, OXPHOS alterations have been associated to neurodegeneration and aging and also to some environmental stresses including tobacco smoking, exposition to herbicides or carbon monoxide. Gentoype/phenotype correlations for the genetic syndromes and the determination of the severity of the clinical conditions in all cases mentioned largely relay on biochemical analyses of the OXPHOS performance.
In this unit, we have presented classical protocols for the polarographic study of oxygen consumption (Basic Protocols 1 and 2) and spectrophotometric analyses of the mitochondrial respiratory chain enzymes (Basic Protocol 3) in whole cells, tissue homogeneates and isolated mitochondria. The protocols presented are based on methods described elsewhere (Barrientos, 2002, Birch-Machin and Turnbull, 2001, Munnich and Rustin, 2001, Rustin et al., 1991, Rustin et al., 1994, Villani et al., 1998).
To standardize respiratory chain polarographic and spectrophotometric assays for clinical diagnostics and research should facilitate comparison of results among clinical centers and research institutes (Chretien et al., 1994, Chretien et al., 1998, Medja et al., 2009).
Critical Parameters and Troubleshooting
Polarography
The optimization of assays to assess OXPHOS function in cultured cells, isolated mitochondria from cells or tissues, or tissue homogenates is essential in both diagnostic and in research studies. The accuracy of the biochemical studies involves an exquisite handle on the biological material and a rational use of inhibitors since all activities must be expressed as sensitive to the specific inhibitor of the enzyme or enzymatic segment assayed.
Polarographic studies in cells rely on the initial conditions of the cultures (the medium should be changed a few hours before collecting the cells) and on the appropriate permeabilization of the cell membranes to the several substrate additions, avoiding over-permeabilization. For this purpose, the time of incubation with digitonin depends on the cell type and a “time/percentage of permeabilized cells” curve should be empirically obtained for each one. Following digitonin incubation, it is advised to check the degree of permeabilization by toluene blue exclusion under optic microscopy. Approximately 95% of the cells should be permeabilized (i.e., the dye accesses the cytoplasm and shows up blue).
The success of all respiratory studies performed in isolated mitochondria relies on the quality of the organellar preparation. Classical laboratory methods and kit-based methods can be confidently used (Birch-Machin and Turnbull, 2001, Hartwig et al., 2009, Pallotti and Lenaz, 2001, Rustin et al., 1994). All reagents should be freshly prepared. The respiratory buffers can be prepared and stored but ADP should always be freshly added. Substrate oxidation should be followed in the presence and absence of ADP and in the presence and absence of an uncoupler. The maintenance and calibration of the polarographic unit, following the recommendations of the manufacturer, is also a must to obtain reproducible results.
Spectrophotometry
Spectrophotometric assays of the OXPHOS enzymes rely on an appropriate disruption of the mitochondrial membranes, which differs depending on the maximal activity of the enzyme to be measured. The reduction of cytochrome c or decylubiquinone should be freshly done and monitored spectrophotometrically to obtain reproducible results. Every enzyme assay is performed in a particular buffer system that allows for optimization of the assay. Most enzymatic activities can be confidently measured in cells and tissue homogeneates, with the exception of complex I activity, for which no reliable method is currently available.
To effectively evaluate the enzymatic data obtained, the results must be normalized by cell number, unit of protein, or the activity of a non-OXPHOS mitochondrial enzyme such as citrate synthase. Additionally, these values should be supplemented with the activity ratios between the various OXPHOS complexes, which are known to be maintained significantly constant among cell types and individuals (Barrientos, 2002, Chretien et al., 1998) as additional parameters to help in discriminating very partial specific enzymatic OXPHOS defects.
Anticipated Results
The interpretation of the polarographic results is as follows. A decreased oxidation rate with NAD+-linked substrates (pyruvate or glutamate), but a normal rates with succinate (complex II substrate) is an indicator of complex I deficiency. The use of two different substrates allows excluding a possible defect in the tricarboxylic acid carrier or in the primary dehydrogenase. Opposite results will be obtained in case of complex II deficiency. In the case of complex III or IV deficiencies, a defective oxidation of both site I and site II substrates is observed. In the case of quinone deficiency, the defect will be pleiotropic but normal rates of oxidation with all substrates will be obtained if the respiratory buffer is supplemented with exogenous ubiquinone. In the case of complex V deficiency, the oxidation of several substrates can be found impaired but it is restored when measured in uncoupling conditions in the presence of CCCP.
The enzymatic defects can be either specific to one complex or affect several complexes. Because the mitochondrial respiratory chain is organized forming supercomplexes or respirasomes (Acin-Perez et al., 2008), alterations primarily affecting the function and activity of one enzyme can pleiotropically affect others. For example, this is the case of some mutations primarily affecting complex III which also produce a complex I deficiency (Acin-Perez et al., 2004).
Reference charts for respiratory chain activities in human tissues have been reported elsewhere (Chretien et al., 1994, Chretien et al., 1998).
Time considerations
Once the biological material has been obtained, the whole set of polarographic and spectrophotometric assays proposed here can be easily performed for 2–3 samples in one day of work. Familiarity with the protocols is key for efficiency and accuracy.
AKCNOWLEDGEMENTS
We thank Dr. Tiziana Lodi and Darryl Horn for critically reading the manuscript. The following foundations are acknowledged for their financial support: NIH (RO1 Grant GM071775A to A.B.), Muscular Disease Association (MDA Research Grant to A.B.). The James and Esther King Biomedical Research Program, Florida Department of Health 08KN-01 (to F.D.).
REFERENCES
- Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Perez-Martos A, Bruno C, Moraes CT, Enriquez JA. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell. 2004;13:805–815. doi: 10.1016/s1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol. Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Barrientos A. In vivo and in organello assessment of OXPHOS activities. Methods. 2002;26:307–316. doi: 10.1016/S1046-2023(02)00036-1. [DOI] [PubMed] [Google Scholar]
- Birch-Machin MA, Turnbull DM. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell. Biol. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- Chretien D, Bourgeron T, Rotig A, Munnich A, Rustin P. The measurement of the rotenone-sensitive NADH cytochrome c reductase activity in mitochondria isolated from minute amount of human skeletal muscle. Biochem. Biophys. Res. Commun. 1990;173:26–33. doi: 10.1016/s0006-291x(05)81016-2. [DOI] [PubMed] [Google Scholar]
- Chretien D, Rustin P, Bourgeron T, Rotig A, Saudubray JM, Munnich A. Reference charts for respiratory chain activities in human tissues. Clin. Chim. Acta. 1994;228:53–70. doi: 10.1016/0009-8981(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Chretien D, Gallego J, Barrientos A, Casademont J, Cardellach F, Munnich A, Rotig A, Rustin P. Biochemical parameters for the diagnosis of mitochondrial respiratory chain deficiency in humans, and their lack of age-related changes. Biochem J. 1998;329:249–254. doi: 10.1042/bj3290249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chretien D, Slama A, Briere JJ, Munnich A, Rotig A, Rustin P. Revisiting pitfalls, problems and tentative solutions for assaying mitochondrial respiratory chain complex III in human samples. Curr. Med. Chem. 2004;11:233–239. doi: 10.2174/0929867043456151. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- Fernandez-Vizarra E, Tiranti V, Zeviani M. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta. 2009;1793:200–211. doi: 10.1016/j.bbamcr.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Hartwig S, Feckler C, Lehr S, Wallbrecht K, Wolgast H, Muller-Wieland D, Kotzka J. A critical comparison between two classical and a kit-based method for mitochondria isolation. Proteomics. 2009 May 4; doi: 10.1002/pmic.200800344. [DOI] [PubMed] [Google Scholar]
- Krahenbuhl S, Talos C, Wiesmann U, Hoppel CL. Development and evaluation of a spectrophotometric assay for complex III in isolated mitochondria, tissues and fibroblasts from rats and humans. Clin. Chim. Acta. 1994;230:177–187. doi: 10.1016/0009-8981(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Medja F, Allouche S, Frachon P, Jardel C, Malgat M, de Camaret BM, Slama A, Lunardi J, Mazat JP, Lombès A. Development and implementation of standardized respiratory chain spectrophotometric assays for clinical diagnosis. Mitochondrion. 2009 May 9; doi: 10.1016/j.mito.2009.05.001. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Munnich A, Rustin P. Clinical spectrum and diagnosis of mitochondrial disorders. Am. J. Med. Genet. 2001;106:4–17. doi: 10.1002/ajmg.1391. [DOI] [PubMed] [Google Scholar]
- Pallotti F, Lenaz G. Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol. 2001;65:1–35. doi: 10.1016/s0091-679x(01)65002-7. [DOI] [PubMed] [Google Scholar]
- Ragan CI, Cottingham IR. The kinetics of quinone pools in electron transport. Biochim. Biophys. Acta. 1985;811:13–31. doi: 10.1016/0304-4173(85)90003-5. [DOI] [PubMed] [Google Scholar]
- Reeve AK, Krishnan KJ, Turnbull D. Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann. N. Y. Acad. Sci. 2008;1147:21–29. doi: 10.1196/annals.1427.016. [DOI] [PubMed] [Google Scholar]
- Rustin P, Chretien D, Bourgeron T, Wucher A, Saudubray JM, Rotig A, Munnich A. Assessment of the mitochondrial respiratory chain. Lancet. 1991;338:60. doi: 10.1016/0140-6736(91)90057-v. [DOI] [PubMed] [Google Scholar]
- Rustin P, Chretien D, Bourgeron T, Gerard B, Rotig A, Saudubray JM, Munnich A. Biochemical and molecular investigations in respiratory chain deficiencies. Clin. Chim. Acta. 1994;228:35–51. doi: 10.1016/0009-8981(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Villani G, Greco M, Papa S, Attardi G. Low reserve of cytochrome c oxidase capacity in vivo in the respiratory chain of a variety of human cell types. J. Biol. Chem. 1998;273:31829–31836. doi: 10.1074/jbc.273.48.31829. [DOI] [PubMed] [Google Scholar]