Abstract
The Sleeping Beauty (SB) transposon system has been used as a somatic mutagen to identify candidate cancer genes. In previous studies, efficient leukemia/lymphoma formation on an otherwise wild-type genetic background occurred in mice undergoing whole-body mobilization of transposons, but was accompanied by high levels of embryonic lethality. To explore the utility of SB for large-scale cancer gene discovery projects, we have generated mice that carry combinations of different transposon and transposase transgenes. We have identified a transposon/transposase combination that promotes highly penetrant leukemia/lymphoma formation on an otherwise wild-type genetic background, yet does not cause embryonic lethality. Infiltrating gliomas also occurred at lower penetrance in these mice. SB-induced or accelerated tumors do not harbor large numbers of chromosomal amplifications or deletions, indicating that transposon mobilization likely promotes tumor formation by insertional mutagenesis of cancer genes, and not by promoting wide-scale genomic instability. Cloning of transposon insertions from lymphomas/leukemias identified common insertion sites at known and candidate novel cancer genes. These data indicate that a high mutagenesis rate can be achieved using SB without high levels of embryonic lethality or genomic instability. Furthermore, the SB system can be used to identify new genes involved in lymphomagenesis/leukemiogenesis.
Keywords: Transposon, leukemia, glioma
Introduction
Forward somatic cell genetic screens in model organisms are a powerful approach for the identification and validation of tumor suppressor genes (tsgs) and oncogenes relevant in human cancer (1–3). Insertional mutagens such as retroviruses and transposable elements are frequently used for this purpose because the mutagen itself serves as a molecular tag, allowing rapid identification of mutagenized genomic loci. Candidate cancer genes are identified by finding regions of the genome that are insertionally mutated in multiple independent tumors, so-called common insertion sites (CISs).
The SB transposon system has been used as such an insertional mutagen. The SB system is bipartite; consisting of the mobilized piece of DNA, the transposon, and the enzyme that catalyzes the transposition reaction, the transposase (4). Different combinations of SB transposon and transposase transgenics have been used for whole-body somatic cell genetic screens in vivo (5, 6). For these studies, different lines of mice harboring multiple copies of the T2/onc transposon in a head-to-tail arrangement in a chromosomally resident concatomer were utilized. Lines harboring about 25 copies of T2/onc in the donor concatomer were designated as low-copy lines (5) while lines harboring greater than 140 copies of T2/onc were designated as high-copy lines (6). Two SB transposase transgenic lines were used to mobilize T2/onc throughout the soma. One transgenic was engineered with the SB11 version of the transposase “knocked” into the Rosa26 locus (Rosa26–SB11) (6) while one transgenic expresses the SB10 version of the transposase under the control of the CAGGS promoter (7) (CAGGS-SB10) (5). Mobilizing T2/onc from low-copy lines by CAGGS-SB10 could not generate tumors on an otherwise wild-type genetic background, yet did accelerate sarcoma formation in mice deficient for the tsg p19Arf (5). T2/onc mobilization from high-copy lines by Rosa26-SB11 on an otherwise wild-type genetic background resulted in high levels of embryonic lethality which limited the number of transposon;transposase doubly transgenic mice that could be generated (6). All mice surviving to birth eventually succumbed to tumors, primarily lymphocytic lymphoma/leukemia, by 120 days. Medulloblastoma and other hyperplasias/neoplasias were also observed at low penetrance. Cloning insertions from 15 lymphoma/leukemias and one medulloblastoma identified 33 CISs at known and candidate cancer genes, only a few of which had been previously identified in retroviral screens for lymphoma/leukemia genes (6).
The SB system is two-component (consisting of both transposons and transposase), so the possibility exists to modify each component individually to determine the effects on tumorigenesis. To this end, we crossed a T2/onc high-copy line to CAGGS-SB10 and two T2/onc low-copy lines to Rosa26-SB11. We have discovered that a rate of mutagenesis sufficient for promoting highly penetrant tumor formation yet insufficient for causing embryonic lethality can be achieved with the SB system. Leukemias/lymphomas predominate the tumor spectrum in mice undergoing whole-body transposon mutagenesis. Gliomas also occur with reduced penetrance, indicating that this tumor type can be modeled using SB mutagenesis. Furthermore, widespread genomic instability is not observed in SB-induced or accelerated tumors, suggesting that transposon insertional mutagenesis and not genomic instability drives tumorigenesis in these models. Transposon insertion sites from SB-induced leukemias/lymphomas identify CISs at both known and candidate novel cancer genes, suggesting that the SB system can reveal a different spectrum of cancer loci than retroviruses.
Materials and Methods
Mice
Mouse work was performed under University of Minnesota IACUC guidelines. All strains have been described (5, 6, 8). At necropsy, tissues were snap frozen for DNA preparation and formalin fixed/paraffin embedded for pathological analysis at the Masonic Cancer Center Histopathology Core and the Mayo Clinic Tissue and Cell Molecular Analysis Shared Resource. Kaplan-Meier survival analysis was performed using Prism software.
Genotyping
Transposase transgenics were PCR genotyped using the following primers: 5’GGACAACAAAGTCAAGGTAT3’ and 5’TAACTTGGGTCAAACGTTTC3’. T2/onc mice were genotyped as described (9).
Flow cytometry
Cell staining and flow cytometry techniques were as described (10, 11). Antibodies used were CY5-conjugated anti-CD4, APC-conjugated anti-CD8, FITC-conjugated anti-B220, P E-conjugated anti-TCRβ, PE-conjugated Gr1 and FITC-conjugated Mac1 (BD Biosciences, San Jose, CA). Data were analyzed using FlowJo software.
Linker-mediated PCR
For tumor DNA, linker-mediated PCR was performed as described (12). PCR products from tumors were shotgun cloned into pCR4-Topo (Invitrogen, Carlsbad, CA). For each PCR, 96 bacterial colonies were robot picked, prepped and sequenced on the ABI 3730 platform. For the dataset from tail DNA, sequencing was performed on the 454 platform as described (9).
Insertion mapping and CIS analysis
Mapping of insertion sites to NCBI 36 build of the mouse genome was performed as described (6, 13). CIS analysis was based on published methods (14). Because of the possibility for transposons to local hop after a prior mobilization, insertions from the same animal were not allowed to solely define a CIS. Insertion data is deposited in the RTCGD (13).
Array CGH
Tumor DNA samples (1 µg each) were labeled with Cy-3-dUTP and control DNA samples from muscle or spleen tissue (from the same animal when possible and from littermates in all other cases) were labeled with Cy-5 d-UTP essentially as described (15), with the omission of the Dpn II digest. Samples were combined and mixed with mouse Cot-1 DNA and hybridized to 1344-element BAC arrays (16) as described. Array images were captured using a CCD camera, and automated spot identification and statistical analysis was carried out using custom software (17) as described (15).
IHC
IHC for transposase was performed using the M.O.M. kit (Vector Laboratories, Burlingame, CA). Anti-transposase antibody (R&D Systems, Minneapolis, MN) was used at 1 µg/ml. Immunostain was developed using the ABC Vectastain peroxidase system (Vector Laboratories, Burlingame, CA), and sections were counterstained with hematoxylin. IHC was performed using anti-GFAP (Dako, Denmark, polyclonal, dilution 1:4000) and anti-synaptophysin antibodies (ICN, Costa Mesa, CA, clone SY38, dilution 1:40) and showed a similar pattern in ten gliomas examined. Primary antibody incubation was performed for 30 minutes, followed by 20 minutes in the Envision+Dual Link detection system on a Dako autostainer.
Results and Discussion
Combining CAGGS-SB10 with high-copy T2/onc does not result in tumor formation due to limited transposase expression
To determine if mobilization of T2/onc from high-copy lines by CAGGS-SB10 is sufficient to induce tumors, mice doubly transgenic for a T2/onc high-copy concatomer located on chromosome 4 (6) and the CAGGS-SB10 transgene (8) were generated. No evidence of embryonic lethality was observed (data not shown). CAGGS-SB10 only controls (n=11) and T2/onc high-copy; CAGGS-SB10 experimental mice (n=9) were aged and monitored for tumor formation for 18 months. No statistical difference in survival was observed (P=.6848, Log rank test) (Figure S1), indicating that the mutagenesis rate achieved by mobilizing T2/onc from a high-copy line by CAGGS-SB10 is insufficient for tumor formation on an otherwise wild-type genetic background.
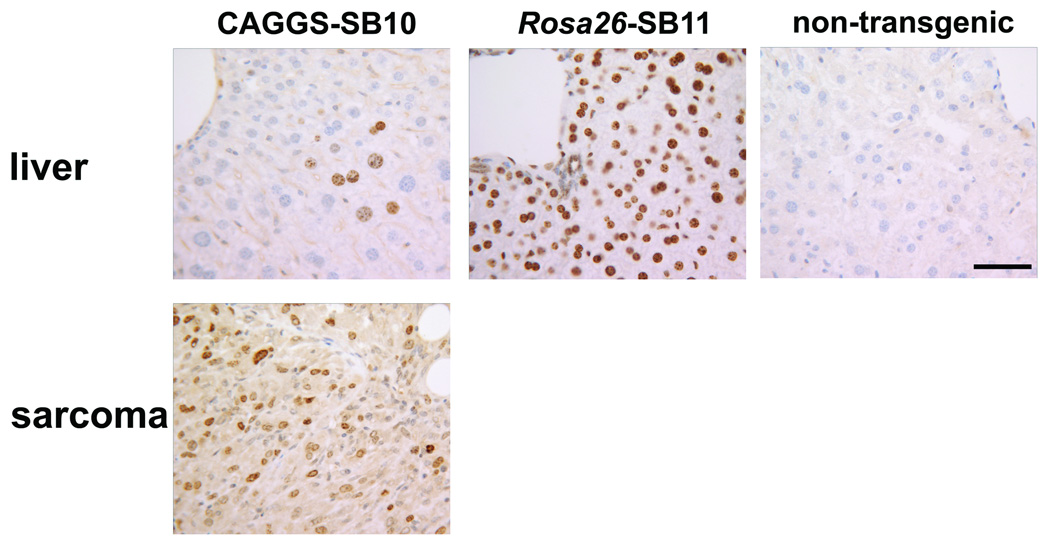
To investigate if transposase expression levels influence mutagenesis rates, immunohistochemistry (IHC) was performed to detect transposase in CAGGS-SB10 and Rosa26-SB11 mice. Normal adult tissues were examined, as was a sarcoma from a p19Arf−/−;CAGGS-SB10;T2/onc low-copy mouse (5). Although transposase was detected in the sarcoma, it was absent from most normal somatic tissues in CAGGS-SB10 mice (Figure 1 and Figure S2). When expression was detected, it occurred in a highly variegated pattern (liver in Figure 1; kidney in Figure S2). In contrast, transposase was robustly expressed in the majority of cell types in Rosa26-SB11 mice (Figure 1 and Figure S2). The low level and variegated expression in CAGGS-SB10 mice is potentially due to epigenetic silencing that is often observed in standard transgenics.
Figure 1. IHC reveals transposase expression in transgenic mice.
Transposase is poorly expressed in somatic tissues in CAGGS-SB10 mice (liver shown), while a sarcoma from a p19Arf−/−;CAGGS-SB10;T2/onc low-copy mouse contains many transposase expressing cells. Most cells in Rosa26-SB11 mice stain positive for transposase (liver shown). Brown indicates antibody staining, nuclei are counter-stained blue. A liver from a non-transposase transgenic mouse demonstrates antibody specificity. Scale bar=50 microns.
The presence of transposase in a p19Arf−/−;CAGGS-SB10;T2/onc sarcoma indicates that transposase is expressed in these mice in an appropriate cell type to promote sarcomagenesis. It could be hypothesized that T2/onc high-copy;CAGGS-SB10 mice could have developed sarcomas on an otherwise wild-type genetic background due to the availability of many T2/onc copies for mutagenesis. However, tumor formation was not observed. In murine models, p19Arf is known to play a role in oncogene-induced senescence (18–20). Therefore, in sarcoma-initiating cells in p19Arf+/+ mice, T2/onc mutagenesis of cancer genes could promote Arf-mediated senescence, providing a block to tumor formation. This experiment suggests that performing SB-screens in tumor-predisposed genetic backgrounds may be necessary for robust tumor formation in certain tissue types.
Combining T2/onc low-copy lines with Rosa26-SB11 does not cause embryonic lethality but promotes tumor formation in otherwise wild-type mice
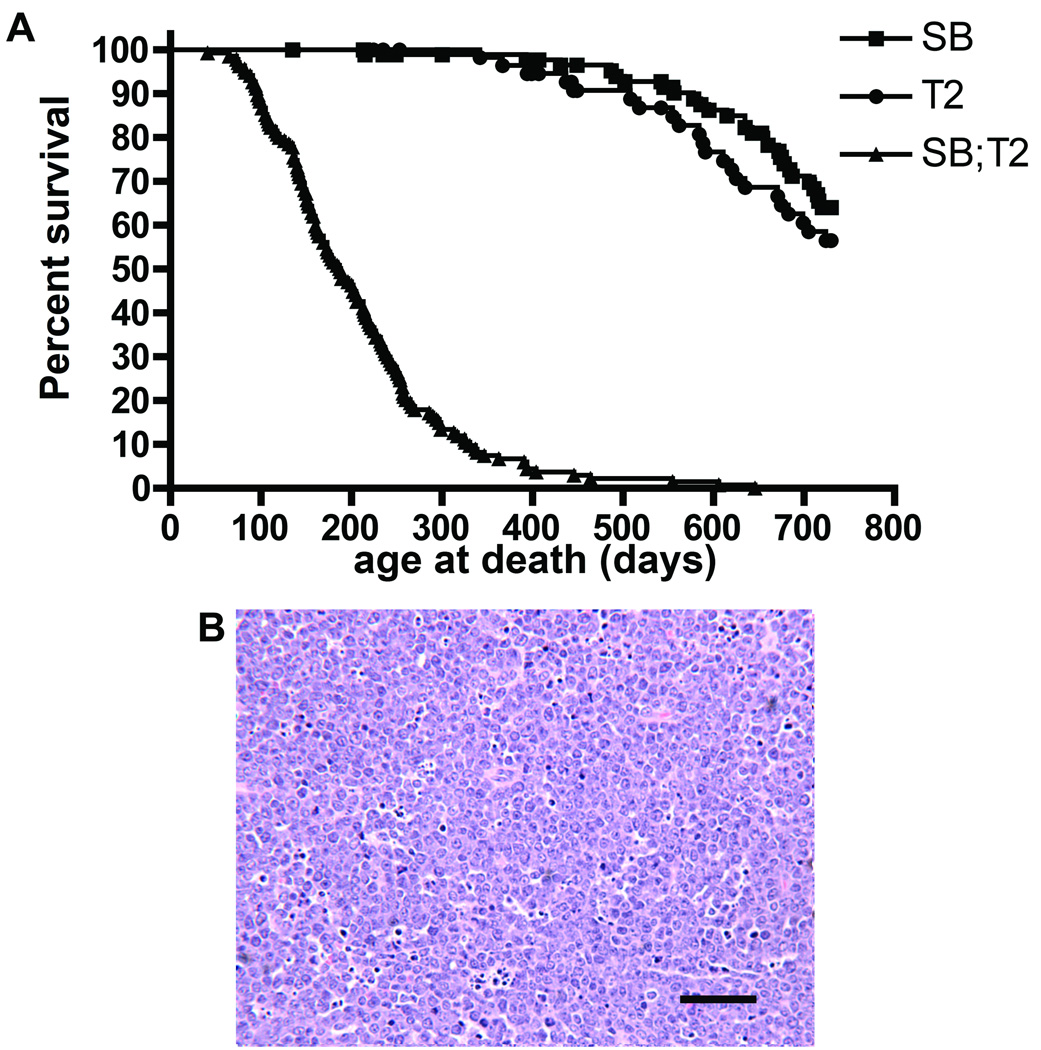
To determine if mobilization from low-copy lines is sufficient for tumor formation, two T2/onc low-copy lines (lines 68 and 76 (5)) were crossed to Rosa26-SB11. Chi square analysis of the resulting progeny (Table 1) revealed no evidence for non-Mendelian inheritance of the transgenes (p= 0.4153, 3 degrees of freedom). A large cohort of doubly transgenic mice and single transgenic littermate controls were therefore generated. T2/onc low-copy;Rosa26-SB11 mice became moribund with an average latency of 187 days while controls had normal lifespans (Figure 2A). Separate analysis of each T2/onc low-copy line revealed that T2/onc low-copy line 68;Rosa26-SB11 mice develop disease much more rapidly than T2/onc low-copy line 76;Rosa26-SB11 mice (Figure S3). However, the tumor spectrum was the same for both lines. At necropsy, 89% (97 of 109) of analyzed doubly transgenic mice had signs of hematopoietic disease including splenomegaly, lymphadenopathy and/or an enlarged thymus. Twenty-seven mice with hematopoietic involvement were analyzed by veterinary pathologists at the Masonic Cancer Center Comparative Pathology core. Nineteen mice were diagnosed with lymphocytic lymphoma/leukemia (Figure 2B), three with hematopoietic neoplasia of undetermined lineage, four with hematopoietic hyperplasia and one with myeloid leukemia. Flow cytometry analysis for cell surface markers on nineteen tumors verified that the majority of leukemias/lymphomas arising in these mice are phenotypically T-cell lymphocytic disease (Table S1).
Table 1.
No evidence for non-Mendelian transgene inheritance in the T2/onc low-copy; Rosa26-SB11 cross.
| genotype (T2/onc, Rosa26-SB11): | +,− | −,+ | +,+ | −,− |
| number of mice: | 123 | 138 | 112 | 129 |
T2/onc low-copy heterozygous mice were crossed to Rosa26-SB11 heterozygous mice and the resulting progeny were genotyped for each transgene. The four possible genotypes and number of mice observed for each genotype are shown.
Figure 2. Rosa26-SB11; T2/onc low-copy mice are tumor prone.
A) Kaplan-Meier survival curve for Rosa26-SB11;T2/onc low-copy (SB;T2, triangles), Rosa26-SB11 (SB, squares), and T2/onc low-copy (T2, circles) mice. Rosa26-SB11;T2/onc low-copy mice become moribund more rapidly than controls (p<.001, Logrank test). B) Hematoxylin and Eosin (H&E) stained example of a lymphocytic leukemia/lymphoma from a Rosa26-SB11;T2/onc low-copy mouse. Scale bar=50 microns.
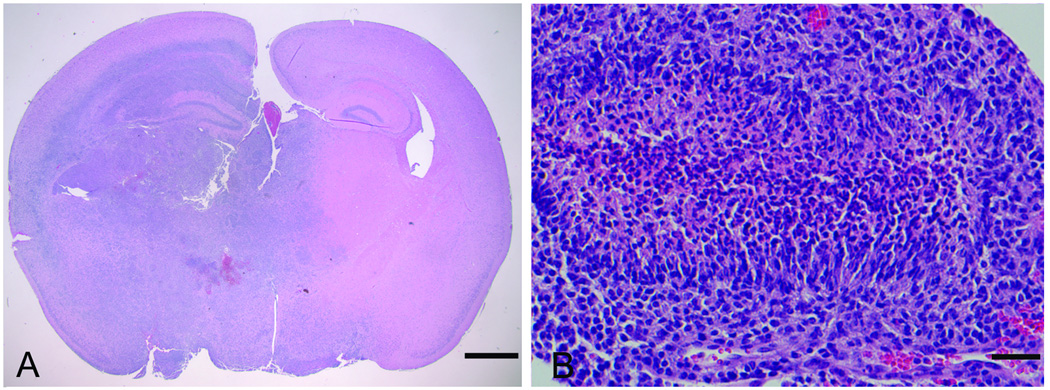
Several mice presented with neurological symptoms at morbidity. Medulloblastomas, a tumor of the cerebellum, occurred at low penetrance in T2/onc high-copy;Rosa26-SB11 doubly transgenic mice (6). To determine if medulloblastomas also occur in T2/onc low-copy;Rosa26-SB11 mice, 82 brains were extensively sectioned for pathology. Fourteen brain tumors were discovered. One tumor was a sarcoma growing on the surface of the brain while one was a glioma of undetermined origin (data not shown). Histopathological analysis determined that the remaining tumors were high-grade astrocytomas (Figure 3A). Pseudopalisading necrosis was present in three cases that were therefore classified as glioblastomas (Figure 3B). IHC for Glial fibrillary acidic protein (GFAP) and was performed on a sub-set of tumors to confirm the diagnosis. Immunoreactivity for GFAP was noted at least focally in all tumors examined but they were negative for synaptophysin (data not shown), supporting the diagnosis of astrocytomas. No gliomas were detected in 28 aged-matched transposon or transposase only mice sacrificed for analysis, indicating that T2/onc mobilization induces high-grade astrocytomas. Molecularly defined hyperproliferative lesions were also found in the prostates of moribund T2/onc low-copy;Rosa26-SB11 mice (21); but no additional overt tumor types were commonly observed despite the fact that transposase is expressed in most cell types. The aggressive nature of the leukemias/lymphomas and gliomas in these mice limits animal survival, and therefore likely prevents the ability of transposon mobilization by Rosa26-SB11 to model more slowly developing tumor types.
Figure 3. Gliomas are present in Rosa26-SB11; T2/onc low-copy mice.
A) Gliomas sometimes involved essentially an entire hemisphere (H&E), scale bar=1mm. B) Pseudopalisading necrosis was evident in a subset of cases which were therefore classified as glioblastoma (H&E), scale bar=50 microns.
In contrast to T2/onc high-copy lines, no embryonic lethality was observed in T2/onc low-copy;Rosa26-SB11 mice. Embryonic lethality was proposed to potentially result from unrepaired DNA damage after transposition (6). The lower number of mobilizing transposons in T2/onc low-copy;Rosa26-SB11 mice could result in fewer double strand breaks for cellular machinery to repair. Another explanation for embryonic lethality in T2/onc high-copy;Rosa26-SB11 double transgenics could be the generation of concatomer-associated rearrangements which can accompany SB germline transposition (22). High transposition rates associated with high-copy concatomers could increase the severity of these rearrangements. Whatever the cause of embryonic lethality, the Mendelian inheritance of Rosa26-SB11 with T2/onc low-copy transgenes indicates that a mutagenic rate sufficient to promote tumor formation but insufficient to interfere with normal development can be achieved.
Brain tumors have been found in mice in which T2/onc is mobilized by Rosa26-SB11. In high-copy lines, the tumors were medulloblastomas (6) while in low-copy lines the tumors were infiltrating gliomas. Differences in tumor latency could contribute to differences in brain tumor type. Medulloblastomas are predominantly found in children, and it is hypothesized that they arise from granule cell precursor cells. It is hypothesized that most granule cell precursors have completed proliferation and differentiation by adulthood, and therefore fewer potential medulloblastoma-initiating cells exist in adults. The high mutagenesis rate in T2/onc high-copy;Rosa26-SB11 mice could allow enough mutations in cancer genes to occur prior to terminal differentiation. Conversely, in T2/onc low-copy;Rosa26-SB11 mice, the mutagenesis rate may be too low to promote tumor formation prior to terminal differentiation of these cells. This slower mutagenesis rate could still promote mutagenesis in the longer-lived glial precursor cell.
Somatic mobilization of transposons does not cause substantial genomic instability
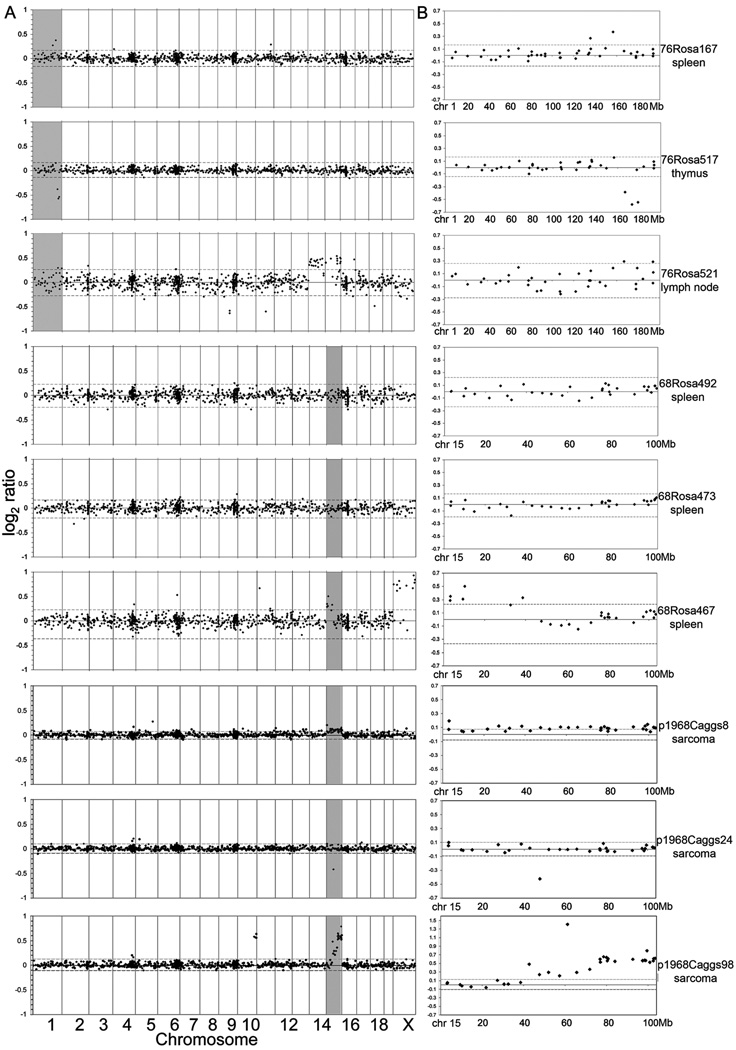
Transposition of DNA transposons involves double strand break formation and repair (23). It is possible that somatic mobilization of SB transposons could cause tumor formation by promoting genomic instability due to illegitimate repair of these breaks. To investigate this possibility, BAC-based array comparative genomic hybridization (CGH) (16) was used to look at genomic copy number changes in six T2/onc low-copy;Rosa26-SB11 lymphomas/leukemias and three sarcomas from p19Arf−/−;CAGGS-SB10;T2/onc low-copy mice (5) using non-tumor DNA as a reference sample (Figure 4). Two spontaneously arising sarcomas in p19Arf−/− mice served to demonstrate the ability of the BAC array platform to detect copy number changes in tumors (Figure S4). Whole chromosome gains or losses were rarely detected in tumors with mobilizing transposons (for example, the gain of chromosomes 14 and 15 in 76Rosa521 lymph node). Deletions or amplifications defined by one or two adjacent probes were occasionally detected. However, for line 76 leukemias/lymphomas, two of three displayed evidence of amplification or deletion on chromosome 1 at probes flanking 166Mb, the approximate location of the T2/onc concatomer in line 76 (5) (Figure 4B). The exact chromosomal location of the line 68 concatomer has not been determined, but FISH and local hopping patterns have placed it at approximately 45Mb on chr. 15. One (68Rosa467 spleen) of 3 leukemias/lymphomas from T2/onc line 68 and all three sarcomas from p19Arf−/−;CAGGS-SB10;T2/onc line 68 mice showed evidence for amplification or deletion on chromosome 15.
Figure 4. Rosa26-SB11;T2/onc low-copy leukemias do not show genome-wide chromosomal amplifications and deletions.
Array CGH profiles from six leukemias/lymphomas from Rosa26-SB11;T2/onc low-copy mice (76Rosa and 68Rosa) and three sarcomas from p19Arf−/−;CAGGS-SB10;T2/onc low-copy mice (p1968Caggs). Each row represents one tumor. A) Genome-wide log2 ratios. Dotted lines represent 3 standard deviations from the central mean of all clones genome wide, indicating cutoffs for gains and losses, respectively. Tumors from 76Rosa167, 76Rosa517, 68Rosa467, p1968Caggs8, p1968Caggs24 and p1968Caggs98 show localized rearrangements in the region surrounding the transposon concatomer on chromosome 1 and 15 (shaded areas). The apparent gain of the X chromosome in the tumor from 68Rosa467 is due to DNA from normal male spleen being used as reference DNA for a tumor from a female littermate mouse. B) Profiles of clones on the donor concatomer chromosome for the tumors profiled in (A).
Previously, chromosomal rearrangements flanking a SB transposon donor concatomer on chromosome 11 have been detected as a result of transposition in the germline and in normal splenocytes (22). The array CGH reported here indicates that this phenomenon is not limited to the concatomer on chromosome 11, and that they can occur in SB-induced tumors. The data suggest that SB-induced tumorigenesis does not promote genome-wide instability, but does frequently cause amplifications and/or deletions flanking the donor concatomer that could contribute to tumor formation if the donor concatomer happens to reside near a tsg or oncogene. As other DNA elements are known to cause genomic rearrangements (24), it will be important to determine if additional transposons proposed to be used as somatic mutagens including piggyBac (25), Tol2 (26) and Minos (27) also promote deletions or amplifications flanking the donor locus.
T2/onc local hops in somatic cells
Cancer gene identification using insertional mutagens relies on performing CIS analysis to identify chromosomal regions where transposons have inserted in tumors at a rate greater than that expected by random chance. In vivo, SB is known to have a preference for re-inserting at loci linked to the starting integration site, a phenomenon termed local hopping (28), which complicates CIS analysis. Previously, an unselected insertion set (n=490) obtained from T2/onc high-copy;Rosa26-SB11 embryos was used to examine local hopping rates from chromosomal concatomers in somatic cells in vivo (6). Although only 6–11% of insertions were found in the 25-megabase region surrounding donor concatomers, 38.6% of insertions were found on the same chromosome as the donor concatomer. However, it is unclear if transposon copy number or the chromosomal location of a concatomer influences local hopping rates.
To determine how local hopping from low-copy concatomers may influence analysis of SB transposon tumor insertion sites, a dataset of unselected SB transposon insertion sites from tail biopsy DNA isolated from 14–21 day old T2/onc low-copy;Rosa26-SB11 mice was generated (9, 29). As no leukemias have been observed in mice this young, the insertions cloned from this material likely represent the SB transposon insertion site profile under un-selected conditions. 16,411 unique SB insertion sites were cloned from 88 doubly transgenic mice using linker-mediated PCR and 454 pyrosequencing methods (Table S2). Of these, 22.8% were located in the 25 Mb surrounding the donor concatomer, indicating that transposons in low-copy concatomers local hop in somatic cells. Furthermore, 49.3% of insertions were located on the same chromosome as the donor concatomer (8095 of 16411). The percentage of insertions on the donor chromosome contrasts with T2/onc high-copy line embryo insertions in which 38.6% of insertions reside on the same chromosome as the donor concatomer. This could potentially be explained by differences in local hopping rates from different copy number concatomers. Nevertheless, both of these datasets indicate that in somatic cells, SB transposons have a local hopping interval that encompasses the whole chromosome on which the concatomer resides.
CIS analysis was performed on this control dataset after removal of insertions mapping to the donor chromosomes (n= 8316 insertions) using criteria described by Mikkers et al(14) at an expected fraction (Efr) of 0.001 (2 insertions in .325 kb, 3 insertions in 14.75 kb and 4 insertions in 62 kb), which is predicted by Monte Carlo simulations to result in approximately 25 CISs being identified by random chance alone. 43 CISs were identified in the control dataset. Clustering criteria using an Efr of 0.005 was also applied (2 insertions in 1.625 kb, 3 in 33.75 kb and 4 in 109.75 kb) which is predicted to result in a total of 124.5 CISs identified based on random chance alone. Using these criteria, 134 CISs were identified (of which 43 also met the criteria used above for an Efr of 0.001) (Table S3).
More CISs were identified in this control dataset than would be predicted by Monte Carlo simulations. This observation could actually be due to random chance, as Monte Carlo simulations predictions of false positive clustering rates are based on averages for an infinite number of experiments. Therefore, 50% of the time a random dataset of 8316 insertions is generated, the number of CISs identified at an Efr of .005 would be ≥ 124.5 and 50% of the time the number of CISs identified would be ≤ 124.5. Alternatively, SB is known to have some insertion preferences for specific sequences or DNA conformations (30, 31), and preferred SB insertion sites may not be randomly distributed through the genome. Five of the CISs in the control dataset are also CISs in leukemias (see below), supporting the hypothesis that there are genomic “hot spots” for SB integration. Interestingly, CISs are found in the control dataset on proximal regions of chrs. 5, 11, 12, 13 and 18; indicating the possibility that SB transposons have affinity for inserting into centromeric regions. The generation and analysis of additional datasets under unselected conditions will help refine the statistical methods used for CIS analysis in SB-induced tumors.
T2/onc identifies candidate genes involved in lymphoma/leukemia formation not identified by MuLV
To determine if T2/onc identifies new lymphoma/leukemia cancer genes, 2296 independent T2/onc insertion sites were cloned from 59 lymphomas/leukemias from 58 T2/onc low-copy;Rosa26-SB11 animals (Table S4). Local hopping was observed as 13.1% of insertions from line 68 occurred within the 25 Mb surrounding the donor concatomer, 18.6% within the 40 Mb surrounding the donor concatomer and 33.6% on the entire donor chromosome. For line 76 the local hopping percentages were similar at 11.2%, 15.6% and 31.5%, respectively.
The local hoping rate in T2/onc low-copy;Rosa26-SB11 tumors is intermediate between those reported for sarcomas from p19Arf−/−;CAGGS-SB10;T2/onc low-copy mice (23% of insertions within the 40Mb surrounding the donor concatomer (5)) and that reported for T2/onc high-copy;Rosa26-SB11 leukemias/lymphomas (“little local hopping” (6)). The chromosomal location and environment of the concatomers could influence local hopping rates. Transposase levels could also influence local hopping rates as differences were observed between p19Arf−/−;CAGGS-SB10;T2/onc low-copy sarcomas and Rosa26-SB11;T2/onc low-copy leukemias/lymphomas. The lower local hopping rates in Rosa26-SB11;T2/onc low-copy leukemias/lymphomas compared to weaning tail biopsies from the same cohort of mice could be due to increased time for transposons to remobilize in tumors from older mice compared to tissue from young mice.
CIS analysis was performed on insertions cloned from 59 Rosa26-SB11;T2/onc low-copy induced leukemias/lymphomas after removal of insertions residing on the donor concatomer chromosome (n=1547 insertions) to identify candidate leukemia/lymphoma genes. Analysis was performed using criteria described by Mikkers et al (14) at an expected fraction (Efr) of 0.001 (2 insertions in 1.95 kb, 3 insertions in 88.5 kb and 4 insertions in 371.5 kb), which is predicted to result in 4.5 CISs being identified by random chance alone. This resulted in the identification of 28 CISs in the leukemia dataset. Clustering criteria of insertions using an Efr of 0.005 was also applied (2 insertions in 9.75kb, 3 in 202kb and 4 in 658.5 kb) which is predicted to result in a total of 22.5 CISs identified based on random chance alone. Using these criteria a total of 49 CISs were identified (of which 28 also met the criteria used above for an Efr of 0.001). Five of these CISs were also CISs in the unselected dataset, indicating that they likely do not tag a locus important for cancer formation. Removal of these resulted in a final total of 44 CISs in leukemias/lymphomas (Table 2).
Table 2.
CISs identified in leukemias/lymphomas from T2/onc;Rosa26-SB11 mice.
| CIS name | Chr | CIS address start |
CIS address end |
# of tumors |
# of independent insertions |
other genes in CIS interval |
|---|---|---|---|---|---|---|
| Myb * | 10 | 20824216 | 20921673 | 4 | 4 | |
| Ube2d1 # | 10 | 70669435 | 70673276 | 2 | 2 | |
| Stab2 # | 10 | 86412419 | 86505987 | 3 | 3 | |
| Ikzf1 * | 11 | 11407716 | 11749020 | 14 | 18 | 4930415F15Rik, 4930512M02Rik, Q3UW08, Fignl1, Ddc |
| Csf2 # | 11 | 54031828 | 54569276 | 4 | 4 | ENSMUSG00000060068, Il3, Acsl6, 4930404A10Rik, Fnip1, Rapgef6, Cdc42se2 |
| Cox10 # | 11 | 63785103 | 63794563 | 2 | 2 | |
| Rab5c *# | 11 | 100370989 | 100543223 | 3 | 3 | Ttc25, Cnp1, Dnajc7, Nkiras2, A930006D11Rik, D11Lgp2e, Gcn5l2, Hspb9 |
| Dusp22 | 13 | 30572119 | 30813368 | 4 | 4 | Irf4 |
| Ibrdc2 # | 13 | 47050885 | 47464428 | 4 | 5 | Tpmt, Aof1, Dek |
| H2afy *# | 13 | 56100124 | 56645908 | 4 | 4 |
BC027057, Neurog1, Cxcl14, Q8CDW6, AU042651, Il9, Fbxl21, Lect2, Q3U1K8, Tgfbi |
| Mef2c * | 13 | 83767562 | 84048492 | 5 | 5 | |
| Cenpk | 13 | 105370547 | 105371252 | 2 | 2 | |
| Zmiz1 * | 14 | 24269629 | 24414687 | 5 | 5 | |
| Heg1 | 16 | 33678449 | 33679145 | 2 | 2 | |
| Btla # | 16 | 44873922 | 45525600 | 4 | 4 | Cd200r3, Ccdc80, Q3UVS9, Slc35a5, Atg3, EG547267, Cd200, Gm609, ENSMUST00000060550, Slc9a10 |
| Erg * | 16 | 95072653 | 95570852 | 20 | 10 | Kcnj6, Kcnj15 |
| Heatr5b # | 17 | 78655675 | 78761809 | 3 | 3 | ENSMUST00000059920, 2310002B06Rik, ENSMUST00000043373, Eif2ak2 |
| Mbd2 * | 18 | 70309981 | 70807502 | 7 | 6 | 2310002L13Rik, ENSMUST00000096551, 4930503L19Rik, Poli, ENSMUST00000031200 |
| Pten | 19 | 32611775 | 32885582 | 7 | 12 | Papss2, Atad1, B430203M17Rik |
| Notch1 * | 2 | 26281337 | 26321310 | 21 | 20 | |
| Zbtb34 | 2 | 33060035 | 33919549 | 5 | 6 | Angptl2, Ralgps1, Lmx1b, C130021I20Rik, ENSMUST00000100174, 2610528K11Rik |
| Rasgrp1 * | 2 | 117030934 | 117031956 | 2 | 2 | |
| Gm414 | 3 | 70203770 | 70205270 | 2 | 2 | |
| Ppp3ca # | 3 | 136626657 | 136865395 | 2 | 6 | |
| Bach2 *# | 4 | 32416902 | 33046612 | 4 | 6 |
XR_001707.1, Q8BQ29, ENSMUST00000093133, Gja10, Casp8ap2, Mdn1 |
| Ptpn12 # | 5 | 20125026 | 21032439 | 5 | 5 | Magi2, Q3U0Y7, Q8CEH7, Phtf2, Tmem60, Rsbn1l, ENSMUST00000053060, EG626903, A530088I07Rik, 4930528G09Rik, Fgl2, AI847670, Fbxl13 |
| Kit/Kdr * | 5 | 75834997 | 76133546 | 4 | 5 | |
| AB041803 | 6 | 31101079 | 31233559 | 4 | 4 | |
| Wnk1 # | 6 | 119698409 | 120263825 | 4 | 4 | Erc1, 3110021A11Rik, ENSMUST00000036010, Rad52, EG406236, mmu-mir-706, Ninj2, B4galnt3, ENSMUSG00000053059 |
| Etv6 | 6 | 134104562 | 134557768 | 4 | 4 | Bcl2l14, Lrp6, Q8BPW4 |
| Akt2 | 7 | 27305138 | 27309525 | 3 | 3 | |
| Klf13 | 7 | 63514893 | 63864073 | 4 | 4 | Otud7a, ENSMUST00000003521 |
| Mctp2 # | 7 | 72254322 | 72263587 | 2 | 2 | |
| Eed | 7 | 89832779 | 89844617 | 2 | 3 | |
| EG209380 # | 7 | 105978304 | 106615569 | 4 | 4 | Gvin1, Q922V0, Q9D303, ENSMUST00000071162, Gm1966, Olfr693-701 |
| Zfp629 | 7 | 127271619 | 127383373 | 4 | 4 | Fbs1, Q8C4A9, D030022P06Rik, Phkg2, Gm166, Rnf40 |
| Dcun1d5 # | 9 | 7186494 | 7196156 | 2 | 2 | |
| Naalad2 # | 9 | 18088306 | 18093482 | 2 | 2 | |
| Fli1 * | 9 | 31723619 | 32229349 | 6 | 5 | Grit, Kcnj5, Kcnj1 |
| BC033915 | 9 | 45938617 | 45993423 | 3 | 4 | Apoa1, Tcea1, Apoc3, Efhc1 |
| 4833427G06Rik | 9 | 50485627 | 50898988 | 4 | 4 | Dixdc1, 2310030G06Rik, Cryab, Hspb2, 1110032A03Rik, D630004A14Rik, Alg9, Ppp2r1b, Snf1lk2, Layn, Btg4, mmu-mir-34b,c |
| Tcf12 *# | 9 | 71786484 | 71892068 | 3 | 3 | |
| Eras *# | X | 7019492 | 7220920 | 3 | 3 | Otud5, Pim2, Slc35a2, Pqbp1, Timm17b, ENSMUST00000085330, Q3UUQ2, Pcsk1n, Hdac6, Gata1, 2010001H14Rik, EG632013, Suv39h1 |
| Tbl1x # | X | 73774303 | 74215530 | 4 | 4 | EG628893, Prkx, Pbsn |
The name of the CIS is presented along with the chromosome (chr), the base pair of the first insertion defining the CIS, the base pair of the last insertion defining the CIS, the number of tumors defining the CIS, the number of insertions in independent TA dinucleotides, additional Ensembl annotated genes found within the bounds of the CIS. CISs previously identified in MuLV mutagenesis studies (13, 32) are marked with an * and those defined by an expected fraction (Efr) of .005 are marked with a #
Of the 44 CISs identified, 15 are CISs in the RTCGD of retroviral screens for lymphoma/leukemia genes or in a recent report analyzing MuLV insertions from over 500 tumors (13, 32) (Table 2). Therefore, the majority of CISs identified by SB have not been previously identified in retroviral screens. Notably, in lymphomas/leukemias resulting from mobilization from T2/onc low-copy lines, a CIS is found in Pten. Although an important tsg in the hematopoietic system and other cancers (33, 34), Pten has not been previously identified as a CIS in retroviral screens (13, 32). This supports the hypothesis that SB can identify cancer genes that are not readily tagged by retroviruses, including tsgs. Only seven CISs were common between the leukemia/lymphoma dataset described here and the dataset from the 15 leukemias/lymphomas generated using high-copy lines (6), indicating that cloning insertions from a larger cohort of tumors increases CIS identification power.
In summary, by combining T2/onc low-copy lines with Rosa26-SB11 we have achieved whole-body mobilization rates that are sufficient to promote penetrant tumorigenesis without the complication of embryonic lethality or genomic instability. Although lymphomas/leukemias predominate the tumor spectrum, whole-body mobilization of T2/onc can also promote glioma formation including glioblastoma, a tumor type in humans with an extremely poor prognosis. In lymphomas/leukemias, T2/onc tags both known and candidate novel cancer genes. Recent reports have demonstrated the ability of T2/onc mobilization by tissue-specific transposase expression to generate liver tumors and intestinal tumors useful for candidate cancer gene discovery (9, 29). In these models, true carcinoma/adenocarcinoma on a wild-type genetic background occurred with long latency and incomplete penetrance, indicating that additional improvements to increase mutagenesis rates are still needed for the SB system. Our data indicate that such mutagenesis rates can be obtained without undesired consequences such as lethality or genome-wide instability and that further development of the SB system is warranted for cancer gene discovery in a wider range of cell types.
Supplementary Material
Acknowledgments
We thank Erin Riley for technical assistance, Paul Marker and Michael Taylor for critical reading of the manuscript and the members of the Center for Genome Engineering for many helpful discussions.
Funding: K01CA122183 and an American Cancer Society pre-doctoral fellowship (LSC), MN Dept of Employment and Economic Development SPAP-05-0013-P-FY06 (DAL and RBJ), R01CA113636-01A1 (DAL), R01NS055750 (WAW), Cancer Research-UK and the Wellcome Trust (DJA).
Footnotes
Conflicts of interest: In the past, SB technology was exclusively licensed to Discovery Genomics Inc. (DGI), which is co-founded by DAL. DAL has an equity interest in and is an unpaid scientific advisor to DGI. DGI is pursuing the use of SB for human gene therapy. Neither DGI money nor personnel were involved in the work reported here. The University of Minnesota has a pending patent on the process of using transposons such as SB for cancer gene discovery. DAL, LSC, AJD, NAJ and NGC are named inventors.
References
- 1.Uren AG, Kool J, Berns A, van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24:7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 2.Collier LS, Largaespada DA. Transforming science: cancer gene identification. Curr Opin Genet Dev. 2006;16:23–29. doi: 10.1016/j.gde.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Callahan R, Smith GH. MMTV-induced mammary tumorigenesis: gene discovery, progression to malignancy and cellular pathways. Oncogene. 2000;19:992–1001. doi: 10.1038/sj.onc.1203276. [DOI] [PubMed] [Google Scholar]
- 4.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 5.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 6.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 7.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. 'Green mice' as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 8.Dupuy AJ, Fritz S, Largaespada DA. Transposition and gene disruption in the male germline of the mouse. Genesis. 2001;30:82–88. doi: 10.1002/gene.1037. [DOI] [PubMed] [Google Scholar]
- 9.Starr TK, Allaei R, Silverstein KA, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim WI, Matise I, Diers MD, Largaespada DA. RAS oncogene suppression induces apoptosis followed by more differentiated and less myelosuppressive disease upon relapse of acute myeloid leukemia. Blood. 2009;113:1086–1096. doi: 10.1182/blood-2008-01-132316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim WI, Wiesner SM, Largaespada DA. Vav promoter-tTA conditional transgene expression system for hematopoietic cells drives high level expression in developing B and T cells. Exp Hematol. 2007;35:1231–1239. doi: 10.1016/j.exphem.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Largaespada DA, Collier LS. Transposon-mediated mutagenesis in somatic cells: identification of transposon-genomic DNA junctions. Methods Mol Biol. 2008;435:95–108. doi: 10.1007/978-1-59745-232-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikkers H, Allen J, Knipscheer P, et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002;32:153–159. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- 15.Hackett CS, Hodgson JG, Law ME, et al. Genome-wide array CGH analysis of murine neuroblastoma reveals distinct genomic aberrations which parallel those in human tumors. Cancer Res. 2003;63:5266–5273. [PubMed] [Google Scholar]
- 16.Hodgson JG, Malek T, Bornstein S, et al. Copy number aberrations in mouse breast tumors reveal loci and genes important in tumorigenic receptor tyrosine kinase signaling. Cancer Res. 2005;65:9695–9704. doi: 10.1158/0008-5472.CAN-05-0755. [DOI] [PubMed] [Google Scholar]
- 17.Jain AN, Tokuyasu TA, Snijders AM, Segraves R, Albertson DG, Pinkel D. Fully automatic quantification of microarray image data. Genome Res. 2002;12:325–332. doi: 10.1101/gr.210902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Stanchina E, McCurrach ME, Zindy F, et al. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zindy F, Eischen CM, Randle DH, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 21.Rahrmann EP, Collier LS, Knutson TP, et al. Identification of PDE4D as a proliferation promoting factor in prostate cancer using a Sleeping Beauty transposon-based somatic mutagenesis screen. Cancer Res. 2009;69:4388–4397. doi: 10.1158/0008-5472.CAN-08-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geurts AM, Collier LS, Geurts JL, et al. Gene Mutations and Genomic Rearrangements in the Mouse as a Result of Transposon Mobilization from Chromosomal Concatemers. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Luenen HG, Colloms SD, Plasterk RH. The mechanism of transposition of Tc3 in C. elegans. Cell. 1994;79:293–301. doi: 10.1016/0092-8674(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 24.Gray YH. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16:461–468. doi: 10.1016/s0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- 25.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Balciunas D, Wangensteen KJ, Wilber A, et al. Harnessing a High Cargo-Capacity Transposon for Genetic Applications in Vertebrates. PLoS Genetics. 2006 doi: 10.1371/journal.pgen.0020169. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zagoraiou L, Drabek D, Alexaki S, et al. In vivo transposition of Minos, a Drosophila mobile element, in mammalian tissues. Proc Natl Acad Sci U S A. 2001;98:11474–11478. doi: 10.1073/pnas.201392398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo G, Ivics Z, Izsvak Z, Bradley A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keng VW, Villanueva A, Chiang DY, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27:264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G, Geurts AM, Yae K, et al. Target-site preferences of Sleeping Beauty transposons. J Mol Biol. 2005;346:161–173. doi: 10.1016/j.jmb.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 31.Geurts AM, Hackett CS, Bell JB, et al. Structure-based prediction of insertion-site preferences of transposons into chromosomes. Nucleic Acids Res. 2006;34:2803–2811. doi: 10.1093/nar/gkl301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uren AG, Kool J, Matentzoglu K, et al. Large-scale mutagenesis in p19(ARF)-and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chow LM, Baker SJ. PTEN function in normal and neoplastic growth. Cancer Lett. 2006;241:184–196. doi: 10.1016/j.canlet.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 34.Maser RS, Choudhury B, Campbell PJ, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.