Abstract.
Human bystin was identified as a cytoplasmic protein directly binding to trophinin, a cell adhesion molecule potentially involved in human embryo implantation. Although the trophinin gene is unique to mammals, the bystin gene (BYSL) is conserved across eukaryotes. Recent studies show that bystin plays a key role during the transition from silent trophectoderm to an active trophoblast upon trophinin-mediated cell adhesion. Bystin gene knockout and knockdown experiments demonstrate that bystin is essential for embryonic stem cell survival and trophectoderm development in the mouse. Furthermore, biochemical analysis of bystin in human cancer cells and mouse embryos indicates a function in ribosomal biogenesis, specifically in processing of 18S RNA in the 40S subunit. Strong evidence that BYSL is a target of c-MYC is consistent with a role for bystin in rapid protein synthesis, which is required for actively growing cells.
Keywords. BYSL, Enp1, ribosome, embryo, trophoblast, placenta, cancer
Introduction
Bystin was originally identified as a cytoplasmic protein that forms a complex with trophinin and tastin in human trophoblastic embryonal carcinoma HT-H cells [1–3]. Trophinin is an intrinsic membrane protein that mediates cell adhesion by homophilic trophinin-trophinin binding. Tastin and bystin are cytoplasmic proteins required for trophinin to function efficiently as a cell adhesion molecule. In humans, trophinin, tastin and bystin are expressed at the uteroplacental interface or at implantation sites [4, 5]. These proteins are expressed in human placenta at early stages of pregnancy but disappear from the placenta after 10 weeks of pregnancy [4].
Genes encoding trophinin and tastin are only found in mammals. Knockout of trophinin in mice revealed that trophinin does not play an essential role in embryo implantation in that species [6]. Nevertheless, analysis of the function of human trophinin, particularly in cases of ectopic pregnancy, supports the hypothesis that trophinin is uniquely involved in human embryo implantation [5].
By contrast, the bystin gene, Bysl, is conserved across a wide range of eukaryotes, including yeast, nematodes, insects, snakes, and mammals [7–11]. The yeast Bysl homolog, ENP1 or essential nuclear protein 1, is required for vegetative growth in budding yeast [7] and functions in pre-ribosomal RNA processing [8]. Biochemical and cell biological analysis of bystin in human cancer cells and mouse embryos indicates that bystin functions in the biogenesis of the 40S ribosome and in cell growth [12, 13].
Involvement of bystin in human embryo implantation
A fertilized mammalian egg autonomously develops into a blastocyst, which must be successfully implanted in the uterus to develop into a fetus. In higher primates including humans, initial adhesion of the blastocyst to the uterus occurs via the trophectoderm and endometrial luminal epithelial cells. Thus the blastocyst, which is composed of a trophectoderm monolayer surrounding embryonic stem cells or the inner cell mass, adheres to the apical surface of the endometrial luminal epithelia at its embryonic pole [14–16]. This morphology of the human embryo implantation site differs significantly from that seen in the mouse. In mouse, an implanting blastocyst is surrounded by endometrium, and the initial adhesion takes place at the abembryonic pole of the mouse blastocyst, which orients the embryo within the implantation chamber [17, 18]. Nonetheless, in all mammals, the initial adhesion of the blastocyst to the uterus occurs at apical membranes of two polarized epithelial cells, whereas, apical cell surfaces of epithelia are generally non-adhesive. In mouse embryo implantation, the ErbB family receptor tyrosine kinase (probably ErbB4) interacts with membrane-bound heparin-binding epidermal growth factor (EGF)-like growth factor (HB-EGF), which is induced in the endometrial epithelia in a spatially restricted manner by an implanting blastocyst [18, 19]. We hypothesized that such spatially and temporally regulated and cell type-specific apical adhesion was mediated by a unique adhesion molecule. We also hypothesized that some cancer cell lines derived from human embryonal carcinomas may exhibit activity of trophectoderm cells at the implantation stage. As human embryonal carcinoma cell lines tend to differentiate into trophoblastic cells [20], we used the human embryonal carcinoma HT-H line, which differentiates spontaneously into trophoblastic cells [21]. When trophoblastic HT-H cells are added to a monolayer of human endometrial epithelial SNG-M cells, HT-H cells adhere rapidly to the upper surface of the SNG-M cells [1]. Using both lines, we identified gene products responsible for apical adhesion by expression cloning. COS cells, which do not adhere to the apical surface of SNG-M cells, were transfected with an HT-H cDNA library constructed in a mammalian expression vector, and cells that then adhered to SNG-M were selected. When a mixture of cDNA clones was transfected, COS cells adhered to SNG-M cells. However, no single cDNA from that pool promoted adhesion. Thus clones were subtracted from the original positive pool to identify potential combinations of cDNAs required for adhesion. This approach identified trophinin (TRO gene product) and tastin (TROAP gene product) as necessary for COS to adhere to SNG-M [1].
We found that trophinin and tastin do not interact directly and subsequently identified within the positive cDNA pool a cytoplasmic protein that bridges the two proteins [3]. This new protein was designated bystin (BYSL gene product), as the gene encoding it is homologous to Drosophila Bys (standing for ‘by S’ because it is next to the gene encoding ribosomal protein S6) [22]. Bystin was likely missed in the initial cloning of trophinin and tastin [1] because it is expressed in COS cells.
In humans, the BYSL gene localizes to chromosome 6, between TRFP encoding a transcription mediater and CCND3 encoding cyclin D3 (Fig. 1a, b). The bystin protein contains many potential protein kinase phosphorylation sites, suggesting an active role of bystin in signal transduction (Fig. 1c). However, no known structural motif is found in the bystin protein.
Figure 1.
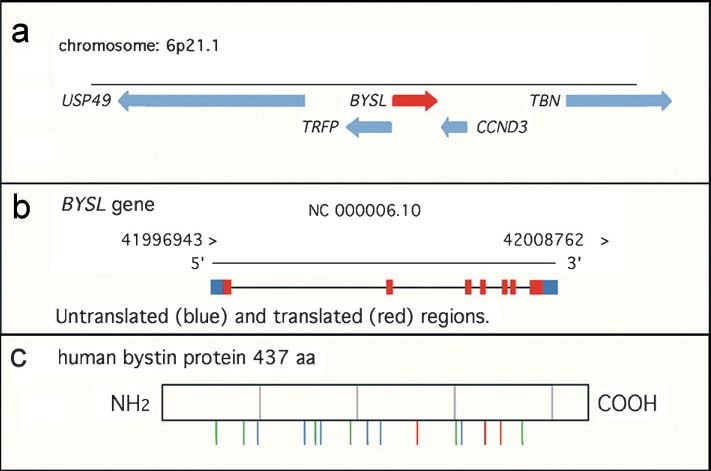
Structures of the human bystin gene BYSL and bystin protein. (a) Genomic structure of the BYSL gene. BYSL maps to chromosome 6, between TRFP encoding transcription mediator and CCND3 encoding cyclin D3. (b) Transcription of the BYSL gene. Untranslated region and translated regions are shown in blue and red, respectively. (c) Human bystin protein. Potential phosphorylation sites of the human bystin protein for protein kinase C, casein kinase II, and tyrosine kinase are shown in green, blue, and red, respectively.
Bystin interacts with trophinin, tastin, and cytokeratins 8 and 18 [3]. Interestingly, interaction of bystin with trophinin or tastin became stronger when cytokeratins 8 and 18 were included [3]. Such results suggest that the trophinin-bystin-tastin complex is stabilized by cytokeratins. In the mouse, cytokeratins 8 (EndoA) and 18 (EndoB) are specific differentiation markers for trophectoderm, which constitutes the first differentiated cells of the fertilized egg [23]. We hypothesize that cytokeratins stabilize the apical localization of the trophinin-bystin-tastin complex in trophectoderm cells prior to implantation.
At the human embryo implantation site, trophinin, bystin, and tastin are strongly expressed in chorionic trophoblasts and endometrial glandular epithelial cells [4]. These three proteins are also expressed at the implantation site in ectopic pregnancy, a condition unique to humans [5]. Their expression patterns also suggest that these proteins function in placental development in humans.
The role of bystin in signal transduction following trophinin-mediated cell adhesion
The histology of human implantation sites shows that trophectoderm cells at the initial adhesion proliferate and invade maternal tissue [14, 24]. By contrast, trophectoderm cells at non-adherent sites remain silent. Clearly, adhesion triggers transformation of silent trophectoderm to an activated trophoblast in vivo.
When HT-H cells were co-cultured on an endometrial epithelial cell monolayer, the morphologies of both cell types were altered [1, 25, 26]. Many microvilli on the apical cell surfaces became flat and disappeared, suggesting that initial apical adhesion mediated by trophinin transduced a signal promoting cytoplasmic reorganization in both cell types.
Because trophinin-mediated cell adhesion is based on homophilic trophinin-trophinin binding, HT-H cells also adhere to themselves [1]. Thus when HT-H cells were detached from tissue culture plates by trypsin treatment and added to an HT-H cell monolayer, they adhered to that monolayer. Immunocytochemistry of the HT-H monolayer after adhesion showed elevated tyrosine phosphorylation [25], suggesting that trophinin-mediated cell adhesion activated HT-H cells in those cells. When the surface of HT-H cells was bound with an anti-trophinin antibody and cross-linked using a second antibody, tyrosine phosphorylation levels and F-actin formation were increased, suggesting that engagement of cell surface trophinin elicits an activation signal to the cytoplasm. To further investigate this mechanism, we screened a peptide-displaying phage library and identified a short GWRQ peptide that mimics trophinin-mediated cell adhesion [25]. HT-H cells cultured in the presence of GWRQ exhibited higher mobility and proliferation than did cells cultured with control peptide or without peptide. When GWRQ was added to monkey blastocysts cultured in vitro, trophectoderm cells migrated out from the blastocyst, and total numbers of cells in the blastocyst, including embryonic stem cells, increased [25].
Using GWRQ peptide as a reagent mimicking trophinin-mediated cell adhesion, we showed that the cytoplasmic domain of trophinin arrests ErbB4, an ErbB family receptor kinase, through bystin in HT-H cells [25, 26]. When trophinin-mediated cell adhesion occurs, which is mimicked by binding of GWRQ peptide to the trophinin extracellular domain, bystin is released from trophinin, allowing activation of ErbB4 and increasing proliferation and motility of trophoblastic cells. These results indicate that bystin plays a key role in switching silent trophectoderm to activated trophoblast.
The role of bystin in pre- and post-implantation stage mouse embryos and in maternal uterine epithelia
When mouse blastocysts are returned to the uterus of ovariectomized and progesterone-injected female mice, blastocysts remain dormant or do not implant [27, 28]. Activation occurs following estrogen injection into host mice. Microarray analysis of dormant and activated mouse blastocysts showed that, among several genes, Bysl expression was elevated in activated blastocysts [29].
Immunohistochemistry of mouse pre-implantation stage embryos indicates that bystin is not expressed or only weakly expressed in the fertilized egg earlier than the blastocyst stage [30]. Bystin protein was not detected in the blastocyst inside the zona pellucida, while bystin expression was first detected during hatching, and was then strongly expressed in the fully expanded blastocyst (Fig. 2). After this stage, bystin protein was down-regulated in trophectoderm cells throughout the entire period of adhesion to the maternal epithelia but reappeared in the epiblast composed of pluripotent embryonic stem cells after implantation. The expression pattern of mouse bystin at peri-implantation [30] is similar to that of mouse trophinin [6]. Bystin-null mouse embryos implanted successfully but died soon after implantation [30], suggesting that bystin is essential for mouse embryo survival after implantation. However, as described below, Bysl gene knockdown experiments show that bystin is also required for survival of preimplantation stage mouse embryos [12]. In the knockout mouse, it is likely that maternally derived Bysl mRNA masks loss of Bysl at pre-implantation stages [30].
Figure 2.
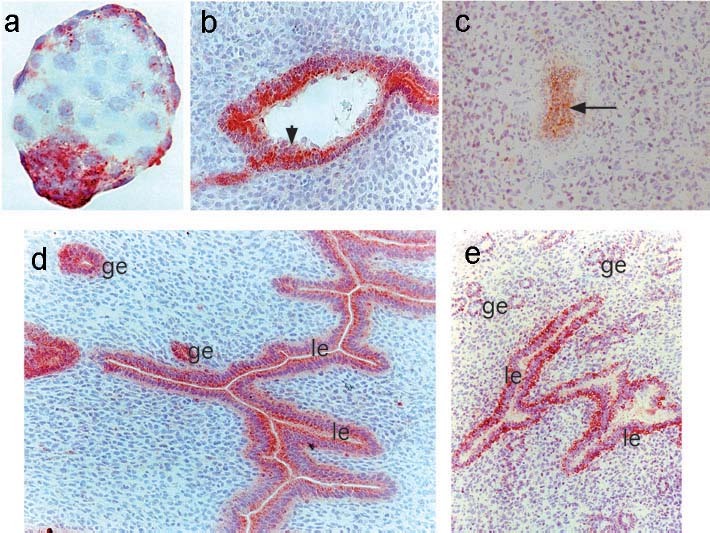
Expression of bystin protein in mouse embryos before, during and after implantation. (a) Immunohistochemistry for bystin in a blastocyst. Bystin protein was found in a blastocyst but not detected in a fertilized mouse egg or embryos earlier than the blastocyst stage [30]. (b) Bystin protein was barely detected in trophectoderm cells (arrowhead) during implantation. (c) After implantation, bystin protein was found in the epiblast including embryonic stem cells (arrow). (d) In the pregnant mouse uterus or in the presence of blastocysts, bystin expression is seen on the apical side of luminal epithelia (le), whereas it is distributed evenly in glandular epithelia (ge). (e) Immunohistochemistry of the uterus of a non-pregnant female mouse. Note that bystin proteins were found at the abluminal side of endometrial luminal epithelia (le) and glandular epithelia (ge). Tissue sections were reacted with rabbit anti-bystin antibody [30], followed by biotinylated antirabbit IgG antibody and peroxidase avidin. The peroxidase substrate 3-amino-9-ethyl carbazole was used to detect immunostaining. Hematoxylin was used for counter-staining.
When Bysl siRNAs were microinjected into fertilized eggs, compaction at the eight-cell stage occurred normally in vitro [12]. Bysl siRNA-injected embryos showed slightly reduced expression of cytokeratin 8 (EndoA), an early trophectoderm marker [31]. While control blastocysts showed assembled cytokeratin structures in the trophectoderm layer, no organized structures were detected in Bysl siRNA-injected embryos. Consequently, blastocyst formation was completely inhibited. These embryos failed to hatch from the zona pellucida and could not outgrow in culture. These results indicate that the bystin functions in trophectoderm differentiation. Bysl knockdown also inhibited embryonic stem cell proliferation [12]. Bystin is expressed in mouse endometrial luminal and glandular epithelial cells throughout hormonal cycles [30]. Interestingly, localization of bystin in the luminal epithelia showed a distinct blastocyst-dependent pattern: in the presence of blastocysts, bystin proteins localized to the apical side of the epithelia, whereas in their absence, bystin protein was localized to the abluminal or basal side of the epithelia (Fig. 2). This observation suggests the existence of an embryonic factor affecting the localization of bystin in the maternal epithelia. The molecular basis underlying apical or basal localization of bystin is presently unknown.
Drosophila bys for growth and cell adhesion
In Drosophila, the bys gene is implicated in cell growth [9, 22, 32] and in cell-cell interaction [9]. Maternal bys is strongly expressed in cells exhibiting high levels of rRNA synthesis and ribosome production [9, 33]. In the Drosophila embryo, bys expression is ubiquitous but relatively weak at early stages (gastrulation, stage 9), but at later stages (germband-extended embryos), bys expression is strong and specifically localized, and in larval imaginal discs, bys expression is restricted to specific patterns: for example, bys is strongly expressed in the dorsal and ventral regions of the wing pouch that will form epithelia of the adult wing. These embryonic and larval expression patterns could indicate a role of bys in cell adhesion. In particular, bys expression is strong in the region of the wing pouch giving rise to two epithelial sheets of the adult wing that adhere to one another after the disc everts.
The role of bystin in ribosomal biogenesis
Complementation analysis of budding yeast identified an essential nuclear protein designated Enp1 [7], the yeast bystin homolog. Atemperature-sensitive ENP1-null mutant showed defective processing of rRNA[8]. Studies of ribosomal biogenesis in yeast indicate that Enp1 is required to synthesize 40S ribosomal subunits by functioning in their nuclear export [34, 35].
While the expression of bystin in human tissues is related to embryo implantation [4, 5], the expression level of this protein is low in normal adult tissues [3]. A publicly available database shows that bystin mRNA levels are relatively low in normal human tissues, but that expression of bystin increases in various tumor cells [13]. Other microarrays analyzing specimens of human breast tumors identify bystin as part of a ‘proliferation cluster’ [36]. The embryonic lethality of the Bysl-null mouse and observation of bystin expression in the epiblast also support the hypothesis that bystin is essential for rapid cell growth in mammals, as both the epiblast and embryonic stem cells should proliferate rapidly after implantation. Bystin expression in the epiblast is consistent with a report showing that Bysl is one of 216 genes commonly expressed in embryonic, neural and hematopoietic stem cells in themouse [37]. Thus Bysl is a specific stem cell marker. Bysl is also included in a gene cluster of stem cell markers found on mouse chromosome 16 [37].
Ribosomal biogenesis differs significantly between prokaryotes and eukaryotes. In eubacteria, functional ribosomes self-assemble and can be reconstituted in vitro [38]. In budding yeast, ribosome synthesis requires more than 150 non-ribosomal proteins, many of which are essential for growth [39, 40]. These proteins have homologs in other species, and some share properties with proteins found in mammalian pre-ribosomal complexes [41]. Some human proteins, including fibrillarin [42], can complement at least partially yeast strains with mutations in their orthologs. Hence, a complex biogenesis system is likely conserved in most eukaryotes, including humans. Eukaryotic ribosome formation occurs predominantly in nucleoli, but late maturation steps occur in both the nucleoplasm and cytoplasm [40, 43].The location of bystin in the cytoplasm during G1 and its nuclear localization prior to mitosis [30] suggest that bystin plays dual roles in cell growth and proliferation in mammalian cells.
Several lines of evidence indicate that bystin functions in ribosome biogenesis in human cells (Fig. 3). First, it is located in the nucleolus, the organelle where ribosomal biogenesis takes place [12, 13]. It is also associated with the cytoplasmic 40S subunit, a component of the 80S monosome, before translation is initiated [13]. Bystin down-regulation delays processing of 18S rRNA or the mature form required for translation, resulting in compromised cell viability [12, 13]. In addition, inhibition of activity of the mammalian target of rapamycin (mTOR) suppresses bystin expression [13]. Furthermore, both nucleolar bystin and bystin associated with the 40S subunit disappear under conditions of nucleolar stress, suggesting that bystins in both locations are functionally linked in human cells [13]. Bystin may be associated with a precursor of the 40S subunit — a pre-40S particle — in the nucleolus and exported with these particles through nuclear pores, where it dissociates from particles at very late phases of subunit synthesis, as has been shown for Enp1 [34]. Given the dependence of cancer cell growth on ribosome biogenesis, high bystin expression in cancer cells may contribute to proliferation, a hypothesis supported by the fact that the bystin gene is amplified in diffuse large B cell lymphoma [44].
Figure 3.
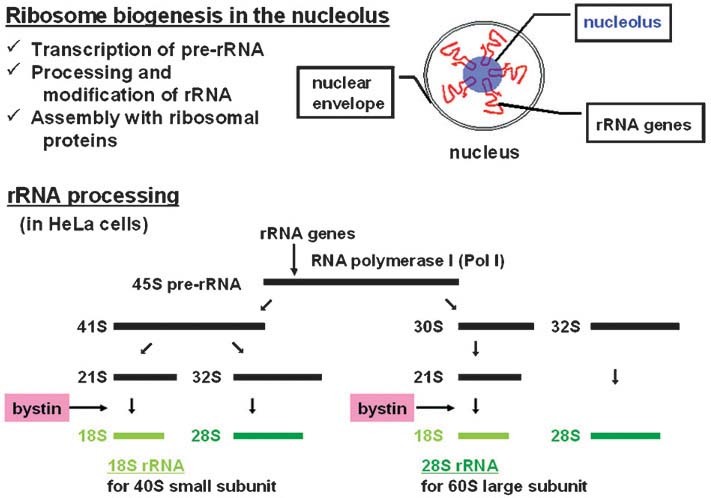
Ribosomal biogenesis and rRNA processing in eukaryotic cells. The initial pre-rRNA transcript is first transcribed from repetitive ribosomal DNA genes by RNA polymerase I (Pol I) in the nucleolus. rRNA precursors are then processed, chemically modified, and folded in the nucleolus, and ribosomal proteins, which are translated in the cytoplasm and imported into this organelle, concomitantly assemble with pre-rRNAs [40, 46]. There are two alternative pathways for rRNA processing in human HeLa cells [56]. Bystin is likely involved in processing of a 21S intermediate, of which the final product, 18S rRNA, is included in the 40S small subunit [13]. The importance of bystin homolog in 18S rRNA processing has also been shown in budding yeast [8] and in mouse [12].
Although bystin exhibits activities similar to Enp1 [8, 34], human bystin cannot rescue the lethal phenotype of a ENP1-null yeast mutant [8], suggesting that ribosomal RNA processing pathways in multicellular organisms differ from those in yeast [45] and that bystin activities may have been modified during evolution. In higher organisms, at early developmental stages, embryos must grow rapidly after implantation, requiring rapid ribosome biosynthesis. Bystin upregulation in the epiblast or in embryonic stem cells [30] is consistent with the idea that bystin is linked to rapid ribosomal biogenesis.
The C-terminal region of bystin/Enp1 sequences is well conserved across eukaryotes [7, 8]. Previous studies showed that human bystin binds trophinin, tastin, and cytokeratin [3]. Human bystin also binds to ErbB4 receptor tyrosine kinase [25]. However, it is not known which part of the bystin polypeptide is responsible for these binding activities. Some deletion mutants of mouse bystin localized to the nucleolus failed to rescue Bysl knockdown phenotypes and suppressed proliferation of transfected cells [12]. However, bystin protein contains no known functional domains linked to these activities. Defining the structure and function relationship of bystin awaits future studies.
The cytoplasmic localization of bystin contrasts with the almost exclusively nuclear localization of Enp1 in yeast cells [7, 8, 34, 35]. Comprehensive proteomics analysis of yeast nuclear proteins indicates that nuclear proteins are not stored in the cytoplasm [34, 35]. Thus newly synthesized ribosomal proteins in the cytoplasm are immediately transported to the nucleolus [46, 47]. Indeed, no cytoplasmic pool of nucleolar fibrillarin and ribosomal S6 has been detected in human cells [13]. Thus, the cytoplasmic localization of bystin may have evolved in higher organisms. Bystin function in cell adhesion during human embryo implantation [3] suggests a cytoplasmic role in cell adhesion and signal transduction [25]. In prostate cancer cells, which adhere to neurons, bystin protein is expressed in a manner suggesting a role in cell-cell contact and cell growth [48].
For higher organisms, it was long believed that rRNA processing is completed within the nucleus. However, maturation of the 40S subunit, including final processing of 18S rRNA, has recently been demonstrated to occur in the cytoplasm in human cells [reviewed in ref. 43], as well as in yeast [40]. Since part of cytoplasmic bystin is associated with the 40S subunit before translation in human cells [13], bystin may also function in the final step of 40S subunit synthesis in the cytoplasm.
We found that bystin associates with undefined nuclear particles following actinomycin D treatment of HeLa cells [13]. Although these particles were detected under conditions of bystin overexpression, nucleolar stress-induced particles appear specific to bystin. As nuclear stress granules can serve as storage sites for transcription factors [49], soluble proteins involved in ribosome biogenesis may shuttle between the nucleolus and nucleoplasm [40, 47]. Given the dependence of cell proliferation on ribosome biogenesis, when biogenesis is halted by nucleolar stress, this system may allow rapid ribosome resynthesis following relief from stress. Future proteomic analysis of particles including tagged-bystin may define characteristics of the bystin-containing nuclear particles as has been shown with other nuclear particles [50, 51]. Cancer progression depends on ribosome biogenesis, as exemplified by regulation of ribosome synthesis by the tumor suppressors, pRb and p53, and RNA-processing factors, including B23/nucleophosmin, are involved in cancer progression [52]. Compounds such as rapamycin, an inhibitor of ribosome biogenesis and translation initiation, are effective anticancer drugs [53]. Therefore, defining factors and pathways involved in ribosome biogenesis is required for cancer therapy. Just as many antibiotics interfere with the formation of the prokaryotic ribosome [54], targeting biogenesis of human ribosomes may antagonize malignant neoplasms.
Bystin as a prominent c-MYC target
In Drosophila embryos, the expression pattern of bys mRNA is almost identical to that of pitchoune (pit) and modulo (mod) genes, which are potential transcriptional targets of Drosophila Myc (dMyc) [9]. As the 5′ region of the Drosophila bys gene contains a canonical E-box Myc-binding site, it is possible that dMyc promotes bys gene expression in Drosophila.
Network reverse engineering, which enables identification of co-regulated genes from genome-wide expression profiles, identified more than 100 first neighbors or genes directly interconnected with the MYC proto-oncogene in human B cells [55]. Intriguingly, BYSL was identified as the highest scored gene co-regulated by MYC. This finding indicates that c-MYC overexpression in human cells could lead to ribosomal biogenesis and cellular overgrowth. Both c-MYC and BYSL are overexpressed in many human cancers, suggesting that bystin expression is associated with malignant phenotypes.
Concluding remarks
In a wide variety of organisms from yeast to humans, bystin and Enp1 play a role in facilitating ribosome biogenesis required for cell growth. Drosophila bys and human bystin are also involved in cell adhesion, suggesting that bys and bystin acquired additional functions during the course of evolution.
In higher organisms, many cellular functions are controlled by cell-cell interactions. Embryo implantation and its subsequent processes for placenta formation involve both cell-cell interactions and cell growth. Although some mechanisms underlying malignant cancers are shared with those in embryo implantation, distinct differences between them are seen in their regulation: embryo implantation is a well orchestrated and regulated process, while cancers grow without regulation. Further studies on bystin will provide a better understanding of these processes and could suggest novel therapeutic strategies against malignant cancers.
Footnotes
Received 30 June 2007; received after revision 7 August 2007; accepted 29 August 2007
References
- 1.Fukuda M. N., Sato T., Nakayama J., Klier G., Mikami M., Aoki D., Nozawa S. Trophinin and tastin, a novel cell adhesion molecule complex with potential involvement in embryo implantation. Genes Dev. 1995;9:1199–1210. doi: 10.1101/gad.9.10.1199. [DOI] [PubMed] [Google Scholar]
- 2.Fukuda M. N., Nozawa S. Trophinin, tastin, and bystin: a complex mediating unique attachment between trophoblastic and endometrial epithelial cells at their respective apical cell membranes. Semin. Reprod. Endocrinol. 1999;17:229–234. doi: 10.1055/s-2007-1016230. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki N., Zara J., Sato T., Ong E., Bakhiet N., Oshima R. G., Watson K. L., Fukuda M. N. A novel cytoplasmic protein, bystin, interacts with trophinin, tastin and cytokeratin, and may be involved in trophinin mediated cell adhesion between trophoblast and endometrial epithelial cells. Proc. Natl. Acad. Sci. USA. 1998;95:5027–5032. doi: 10.1073/pnas.95.9.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki N., Nakayama J., Shih I. M., Aoki D., Nozawa S., Fukuda M. N. Expression of trophinin, tastin, and bystin by trophoblast and endometrial cells in human placenta. Biol. Reprod. 1999;60:621–627. doi: 10.1095/biolreprod60.3.621. [DOI] [PubMed] [Google Scholar]
- 5.Nakayama J., Aoki D., Suga T., Akama T. O., Ishizone S., Yamaguchi H., Imakawa K., Nadano D., Fazleabas A. T., Katsuyama T., Nozawa S., Fukuda M. N. Implantation-dependent expression of trophinin by maternal fallopian tube epithelia during tubal pregnancies: possible role of human chorionic gonadotrophin on ectopic pregnancy. Am. J. Pathol. 2003;163:2211–2219. doi: 10.1016/S0002-9440(10)63579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nadano D., Sugihara K., Paria B. C., Saburi S., Copeland N. G., Gilbert D. J., Jenkins N. A., Nakayama J., Fukuda M. N. Significant differences between mouse and human trophinins are revealed by their expression patterns and targeted disruption of mouse trophinin gene. Reprod. 2002;66:313–321. doi: 10.1095/biolreprod66.2.313. [DOI] [PubMed] [Google Scholar]
- 7.Roos J., Luz J. M., Centoducati S., Sternglanz R., Lennarz W. J. ENP1, an essential gene encoding a nuclear protein that is highly conserved from yeast to humans. Gene. 1997;185:137–146. doi: 10.1016/S0378-1119(96)00661-0. [DOI] [PubMed] [Google Scholar]
- 8.Chen W., Bucaria J., Band D. A., Sutton A., Sternglanz R. Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res. 2003;31:690–699. doi: 10.1093/nar/gkg145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart M. J., Nordquist E. K. Drosophila Bys is nuclear and shows dynamic tissue-specific expression during development. Dev. Genes Evol. 2005;215:97–102. doi: 10.1007/s00427-004-0447-8. [DOI] [PubMed] [Google Scholar]
- 10.Sheng J., Yang S., Xu L., Wu C., Wu X., Li A., Yu Y., Ni H., Fukuda M., Zhou J. Bystin as a novel marker for reactive astrocytes in the adult rat brain following injury. Eur. J. Neurosci. 2004;20:873–884. doi: 10.1111/j.1460-9568.2004.03567.x. [DOI] [PubMed] [Google Scholar]
- 11.Pack S. D., Pak E., Tanigami A., Ledbetter D. H., Fukuda M. N. Assignment of the bystin gene BYSL to human chromosome band 6p21.1 by in situ hybridization. Cytogenet. Cell Genet. 1998;83:76–77. doi: 10.1159/000015131. [DOI] [PubMed] [Google Scholar]
- 12.Adachi K., Soeta-Saneyoshi C., Sagara H., Iwakura Y. Crucial role of Bysl in mammalian preimplantation development as an integral factor for 40S ribosome biogenesis. Mol. Cell. Biol. 2007;27:2202–2214. doi: 10.1128/MCB.01908-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyoshi M., Okajima T., Matsuda T., Fukuda M. N., Nadano D. Bystin in human cancer cells: intracellular localization and function in ribosome biogenesis. Biochem. J. 2007;404:373–381. doi: 10.1042/BJ20061597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindenberg S. Ultrastructure in human implantation: transmission and scanning electron-microscopy. Baillieres Clin. Obstet. Gynaecol. 1991;5:1–14. doi: 10.1016/S0950-3552(05)80067-7. [DOI] [PubMed] [Google Scholar]
- 15.Enders A. C., Blankenship T. N. Comparative placental structure. Adv..Drug Deliv. Rev. 1999;38:3–15. doi: 10.1016/S0169-409X(99)00003-4. [DOI] [PubMed] [Google Scholar]
- 16.Enders A. C., Lantz K. C., Peterson P. E., Hendrick A. G. From blastocyst to placenta: the morphology of implantation in the baboon. Human Reprod. Update. 1997;3:561–573. doi: 10.1093/humupd/3.6.561. [DOI] [PubMed] [Google Scholar]
- 17.Wang H., Dey S. K. Roadmap to embryo implantation: clues from mouse models. Nat. Rev. Genet. 2006;7:185–199. doi: 10.1038/nrg1808. [DOI] [PubMed] [Google Scholar]
- 18.Paria B. C., Reese J., Das S. K., Dey S. K. Deciphering the cross-talk of implantation: advances and challenges. Science. 2002;296:2185–2188. doi: 10.1126/science.1071601. [DOI] [PubMed] [Google Scholar]
- 19.Paria B. C., Elenius K., Klagsbrun M., Dey S. K. Heparin-binding EGF-like growth factor interacts with mouse blastocysts independently ofErbB1: a possible role for heparan sulfate proteoglycans and ErbB4 in blastocyst implantation. Development. 1999;126:1997–2005. doi: 10.1242/dev.126.9.1997. [DOI] [PubMed] [Google Scholar]
- 20.Andrews P.W., Bronson D. L., Benham F., Strickland S., Knowles B. B. A comparative study of eight cell lines derived from human testicular teratocarcinoma. Int. J. Cancer. 1980;26:269–280. doi: 10.1002/ijc.2910260304. [DOI] [PubMed] [Google Scholar]
- 21.Izhar M., Siebert P., Oshima R. G., DeWolf W. C., Fukuda M. N. Trophoblastic differentiation of human teratocarcinoma cell line HT-H. Dev. Biol. 1986;116:510–518. doi: 10.1016/0012-1606(86)90151-X. [DOI] [PubMed] [Google Scholar]
- 22.Stewart M. J., Denell R. Mutations in the Drosophila gene encoding ribosomal protein S6 cause tissue overgrowth. Mol. Cell. Biol. 1993;13:2524–2535. doi: 10.1128/mcb.13.4.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oshima R. G. Identification and immunoprecipitation of cytoskeletal proteins from murine extra-embryonic endothelial cells J. Biol. Chem. 1981;256:8124–8133. [PubMed] [Google Scholar]
- 24.Knoth M., Larsen J. F. Ultrastructure of a human implantation site. Acta Obstet. Gynecol. Scand. 1972;51:385–393. doi: 10.3109/00016347209156879. [DOI] [PubMed] [Google Scholar]
- 25.Sugihara K., Sugiyama D., Byrne J., Wolf D. P., Lowitz K. P., Kobayashi Y., Kabir-Salmani M., Nadano D., Aoki D., Nozawa S., Nakayama J., Mustelin T., Ruoslahti E., Yamaguchi N., Fukuda M. N. Trophoblast cell activation by trophinin ligation is implicated in human embryo implantation. Proc. Natl. Acad. Sci. USA. 2007;104:3799–3804. doi: 10.1073/pnas.0611516104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukuda M. N., Sugihara K. Signal transduction in human embryo implantation. Cell Cycle. 2007;6:1153–1156. doi: 10.4161/cc.6.10.4266. [DOI] [PubMed] [Google Scholar]
- 27.Yoshinaga K., Adams C. E. Delayed implantation in the spayed, progesterone treated adult mouse. J. Reprod. Fertil. 1966;12:593–595. doi: 10.1530/jrf.0.0120593. [DOI] [PubMed] [Google Scholar]
- 28.Paria B. C., Huet-Hudson Y. M., Dey S. K. Blastocyst's state of activity determine the window of implantation in the receptive mouse uterus. Proc Natl. Acad. Sci.USA. 1993;90:10159–10162. doi: 10.1073/pnas.90.21.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamatani T., Daikoku T., Wang H., Matsumoto H., Carter M. G., Ko M. S., Dey S. K. Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation. Proc. Natl. Acad. Sci. USA. 2004;101:10326–10331. doi: 10.1073/pnas.0402597101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aoki R., Suzuki N., Paria B. C., Sugihara K., Akama T. O., Raab G., Miyoshi M., Nadano D., Fukuda M. N. The Bysl gene product, bystin, is essential for survival of mouse embryos. FEBS Lett. 2006;580:6062–6068. doi: 10.1016/j.febslet.2006.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshima R. G., Millan J. L., Cecena G. Comparison of mouse and human keratin 18: a component of intermediate filaments expressed prior to implantation. Differentiation. 1986;33:61–68. doi: 10.1111/j.1432-0436.1986.tb00411.x. [DOI] [PubMed] [Google Scholar]
- 32.Watson K. L., Konrad K. D., Woods D. F., Bryant P. J. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc. Natl. Acad. Sci. USA. 1992;89:11302–11306. doi: 10.1073/pnas.89.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin H., Spradling A. C. Germline stem cell division and egg chamber development in transplanted Drosophila germaria. Dev. Biol. 1993;159:140–152. doi: 10.1006/dbio.1993.1228. [DOI] [PubMed] [Google Scholar]
- 34.Schafer T., Strauss D., Petfalski E., Tollervey D., Hurt E. The path from nucleolar 90S to cytoplasmic 40S preribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schafer T., Maco B., Petfalski E., Tollervey D., Bottcher B., Aebi U., Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- 36.Perou C. M., Sorlie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Rees C. A., Pollack J. R., Ross D. T., Johnsen H., Akslen L. A., Fluge O., Pergamenschikov A., Williams C., Zhu S. X., Lonning P. E., Borresen-Dale A. L., Brown P. O., Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 37.Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R. C., Melton D. A. 'stemness': transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 38.Nomura M. Assembly of bacterial ribosomes. Science. 1973;179:864–873. doi: 10.1126/science.179.4076.864. [DOI] [PubMed] [Google Scholar]
- 39.Warner J. R. Nascent ribosomes. Cell. 2001;107:133–136. doi: 10.1016/S0092-8674(01)00531-1. [DOI] [PubMed] [Google Scholar]
- 40.Dez C., Tollervey D. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 2004;7:631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi N., Yanagida M., Fujiyama S., Hayano T., Isobe T. Proteomic snapshot analyses of preribosomal ribonucleoprotein complexes formed at various stages of ribosome biogenesis in yeast and mammalian cells. Mass Spectrom. Rev. 2003;22:287–317. doi: 10.1002/mas.10057. [DOI] [PubMed] [Google Scholar]
- 42.Jansen R. P., Hurt E. C., Kern H., Lehtonen H., Carmo-Fonseca M., Lapeyre B., Tollervey D. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J. Cell Biol. 1991;113:715–729. doi: 10.1083/jcb.113.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zemp I., Kutay U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 2007;581:2783–2793. doi: 10.1016/j.febslet.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 44.Kasugai Y., Tagawa H., Kameoka Y., Morishima Y., Nakamura S., Seto M. Identification of CCND3 and BYSL as candidate targets for the 6p21 amplification in diffuse large B-cell lymphoma. Clin. Cancer Res. 2005;11:8265–8272. doi: 10.1158/1078-0432.CCR-05-1028. [DOI] [PubMed] [Google Scholar]
- 45.Gerbi S. A., Borovjagin A. V. Pre-ribosomal RNA processing in multicellular organisms. In: Olson M. O.J., editor. The Nucleolus. New York: Kluwer Academic; 2004. pp. 170–198. [Google Scholar]
- 46.Aitchison J. D., Rout M. P. The road to ribosomes: filling potholes in the export pathway. J. Cell Biol. 2000;151:F23–F26. doi: 10.1083/jcb.151.5.F23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lam Y.W., Lamond A. I., Mann M., Andersen J. S. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ayala G. E., Dai H., Li R., Ittmann M., Thompson T. C., Rowley D., Wheeler T. M. Bystin in perineural invasion of prostate cancer. Prostate. 2006;66:266–272. doi: 10.1002/pros.20323. [DOI] [PubMed] [Google Scholar]
- 49.Sandqvist A., Sistonen L. Nuclear stress granules: the awakening of a sleeping beauty? J. Cell Biol. 2004;164:15–17. doi: 10.1083/jcb.200311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersen J. S., Lyon C. E., Fox A. H., Leung A. K., Lam Y.W., Steen H., Mann M., Lamond A. I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/S0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 51.Gavin A. C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Hofert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 52.Ruggero D., Pandolfi P. P. Does the ribosome translate cancer? Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 53.Pandolfi P. P. Aberrant mRNA translation in cancer pathogenesis: an old concept revisited comes finally of age. Oncogene. 2004;23:3134–3137. doi: 10.1038/sj.onc.1207618. [DOI] [PubMed] [Google Scholar]
- 54.Champney W. S. Bacterial ribosomal subunit assembly is an antibiotic target. Curr. Top. Med. Chem. 2003;3:929–947. doi: 10.2174/1568026033452186. [DOI] [PubMed] [Google Scholar]
- 55.Basso K., Margolin A. A., Stolovitzky G., Klein U., Dalla-Favera R., Califano A. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 56.Hadjiolova K. V., Nicoloso M., Mazan S., Hadjiolov A. A., Bachellerie J. P. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. J. Biochem. 1993;212:211–215. doi: 10.1111/j.1432-1033.1993.tb17652.x. [DOI] [PubMed] [Google Scholar]


