Abstract
Fisher [1925] was the first to suggest a method of combining the p-values obtained from several statistics and many other methods have been proposed since then. However, there is no agreement about what is the best method. Motivated by a situation that now often arises in genetic epidemiology, we consider the problem when it is possible to define a simple alternative hypothesis of interest for which the expected effect size of each test statistic is known and we determine the most powerful test for this simple alternative hypothesis. Based on the proposed method, we show that information about the effect sizes can be used to obtain the best weights for Liptak’s method of combining p-values. We present extensive simulation results comparing methods of combining p-values and illustrate for a real example in genetic epidemiology how information about effect sizes can be deduced.
Keywords: Fisher, Liptak, effect size
1. INTRODUCTION
Since the first approach proposed by Fisher1, several other approaches2–5 have been suggested for combining p-values. Combining p-values is usually required in one of two situations: (1) when either the values of the actual statistics that need to be combined or the forms of their distributions are unknown, or (2) this information is available, but the distributions are such that there is no known or reasonably convenient method available for constructing a single overall test6. In addition, in practical situations, combining p-values gives the statistician flexibility to weight the individual statistics according to how informative they are and allows the designs of complex experiments to be determined independently of each other.
Combining p-values has usually been used for multi-stage analyses, in which inferences are pooled using the same statistic from different samples. However, another situation has recently arisen in genetic epidemiology, which we here call multi-phase analysis. Multi-phase analysis is the process of drawing similar inferences using different statistics calculated from the same sample7–9. The null hypothesis, stated in genetic terms, is simply that a particular genomic region is not associated with the presence of disease. If this hypothesis is rejected, there is reason to seek a causal mechanism experimentally, for example using cell lines or an animal model. There are two different types of multi-phase analysis: the independent (or predictor) variables can be biologically either the same or different, and correspondingly the statistical tests will be quite different or of the same form. For example, in genetic epidemiology, association between a marker locus and a disease can be confirmed by differences between cases and controls either in marker allele frequencies or in parameters for Hardy-Weinberg disequilibrium7, 9; in this case the same genetic marker can be the biological predictor, but the two statistics that test for association are different in form, each testing a different aspect of the distribution of marker genotypes (i.e. we have different statistics for the same biological predictor marker). Alternatively, several different genetic markers near a disease locus may be associated with the disease of interest and we perform tests of allele frequency difference between cases and controls for the alleles at each of the marker loci7, 10–14; in this case each marker locus is a different biological predictor (i.e. we use the same type of statistic to test for association with each of the marker loci). The importance of both kinds of multi-phase analysis is related to power, because power can be improved by combining the p-values of the different tests. The methods for multi-phase analysis in genetic epidemiology, for example, have so far not considered the expected genetic effects, 7–9, 13, 14 even though the optimal method of combining p-values depends on the magnitude of the genetic effects to be expected, and theoretical investigations on detecting genetic association has shed light on the genetic effect size expected under alternative hypotheses 15, 16. Thus multi-phase analysis should be performed using this information which, because it can be determined a priori, allows us to choose the most powerful method for combining the p-values that these tests produce.
fter Fisher introduced his χ2-based method, Pearson suggested an approach that has a similar, but different, rejection function. Let Uj be the p-value resulting from the j-th of P independent statistics. Whereas Fisher’s method rejects the null hypothesis if and only if U1·U2· ⋯ UP ≤ c, Pearson’s method rejects it if and only if (1 – U1)·(1 – U2)⋯(1 – UP) ≥ c, where in each case c is a predetermined constant corresponding to the desired overall significance level. Wilkinson5 suggested a method in which the null hypothesis is rejected if and only if U j ≤ c for r or more of the Uj, where r is a predetermined integer, 1 ≤ r ≤ P. The approaches of Fisher and Pearson were also generalized by using the inverse of a cumulative normal distribution, and extended by Liptak to allow each test to have different weights wj, where , using the combined statistic . This, if Φ is the cumulative standard normal distribution, follows the standard normal distribution under the null hypothesis3, 17. Either the inverse of the standard error or the square root of the sample size has been suggested for the weight wj, but we shall see that neither of these may be appropriate. Goods18 suggested another function for weighting p-values, (1 – Uj) is the (1 – Uj)-th quantile of the chi-square distribution with kj degrees of freedom (DF). Lancaster19 suggested for when the k-th test has kj DF. Koziol20 showed the asymptotic equivalence of Lancaster’s and Liptak’s tests when . Furthermore, there have been extensions to allow for the statistics to be correlated 13, 21–23. However, so far little work has been done to find the most powerful (MP) way of combining p-values and this is now becoming of increasing interest.
For a method of combining p-values to be optimal, the method needs to be uniformly MP (UMP). However, it has been shown that a UMP test does not exist because the MP test is different according to the situation6. In view of this, admissibility – which is satisfied by many of the methods and is always preserved in the UMP method – can be considered as a minimum requirement for the method to be valid. For a method of combining p-values to be admissible, if the null hypothesis is rejected for any given set of p-values uj, then it must also be rejected for all sets of νj such that νj ≤ uj6. Also, even though Fisher’s method was shown to be approximately most efficient in terms of Bahadur relative efficiency24, Naik25 and Zaykin et al.13, 14 found that Wilkinson’s5 method can be better than Fisher’s. Here we shall show that an admissible MP test can be found for a particular situation that occurs in practice, even though there is no UMP test.
In many practical situations, the parameter spaces for both the null (H0) and alternative (H1) hypotheses can be considered simple, because the effect size is naturally assumed to be zero under the null hypothesis and there is an expected effect, or at least a minimum magnitude of effect that we would wish to detect, under the alternative hypothesis. In this situation, when the alternative hypothesis is simple, it can be shown that there is a MP test. Here we derive the MP test for a simple alternative hypothesis when we can specify this expected effect size for each alternative, and also an approximation to this test if only their ratios are available. We compare the method we derive for this situation with the previously suggested methods and show that it has optimal power as long as the prerequisites are satisfied. In section 2 we show theoretically that the most powerful method for combining p-values can be approximately achieved with information about the effect sizes; and that the parameters that are needed for existing methods of combining p-values, such as the weights in Liptak’s method, should be chosen using the expected effect sizes. In section 3 we give detailed simulation results comparing the various methods in different situations, and illustrate how the information about effect sizes can be deduced for a particular type of genetic association analysis. Finally, in Section 4, we discuss extensions, including the case of correlated tests, and suggest a general strategy for combining p-values.
2. MOST POWERFUL REJECTION REGION
Suppose we want to combine the p-values from P tests. Let be the null and alternative hypotheses for each test, respectively. The null and alternative hypotheses for combining the p-values are
If we restrict the parameter space for the alternative hypothesis to the simple case, then the alternative is
where it should be noted that some of the can be in because the alternative hypothesis is that at least one of the is rejected. As usual, the rejection region for any test, ϕ, for combining p-values should be admissible, i.e. if H0 is rejected for any given set of Uj = uj, then it will also be rejected for all sets of νj such that νj ≤ uj for each j6 However, ϕ is different from the usual hypothesis testing paradigm for a single parameter because, if we let the p-values from each test be U1, U2, … and UP and they are independent, the density function of U1, U2, … and UP under H0 must be 1. Then the Neyman-Pearson lemma results in the following, if we let ϕ = 1 when H0 is rejected and ϕ = 0 otherwise, and let fA(U1, … , UP), Tj and Aj(Uj) be respectively the density function of U1, U2, … and UP under H1 , the statistics for Uj, and a region that results in the p-value Uj for Tj:
Given α, where 0 ≤ α ≤ 1, there exists k such that
where independence of the statistics implies that and fA,j(Uj) is . It should be noted that fA,j(Uj) is 1 if . Admissibility requires that fA,j(Uj) be a monotonic decreasing function of Uj for all j, by the following lemma:
If we let be the rejection region of ϕ at the significance level α, is admissible if all the fA,j(Uj) are positive monotonic decreasing functions of Uj. The proof of this lemma is trivial. As a result, by the Neyman-Pearson lemma, as long as fA,j(Uj) is a monotonic decreasing function of Uj we can find an optimal test.
In general, we can define the functions P(Tj ∈ Aj(Uj); θj) and fA,j(Uj) as functions of the quantiles of the distribution of p-values. For example, if Tj is a statistic for a two-tail test that estimates a difference in means, sej is its standard error, and the sample size is large enough, then, for some positive real constant c and denoting the cumulative standard normal distribution Φ,
where Sj is the standardized expected difference in means (i.e. the expected difference in means under H1, divided by sej - or sêj if sej is unknown), and ZU is the U- th quantile of the standard normal distribution. For a one-tail test, fA,j(Uj)=c′exp(tjZ1-UjSj), where tj is 1 for testing positive effects and −1 for testing negative effects. Thus, the test ϕ for two-tail tests becomes, by the Neyman-Pearson lemma,
If the Tj include both one-tail and two-tail tests, ϕ is a product of both types of fA,j(Uj). These results can be applied in a practical situation using the following Monte-Carlo algorithm, provided we have information about the expected effect sizes.
algorithm 1:
Generate z 1, … , and z p from a standardized normal distribution.
Confirm whether gA(z1, … , zp) is larger than gA(Z1-U1/2, … , Z1-Up/2)(gA(Z1-U1, … , Z1-Up) if the Tj are one-side tests) and, if it is larger, add 1 to C.
After N iterations, the p-value C/N, where gA(z1, … , zP) = Πj gA,j(zj), gA,j(zj) = exp(zj·Sj) + exp(−zj·Sj) for a two-tail test and gA,j(zj) = exp(zj·Sj) (gA,j(zj) = exp(−zj·Sj)) for a one-tail test of positive (negative) effect. This approach can be shown to be the same as Liptak’s method when p-values from one-tail tests are combined if Sj is used for wj, because
where , tj is 1 for testing a positive effect and −1 for testing a negative effect, and z′ ~ N(0,1). In addition, if only ratios of the expected differences are available, we have the following approximation:
This can also be implemented using a Monte-Carlo algorithm, as follows.
algorithm 2:
Generate z 1, … , and z p from a standardized normal distribution.
Confirm whether and, if it is larger, add 1 to C.
After N iterations, the p-value C/N, Thus, the rejection region that results in the predefined significance level is similar to that of Liptak’s method if Liptak’s method uses as weight the standardized effect size instead of the square root of the sample size or the inverse of the standard error.
Also, with a slight modification, the method can be applied to statistics with other distributions, such as chi-square or F distributions. For example, if Tj follows a chi-square distribution with kj DF, then
where Sj is the non-centrality parameter for Tj, and Ia(·) and Γ(·) are respectively a modified Bessel function of the first kind and a gamma function.
However, the Monte-Carlo algorithm usually requires intensive computation, which can be alleviated as follows when all the statistics are normally distributed. The combined p-value for the p-values (U1, …, UP) is the hyper-volume of the region where gA(z1,…zp) is larger than gA(Z1-U1/2,…,Z1-Up/2) and, for any z2,…,zP that are between 0 and 1, if ν1 satisfies the following inequality, then gA(z1,…zp) > gA(Z1-U1/2,…,Z1-Up/2):
where . Thus, the calculation only involves being able to calculate the cumulative normal distribution and numerical integration over the above region.
3. COMPARISON WITH PREVIOUS METHODS
3.1. Rejection regions
Several approaches to combine p-values from independent tests have been suggested but their power has not been compared. Because algorithm 1 always results in the most powerful rejection region, we compare it with the following previously suggested approaches for P = 2 only:
Minimum p-value method: reject H 0 if and only if min (U 1,U 2)<c
Cutoff-based method: reject H 0 if and only if U 1 < c 1 and U 2 < c 2
Pearson's method : reject H 0 if and only if (1–U 1)(1–U 2)≥c
Fisher's Method : reject H 0 if and only if U 1 U 2≤c
Liptak's method : reject H 0 if and only if where Zw~N(0,1).
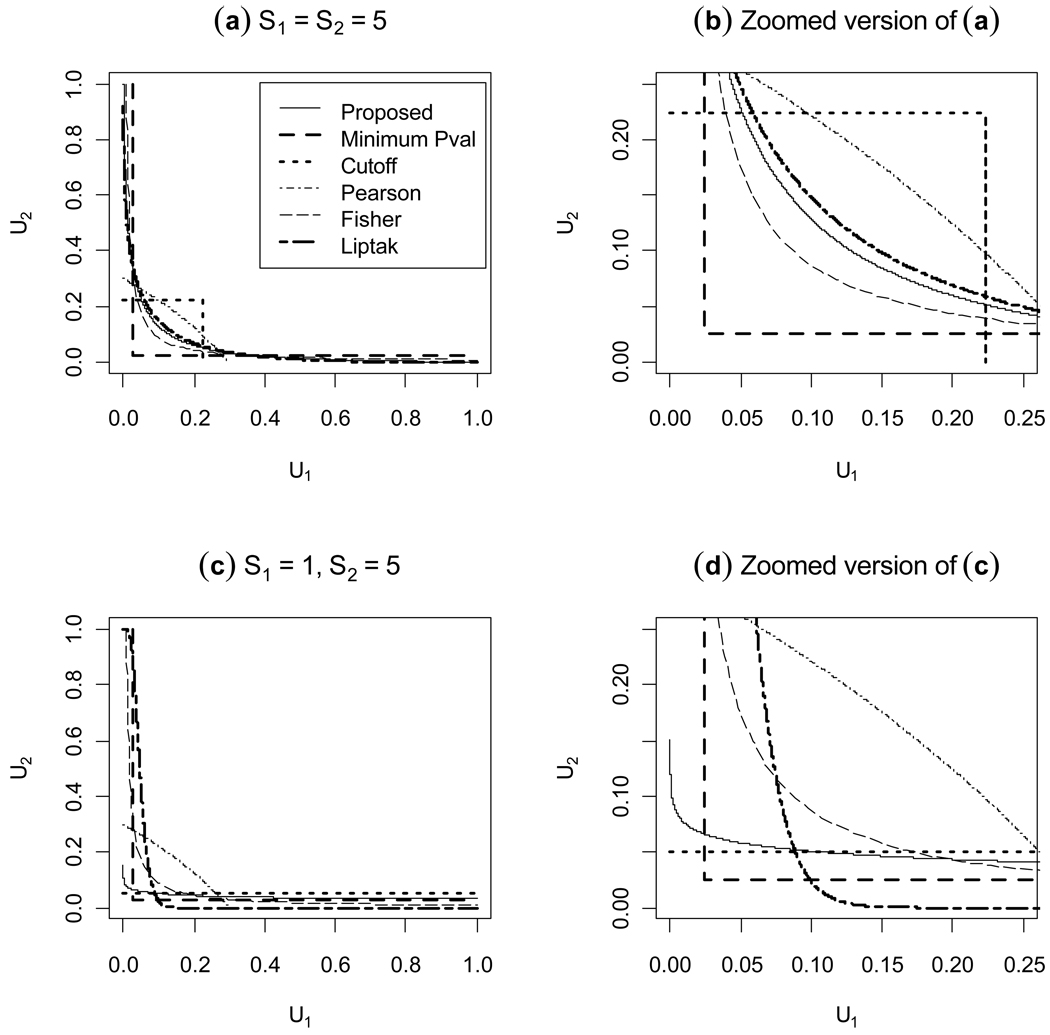
In each case c is determined by the desired value of α. Figure 1 shows results for two different cases: S1 = S2 = 5 for (a) and (b), and S1 = 1, S2= 5 for (c) and (d). First, when S1 and S2 are equal, the MP region is expected to be symmetric and this is seen to be the case for the rejection region of our proposed method. Also, the MP region is fairly similar to the region given by Liptak’s method using S1 and S2 as weights in this case, and Liptak’s approach is second best. Investigation shows that which method is second best depends on the size of S1 and S2; if it is less than about 3, Fisher’s method is better than Liptak’s method, but otherwise Liptak’s is better. Second, when S1 and S2 are unequal, the MP region is expected to be asymmetric and our results confirm this. For Liptak’s method, we used the weights because the ratio between S1 and S2 is 1:5. The plot (c) and (d) in Figure 1 shows that the cutoff-based approach with c1=1 and c2=0.05 is the closest to the MP region, though Liptak’s rejection region is very close to the MP region. Here the results indicate that using a more powerful statistic alone, T2 in this case, is better than combining the two together using Liptak’s method.
Figure 1. Rejection regions for five methods of combining two P-values at the 0.05 significance level.
Two different cases are considered: the standardized effect sizes under H1 are equal (a) and unequal (c). Because the proposed method is the most powerful, we can compare the methods that have been suggested by comparing the closeness of their rejection regions to that of our proposed method.
3.2. Simulation when the standardized expected difference is known
We applied our approach to the mean difference test of two samples when the standardized expected difference is known. For two different test statistics, we generated Xi1, Xi2, Yk1 and Yk2 (i = 1, 2, … , N1 and k = 1, … , N2) as follows:
where μX and μY are either 0.1 or 0.02, εil and εil′ (l = 1, 2) independently follow normal distribution with mean 0 and variance 0.5. The t-statistic approximately follows a normal distribution if the sample size is large enough. Thus, we obtain the following function for our suggested approach:
where .
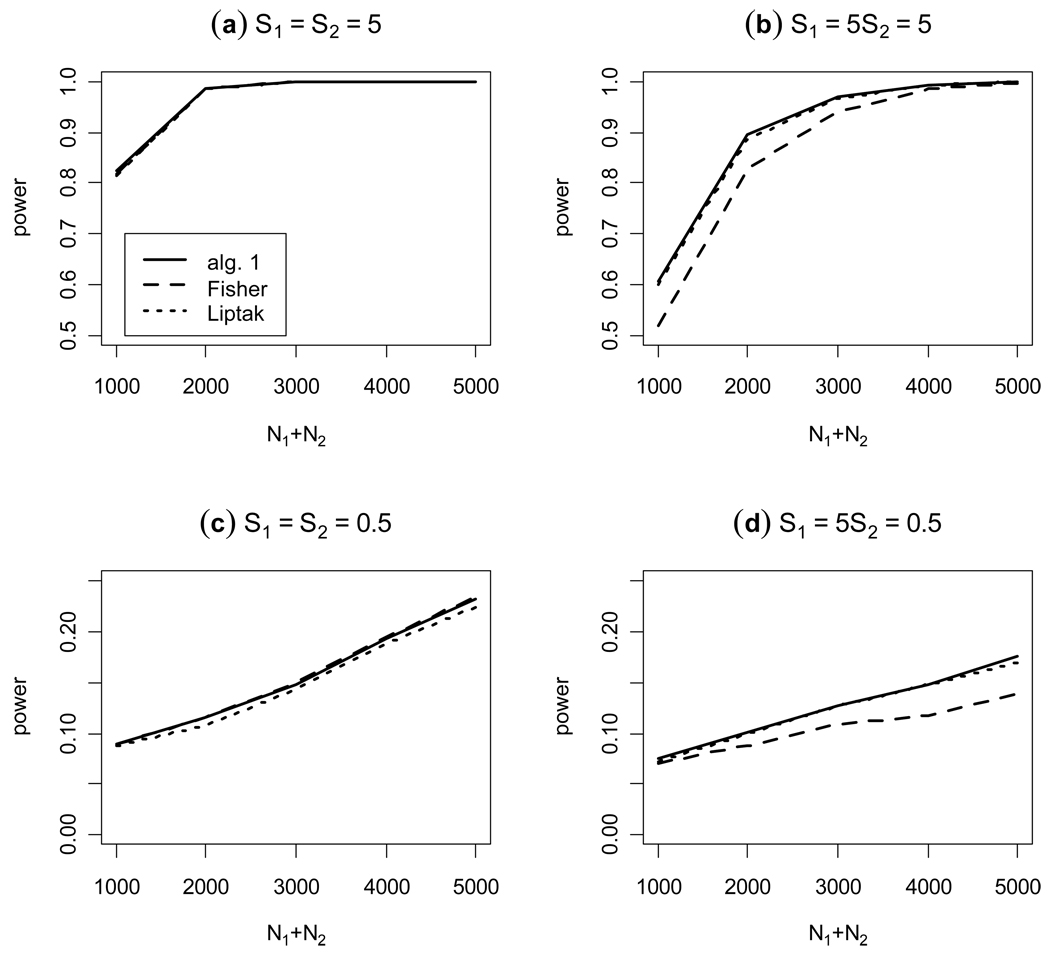
Figure 2 shows the power results of a simulation with various sample sizes for four different cases and they are calculated from 5000 replicate samples: (a) μX = μY = 0.1 and N1 = N2, (b) μX = μY = 0.1 and N1 = 5N2, (c) μX = μY = 0.02 and N1 = N2 and (d) μX = μY = 0.02 and N1 = 5N2. In each case we compare our proposed method with Fisher’s and Liptak’s methods. First, even though the three approaches have similar power when S1 = S2, the proposed method has the best empirical power, followed by Liptak’s method and then by Fisher’s for case (a), but followed by Fisher’s method and then by Liptak’s for case (c). As we mentioned before, Liptak’s method is better than Fisher’s if the standardized expected effect size is larger than about 3, and otherwise Fisher’s is better; the range of Sj considered for the simulation is from 2.2 to 5 for case (a) and from 0.4 to 1 for case (c). Second, if S1 and S2 are different (we considered the ratio 5:1), Fisher’s is the worst method and Liptak’s method using S1 and S2 as weights is approximately equal to our proposed method.
Figure 2. Empirical power as a function of the sum of the sample sizes for two p-values.
The empirical power at the 0.01 significance level is calculated, in each case for total sample sizes 1000, 2000, 3000, 4000, and 5000, obtained from a simulation of 5000 replicate samples, as a function of the sum of the sample sizes; for algorithm 1, 100,000 Monte Carlo replicates were used. Two different cases are considered: the standardized effect sizes under H1 are equal, (a) and (c), and unequal (b) and (d).
3.3. Simulation for a multi-stage analysis with estimated standard error
As mentioned above, methods for combining p-values can be used for both multi-stage analysis and multi-phase analysis. We applied our approach to testing the mean difference between two samples for multi-stage analysis, i.e. pooling the results of using the same statistic from different samples, when the standard deviation is unknown. In general, because multi-stage analyses (including meta-analysis) are usually for the same hypothesis, we can assume the same expected difference under the alternative hypothesis but the sample sizes or variances could be different. We therefore applied our approach to test a mean difference from simulated samples with different sample sizes and with different variances, using numerical integration for the proposed method of combing the p-values. For two different test statistics, we generated Xi1, Xi2, Yk1 and Yk2 (i = 1, 2, … , N1 and k = 1, … , N2) as follows:
where εil and εil′ (l = 1, 2) are independent. Because the t-statistic approximately follows a normal distribution if the sample size is large enough, we obtain the following functions:
where S1 = μ′ /sê1, S2 = μ′/sê2, and μ′ is the expected difference under the alternative hypothesis. It should be noted that in this case wj depends only on the standard error. We also applied Fisher’s method and Liptak’s method using as weights the inverse of the standard error (Liptak1) and the square root of the sample size (Liptak2).
Table 1 shows the empirical type I error from 10,000 replicate samples as a function of N2 when we assume and N1 is 1000. μ and μ′ are assumed to be 0 and 0.05 respectively. The results show that all methods preserve type I error well at the significance level 0.05. Table 2 and Table 3 show the empirical power based on 5,000 replicate samples when equal variances but different sample sizes are assumed, and when equal sample sizes but different variances are assumed. In both cases, μ and μ′ are equal to 0.05. For Table 2, we assume that and N2 = 600, 700, 800, …, 1900 and 2000 while N1 is fixed at 1000, and for Table 3, N1=N2=1000 and is 1. In Table 2, algorithm 1 shows the best result, followed by algorithm 2. Fisher’s method is better than the Liptak methods when N2 is similar to N1 but otherwise the Liptak methods are better. Liptak 1 and Litpak 2 have similar empirical power because the weights for both are similar. In Table 3, we find similar results except that Liptak 1 is much better than Liptak 2 because the weights for Liptak 2 are not close to the standardized expected differences. Thus, we conclude that algorithm 1 is always the most powerful when we have information about the expected differences, but no information about the variances, and the same power can be approximately achieved with only their ratios using algorithm 2.
Table 1. Empirical type I error for multi-stage analysis.
The empirical type I error at the significance level 0.05 is calculated from 10,000 replicate samples, in each case for N2=600, 200, 300, … , 1900 and 2000. For algorithm 1, μ′ is assumed to be 0.05 for Sj, and for simulating Xil and Ykl μ is assumed to be 0. Liptak’s method uses as weights the inverse of the standard error (Liptak1) and the square root of the sample size (Liptak2); for algorithms 1 and 2, numerical integration was used.
| N2 | Fisher | Liptak1 | Liptak2 | alg. 1 | alg. 2 |
|---|---|---|---|---|---|
| 600 | 0.052 | 0.0524 | 0.0524 | 0.0526 | 0.0532 |
| 700 | 0.0531 | 0.0479 | 0.0502 | 0.0513 | 0.0518 |
| 800 | 0.0471 | 0.0507 | 0.0479 | 0.0472 | 0.0471 |
| 900 | 0.0512 | 0.0501 | 0.0502 | 0.0499 | 0.05 |
| 1000 | 0.0505 | 0.052 | 0.0522 | 0.0499 | 0.0498 |
| 1100 | 0.0528 | 0.0503 | 0.0522 | 0.0532 | 0.0526 |
| 1200 | 0.0508 | 0.0521 | 0.0502 | 0.0505 | 0.0499 |
| 1300 | 0.053 | 0.049 | 0.0518 | 0.0534 | 0.0529 |
| 1400 | 0.0507 | 0.0499 | 0.049 | 0.0498 | 0.0503 |
| 1500 | 0.0505 | 0.0521 | 0.0503 | 0.051 | 0.0497 |
| 1600 | 0.0542 | 0.05 | 0.052 | 0.0523 | 0.0519 |
| 1700 | 0.496 | 0.0501 | 0.0501 | 0.0514 | 0.0504 |
| 1800 | 0.473 | 0.05 | 0.0499 | 0.497 | 0.0489 |
| 1900 | 0.497 | 0.0485 | 0.0482 | 0.474 | 0.478 |
| 2000 | 0.502 | 0.0505 | 0.0508 | 0.0488 | 0.495 |
Table 2. Empirical power for multi-stage analysis for different sample sizes.
The empirical power at the significance level 0.05 is calculated from 5,000 replicate samples, for N2=600, 200, 300, … , 1900 and 2000. For algorithm 1, both μ and μ′ are assumed to be 0.05 for simulating Xil and Ykl, and Sj in algorithm 1; for algorithms 1 and 2, numerical integration was used. Liptak’s method uses as weights the inverse of the standard error (Liptak1) and the square root of the sample size (Liptak2).
| N2 | Fisher | Liptak1 | Liptak2 | alg. 1 | alg. 2 |
|---|---|---|---|---|---|
| 600 | 0.2338 | 0.2332 | 0.2316 | 0.2376 | 0.234 |
| 700 | 0.2392 | 0.235 | 0.2356 | 0.2412 | 0.2402 |
| 800 | 0.2454 | 0.2414 | 0.2418 | 0.2488 | 0.2464 |
| 900 | 0.2574 | 0.2474 | 0.2468 | 0.2574 | 0.2522 |
| 1000 | 0.2834 | 0.2792 | 0.2796 | 0.283 | 0.2822 |
| 1100 | 0.291 | 0.2872 | 0.287 | 0.2946 | 0.29 |
| 1200 | 0.3042 | 0.302 | 0.303 | 0.3078 | 0.3074 |
| 1300 | 0.3158 | 0.3146 | 0.3158 | 0.3234 | 0.3214 |
| 1400 | 0.3266 | 0.326 | 0.3262 | 0.329 | 0.328 |
| 1500 | 0.3362 | 0.3354 | 0.3354 | 0.343 | 0.3428 |
| 1600 | 0.3356 | 0.3334 | 0.3328 | 0.3468 | 0.3434 |
| 1700 | 0.3598 | 0.3592 | 0.3592 | 0.3662 | 0.3652 |
| 1800 | 0.3694 | 0.3688 | 0.3688 | 0.3824 | 0.3804 |
| 1900 | 0.3842 | 0.3876 | 0.3862 | 0.3954 | 0.3976 |
| 2000 | 0.3998 | 0.4074 | 0.4064 | 0.4144 | 0.415 |
Table 3. Empirical power for multi-stage analysis with different variances.
The empirical power at the significance level 0.05 is calculated from 5,000 replicate samples, for and 10. For algorithm 1, both μ and μ′ are assumed to be 0.05 for simulating Xil and Ykl, and Sj in algorithm 1; for algorithms 1 and 2, numerical integration was used. Liptak’s method uses as weights the inverse of the standard error (Liptak1) and the square root of the sample size (Liptak2).
| Fisher | Liptak1 | Liptak2 | alg. 1 | alg. 2 | ||
|---|---|---|---|---|---|---|
| 1 | 0.2846 | 0.2716 | 0.2726 | 0.283 | 0.2798 | |
| 2 | 0.1966 | 0.195 | 0.186 | 0.2088 | 0.2016 | |
| 3 | 0.1848 | 0.1974 | 0.1824 | 0.2046 | 0.2018 | |
| 4 | 0.1778 | 0.1876 | 0.1644 | 0.2006 | 0.1924 | |
| 5 | 0.1818 | 0.2016 | 0.1676 | 0.2126 | 0.2066 | |
| 6 | 0.174 | 0.2004 | 0.1604 | 0.2058 | 0.2054 | |
| 7 | 0.1628 | 0.19 | 0.1486 | 0.1988 | 0.1926 | |
| 8 | 0.1702 | 0.189 | 0.152 | 0.1932 | 0.192 | |
| 9 | 0.1716 | 0.201 | 0.1584 | 0.2092 | 0.2068 | |
| 10 | 0.1738 | 0.2062 | 0.154 | 0.2134 | 0.2096 |
3.4. Simulation for multi-phase analysis with estimated standard error
We also applied our approach to the mean difference test of two samples for multi-phase analysis when the standard deviation is unknown. Here we assume that the expected differences are known either from previous studies or on the basis of theoretical results that allow us to know their ratios, as can occur in genetic epidemiology 15,16 . Again for two different test statistics, we generated Xi1, Xi2, Yk1 and Yk2 (i = 1, 2, … , N1 and k = 1, … , N2) as follows:
where εil and εil′ (l = 1, 2) independently follow normal distributions with mean 0 and variance 1. For large sample sizes, we obtain the following function for our proposed approach:
where S1 = μX′/sê1, S2 = μY′/sê2, wj = Sj, and μX′ and μY′ are the expected differences under the alternative hypothesis. For Liptak’s method, we again used as weights these wj (Liptak1) and the inverse of the standard error (Liptak2). For both algorithms, the combined p-values were calculated using numerical integration.
Table 4 shows the results of a simulation with 10,000 replicate samples when N1=N2=1,000 and . For empirical type I error, μX and μY are assumed to be 0 for simulating Xil and Yk, and the empirical type I errors calculated at the nominal 0.05 significance level. For S1 and S2, μX′ is assumed to be 0.05 and we consider 0.01, 0.02, … and 0.15 for μY′. The results show that both algorithm 1 and algorithm 2 preserve the type I error well, as in multi-stage analysis. Table 5 shows the results of a simulation with 5,000 replicate samples at the 0.05 significance level when N1=N2=1,000 and . For empirical power, μX and μY were assumed equal to μX′ and μY′, respectively, μX was assumed to be 0.05 and we considered 0.01, 0.02, …, 0.15 for μY. The results show that algorithm 1 generally has the best power, followed by algorithm 2. Also, the proposed algorithms are always better than the Liptak methods and Fisher’s method, though the difference is not large. Finally, Liptak’s method is better than Fisher’s method if the Sj are used as weights and μX and μY are not similar, which confirms first that Fisher’s method is usually good when the standardized expected differences are similar and small, and second that Liptak’s method should use the standardized expected differences as weights, as in algorithm 2.
Table 4. Empirical type I error for multi-phase analysis.
The empirical type I error at the significance level 0.05 is calculated from 10,000 replicate samples for multi-phase analysis, for μY′ = 0.01, 0.02, 0.03, … , 0.14 and 0.15. Both μX and μY are assumed to be 0 for simulating Xil and Ykl, and μX′ is equal to 0.05 for algorithm 1. Liptak’s method uses as weights the standardized expected difference with the standard error estimated (Liptak1) and the inverse of the standard error (Liptak2); for algorithms 1 and 2, numerical integration was used.
| μY′ | Fisher | Liptak1 | Liptak2 | alg. 1 | alg. 2 |
|---|---|---|---|---|---|
| 0.01 | 0.0512 | 0.0515 | 0.052 | 0.0514 | 0.0531 |
| 0.02 | 0.0489 | 0.0497 | 0.0481 | 0.0496 | 0.0508 |
| 0.03 | 0.0505 | 0.0501 | 0.0512 | 0.0498 | 0.0496 |
| 0.04 | 0.0541 | 0.0545 | 0.0551 | 0.0555 | 0.0539 |
| 0.05 | 0.0495 | 0.0485 | 0.0483 | 0.0483 | 0.0479 |
| 0.06 | 0.0483 | 0.0477 | 0.0481 | 0.0476 | 0.0483 |
| 0.07 | 0.047 | 0.0461 | 0.046 | 0.0469 | 0.0466 |
| 0.08 | 0.0483 | 0.0494 | 0.0502 | 0.0495 | 0.0501 |
| 0.09 | 0.0504 | 0.0473 | 0.0503 | 0.0483 | 0.048 |
| 0.10 | 0.0477 | 0.048 | 0.0492 | 0.0481 | 0.0476 |
| 0.11 | 0.0498 | 0.0516 | 0.0512 | 0.0513 | 0.0524 |
| 0.12 | 0.0482 | 0.0471 | 0.0466 | 0.049 | 0.0467 |
| 0.13 | 0.0467 | 0.0462 | 0.0486 | 0.0481 | 0.0476 |
| 0.14 | 0.0543 | 0.0522 | 0.0534 | 0.0518 | 0.0519 |
| 0.15 | 0.0524 | 0.0523 | 0.0508 | 0.0516 | 0.0521 |
Table 5. Empirical power for multi-phase analysis.
The empirical power at the significance level 0.05 is calculated from 5,000 replicate samples for μY′=0.01, 0.02, 0.03, … , 0.14 and 0.15. Both μX and μX′ are assumed to be 0.05 and μY′ is equal to μY for algorithm 1; for algorithme 1 and 2, numerical integration was used. Liptak’s method uses as weights the standardized expected differences with the standard error estimated (Liptak1) and the inverse of the standard error (Liptak2).
| μY′ | Fisher | Liptak1 | Liptak2 | alg. 1 | alg. 2 |
|---|---|---|---|---|---|
| 0.01 | 0.157 | 0.201 | 0.141 | 0.202 | 0.203 |
| 0.02 | 0.166 | 0.187 | 0.158 | 0.198 | 0.189 |
| 0.03 | 0.203 | 0.207 | 0.200 | 0.211 | 0.209 |
| 0.04 | 0.233 | 0.230 | 0.227 | 0.242 | 0.236 |
| 0.05 | 0.275 | 0.264 | 0.266 | 0.273 | 0.270 |
| 0.06 | 0.331 | 0.328 | 0.322 | 0.340 | 0.334 |
| 0.07 | 0.384 | 0.386 | 0.372 | 0.395 | 0.390 |
| 0.08 | 0.469 | 0.475 | 0.444 | 0.493 | 0.489 |
| 0.09 | 0.535 | 0.559 | 0.506 | 0.575 | 0.570 |
| 0.10 | 0.602 | 0.637 | 0.565 | 0.648 | 0.646 |
| 0.11 | 0.671 | 0.703 | 0.612 | 0.719 | 0.717 |
| 0.12 | 0.736 | 0.772 | 0.673 | 0.786 | 0.781 |
| 0.13 | 0.796 | 0.838 | 0.717 | 0.847 | 0.846 |
| 0.14 | 0.841 | 0.876 | 0.768 | 0.882 | 0.881 |
| 0.15 | 0.893 | 0.920 | 0.817 | 0.923 | 0.951 |
3.5 A genetic multi-phase example
The simulation results in sections 3.3 and 3.4 demonstrate the increase in power possible, but do not illustrate exactly how the effect sizes could be obtained in practice, nor do they examine the sensitivity of the method to assumptions made about those effect sizes. We illustrate this here for one particular genetic example of multi-phase analysis.
In genetic epidemiology, the Cochran-Armitage (CA) trend is usually used for association analysis in a case-control design assuming the two alleles of a diallelic marker act in an additive manner on disease susceptibility26. The test for Hardy Weinberg proportions (HWP) in cases has been combined with the CA test in an attempt to improve the statistical power for association analysis by using either Fisher’s method7 or a cutoff-based method9, which was called self-replication. However, combining these two tests sometimes leads to a reduction in power11. We now show that if we use the expected effect size information the power is improved with either algorithm 2 or Liptak’s method, and explain why, without using this information, combining these two tests sometimes fails to increase power.
For a disease locus D with disease allele D1 and normal allele D2, let PDk|case and PDk|cont be the frequencies of allele Dk in the case group and control group, respectively, and let PDkDk'|case and PDkDk'|cont be the analogous frequencies of the genotype DkDk'. As a measure of Hardy Weinberg disequilibrium (HWD) in cases, let dA|case ≡ PA1A1|case – (PA1|case)2. If we let ϕ and ϕl be the disease prevalence and the penetrance of disease genotype l at the disease location, the expected sizes of these quantities are16, 27:
If the disease genotype effect is small, we can assume that ϕD2D2/ϕ ≈ 1 and then, for a rare disease with known mode of inheritance we have
and , where λ1 is the heterozygous disease genotype relative risk and λ2 is the homozygous disease genotype relative risk (i.e. relative to the homozygous genotype containing no disease predisposing allele). Thus, for a recessive disease we have
Also, we have (PD1|case – PD1|cont):dD|case ≈ 1: λ2PD1 ≈ 1:PD1 and dD|case = 0 respectively, for dominant and multiplicative modes of inheritance. Because additive and multiplicative disease inheritance have similar λ1, we can conclude that the HWP test in cases is non-informative for additive and multiplicative diseases, but can improve the CA test for dominant and recessive diseases, with the above expected ratios of effect sizes.
Figure 3 and Figure 4 show the empirical power from 10,000 replicate samples at the significance level 0.05 when the disease modes of inheritance are dominant and recessive, respectively. We assume the disease allele frequency is 0.2, and the numbers of cases and controls are equal. For the weights, w1 and w2, in algorithm 2 and Liptak’s method, we used
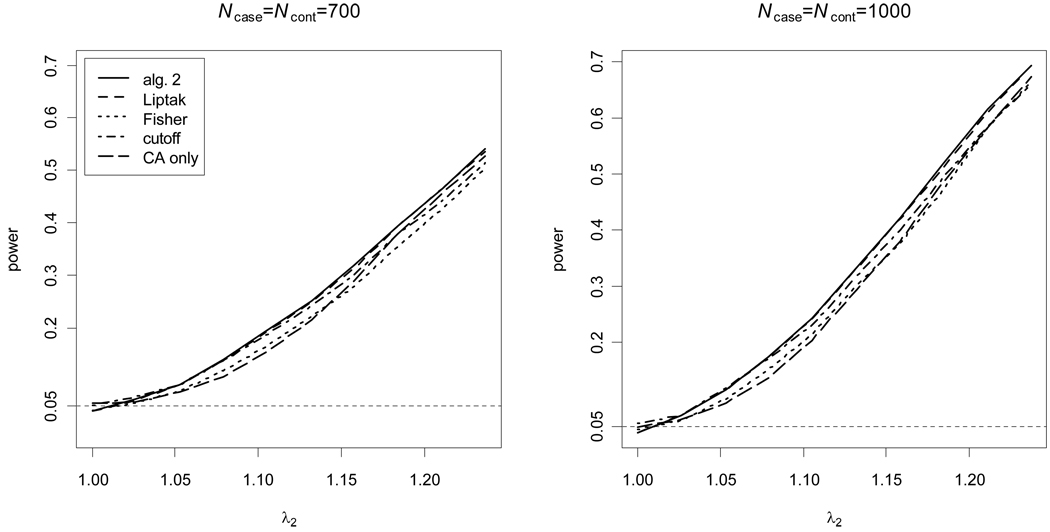
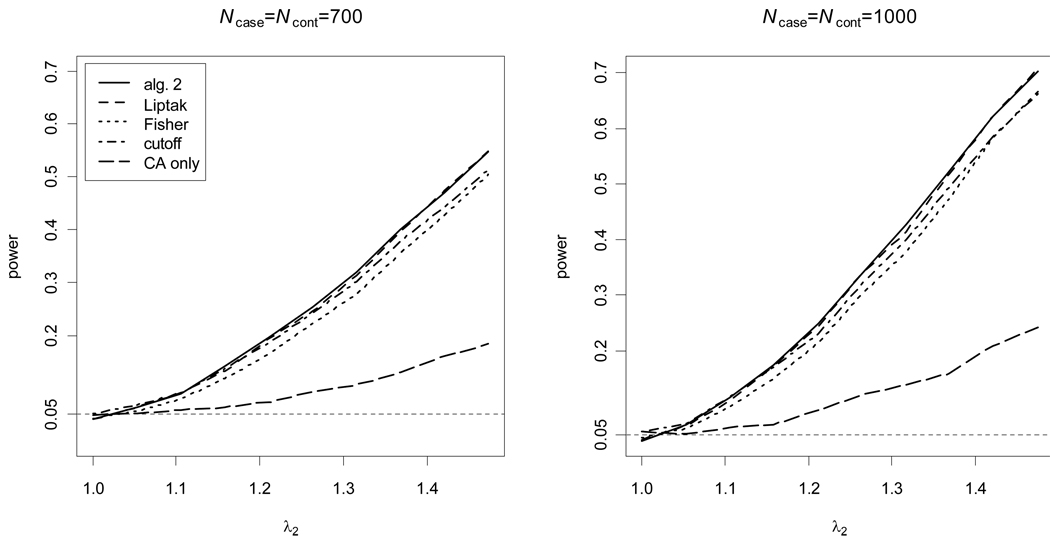
divided by their estimated standard errors; and algorithm 1 is not considered because only ratios of the effect sizes are available. For the cutoff-based method, the value of c1 that results in the maximal empirical power (among the values 0.05, 0.1, … , 0.95) was used. A one-tail test was applied for the HWP test because it is known, if the disease-predisposing allele is the less common allele, that dD|case > 0 for a recessive disease and dD|case < 0 for a dominant disease28. The results show that algorithm 2 and Liptak’s method are similar, because algorithm 2 is equal to Liptak’s method in the case of a one-tail test. However, the results also confirm that the other methods that have been used to combine the CA and HWP tests are not as powerful as the proposed methods. Also, it should be noted that algorithm 2 and Liptak’s method using the proposed weights work well even though the proposed weights are not the true effect sizes, and the CA and HWP tests are not strictly independent under the alternative hypothesis. In particular, the empirical power at λ2 = 1 is equivalent to the empirical type I error at significance level 0.05, and it is seen that both algorithm 2 and Liptak’s method using the proposed weights preserve this type I error.
Figure 3. Empirical power for a dominant disease.
The CA and HWP test p-values are combined using algorithm 2, Fisher’s method, Liptak’s method, and the best cutoff-based method, compared to the CA test alone. The empirical power at the significance level 0.05 is calculated from 10,000 replicate samples; for algorithm 2, 50,000 Monte Carlo replicates were used.
Figure 4. Empirical power for a recessive disease.
The CA and HWP test p-values are combined using algorithm 2, Fisher’s method, Liptak’s method, and the best cutoff-based method, compared to the CA test alone. The empirical power at the significance level 0.05 is calculated from 10,000 replicate samples; for algorithm 2, 50,000 Monte Carlo replicates were used.
4. DISCUSSION
Over the last few decades, the UMP region for combining p-values has been sought and it has been proved that there is no UMP test. Because of this, the various combination methods were compared empirically instead of by further theoretical investigation. However, all these investigations failed to find any practically MP region. Here we have shown that a MP test can be found if we specify the expected effect sizes, or it can be approximated if we only know their ratios. Also, our results show that this proposed method always has the best power, though the power may not be substantially larger than that of other methods. We have illustrated the method with a genetic example that demonstrated moderately increased power, even when the ratio of effect sizes was misspecified.
Although the proposed algorithms described here are for independent tests with underlying normal distributions, they can also be extended to other cases, such as the Tj follow different distributions or, for example, a multivariate normal distribution with correlations. If the statistic Tj for each p-value follows a different distribution, a factor gAj(·) appropriate for each Tj should be used in the Monte-Carlo algorithm, instead of only factors of the form exp(zj·Sj) + exp(−zj·Sj). In addition, when the statistics Tj for each p-value follow a multivariate normal distribution (MVN), the gA(·) for the MVN density should be used in step (2) after sampling from the standard MVN distribution with appropriate correlations between the Tj under the null hypothesis. Thus, with some slight modification the same approach can be extended to complex cases.
Though the proposed method improves power, the need to have information about the expected effect sizes could limit its application. However, hypothesis testing for combining p-values should not be understood in the same way as for testing parameters because there is no uniformly most powerful method and the statistical power can be substantially different according to the situation. Instead, it would be better to use information about effect sizes, something that is often available, especially – as we have shown – in genetic epidemiology. The proposed method suggests the following general strategy for its application in practice:
If the effect sizes are known, the proposed method using the expected differences (algorithm 1) will give the best power.
If only the ratios between the effect sizes are available, the proposed method using their ratios (algorithm 2) should be considered. To avoid excess computation, Liptak’s method using as weights ratios of the standardized effect sizes can be used when the expected differences are different.
If precise information is not available but the effect sizes are expected to be small (large), Fisher method (Liptak’s method using equal weights) should be used.
Alternatively, if we consider other methods, such as a cutoff-based method (which is sometimes called self-replication or screening 8, 9), instead of Liptak’s method or the proposed method, c1 and c2 should be determined by the ratios of the standardized effect sizes. Sometimes nothing is available about effect sizes, but the effect sizes can nevertheless be expected to be equal. For example, in a meta-analysis, we usually want to combine results from several studies for the same hypothesis. Then we suggest using inferences based on ratios between the standard errors because S1:S2=1/σ̂1:1/σ̂2. Finally, it should be remembered that Liptak’s method with appropriate weights is the same as our proposed method if the p-values are from one-tail tests.
Acknowledgements
This work was supported in part by a U.S. Public Health Research grant (GM28356) from the National Institute of General Medical Sciences, Cancer Center Support grant (P30CAD43703) from the National Cancer Institute, and Training grant (HL07567) from the National Heart, Lung and Blood Institute.
Reference List
- 1.Fisher RA. Statistical Methods for Research Workers. London: Oliver & Boyd; 1950. pp. 99–101. [Google Scholar]
- 2.George EO, Mudholkar GS. On the Convolution of Logistic Random Variables. Metrika. 1983;30:1–14. [Google Scholar]
- 3.Liptak T. On the combination of independent tests. Magyar Tud.Akad.Mat.Kutato’ Int.Ko”zl. 1958;3:171–197. [Google Scholar]
- 4.Pearson ES. The probability integral transformation for testing goodness of fit and combining independent tests of significance. Biometrika. 1938;30(1):134–148. [Google Scholar]
- 5.Wilkinson B. A statistical consideration in psychological research. Psychological Bulletin. 1951;48:156–157. doi: 10.1037/h0059111. [DOI] [PubMed] [Google Scholar]
- 6.Birnbaum A. Combining independent tests of significance. J Am Stat Assoc. 1954;49(267):559–574. [Google Scholar]
- 7.Hoh J, Wille A, Ott J. Trimming, weighting, and grouping SNPs in human case-control association studies. Genome Res. 2001;11(12):2115–2119. doi: 10.1101/gr.204001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Steen K, McQueen MB, Herbert A, Raby B, Lyon H, Demeo DL, Murphy A, Su J, Datta S, Rosenow C, Christman M, Silverman EK, Laird NM, Weiss ST, Lange C. Genomic screening and replication using the same data set in family-based association testing. Nat.Genet. 2005;37(7):683–691. doi: 10.1038/ng1582. [DOI] [PubMed] [Google Scholar]
- 9.Zheng G, Song K, Elston RC. Adaptive two-stage analysis of genetic association in case-control designs. Hum.Hered. 2007;63(3–4):175–186. doi: 10.1159/000099830. [DOI] [PubMed] [Google Scholar]
- 10.Dudbridge F, Koeleman BP. Rank truncated product of P-values, with application to genomewide association scans. Genet.Epidemiol. 2003;25(4):360–366. doi: 10.1002/gepi.10264. [DOI] [PubMed] [Google Scholar]
- 11.Hao K, Xu X, Laird N, Wang X, Xu X. Power estimation of multiple SNP association test of case-control study and application. Genet.Epidemiol. 2004;26(1):22–30. doi: 10.1002/gepi.10293. [DOI] [PubMed] [Google Scholar]
- 12.Yang HC, Lin CY, Fann CS. A sliding-window weighted linkage disequilibrium test. Genet.Epidemiol. 2006;30(6):531–545. doi: 10.1002/gepi.20165. [DOI] [PubMed] [Google Scholar]
- 13.Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS. Truncated product method for combining P-values. Genet.Epidemiol. 2002;22(2):170–185. doi: 10.1002/gepi.0042. [DOI] [PubMed] [Google Scholar]
- 14.Zaykin DV, Zhivotovsky LA, Czika W, Shao S, Wolfinger RD. Combining p-values in large-scale genomics experiments. Pharm.Stat. 2007;6(3):217–226. doi: 10.1002/pst.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66(1):279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Won S, Elston RC. The power of independent types of genetic information to detect association in a case-control study design. Genet.Epidemiol. 2008 doi: 10.1002/gepi.20341. In press. [DOI] [PubMed] [Google Scholar]
- 17.Zheng G, Milledge T, George EO, Narasimhan G. Lecture Notes in Computer Science. Springer: Berlin Heidelberg. Vol. 3992. 2006. Pooling Evidence to Identify Cell Cycle-Regulated Genes; pp. 694–701. [Google Scholar]
- 18.Goods IJ. On the Weighted Combination of Significance Tests. J R Stat Soc B. 1955;17(2):264–265. [Google Scholar]
- 19.Lancaster HO. The combination of probabilities: an application of orthonormal functions. Austral.J.Stat. 1961;3:20–33. [Google Scholar]
- 20.Koziol JA. A note on Lancaster’s procedure for the combination of independent events. Biom.J. 1996;38(6):653–660. [Google Scholar]
- 21.Delongchamp R, Lee T, Velasco C. A method for computing the overall statistical significance of a treatment effect among a group of genes. BMC.Bioinformatics. 2006;7 Suppl 2:S11. doi: 10.1186/1471-2105-7-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudbridge F, Koeleman BP. Efficient computation of significance levels for multiple associations in large studies of correlated data, including genomewide association studies. Am J Hum Genet. 2004;75(3):424–435. doi: 10.1086/423738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin DY. An efficient Monte Carlo approach to assessing statistical significance in genomic studies. Bioinformatics. 2005;21(6):781–787. doi: 10.1093/bioinformatics/bti053. [DOI] [PubMed] [Google Scholar]
- 24.Littell RC, Folks L. Asymptotic Optimality of Fisher’s method of combining independent tests. J Am Stat Assoc. 1971;66(336):802–806. [Google Scholar]
- 25.Naik UD. The equal probability tests and its applications to some simulataneous inference problems. J Am Stat Assoc. 1969;64(327):986–998. [Google Scholar]
- 26.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielson DM, Ehm MG, Weir BS. Detecting marker-disease association by testing for Hardy-Weinberg disequilibrium at a marker locus. Am J Hum Genet. 1999;63(5):1531–1540. doi: 10.1086/302114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng G, Ng HK. Genetic model selection in two-phase analysis for case-control association studies. Biostatistics. 2008;9(3):391–399. doi: 10.1093/biostatistics/kxm039. [DOI] [PMC free article] [PubMed] [Google Scholar]