Abstract
Drosomycin (Drs) encoding an inducible 44-residue antifungal peptide is clustered with six additional genes, Dro1, Dro2, Dro3, Dro4, Dro5, and Dro6, forming a multigene family on the 3L chromosome arm in Drosophila melanogaster. To get further insight into the regulation of each member of the drosomycin gene family, here we investigated gene expression patterns of this family by either microbe-free injury or microbial challenges using real time RT-PCR. The results indicated that among the seven drosomycin genes, Drs, Dro2, Dro3, Dro4, and Dro5 showed constitutive expressions. Three out of five, Dro2, Dro3, and Dro5, were able to be upregulated by simple injury. Interestingly, Drs is an only gene strongly upregulated when Drosophila was infected with microbes. In contrast to these five genes, Dro1 and Dro6 were not transcribed at all in either noninfected or infected flies. Furthermore, by 5′ rapid amplification of cDNA ends, two transcription start sites were identified in Drs and Dro2, and one in Dro3, Dro4, and Dro5. In addition, NF-κB binding sites were found in promoter regions of Drs, Dro2, Dro3, and Dro5, indicating the importance of NF-κB binding sites for the inducibility of drosomycin genes. Based on the analyses of flanking sequences of each gene in D. melanogaster and phylogenetic relationship of drosomycins in D. melanogaster species-group, we concluded that gene duplications were involved in the formation of the drosomycin gene family. The possible evolutionary fates of drosomycin genes were discussed according to the combining analysis of gene expression pattern, gene structure, and functional divergence of these genes.
1. Introduction
Antimicrobial peptides are the major humoral immune factors in the insect innate immune system. Drosophila melanogaster has emerged as a powerful model system for the study of innate immunity, especially for the antimicrobial peptides studies, because of its flexible genetics [1]. To date, seven classes of the inducible antimicrobial peptides (or peptides families), drosomycins, metchnikowin, defensin, attacins, cecropins, drosocin, and diptericins have been characterized in the D. melanogaster, each with a specific spectrum of activity [2]. Among them, defensin [3], drosocin [4, 5], and metchnikowin [6] have only one single copy in the genome of D. melanogaster. Others are encoded by multigene families.
Drosomycin (Drs) is an inducible insect antifungal peptide from D. melanogaster. It exhibits potent activity against filamentous fungi but no obvious activity to bacteria and yeast [7]. The mature peptide of Drs is consisting of 44 amino acid residues with 8 cysteine residues to form 4 intramolecular disulfide bridges [8]. It involves an α-helix and a twisted three-stranded β-sheet, that is, Cysteine-stabilized αβ motif [9]. In the genome of D. melanogaster, there are six additional mRNAs with high sequence homology to Drs gene, Dro1, Dro2, Dro3, Dro4, Dro5, and Dro6 (resp., equivalent to Drs-lC, Drs-lD, Drs-lE, Drs-lF, Drs-lG, and Drs-lI). These seven genes are clustered into a drosomycin multigene family on 3L chromosome arm [10, 11]. Similar to Drs, the predicted amino acid sequences of Dro1, Dro2, Dro3, Dro4, Dro5, and Dro6 also contain eight cysteines and four conservative residues (Ser4, Gly9, Glu26, and Gly31), which have been reported to be involved in protein structure stabilization or in the protein folding pathway [9]. To experimentally determine the antifungal function of the six isoforms, Yang et al. [11] cloned all the seven genes of the drosomycin gene family into pET 3c and expressed in Escherichia coli. All purified expression products showed different antifungal activity against tested fungal strains with the exception of Dro6 which showed no activity against all the tested fungal strains [11]. These experiments provided the evidence of the significant functional divergence in the drosomycin family.
Genes coding for antibacterial and antifungal peptides were differentially induced after injection of various classes of microorganisms [12]. In the drosomycin family, although the Drs was constitutively expressed in the larvae and adult of D. melanogaster, it was strongly upregulated by infection of fungi and Gram-positive bacteria and weakly upregulated by Gram-negative bacteria [7, 12–14]. Previous results from the genomewide analysis of the Drosophila immune response genes indicated that Dro5 (Drosomycin B), in addition to Drs, was also significantly upregulated by the immunization of fungi or mixed bacteria [15]. Recent studies have demonstrated that Dro2 and Drs were expressed in larvae, pupae, and adult, Dro3, Dro4, and Dro5 expressed in larvae and adult, whereas the transcripts of Dro1 and Dro6 were not detected in all the stages of insect development [16]. However, the expression and regulation of each member of drosomycin family in response to microbial infection and injury stimulation has not yet been fully elucidated. In the present study, we investigated the expression patterns of seven members of the drosomycin family by simple injury and the various microbial challenges in the adults of D. melanogaster. In addition, we identified the transcription start sites and predicted the cis-regulatory elements at 5′-flanking region of individual gene in the drosomycin family. Moreover, the evolutionary history of this gene family was predicted. These studies will shed light on the gene expression divergence and the functional evolution of antimicrobial peptides multigene family.
2. Materials and Methods
2.1. Drosophila Strain
Wild-type strain D. melanogaster Oregon-R was obtained from Drosophila genetic resource center, Kyoto Institute of Technology, Japan. Flies were maintained on the cornmeal-malt medium at 25°C under continuous light.
2.2. Collection of Bacteria or Fungal Spores
Bacteria strains, including Gram-negative Escherichia coli K12D31 and Pseudomonas aeruginosa and Gram-positive Staphylococcus aureus and Bacillus subtilis, were grown in Luria broth (LB) medium at 37°C to an OD600 of approximate 0.6. The cells were harvested by centrifugation at 5000 g for 5 minutes and resuspended in sterile Ringer's solution at volume of 1/10.
Fungi strains including Fusarium oxysporum, Fusarium culmorum (W. G. Smith), Beauveria bassiana (vuill), Botrytis cinerea, Penicillium digitatum, and Aspergillus niger were grown on potato dextrose agar (PDA) medium. Spores from agar plates were resuspended in sterile Ringer's solution. After filtering through 8 layers of sterile cheese-cloth, the number of spores was counted under a light microscopy, and the final concentration of the spore solution was adjusted into 109 cells per milliliter.
2.3. Immunization of Flies
Two-day-old adult flies were anesthetized with ethyl ether, then individually pricked into the thorax with a thin needle which was previously dipped into either bacterial culture (OD600 ≈ 6) or fungal spore suspension (109 cells/mL). The immunized flies were kept on the cornmeal-malt medium at 28°C. The survival flies at the different times of infection were immediately frozen in liquid Nitrogen and stored at −70°C until RNA extraction.
2.4. RNA Preparation, Regular RT-PCR, and Quantitative Real Time RT-PCR
The immunized flies were ground to a fine powder under liquid nitrogen. Total RNA was extracted with the Trizol reagents (Invitrogen, Carlsbad, USA) according to the manufacturer's instructions, and contaminating genomic DNA was removed by incubation with DNase I (Takara, Dalian, China). RNA (1 μg) was reverse-transcribed using the ReverTraAce reverse transcription kit (Toyobo, Tokyo, Japan), and the resulting first strand cDNA was used as a template for RT-PCR with primers specific to drosomycin genes (Table 1). PCR conditions were 94°C, 1 minute; 94°C, 30 seconds; 55°C, 30 seconds and 72°C, 30 seconds for 35 cycles; and 72°C for 7 minutes. RT-PCR fragments were cloned to pTA2 vector with a TA cloning kit (Toyobo, Tokyo, Japan) for sequencing to check the specificity of the primer pairs.
Table 1.
Primers for RT-PCR and real-time PCR.
| Genes | Primer sequence | Product size (bp) |
|---|---|---|
| Drs | forward: 5′ CCCTCTTCGCTGTCCTGA 3′ | 137 |
| reverse: 5′ GCGTCCCTCCTCCTTGC 3′ | ||
| Dro1 | forward: 5′ TGTCCGCTGTCTTGATG 3′ | 135 |
| reverse: 5′ TTCGCCCTTCCCTCT 3′ | ||
| Dro2 | forward: 5′ TCAAATTCCTTTTCGTCTT 3′ | 130 |
| reverse: 5′ CGTCGGCACATCTCGT 3′ | ||
| Dro3 | forward: 5′ GCACACTGTTTTGGCACG 3′ | 80 |
| reverse: 5′ GGCGGCACTTTTCTCC 3′ | ||
| Dro4 | forward: 5′ ATGGCTCAAATTAAAGGATT 3′ | 148 |
| reverse: 5′ AGAGGCGACGGCACT 3′ | ||
| Dro5 | forward: 5′ ACCTCTTCCTGGCTGT 3′ | 87 |
| reverse: 5′ AGGGTCCTCCGTATCT 3′ | ||
| Dro6 | forward: 5′ TGTTCACCTTCCTCGCTCTG 3′ | 156 |
| reverse: 5′ CACTCACTCGTCCTCGTCCC 3′ | ||
| rp49 | forward: 5′ CGTTTACTGCGGCGAGAT 3′ | 102 |
| reverse: 5′ CCGTTGGGGTTGGTGAG 3′ |
To quantify the expression levels of drosomycin genes, fluorescence real-time PCR was performed using an SYBR Green methodology (Applied Biosystems) under the following conditions: 25 μL reaction mixture contained 12.5 μL SYBR Green Real-time PCR Master Mix, 0.5 μmol/L of forward primer, 0.5 μmol/L of reverse primer, and 0.5 μL of cDNA (corresponding to 25 ng of total RNA) template. The real time RT-PCR was run with the 96-well plate on ABI Prism 7300 Real Time PCR system (Applied Biosystems, Foster City, USA) with the program of 95°C for 1 minute, 40 cycles of 95°C for 15 seconds, 55°C for 15 seconds, and 72°C for 45 seconds. The ribosome protein rp49 gene was used as an internal control. All samples were analyzed in triplicate and normalized against rp49 gene.
2.5. Rapid Amplification of 5′ cDNA Ends (5′RACE)
Total RNA was isolated from adult Drosophila infected with S. aureus. The transcription start site was determined using the 5′ RACE kit (Invitrogen, Carlsbad, USA) according to the manufacturer's instructions. Briefly, first strand cDNA was generated from total RNAs with GSP1, a common primer annealed to all the 7 drosomycin mRNAs. After reverse transcription, the cDNAs were purified using columns supplied in 5′ RACE kit. The resulting purified cDNAs were then oligo-dC-tailed at its 3′ end by TdT (Terminal deoxynucleotidyl transferase). PCR products were amplified from dC-tailed cDNAs using an abridged anchor primer (AAP) and a nested gene-specific primer GSP2 (Table 2). To obtain specific RACE products of Dro2, Dro3, and Dro4, diluted primary PCR products were reamplified by using an abridged universal amplification primer (AUAP) and a nested gene-specific primer GSP3 (Table 2). The PCR products were purified by E. N. Z. A. Cycle-pure kit (Omega, USA) and subsequently cloned into a pTA2 vector (Toyobo, Tokyo, Japan). About 20 to 30 positive clones were selected for sequencing using T7 or M13R primers.
Table 2.
Primers for 5′ RACE reactions. The GSP1 sequence was corresponded to positions 190 ~ 208 in the ORFs of Dro1 and Dro5, 193 ~ 211 in Dro2 and Drs, 196 ~ 214 in Dro3 and Dro4, and 199 ~ 208 in Dro6.
| Primers | 5′ to 3′ sequence |
|---|---|
| GSP1 | AGCATCCTTCGCACCAGCA |
| Dro2-GSP2 | AAATACGTCGGCACATCTCG |
| Dro2-GSP3 | CGGCATCAGCCATATTGGCGG |
| Dro3-GSP2 | GACGGCGGCACTTTTCTCC |
| Dro3-GSP3 | CAATCACGTGCCAAAACAGT |
| Dro4-GSP2 | CCTCCCTGCAGAGGCGACG |
| Dro4-GSP3 | CCAGATGGGCAATCCACGGC |
| Dro5-GSP2 | CGCAGGGTCCTCCGTATCT |
| Drs-GSP2 | CAGCATCAGGACAGCGAAGA |
2.6. Prediction of Promoters and Regulatory Elements
The 5′ flanking sequences of the 7 drosomycin genes were loaded to predict the core promoter sequences by NNPP version 2.2 [17] (http://www.fruitfly.org/seq_tools/promoter.html). Binding sites of regulatory factors were analyzed by pDRAW32 software (http://www.acaclone.com/) within the 1 kb 5′-flanking region of drosomycin genes. The putative regulatory elements involved in the immune response were NF-κB/Rel sites with consensus sequence of GGGRAYYYYY in Drosophila [18], GATA sites with WGATAR [19], IL6-RE (interleukin-6 response element) sites with TKNNGNAAK [20], and ICRE (interferon consensus element) sites with GGAAANN [21].
2.7. Evolutionary Analysis of the Drosomycin Gene Family
The 5′ and 3′-flanking sequences of drosomycin genes in D. melanogaster were obtained from the Flybase (http://www.flybase.org). A Blast search of the open reading frames (ORFs) corresponding to drosomycins in D. melanogaster was applied to identify the drosomycin genes in 11 Drosophila species other than D. melanogaster at Flybase and UCSC genome websites (http://genome.ucsc.edu/cgi-bin/hgBlat).
Phylogenetic analysis of ORFs sequences of drosomycins in Drosophila species was performed using the Neighbor-Joining method with 1000 bootstrap replicates, as implemented by the Mega 4 programme [22]. Gaps were pairwise deleted and the Maximum Composite Likelihood model was applied to estimate the branch length.
To predict the gene duplication events of drosomycin gene family, repetitive sequences of the flanking regions were identified by RepeatMasker (http://www.repeatmasker.org/). Transposable elements were recognized by BLAST in D. melanogaster transposable element database at Flybase.
3. Results
3.1. Transcriptional Profile of the Drosomycin Gene Family
To detect the primary transcriptional profiles of 7 members of the drosomycin gene family, we used the regular RT-PCR with specific primers (Table 1) to identify the transcriptional products from adult flies after challenging to either bacteria or fungi as indicated in Section 2. The RT-PCR bands were amplified with primers specific to Dro2, Dro3, Dro4, Dro5, and Drs when templates of both the immunized and the native control flies were used. In contrast, no visible RT-PCR bands were observed with primers specific to Dro1 and Dro6 for all the tested templates (data not shown). These results suggested that the expressions of Dro2, Dro3, Dro4, Dro5, and Drs were constitutive, whereas Dro1 and Dro6 were not transcribed at all even in the adult flies that challenged to microbial infections. We cloned and sequenced the RT-PCR fragments including PCR products of Dro1 and Dro6 using the genomic DNA as template, confirming that all primers were gene-specific.
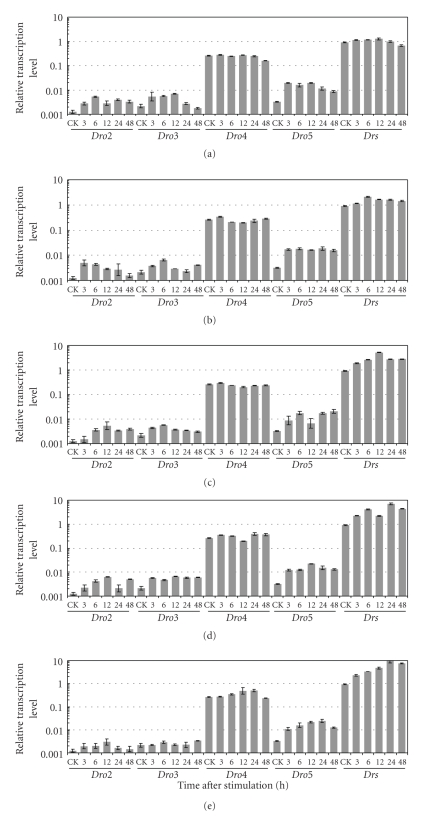
We next examined the time-course expression of Dro2, Dro3, Dro4, Dro5, and Drs in adult flies infected by fungi (F. oxysporum or B. bassiana) or bacteria (E. coli K12D31 or S. aureus) for 3 to 48 hours. The native flies and microbial-free pricked flies were used as controls. The survival rate of microbial-free pricked flies and infected flies with various microorganisms was shown in Supplementary Figure 1. The results of the quantitative real time RT-PCR revealed that (i) in native flies, the transcription activity of Dro2 was the weakest among five genes, only 1/1000 of control rp49. Dro3 and Dro5 expressions were 2 ~ 3-folds higher than that of Dro2. The expression of Dro4 showed even higher, about 1/4 of that of rp49. The transcription activity of Drs was the highest, reaching an mRNA levels similar to rp49 (Figure 1(a)), equivalent to about 80% of the total transcription of all the drosomycin genes. (ii) In the simple injured flies, with the microbe-free pricking treatment, the transcription activity of Dro2, Dro3, and Dro5, but not Dro4 and Drs, was significantly upregulated (Figure 1(a)). (iii) In the microorganism-pricked flies, the transcription activity of Drs was significantly upregulated under stimulation by Gram-positive bacteria and fungi (Figures 1(c)–1(e)) but only increased about 1.5-folds after infection of Gram-negative bacteria (Figure 1(b)), compared to the control flies, suggesting that Gram-negative bacterium was a weak inducer. Taking all these results together, we concluded that of the 7 members, only Drs was greatly triggered by microbial infection. In other words, the microorganisms did not induce the expression of Dro2, Dro3, and Dro5 above the level of simple injury and did not induce Dro4 above the constitutional expression level.
Figure 1.
The transcription activity of drosomycin gene family in D. melanogaster stimulated by microbe-free injury and various microorganisms. Quantitative real time RT-PCR was performed for the detection of the expression levels of drosomycin genes in wild-type Oregon-R flies with SYBR Green. The expression levels of drosomycin genes were normalized to rp49 in the samples. The experiments were done in triplicate and the error bars represented standard deviation. CK: native flies. (a) The 2-day-old adult flies were pricked by a sterile needle (microbe-free injury). (b) to (e) The 2-day-old adult flies were infected with E. coli K12D31, Staphylococcus aureus, Fusarium oxysporum, and Beauveria bassiana, respectively.
3.2. Analysis of the Promoter Regions of Drosomycin Genes
Firstly, we analyzed the promoter sequences of the drosomycin genes for the prediction of core promoter sequence by NNPP software. The results showed that almost all of the drosomycin genes contained one or two typical core promoter sequences within 150 bp upstream of the ORFs with the exception of Dro1. The absence of a core promoter sequence in the Dro1 may explain why this gene has no transcription activity.
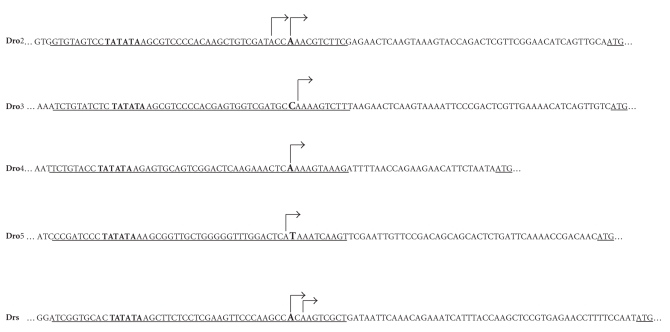
Secondly, we determined the position of transcription start sites of drosomycin genes by 5′ RACE. We initially synthesized the cDNA by primer GSP1 (Table 2) and then amplified these specific 5′ RACE products with the primers GSP2 and AAP in PCR reaction, respectively. A single 5′ RACE band about 200 bp for Dro5 and 120 bp for Drs was obtained by one turn of PCR reaction (Figures 2 (d), 2 (e)). In the second round of PCR reaction, a single specific 5′ RACE band for Dro2, Dro3, and Dro4 was obtained with the primers GSP3 and AUAP, respectively (Figures 2 (a)–2 (c)). The 5′ RACE products were subsequently purified and cloned into a T-vector for sequencing. DNA sequence analysis revealed that Dro3, Dro4, or Dro5 contain one transcription start site, which located at −55A (1 bp downstream of the predicted TSS by NNPP), −36A (same to the predicted site), and −54A (1bp upstream of the predicted TSS) of the genome of D. melanogaster, respectively. Two transcription start sites were identified from the 5′ RACE sequences of Drs and Dro2. The transcription of Dro2 initiated at −55A or −58A, and Drs initiated at −59A or −61A (Figure 3).
Figure 2.
Detection of 5′ RACE products of drosomycin genes. The fragments were visualized by staining with ethidium bromide after separation in 2% agarose gel. M. DNA marker. Both lanes 1 and 2 were the 5′ RACE PCR products.
Figure 3.
Identification of transcription start sites of drosomycin genes by 5′ RACE. The underlined ATG indicated the start of open reading frames (ORFs). Arrows indicated the transcription start sites and transcriptional directions. The putative core promoters predicted by NNPP software were underlined. And the predicated transcription start sites were indicated in larger fonts. The TATA boxes in the promoter were shown in bold.
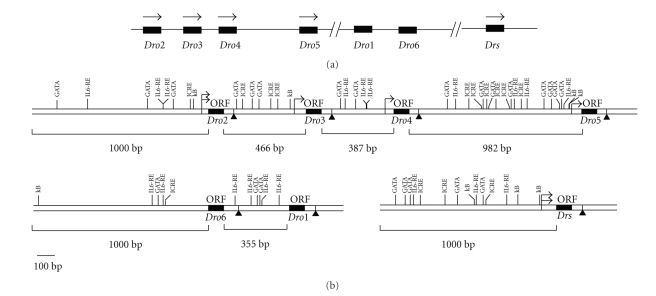
Thirdly, we scanned the potential binding sites for the regulatory factors, such as NF-κB/Rel, GATA, IL-6RE, and ICRE, by pDRAW32 software, as these DNA motifs were known to be important for regulation of antimicrobial gene expression. As indicated in Figure 4, GATA sites were found in the promoter regions of all the drosomycin genes. In addition, IL6-RE sites were identified in most of the drosomycin genes with the exception of Dro3. The putative ICRE sites were also present in most of the drosomycin genes except Dro1 and Dro4. However, several ICRE sites would be identified at the 5′-flanking region of Dro1 and Dro4 when sequence GAAANN [23] was used instead of GGAAANN [21] for the prediction. It was noteworthy to point out that there was one NF-κB site in the promoter region of Dro2 and Dro3. Dro5 contained two overlapping sequences of NF-κB sites and Drs contained three NF-κB sites. Interestingly, Dro2, Dro3, Dro5, and Drs contained at least one NF-κB sites near the transcription start site(s). However, no NF-κB site was found in Dro4 and Dro1. A putative NF-κB site was identified at around 1 kb upstream from the translation start site of Dro6, much farther than the general distance of functional NF-κB sites as previously reported [24]. The presence of the potential cis-regulatory modules in the drosomycin genes suggested that they might be involved in the regulation of the differential expression pattern of drosomycin genes. NF-κB site, in particular, was important for the inducibility of drosomycin genes.
Figure 4.
The schematic organization of the transcription factor binding sites upstream drosomycin genes in D. melanogaster. (a) General position of drosomycin genes on the 3L chromosome. (b) Organization of binding sites for putative transcription factors at the upstream region of drosomycin genes. κB: NF-κB site, GGGRAYYYYY of consensus sequence (there was no sites with perfectly matched sequence of GGGRAYYYYY. Sites with one nucleotide mismatch were given in the figure); GATA: binding site of the GATA factor, WGATAR of consensus sequence; IL-6 RE: sites for interleukin-6 response element with TKNNGNAAK of consensus sequence; ICRE: sites for interferon consensus element with GGAAANN of consensus sequence. Black boxes indicated ORFs of drosomycin genes. Arrows indicated transcription start sites (TSSs) and arrows with double heads represented double TSSs. Bolded triangle (▲) indicated polyadenylation signal.
3.3. The Evolutionary History of Drosomycin Gene Family
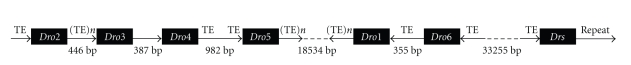
We searched the drosomycins homologs from the 12 sequenced Drosophila genomes by reciprocal Blast with seven members of drosomycins in D. melanogaster. From the Blast results, we found that homologs were present only in the Drosophila melanogaster species-group of Sophophora subgenus: 4 homologs in D. ananassae of ananassae species-subgroup, 6 homologs in the D. sechellia and D. yakuba, and 7 homologs in D. simulans and D. erecta of melanogaster species-subgroup. In melanogaster species-subgroup, Dro3 was lost in D. yakuba and Dro4 was lost in D. sechelia. To predict the possibility of gene duplication involving in the formation of the drosomycin family, we investigated the TEs and repetitive sequences in the flanking regions of each drosomycin gene in D. melanogaster. As shown in Figure 5, there are TEs in flanking regions of each gene, except the 387 bp region between Dro3 and Dro4 and the 3′-flanking region of Drs. There is a repetitive sequence in the 3′-flanking region of Drs (Figure 5). These results suggested that gene duplications took place in the formation of drosomycin family. On the basis of Nei's birth-and-death model of evolution that some duplicate genes are maintained in the genome for a long time but others are deleted or become nonfunctional by deleterious mutations [25], the loss of Dro3 and Dro4 and the existence of other genes in D. yakuba and D. sechelia suggested that the drosomycin family underwent the birth-and-death model of evolution in melanogaster species-subgroup.
Figure 5.
The distribution of TEs and the repetitive sequence in the flanking regions of drosomycin family in D. melanogaster. ORFs of drosomycins were indicated by black squares and the transcription directions were indicated by arrows. Number below the arrows indicated the intergenic length. The location of TEs and repetitive sequence was marked above the arrows. (TE)n meant that there was more than one TE in the region.
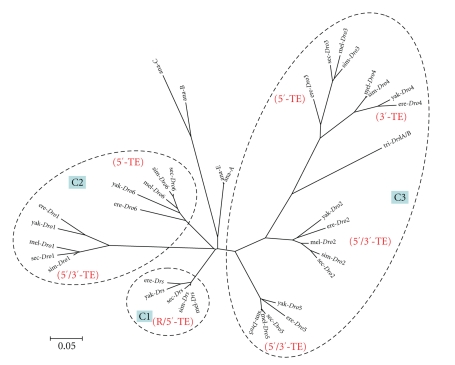
The phylogenetic relationship as described in the Figure 6 indicated that D. ananassae species-subgroup might share the common drosomycin ancestor with D. melanogaster before the speciation event which led to two separate species-subgroups. After splitting of two species-subgroups, the gene duplications happened and from the drosomycin common ancestor to 4 genes in D. ananassae; from the drosomycin common ancestor to 6-7 paralogs in each species of melanogaster species-subgroup, and the gene duplications took place before the speciation events of the melanogaster species-subgroup.
Figure 6.
Neighbor-Joining tree of drosomycin gene family in Drosophila species. The tree was reconstructed by the NJ method implemented in Mega 4 with 1000 bootstrap replicates. Gaps were pairwise deleted, and the Maximum Composite Likelihood model was applied to estimate the branch length. Dot cycles with the light blue green square enclosed genes in the same cluster: the C1 is the cluster 1 at the 63D2 of 3L; C2 and C3 are the cluster 2 and cluster 3 at the 63D1 of 3L. TEs and repetitive sequence in D. melanogaster were shown in red beside each branch: R indicated the repetitive sequence, and 5′-TE was the TE at the 5′-flanking region, 3′-TE was the TE at the 3′-flanking region, and 5′/3′-TE was TEs at both flanking regions. Abbreviation for species: mel: D. melanogaster; sim: D. simulans; sec: D. sechellia; yak: D. yakuba; ere: D. erecta; ana: D. ananassae; tri: D. triauraria.
4. Discussion
The drosomycin gene family in D. melanogaster comprises seven genes. For such a multigene family, expression patterns of individual genes are extremely difficult to be analyzed because of the high degree of sequence identity. In the present study, we used the real time RT-PCR with designed specific primers to uncover the transcriptional pattern of each gene after microbial challenges and physical stimulation. We provide evidence that the drosomycin genes may play a critical role in response not only to microbial infections but also to physical injury in the adult stage. Of particular interest, Drs was the only gene that could be significantly upregulated by microbial challenges, especially by fungi and Gram-positive bacteria (Figures 1(b)–1(e)). Further promoter analysis revealed that there were two transcription start sites and three putative NF-κB sites in 5′-flanking region of the ORF of Drs gene. Such motifs should be important for response following microbial infections. In contrast to Drs, the expressions of Dro2, Dro3, and Dro5 appeared to be upregulated by simple injury but not by microbial infection (Figure 1). As a matter of fact, the NF-κB site was also found in 5′-flanking region of Dro2, Dro3, and Dro5, and we propose that the physical injury may also activate a signal transduction pathway that is related to the Rel family. Lacking of NF-κB site in Dro4 may explain why this gene with high level of constitutive expression is noninducible by both microbe-free injury and microbial infection. Moreover, the gene expression of Dro1 and Dro6 could not be detected in either control flies or microbe-free injured or microbial infected flies. The analysis of promoter sequence indicated that NF-κB site and transcription start sites were not found at the upstream of Dro1 and Dro6.
Genomic analyses of model organisms have shown that over one-third of all protein-coding genes belong to multigene family [26, 27] originating from the gene duplication [28]. In Drosophila genomes, genes can be duplicated via retroposition and DNA-based duplication [28, 29]. In D. melanogaster, most of genes coding for AMPs, for example, cecropins [30], attacins [31], diptericins [32], lysozymes [33], and drosomycins [11], were known to be clustered into multigene families. In the drosomycin gene family of D. melanogaster, one repetitive sequence at the 3′-flanking region of Drs and TEs in flanking regions of most genes (Figure 5) suggested that gene duplications were involved in the formation of drosomycin multigene family. The gene duplication might occur during 12.8–44.2 million years ago, because species within the melanogaster species-subgroup diverged 12.8 million years ago, and the melanogaster species-subgroup and the ananassae species-subgroup separated from each other 44.2 million years ago [34]. At this time, the ancestor gene evolved into the Drs at the 63D2 of 3L in the D. melanogaster, and then gene duplication led to form the cluster 2 and cluster 3 in the 63D1 of 3L. The gene arrangement of cluster 3 indicated that the intergenic length of these genes were very small that from 387 bp to 982 bp, the tandem gene duplication might give rise to these genes.
The evolutionary fates of these duplication genes were very different. In the ORF of Dro1, 10% of DNA sequences in the populations of D. melanogaster mutated resulting in the loss of a disulphide bridge; 5% of them had an internal stop codon [10]. The expression product generated from Dro6 gene with no antifungal activity could be due to the insert of two amino acid residues [11]. From the above evidences, we propose that Dro1 and Dro6 of cluster 2 have evolved as the pseudogenes. This is one more example of nonfunctionalization of duplicate immune genes. The similar event of nonfunctionalization of duplicative genes also happened to cecropin and attacin gene family in D. melanogaster [29, 32]. For the Dro2, Dro3, Dro4, and Dro5 of cluster 3, their expression product displayed antifungal activity against 3–5 strains of tested fungi [11]. Therefore, we proposed that the fate of Dro2, Dro3, Dro4, and Dro5 was of subfunctionilization.
Neofunctionilization is another fate of duplication gene [33]. The Bmglv2-4, which duplicated from the ancestor gene Bmglv1 in the gloverin gene family of silkworm (Bombyx mori.), gained a new function in the embryonic stage due to an intron loss [34]. Does the high level of constitutive transcription of Dro4 identified in present study suggest a new function beyond the immune defense? Or are the high expression levels of Drs and Dro4 in the native flies just for battling pathogens? How other genes in the drosomycin gene family perform the synergistic effect with the predominate gene Drs to combat with the invading pathogens or exoteric stimulations? These questions are worth further investigation.
Supplementary Material
Supplementary Figure 1 shows the percent survival of adult flies following injury and infection with the microorganisms used in this study.
Acknowledgments
This work was supported by Grants of the 973 National Basic Research Program of China (no. 2005CB121000), the 863 National Important Project of China (no. 2006AA10A119), and the project of transformation fund for achievement of science and technology of agriculture (2007GB2E000240).
Abbreviations
- AAP:
Abridged anchor primer
- AMPs:
Antimicrobial peptides
- AUAP:
Abridged universal amplification primer
- Drs:
Drosomycin
- GSP:
Gene-specific primer
- ICRE:
Interferon consensus element
- IL6-RE:
Interleukin-6 response element
- LB:
Luria broth
- ORF:
Open reading frame
- PDA:
Potato dextrose agar
- rp49:
Ribosome protein 49
- RACE:
Rapid amplification of cDNA ends
- RT-PCR:
Reverse transcription-polymerase chain reaction
- TdT:
Terminal deoxynucleotidyl transferase
- TSS:
Transcription start site.
References
- 1.Irving P, Troxler L, Heuer TS, et al. A genome-wide analysis of immune responses in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(26):15119–15124. doi: 10.1073/pnas.261573998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426(6962):33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 3.Dimarcq JL, Hoffmann D, Meister M, et al. Characterization and transcriptional profiles of a Drosophila gene encoding an insect defensin. A study in insect immunity. European Journal of Biochemistry. 1994;221(1):201–209. doi: 10.1111/j.1432-1033.1994.tb18730.x. [DOI] [PubMed] [Google Scholar]
- 4.Bulet P, Dimarcq J-L, Hetru C, et al. A novel inducible antibacterial peptide of Drosophila carries an O-glycosylated substitution. The Journal of Biological Chemistry. 1993;268(20):14893–14897. [PubMed] [Google Scholar]
- 5.Charlet M, Lagueux M, Reichhart JM, Hoffmann D, Braun A, Meister M. Cloning of the gene encoding the antibacterial peptide drosocin involved in Drosophila immunity: expression studies during the immune response. European Journal of Biochemistry. 1996;241(3):699–706. doi: 10.1111/j.1432-1033.1996.00699.x. [DOI] [PubMed] [Google Scholar]
- 6.Levashina EA, Ohresser S, Bulet P, Reichhart JM, Hetru C, Hoffman JA. Metchnikowin, a novel immune-inducible proline-rich peptide from Drosophila with antibacterial and antifungal properties. European Journal of Biochemistry. 1995;233(2):694–700. doi: 10.1111/j.1432-1033.1995.694_2.x. [DOI] [PubMed] [Google Scholar]
- 7.Fehlbaum P, Bulet P, Michaut L, et al. Insect immunity: septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. The Journal of Biological Chemistry. 1994;269(52):33159–33163. [PubMed] [Google Scholar]
- 8.Michaut L, Fehlbaum P, Moniatte M, Van Dorsselaer A, Reichhart J-M, Bulet P. Determination of the disulfide array of the first inducible antifungal peptide from insects: drosomycin from Drosophila melanogaster. FEBS Letters. 1996;395(1):6–10. doi: 10.1016/0014-5793(96)00992-1. [DOI] [PubMed] [Google Scholar]
- 9.Landon C, Sodano P, Hetru C, Hoffmann J, Ptak M. Solution structure of drosomycin, the first inducible antifungal protein from insects. Protein Science. 1997;6(9):1878–1884. doi: 10.1002/pro.5560060908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiggins FM, Kim KW. The evolution of antifungal peptides in Drosophila. Genetics. 2005;171(4):1847–1859. doi: 10.1534/genetics.105.045435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang WY, Wen SY, Huang YD, et al. Functional divergence of six isoforms of antifungal peptide Drosomycin in Drosophila melanogaster. Gene. 2006;379:26–32. doi: 10.1016/j.gene.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(26):14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann JA, Bulet P. Differential display of peptides induced during the immune response of Drosophila: a matrix-assisted laser desorption ionization time-of-flight mass spectrometry study. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(19):11342–11347. doi: 10.1073/pnas.95.19.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy F, Rabel D, Charlet M, Bulet P, Hoffmann JA, Ehret-Sabatier L. Peptidomic and proteomic analyses of the systemic immune response of Drosophila. Biochimie. 2004;86(9-10):607–616. doi: 10.1016/j.biochi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 15.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(22):12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian C, Gao B, Rodriguez MdC, Lanz-Mendoza H, Ma B, Zhu S. Gene expression, antiparasitic activity, and functional evolution of the drosomycin family. Molecular Immunology. 2008;45(15):3909–3916. doi: 10.1016/j.molimm.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 17.Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Computers and Chemistry. 2001;26(1):51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 18.Hultmark D. Immune reactions in Drosophila and other insects: a model for innate immunity. Trends in Genetics. 1993;9(5):178–183. doi: 10.1016/0168-9525(93)90165-e. [DOI] [PubMed] [Google Scholar]
- 19.Kadalayil L, Petersen UM, Engström Y. Adjacent GATA and κB-like motifs regulate the expression of a Drosophila immune gene. Nucleic Acids Research. 1997;25(6):1233–1239. doi: 10.1093/nar/25.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X, Nguyen T, Kimbrell DA. Identification and characterization of the Cecropin antibacterial protein gene locus in Drosophila virilis. Journal of Molecular Evolution. 1997;44(3):272–281. doi: 10.1007/pl00006144. [DOI] [PubMed] [Google Scholar]
- 21.Levashina EA, Ohresser S, Lemaitre B, Imler J-L. Two distinct pathways can control expression of the gene encoding the Drosophila antimicrobial peptide metchnikowin. Journal of Molecular Biology. 1998;278(3):515–527. doi: 10.1006/jmbi.1998.1705. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 23.Dushay MS, Roethele JB, Chaverri JM, et al. Two attacin antibacterial genes of Drosophila melanogaster. Gene. 2000;246(1-2):49–57. doi: 10.1016/s0378-1119(00)00041-x. [DOI] [PubMed] [Google Scholar]
- 24.Senger K, Armstrong GW, Rowell WJ, Kwan JM, Markstein M, Levine M. Immunity regulatory DNAs share common organizational features in Drosophila. Molecular Cell. 2004;13(1):19–32. doi: 10.1016/s1097-2765(03)00500-8. [DOI] [PubMed] [Google Scholar]
- 25.Nei M, Gu X, Sitnikova T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(15):7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meisel RP. Repeat mediated gene duplication in the Drosophila pseudoobscura genome. Gene. 2009;438(1-2):1–7. doi: 10.1016/j.gene.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 27.Clark AG, Wang L. Molecular population genetics of Drosophila immune system genes. Genetics. 1997;147(2):713–724. doi: 10.1093/genetics/147.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazzaro BP, Clark AG. Evidence for recurrent paralogous gene conversion and exceptional allelic divergence in the Attacin genes of Drosophila melanogaster. Genetics. 2001;159(2):659–671. doi: 10.1093/genetics/159.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hedengren M, Borge K, Hultmark D. Expression and evolution of the Drosophila attacin/diptericin gene family. Biochemical and Biophysical Research Communications. 2000;279(2):574–581. doi: 10.1006/bbrc.2000.3988. [DOI] [PubMed] [Google Scholar]
- 30.Daffre S, Kylsten P, Samakovlis C, Hultmark D. The lysozyme locus in Drosophila melanogaster: an expanded gene family adapted for expression in the digestive tract. Molecular and General Genetics. 1994;242(2):152–162. doi: 10.1007/BF00391008. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Molecular Biology and Evolution. 2004;21(1):36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- 32.Ramos-Onsins S, Aguadé M. Molecular evolution of the Cecropin multigene family in Drosophila: functional genes vs. pseudogenes. Genetics. 1998;150(1):157–171. doi: 10.1093/genetics/150.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 34.Mrinal N, Nagaraju J. Intron loss is associated with gain of function in the evolution of the gloverin family of antibacterial genes in Bombyx mori. The Journal of Biological Chemistry. 2008;283(34):23376–23387. doi: 10.1074/jbc.M801080200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 shows the percent survival of adult flies following injury and infection with the microorganisms used in this study.