Abstract
The extracellular matrix is an insoluble aggregate of large proteins and glycosoaminoglycans that comprises the microenvironment of cells in tissue. The matrix displays a host of ligands that interact with cell-surface receptors to mediate the attachment and spreading of cells and regulate signaling processes. Studies of cell–matrix interactions and downstream signaling processes commonly employ substrates having an adsorbed layer of protein and are challenging because of the difficulty in controlling the structure and activity of the immobilized protein. Significant effort has been directed towards the development of model substrates that present adhesion ligands in defined densities, orientations and environments. Among these approaches, self-assembled monolayers of alkanethiolates on gold offer a high level of control over the molecular structure of the surface and are well-suited to studies of cell adhesion. This review describes the design and use of monolayers for applications in cell biology, including the use of monolayers to evaluate the roles of peptide and protein ligands in cell–matrix interactions, the development of methods to pattern ligands on monolayers and applications to cell biology, the development of dynamic monolayers that can switch the activities of ligands presented to an adherent cell, and the rewiring of interactions between a cell and its substrate. These examples illustrate the flexibility inherent to monolayers for applications in cell biology.
Keywords: cell–matrix interactions, self-assembled monolayers, modelling
1. Introduction
Most cells are adherent and must attach to and spread on a surface in order to survive, proliferate and function. In tissue, this surface is the extracellular matrix (ECM), an insoluble scaffold formed by the assembly of several large proteins—including fibronectin, the laminins and collagens and others—that provide a wide range of biochemical and mechanical cues to cells [1,2]. Studies of cellular processes in the laboratory routinely use protein-coated dishes to mimic the in vivo environment and to elucidate the functions of the matrix in regulating cellular processes. The adsorption of protein, however, is complicated and often proceeds with a lack of control over the orientation and conformation of proteins at the surface. As a result, it remains difficult to control the biological activities of proteins that are adsorbed to man-made materials, and in turn compromises the use of these substrates as models of the ECM.
This realization has motivated a significant effort over the past two decades to develop materials that present well-defined biological motifs for use as mimics of the ECM. Many of the approaches have used substrates modified with polymeric materials or monolayer chemistries that can be modified with cell adhesion motifs. These important approaches have been reviewed elsewhere [3–5]. The present review specifically focuses on the use of self-assembled monolayers of alkanethiolates on gold for this application. These surfaces are a recent addition to the strategies that are now used and offer a set of characteristics that make them well-suited to certain classes of problems in studies of cell–ECM interactions. These characteristics and early applications of the monolayers are described in the following pages.
2. Self-assembled monolayers
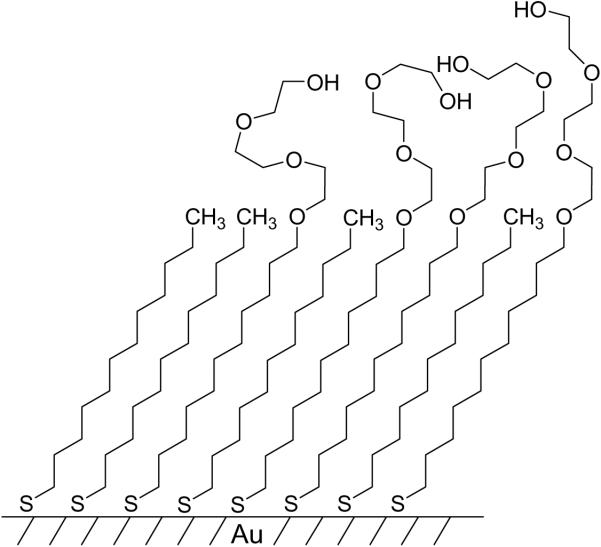
Self-assembled monolayers (SAMs) of alkanethiolates on gold are currently the best available class of model organic surfaces that permit control over interfacial structure and properties [6]. SAMs form spontaneously upon immersion of a gold-coated substrate into a solution of alkanethiols or corresponding disulfide reagents (Fig. 1). The monolayers are structurally well-ordered, though the details of the arrangement of thiolates on the gold surface have been debated [7]. In any event, the sulfur atoms coordinate to the gold(111) surface to give a densely packed and ordered hexagonal array of long chain molecules that are in an extended conformation. The key feature of this system is that the properties of the surface are determined by the terminal functional group of the precursor alkanethiol. Even complex and reactive groups can be introduced—either before or after the monolayer is formed—through straightforward synthetic procedures.
Figure 1.
Representation of a self-assembled monolayer (SAM) of alkanethiolates on the surface of gold. The sulfur atoms of the alkanethiolates coordinate to the gold surface and arrange the alkyl chains in a close-packed layer. The properties of the SAM are determined by the terminal functional group of the precursor alkanethiol. The figure shows alkanethiolates that are terminated in the methyl group and in the tri(ethylene glycol) group. The latter have proven important because they are effective at preventing the non-specific adsorption of protein.
It is also straightforward to introduce two or more different groups onto the substrate, simply by preparing the monolayer from a solution of a mixture of terminally substituted alkanethiols. Hence, monolayers can be used to prepare complex substrates that present a variety of ligands, with good control over the densities and patterns of each ligand. There is the possibility, however, that the alkanethiolates can form phase-separated domains in the monolayer, giving rise to structural heterogeneities. In practice, these domains are usually not present when the density of the ligand-terminated alkanethiolate is kept below 1% relative to the total alkanethiolate. SAMs have several properties that make them useful in cell biology: they are stable in air for periods of months; they are stable in cell culture for periods of several days; when supported on gold 100 Å in thickness they are optically transparent and compatible with immunofluorescence staining [8]. Even these thin films of gold are electrically conductive and therefore compatible with methods that use electrochemistry to control the biological properties of the substrates.
3. Protein adsorption to materials
Most man-made materials, when placed in solutions containing proteins, are rapidly coated with an adsorbed layer of protein [9]. This property is exploited in the routine preparation of substrates for cell culture. For example, polystyrene substrates are often treated with a solution of fibronectin prior to seeding with cells. The resulting protein film is heterogeneous in structure—in that the proteins are presented in a range of orientations and denaturated states—but still presents the cell-binding motifs at sufficient density and in a functional conformation to promote attachment, spreading and migration. In practice, the conditions for adsorbing protein are identified empirically and depend on the structures of the substrates in ways that remain poorly understood. One study, for example, used atomic force microscopy and surface plasmon resonance spectroscopy to characterize fibrinogen and fibronectin that had adsorbed to monolayers terminated either in methyl or carboxyl groups and found that the densities and conformations of the adsorbed proteins strongly depended on surface chemistry [10].
The tendency for proteins to adsorb non-specifically to materials has also interfered with efforts to use model surfaces that present defined cell-binding motifs. When the slide is placed in a suspension of cells, for example, proteins in the medium can rapidly adsorb to the surface, thereby physically preventing access to the immobilized ligands or introduce additional ligands that mediate cell attachment. The common approach to this problem has relied on treating the substrate with a solution of a “blocking” protein—often bovine serum albumin—prior to cell attachment with the expectation that “sticky” sites on the surface are passivated, now allowing the ligands to mediate cell adhesion. But the large fraction of the surface that is occupied by the blocking protein and the substantially larger size of the protein relative to the ligand make this approach ineffective in studies that require molecular control over the surface.
4. Bio-specific recognition at monolayers
A turning point in the use of model substrates came with the development of so-called inert surfaces—i.e. surfaces that prevent the non-specific adsorption of protein. The majority of approaches rely on the use of poly(ethylene glycol), which excludes protein adsorption through mechanisms that are thought to depend on the conformational properties of highly solvated polymer layers [11]. A report by Prime and Whitesides 15 years ago demonstrated that monolayers tailored with short oligomers of the ethylene glycol group were very effective at preventing the non-specific adsorption of protein and later work used surface plasmon resonance spectroscopy—which provides a real-time, or in situ, measure of adsorption—to show that these surfaces even prevented weak, or reversible, adsorption [12,13]. This result was in part surprising because it was believed that the ethylene glycol chains should be present at low density to prevent protein binding [14]. Nonetheless, this finding quickly motivated the development of monolayers to studies of biological problems. Early work demonstrated that monolayers prepared from oligo(ethylene glycol)-terminated alkanethiols, but where approximately 1% of the chains presented a covalently attached ligand, displayed selective interactions with proteins. For example, surface plasmon resonance spectroscopy was used to characterize the selective interaction of carbonic anhydrase with a monolayer presenting the benzenesulfonamide ligand [15]. Subsequent work applied this approach to a range of other protein–ligand pairs including the binding of vancomycin to the D–Ala–D–Ala dipeptide and proteins to carbohydrates [16–18].
5. Monolayers for cell adhesion
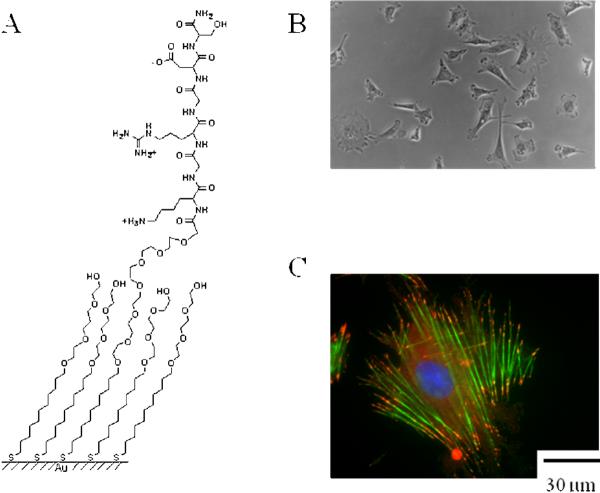
This same approach has been validated for studying the roles of peptide ligands in cell adhesion and migration. In an early example, we had prepared monolayers that presented the peptide Gly–Arg–Gly–Asp–Ser against a background of tri(ethylene glycol) groups [19]. This peptide is found in fibronectin and other ECM proteins and is a ligand for approximately one-half of the integrin family cell-surface receptors, which are an important class of receptors found on all cell surfaces and that mediate the attachment of cells to ECM [20]. We found that Swiss 3T3 fibroblasts attached and spread efficiently to the monolayer and immunostaining showed that the adherent cells assembled normal focal adhesion complexes—a cluster of integrin receptors that forms strong attachments to the substrate and initiates adhesion signals—and actin stress filaments (Fig. 2). Control experiments established that the adhesion of cells could be blocked with a soluble RGD peptide and that cells failed to attach to a monolayer presenting a scrambled form of this peptide. Hence, this work validated the use of monolayers to control the ligand receptor interactions that mediate the adhesion of cells and it also showed that the ligand RGD alone could support cell attachment, spreading and a proper organization of the cytoskeleton.
Figure 2.
(a) Structure of a monolayer that presents the peptide Arg–Gly–Asp mixed with tri(ethylene glycol) groups. (b) An optical micrograph that shows 3T3 fibroblasts attached to a monolayer wherein 0.5% of the alkanethiolates present the peptide ligand. (c) A fluorescent micrograph of a cell that was adherent on these monolayers for 12 h, fixed and stained with phalloidin-rhodamine (green) to reveal the actin cytoskeleton and with anti-vinculin (red) to reveal focal adhesion complexes. The cells assembled stress fibers that were indistinguishable from those found in cells adherent on fibronectin.
The monolayers have also enabled mechanistic studies that evaluated the dependence of cell adhesion on the density of adhesion ligand. We prepared monolayers presenting the peptide at densities ranging from 0.01 to 1.0% (relative to total alkanethiolate) and characterized the adhesion of cells. As expected, the attachment of cells was less efficient on monolayers presenting ligand at lower densities, as was the degree of spreading of adherent cells. We went on to demonstrate that not only the density of the RGD peptide, but also its microenvironment at the surface, had an important role in determining the extent of cell attachment and spreading. Cells attached and spread efficiently on monolayers that presented the peptide at a density of 0.5% mixed with tri(ethylene glycol) groups; when the peptide was presented at the same density but mixed with the more sterically demanding hexa(ethylene glycol) group, fewer cells attached and those that did remained in a rounded morphology. Control experiments were used to show that the affinity of the RGD peptide for the integrin receptor decreased as the ligand was more crowded by the surface. A key point from these studies is that the monolayer substrates are sufficiently structurally ordered that they can be used to study the influence of both ligand density and affinity on cell attachment.
6. Studies of cell–ECM interactions
The application of monolayer substrates to understanding the roles of ligands in fibronectin illustrates the benefits of employing structurally well-defined substrates. Fibronectin is a primary ECM protein that is found in many tissues. This protein comprises more than 30 globular domains that include the type III domains important for cell adhesion. The RGD peptide is found within the 10th type III repeat and was among the earliest ligands discovered to mediate cell adhesion by interacting with integrin receptors [21]. Recent work has implicated a peptide, having sequence PHSRN, residing in the 9th type III domain in the adhesion of cells. A series of studies by Yamada and Grant and coworkers led to the proposal that PHSRN is a synergy peptide that cooperates with RGD to enhance the attachment and spreading of cells but that fails to support cell attachment when present alone [22,23]. Mardon suggested a mechanism wherein the RGD and PHSRN peptides in adjacent domains of fibronectin simultaneously interact with separate binding sites on opposite sides of the integrin receptor [24]. This model was based on studies of cell adhesion to recombinant proteins having scrambled RGD and PHSRN sequences and on a crystal structure of a fragment of fibronectin, which shows that the two peptides are directed towards the same region of space and are separated by a distance that matches the width of the receptor. Yet studies that use protein-coated substrates to study cell adhesion are complicated by the inability to establish that peptide sequences in the proteins are available to interact with cellular receptors once the proteins are adsorbed to tissue culture plastic.
Monolayers erase this ambiguity and were used to revise the mechanistic understanding of the synergy peptide [25]. First, Chinese hamster ovary (CHO) cells attached with similar efficiency to monolayers presenting either the RGD or the PHSRN peptide, although cells failed to spread on the latter substrate. Second, the adhesion of cells could be inhibited by either peptide, i.e. either RGD or PHSRN could block the attachment of cells to a monolayer presenting RGD (and in the same sense, to a monolayer presenting PHSRN). These results require that the two peptides either share a binding site on the integrin receptor or bind to separate sites that are allosterically connected. In either case, these observations are inconsistent with the standing model invoking a cooperative two-point binding of peptides. Indeed, the ability to present defined ligands at controlled densities and in a regular environment (to ensure that all of the peptides are active) against an otherwise non-interacting background represents a powerful tool for establishing the function of ECM ligands and addressing the relationship between distinct ligands.
7. Model substrates for stem cell culture
Kiessling and coworkers have applied monolayers to identify defined culture systems for applications in stem cell biology. By constructing an array that presented several peptide fragments from the ECM protein laminin, this group could apply embryonic stem cells to the substrate and identify those regions that supported the growth and self-renewal of the stem cell cultures [26]. This work identified a peptide that could specifically mediate cell adhesion—though the cell-surface receptor for this ligand has not yet been identified—and went on to create a three-dimensional matrix that presented this peptide for the defined culture of the stem cells. Importantly, this work illustrates that monolayers presenting arrays of peptide motifs can be used to efficiently identify motifs that are biologically active [27].
8. Model substrates that present protein motifs
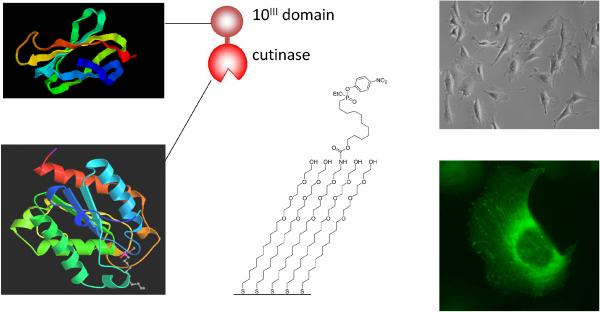
While model substrates that present short peptide ligands are often effective mimics of the ECM—with cell adhesion to RGD serving as the prototypical example—they are not useful for studying the properties of peptide ligands that require a structure enforced by the protein fold, or that are based on a discontinuous epitope in the protein. In these cases, it is necessary to prepare model substrates that present whole protein domains. But many of the common methods for immobilizing proteins offer less control over the orientation and density than is routine with the methods for attaching low molecular weight ligands to materials. The best methods for site-specific immobilization of protein—e.g. the use of His-tagged proteins—do not give linkages that are stable over long periods of time, while methods that employ covalent bonds in the immobilization can be difficult to implement [28,29]. We developed a route that is based on a protein domain that selectively interacts with an irreversible inhibitor to give a covalent adduct [30]. In one example, we expressed a recombinant protein that had the 10th type III repeat from fibronectin fused to cutinase, a serine esterase that is covalently inhibited by phosphonate ligands (Fig. 3). Application of a solution of the protein to a monolayer presenting the ligand resulted in selective immobilization of the cutinase domain to the monolayer, to give an oriented display of the cell adhesion domain [31]. These surfaces were active for cell adhesion and have been applied to the study of multiple ligands within fibronectin. The method has the advantage that it does not require the protein to be purified—since the interaction of cutinase with the irreversible ligand is selective—and the density and pattern of protein can be controlled. This, and related, methods should enable the preparation of model substrates that present complex ligands and therefore that more closely mimic the properties of the ECM [32].
Figure 3.
Strategy for immobilization of proteins containing cell-adhesive ligands. The fibronectin 10th type III domain is fused to cutinase, which specifically binds a suicide ligand of the substrate to give covalent immobilization of proteins while maintaining activity and orientation. An optical micrograph of swiss 3T3 cells adherent to the model substrate (upper right). The cells displayed normal cytoskeleton and focal adhesion structures (lower right).
9. Patterning the attachment of cells
Monolayers that are patterned with cell adhesive ligands allow control over the shapes, sizes and positions of adherent cells, and have proved important in several areas of basic and applied biology. Indeed, the development of patterning methods has been an active field and has provided numerous strategies for patterning the adhesion of cells [33]. The simplest and still most important patterning method is microcontact printing (μCP), developed by the Whitesides group and which uses a rubber stamp to print a monolayer in a defined pattern on a gold substrate [34]. The procedure begins with photolithography, where a silicon wafer coated with a layer of photoresist is exposed to ultraviolet light through a mask to degrade the photoresist in the illuminated regions. The resulting “master” is used to cast an elastomeric “stamp” that is inked with an alkanethiol, brought into conformal contact with a gold-coated substrate for 30 s. Monolayers assemble at the regions where the stamp contacts the surface and after removal of the stamp the substrate is immersed in a solution of an oligo(ethylene glycol)-terminated alkanethiol to render the non-stamped regions of the surface biologically inert. In many applications, the patterned monolayer is treated with a solution of ECM protein to adsorb matrix proteins onto the stamped regions. The μCP method is simple, can pattern feature sizes of 1 μm and can pattern areas several square cm in size with edge resolution of the features better than 100 nm.
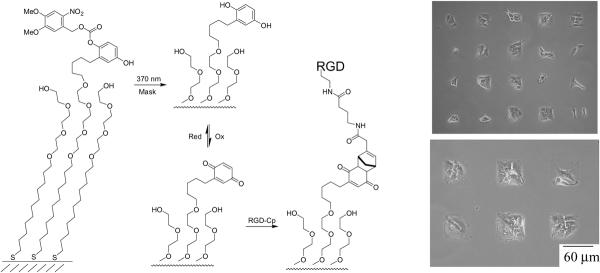
In an early application, the patterned monolayers were used to identify a correlation between cell spreading and apoptosis. Human capillary endothelial cells were allowed to attach to a monolayer patterned with square features ranging in size from 75 to 3000 μm2 [35]. Cells that were restricted to small patterns underwent apoptosis, with progressively less cell death as the spreading was allowed to increase. Further experiments that allowed cells to spread on an array of circular features revealed that it was the extent of cell spreading, and not the total matrix protein presented to the cell, that influenced apoptosis of the cells. A series of papers by several groups have used patterned substrates to control the cytoskeleton structure in adherent cells [36–39]. This work has revealed that shapes having concave features promote the assembly of contractile stress filaments, features having convex shapes promote the assembly of lamellipodia and punctuate features promote the assembly of strong focal adhesions. This work makes it possible to promote a defined cytoskeletal structure in adherent cells. A recent example has also applied the patterned substrates to the differentiation of stem cells [40]. Chen and coworkers found that mesenchymal stem cells adherent to square patterns favored differentiation to bone cells for large squares and to fat cells for smaller squares. The patterned substrates have also been important to the development of cell-based assays in drug discovery [41], where the use of a cell population that shares a common shape reduces the heterogeneity in cellular response. Finally, patterning methods based on photochemical modification of pre-formed monolayers are now providing access to substrates that are patterned with peptides, in gradients and with multiple ligands (Fig. 4) [42].
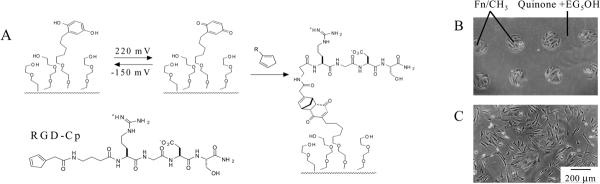
Figure 4.
A photochemical strategy to pattern the immobilization of peptide ligands begins with a monolayer presenting NVOC-protected hydroquinone groups and glycol groups (left). Illumination of the monolayer with light at 370 nm through a mask that had a square pattern revealed the hydroquinone groups, which were converted to the quinones by rinsing with a solution of tetramethylbenzoquinone. The substrate was treated with a conjugate of RGD and cyclopentadiene (RGD-Cp) to immobilize the peptide in illuminated regions. (Right) An optical micrograph shows 3T3 fibroblasts patterned on the substrate.
Microfluidic devices have provided another tool for patterning surfaces and spatially controlling the environment of adherent cells [43]. Whitesides and coworkers have prepared networks by applying an elastomeric mold to a monolayer substrate. The relief pattern of the mold creates a set of microfluidic channels that flow over the monolayer. The channels have been used to pattern the adsorption of ECM proteins and the covalent immobilization of ligands [44]. Of greater significance, the microfluidic networks can be designed so that gradients of ligands are flowed over the surface, resulting in an immobilized gradient [45]. In one example, we created an array of square features, with a gradient of the RGD peptide in each feature. Swiss 3T3 fibroblasts attached and spread to completely fill the square, but had a polarized cytoskeleton in response to the gradient. Whitesides and coworkers showed that the medium surrounding a cell could be patterned such that one side of the cell would be exposed to a drug or protein [46]. These examples reveal the unique capabilities that microfluidic devices offer to cell biology and point to future applications of these microtechnologies.
10. Dynamic substrates
The self-assembled monolayers have enabled models of the ECM that are dynamic, and that allow the activities of immobilized ligands to be switched on and off during the course of cell culture [47]. The approach exploits the conductivity of the gold film that supports the monolayer and follows from extensive work that has shown that electroactive molecules that are tethered to the monolayer can be reduced or oxidized by applying electrical potentials to the gold film [48]. By designing monolayers that incorporate molecular groups that undergo oxidation or reduction and subsequent reactions, it is possible to engineer surfaces that dynamically inactivate or activate ligands that interact with cell-surface receptors. In one example, the RGD peptide was immobilized to a monolayer by way of a tether that incorporated a propanate-benzoquinone fragment [49]. The peptide ligand mediated the attachment and spreading of cells and was stable for several days. Application of a negative potential, however, resulted in reduction of the benzoquinone group and subsequent lactonization to release the peptide from the monolayer. As a result, the adherent cells assumed a rounded morphology and detached from the substrate. This example illustrates the molecular-level design of a monolayer that could selectively release ligands that were tethered by way of the redox-active moiety and in this case non-invasively release an adherent cell culture. We have also developed a tether that released immobilized ligand in response to a positive electrical potential [50]. By combining these two strategies with microelectrode arrays, it is possible to prepare adherent cell cultures and selectively release sub-populations of cells at different times [51]. Another notable approach to the development of dynamic substrates has used polymeric gels that undergo a thermally induced phase transition to release cells [52]. The approach of using physical organic chemistry to design molecules or polymers that undergo redox-active reactions to manipulate the activities of ligands on a monolayer is general and can be applied to the preparation of dynamic substrates having a range of activities.
In another example, we developed a monolayer that could be electrically switched to permit the immobilization of cell adhesive ligands [53]. A monolayer presenting the hydroquinone group against a background of tri(ethylene glycol) groups is inert and does not support the attachment of cells. Application of a positive electrical potential promotes the oxidation of the hydroquinone group to the corresponding benzoquinone. The latter is a reactive dienophile and selectively adds to a cyclopentadiene group to form the Diels–Alder adduct. By conjugating the diene to the RGD peptide ligand, we could selectively switch on the immobilization of peptide by applying an electrical potential to the surface. This dynamic substrate was used to create an assay for cell migration (Fig. 5). Fibroblast cells were first patterned to an array of circular features that were surrounded by the electroactive monolayer. Cells remained confined to the pattern but the application of an electrical potential resulted in immobilization of the peptide and a rapid migration of cells from the pattern. This approach has the benefits over the traditional wound migration assay that the composition of the matrix on which cells migrate can be controlled. These surfaces have also been applied to the preparation of cocultures of two different cell types by allowing a first population of cells to attach to a patterned monolayer and then activating a second pattern for attachment of a second population [54]. The dynamic substrates offer an unprecedented opportunity to control the cellular microenvironment with spatial and temporal control and to assemble complex cell cultures from multiple cell types.
Figure 5.
Design of substrates that can turn on cell migration and growth. (a) Monolayers that present hydroquinone groups mixed with penta(ethylene glycol) groups are inert to cell attachment. Electrochemical oxidation converts the hydroquinone groups to quinone groups, which then undergo Diels–Alder reaction with a diene–Arg–Gly–Asp conjugate (RGD–Cp) to introduce the peptide onto the surface. The resulting monolayers support the attachment and spreading of cells. Cells attached only to the protein-coated regions (b), but after oxidation of the substrate in the presence of RGD–Cp, cells were able to migrate and then grow to fill the entire monolayer (c).
11. Complimentary engineering of the cell–material interface
The properties of monolayers described in this review make this class of model surfaces among the best available for controlling, at the molecular level, the interactions of cells with surfaces. By combining the monolayers with biological strategies to engineer the cell surface, it is possible to rewire the molecular interactions between a cell and substrate and thereby enable the preparation of cell-based devices for applications in drug discovery, sensors, and microtechnologies. One example engineered CHO cells with a chimeric integrin that had the RGD-binding domain replaced with a carbonic anhydrase domain, which binds benzenesulfonamide ligands with micromolar affinity [55]. The chimeric receptor retained the cytoplasmic tail of the β1 integrin and therefore maintained the ability to cluster and signal through focal contacts (Fig. 6). We found that cells transfected with the chimeric receptor efficiently attached and spread on monolayers presenting the synthetic ligand and that these cells were able to migrate on the monolayers with rates that were comparable to that of wild-type cells migrating on RGD-presenting monolayers.
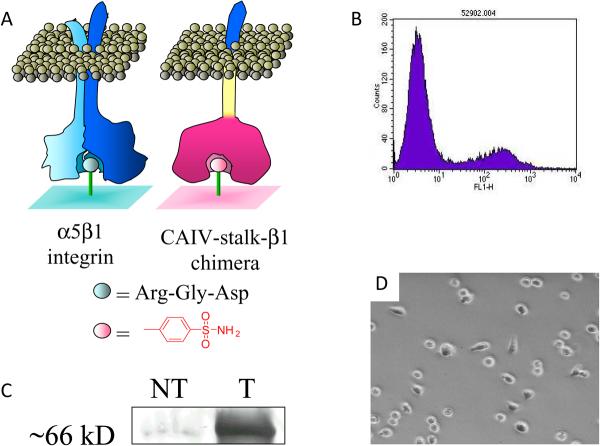
Figure 6.
Strategy for rewiring receptor-mediated cell adhesion. (a) The β1 integrin was engineered to present carbonic anhydrase as the ligand binding domain. (b) FACS analysis was used to sort the expressing CHO cells. (c) Western analysis confirms expression of full-length receptor in CHO cells. (d) Optical micrograph of transfected cells adherent to a monolayer presenting a benzenesulfonamide ligand.
A second example demonstrated an interface that could transducer biological activities in the cell to an electrical signal in the substrate [56]. The approach is based on the expression on the cell surface of an enzyme that can convert an immobilized substrate from a redox-inactive to active form. We used the serine esterase cutinase which hydrolyzes acylated hydroquinones. A monolayer presenting the substrate is not redox-active, whereas addition of the cutinase enzyme converts the substrate to the redox-active hydroquinone that can then be detected electrically [57]. We engineered CHO cells with chimeric integrin receptors that replaced the RGD-binding domain with a cutinase domain. Cells were then allowed to attach to a surface that presented both the RGD adhesion peptide and the acylhydroquinone substrate. Cells that were transfected with the cutinase chimera, but not wild-type cells, resulted in an electrical activity in the monolayer. This example harnessed a complemintary engineering of both the cell surface and the substrate to install a novel interaction between the cell and ECM mimic.
12. Summary and outlook
This review provides a perspective of work over the past decade to develop self-assembled monolayers as model substrates for studies of cell adhesion. Several characteristics inherent to the monolayers make them well-suited for preparing mimics of the ECM. These points include:
Well-defined structure. The regular structure of the monolayers enables wide flexibility in tailoring the surface with ligands and other functional groups. Ligands can be presented with excellent control over their densities and in a uniform environment. This flexibility also enables the preparation of dynamic substrates that can manipulate the presentation of ligands.
Inert surfaces. Self-assembled monolayers that present oligo(ethylene glycol) groups are highly effective at preventing the non-specific adsorption of protein. These surfaces maintain this property in complex solutions, including serum-containing cell culture media and void the need for blocking.
Immobilization schemes. A full portfolio of immobilization chemistries that can be used to tether ligands to monolayers, and that provide for a defined orientation of the ligands and a rigorous control over the density, is available.
Analytical methods. Monolayers are compatible with multiple analytical methods used in characterizing biochips, including surface plasmon resonance spectroscopy, fluorescence imaging, radioisotope detection, and mass spectrometry [58].
Patterning methods. The availability of several patterning methods—and specifically the microcontact printing method—provides routine access to substrates that can control the shapes, sizes and positions of cells. These substrates enable studies of cytoskeleton function in cells and cell-based technologies.
Proven performance. There are hundreds of publications that describe the use of monolayers in biological and bioanalytical applications. These systems are well-suited to experiments involving attached cell cultures and are used commercially in bioanalytical devices.
The ECM is complex and consequently experimental studies benefit from a range of methods and tools that bring insights to the structures and functions of the matrix. The self-assembled monolayers represent one important component of this toolbox and are significant because they offer a straightforward approach to prepare structurally well-defined mimics of the matrix. The tailored substrates are admittedly simple mimics of the matrix but they allow unambiguous studies of the roles that discrete motifs play in mediating cell adhesion and regulating downstream signaling processes. Early work with the monolayers has been important for addressing the roles of matrix ligands, understanding the relationships between cell shape and function, and enabling a class of dynamic substrates that can modulate, in real time, the activities of immobilized ligands. Future work will see an increased use of the monolayers for current problems in cell adhesion and matrix biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berrier AL, Yamada KM. Cell-matrix adhesion. Journal of Cellular Physiology. 2007;213:565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 2.Daley WP, Peters SB, Larsen M. Extracellular matrix dynamics in development and regenerative medicine. Journal of Cell Science. 2008;121:255–264. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 3.Garcia AJ. Interfaces to control cell-biomaterial adhesive interactions. Polymers For Regenerative Medicine. 2006:171–190. [Google Scholar]
- 4.Jagur-Grodzinski J. Polymers for tissue engineering, medical devices, and regenerative medicine. Concise General Review of Recent Studies. Polym. Adv. Technol. 2006;17:395–418. [Google Scholar]
- 5.Mano JF. Stimuli-responsive polymeric systems for biomedical applications. Adv. Eng. Mat. 2008;10:515–527. [Google Scholar]
- 6.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem. Rev. 2005;105:1103–1169. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 7.Cossaro A, Mazzarello R, Rousseau R, Casalis L, Verdini A, Kohlmeyer A, Floreano L, Scandolo S, Morgante A, Klein ML, Scoles G. X-ray diffraction and computation yield the structure of alkanethiols on gold(111) Science. 2008;321:943–946. doi: 10.1126/science.1158532. [DOI] [PubMed] [Google Scholar]
- 8.Mrksich M, Whitesides GM. Using self-assembled monolayers to understand the interactions of man-made surfaces with proteins and cells. Ann. Rev. Biophys. Biomol. Struct. 1996;25:55–78. doi: 10.1146/annurev.bb.25.060196.000415. [DOI] [PubMed] [Google Scholar]
- 9.Ratner BD, Bryant SJ. Biomaterials: where we have been and where we are going. Annual Review of Biomedical Engineering. 2004;6:41–75. doi: 10.1146/annurev.bioeng.6.040803.140027. [DOI] [PubMed] [Google Scholar]
- 10.Servoli E, Maniglio D, Aguilar MR, Motta A, Roman JS, Belfiore LA, Migliaresi C. Quantitative analysis of protein adsorption via atomic force microscopy and surface plasmon resonance. Macromolecular Bioscience. 2008;8 doi: 10.1002/mabi.200800110. in press. [DOI] [PubMed] [Google Scholar]
- 11.Harris JM. Poly(ethylene glycol) chemistry: biotechnical and biomedical applications. Plenum Press; New York: 1992. [Google Scholar]
- 12.Prime KL, Whitesides GM. Self-assembled organic monolayers—model systems for studying adsorption of proteins at surfaces. Science. 1991;252:1164–1167. doi: 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- 13.Mrksich M, Sigal GS, Whitesides GM. Surface plasmon resonanvance permits in situ measurement of protein adsorption on self-assembled monolayers of alkanethiolates on gold. Langmuir. 1995;11:4383–4385. [Google Scholar]
- 14.Jeon SI, Lee JH, Andrade JD, DeGennes PG. Protein-surface interactions in the presence of polyethylene oxide: a simplified theory. J. Colloid Interface Sci. 1991;142:149–158. [Google Scholar]
- 15.Mrksich M, Grunwell JR, Whitesides GM. Bio-specific adsorption of carbonic anhydrase to self-assembled monolayers of alkanethiolates that present benzenesulfonamide groups on gold. J. Am. Chem. Soc. 1995;117:12009–12010. [Google Scholar]
- 16.Rao JH, Yan L, Lahiri J, Whitesides GM GM, Weis RM, Warren HS. Binding of a dimeric derivative of vancomycin to L–Lys–D–Ala–D–Lactate in solution and at a surface. Chemistry & Biology. 1999;6:353–359. doi: 10.1016/S1074-5521(99)80047-7. [DOI] [PubMed] [Google Scholar]
- 17.Horan N, Yan L, Isobe H, et al. Nonstatistical binding of a protein to clustered carbohydrates. Proc. Natl. Acad. Sci., USA. 1996;96:11782–11786. doi: 10.1073/pnas.96.21.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houseman BT, Mrksich M. Carbohydrate arrays for the evaluation of protein binding and enzyme activity. Chem. & Biol. 2002;9:443–454. doi: 10.1016/s1074-5521(02)00124-2. [DOI] [PubMed] [Google Scholar]
- 19.Houseman BT, Mrksich M. Environment of Arg–Gly–Asp peptide ligands immobilized on self-assembled monolayers of alkanethiolates on gold influences the adhesion of 3T3 fibroblasts. Biomaterials. 2001;22:943–955. doi: 10.1016/s0142-9612(00)00259-3. [DOI] [PubMed] [Google Scholar]
- 20.Arnaout MA, Goodman SL, Xiong JP. Structure and mechanics of integrin-based cell adhesion. Curr. Op. Cell Biol. 2007;19:495–507. doi: 10.1016/j.ceb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruoslahti E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 22.Aota S, Nagai T, Yamada KM. Characterization of regions of fibronectin besides the arginine-glycine-aspartic acid sequence required for adhesive function of the cell-binding domain using site-directed mutagenesis. J Biol Chem. 1991;266:15938–15943. [PubMed] [Google Scholar]
- 23.Mardon HJ, Grant KE. The role of the ninth and tenth type III domains of human fibronectin in cell adhesion. FEBS Lett. 1994;340:197–201. doi: 10.1016/0014-5793(94)80137-1. [DOI] [PubMed] [Google Scholar]
- 24.Grant RP, Spitzfaden C, Altroff H, Campbell ID, Mardon HJ. Structural requirements for biological activity of the ninth and tenth FIII domains of human fibronectin. J Biol Chem. 1997;272:6159–6166. doi: 10.1074/jbc.272.10.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng Y, Mrksich M. The adhesion ligand Arg-Gly-Asp and the synergy peptide Pro-His-Ser-Arg-Asn mediate cell adhesion via a common site on integrin receptors. Biochemistry. 2004;43:15811–15821. doi: 10.1021/bi049174+. [DOI] [PubMed] [Google Scholar]
- 26.Derda R, Li L, Orner BP, Lewis RL, Thomson JA, Kiessling LL. Defined substrates for human embryonic stem cell growth identified from surface arrays. ACS Chem. Biol. 2007;2:347–355. doi: 10.1021/cb700032u. [DOI] [PubMed] [Google Scholar]
- 27.Orner BP, Derda R, Lewis RL, Thomson JA, Kiessling LL. Arrays for the combinatorial exploration of cell adhesion. J. Am. Chem. Soc. 2004;126:10808–10809. doi: 10.1021/ja0474291. [DOI] [PubMed] [Google Scholar]
- 28.Sigal GB, Bamdad C, Barberis A, et al. A self-assembled monolayer for the binding and study of histidine tagged proteins by surface plasmon resonance. Anal. Chem. 1996;68:490–497. doi: 10.1021/ac9504023. [DOI] [PubMed] [Google Scholar]
- 29.Lundstrom I, Ivarsson B, Jonsson U, Elwing H. Protein adsorption and interaction at solid surfaces. In: Feast WJ, Munro HS, editors. Polymer Surfaces and Interfaces. Vol. 1. John Wiley; New York: 1987. pp. 201–230. [Google Scholar]
- 30.Hodneland CD, Lee Y-S, Min D-H, Mrksich M. Selective immobilization of protein to self-assembled monolayers presenting active site directed capture ligands. Proc. Natl. Acad. Sci., USA. 2002;99:5048–5052. doi: 10.1073/pnas.072685299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy WL, Mercurius KO, Koide S, Mrksich M. Substrates for cell adhesion prepared via active site-directed immobilization of a protein domain. Langmuir. 2004;20:1026–1030. doi: 10.1021/la035733m. [DOI] [PubMed] [Google Scholar]
- 32.Camarero JA. Recent developments in the site-specific immobilization of proteins onto solid supports. Biopolymers. 2008;90:450–458. doi: 10.1002/bip.20803. [DOI] [PubMed] [Google Scholar]
- 33.Jung DR, Kapur R, Adams T, Giuliano KA, Mrksich M, Craighead HG, Taylor DL. Topographical and physicochemical modification of material surface to enable patterning of living cells. Crit. Rev. in Biotech. 2001;21:111–154. doi: 10.1080/20013891081700. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz SA, Chen CS. Microcontact printing: a tool to pattern. Soft Matter. 2007;3:168–177. doi: 10.1039/b613349e. [DOI] [PubMed] [Google Scholar]
- 35.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1345–1347. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 36.Thery M, Pepin A, Dressaire E, Chen Y, Bornens M. Cell distribution of stress fibres in response to the geometry of the adhesive environment. Cell Mot. and the Cytoskel. 2006;63:341–355. doi: 10.1002/cm.20126. [DOI] [PubMed] [Google Scholar]
- 37.Thery M, Racine V, Piel M, Pepin A, Dimitrov A, Chen Y, Sibarita JB, Bornens M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci., USA. 2006;103:19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia N, Thodeti CK, Hunt TP, Xu QB, Ho M, Whitesides GM, Westervelt R, Ingber DE. Directional control of cell motility through focal adhesion positioning and spatial control of Rac activation. FASEB Journal. 2008;22:1649–1659. doi: 10.1096/fj.07-090571. [DOI] [PubMed] [Google Scholar]
- 39.Maduram JH, Goluch E, Liu C, Mrksich M. Subcellular curvature at the perimeter of micropatterned cells influences lamellipodial distribution and cell polarity. Cell Motility and the Cytoskeleton. 2008 doi: 10.1002/cm.20305. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and rhoa regulate stem cell lineage commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 41.Kapur R, Giuliano KA, Campana M, Adams T, Olson K, Jung D, Mrksich M, Vasudevan C, Taylor DL. Streamlining the drug discovery process by integrating miniaturization, high throughput screening, high content screening, and automation on the CellChip system. Biomedical Microdevices. 1999;2:99–109. [Google Scholar]
- 42.Dillmore WS, Yousaf MN, Mrksich M. A photochemical method for patterning the immobilization of ligands and cells to self-assembled monolayers. Langmuir. 2004;20:7223–7231. doi: 10.1021/la049826v. [DOI] [PubMed] [Google Scholar]
- 43.Weibel DB, Whitesides GM. Applications of microfluidics in chemical biology. Curr. Op. In Chem. Biol. 2006;10:584–591. doi: 10.1016/j.cbpa.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 44.Chen CS, Jiang XY, Whitesides GM. Microengineering the environment of mammalian cells in culture. MRS Bulletin. 2005;30:194–201. [Google Scholar]
- 45.Petty RT, Li H-W, Maduram J, Ismagilov R, Mrksich M. Attachment of cells to islands presenting gradients of adhesion ligands. J. Am. Chem. Soc. 2007;129:8966–8967. doi: 10.1021/ja0735709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takayam S, Ostuni E, LeDuc P, Naruse K, Ingber DE, Whitesides GM. Selective chemical treatment of cellular microdomains using multiple laminar streams. Chemistry & Biology. 2003;10:123–130. doi: 10.1016/s1074-5521(03)00019-x. [DOI] [PubMed] [Google Scholar]
- 47.Yousaf MN, Mrksich M. Dynamic substrates: modulating the behaviors of attached cells. In: Wilson E, et al., editors. New Technologies for Life Sciences: A Trends Guide. Elsevier; Amsterdam: 2000. pp. 28–35. [Google Scholar]
- 48.Murray RW. Nanoelectrochemistry: metal nanoparticles, nanoelectrodes, and nanopores. Chem. Rev. 2008;108:2688–2720. doi: 10.1021/cr068077e. [DOI] [PubMed] [Google Scholar]
- 49.Hodneland CD, Mrksich M. Biomolecular surfaces that release ligands under electrochemical control. J. Am. Chem. Soc. 2000;122:4235–4236. [Google Scholar]
- 50.Yeo W-S, Yousaf MN, Mrksich M. Dynamic interfaces between cells and surfaces: electroactive substrates that sequentially release and attach cells. J. Am. Chem. Soc. 2003;125:14994–14995. doi: 10.1021/ja038265b. [DOI] [PubMed] [Google Scholar]
- 51.Yeo W-S, Mrksich M. Electroactive self-assembled monolayers that permit orthogonal control over the adhesion of cells to patterned substrates. Langmuir. 2006;22:10816–10820. doi: 10.1021/la061212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J, Yamato M, Okano T. Cell-sheet engineering using intelligent surfaces. MRS Bulletin. 2005;30:189–193. [Google Scholar]
- 53.Yousaf MN, Houseman BT, Mrksich M. Turning on cell migration with electroactive substrates. Angew. Chem. Int. Ed. 2001;40:1093–1096. [PubMed] [Google Scholar]
- 54.Yousaf MN, Houseman BT, Mrksich M. Using electroactive substrates to pattern the attachment of two different cell types. Proc. Natl. Acad. Sci. USA. 2001;98:5992–5996. doi: 10.1073/pnas.101112898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato M, Mrksich M. Rewiring cell adhesion. J. Am. Chem. Soc. 2004;126:6504–6505. doi: 10.1021/ja039058e. [DOI] [PubMed] [Google Scholar]
- 56.Collier JH, Mrksich M. Engineering a bio-specific communication pathway between cells and electrodes. Proc. Natl. Acad. Sci., USA. 2006;103:2021–2025. doi: 10.1073/pnas.0504349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeo W-S, Mrksich M. Self-assembled monolayers that transduce enzymatic activities to electrical signals. Angew. Chem. Int. Ed. 2003;42:3121–3124. doi: 10.1002/anie.200250862. [DOI] [PubMed] [Google Scholar]
- 58.Mrksich M. Mass spectrometry of self-assembled monolayers: a new tool for molecular surface science. ACS Nano. 2008;2:7–18. doi: 10.1021/nn7004156. [DOI] [PMC free article] [PubMed] [Google Scholar]