Abstract
The 26S proteasome is the most downstream element of the ubiquitin-proteasome pathway of protein degradation. It is composed of the 20S core particle (CP) and the 19S regulatory particle (RP). The RP consists of 6 AAA-ATPases and at least 13 non-ATPase subunits. Based on a cryo-EM map of the 26S proteasome, structures of homologs, and physical protein-protein interactions we derive an atomic model of the AAA-ATPase-CP sub-complex. The ATPase order in our model (Rpt1/Rpt2/Rpt6/Rpt3/Rpt4/Rpt5) is in excellent agreement with the recently identified base-precursor complexes formed during the assembly of the RP. Furthermore, the atomic CP-AAA-ATPase model suggests that the assembly chaperone Nas6 facilitates CP-RP association by enhancing the shape complementarity between Rpt3 and its binding CP alpha subunits partners.
Keywords: Proteasome, Protein degradation, Modeling, assembly, cryo-EM, Proteasome, AAA-ATPase
Introduction
The ubiquitin-proteasome pathway is the major route used by eukaryotic cells for the disposal of misfolded or damaged proteins and for controlling the life span of regulatory proteins [1; 2; 3]. The 26S proteasome is a molecular machine of approximately 2.5 MDa, which targets poly-ubiquitylated proteins. It comprises two sub-complexes, the 20S core particle (CP) and one or two asymmetric 19S regulatory particles (RPs), which bind to the end(s) of the barrel-shaped CP.
The CP harbors the active sites confining proteolysis to a nano-compartment sequestered from the cytosol, while the RPs recruit substrates and prepare them for translocation into the CP. This preparation includes binding of poly-ubiquitylated substrates, their de-ubiquitylation, substrate unfolding, and opening of the ‘gate’ to the CP. The AAA-ATPase hexamer formed by the proteasome subunits Rpt1-6 is responsible for unfolding and gating [1].
While the structure of the CP was determined by X-ray crystallographic studies more than a decade ago [4; 5], insights into the high-resolution structure of the AAA-ATPases have only become available recently from the archaeal homolog ‘Proteasome Activating Nucleotidase’ (PAN) [6; 7]: each monomer consists of coiled-coils protruding from an OB-fold (PAN-N), and an AAA-fold, which have been crystallized independently. The coiled-coils and OB folds, as well as the AAA-folds assemble into hexameric rings (N-ring and AAA-ring, respectively), but the relationship between both rings with respect to each other is not known.
The order of the AAA-ATPase subunits Rpt1-6 in the hetero-hexamer was suggested to be Rpt1/2/6/4/5/3, based on cross-linking experiments [8]. However, the PAN-N structure challenges this model: only Rpt2, 3, and 5 possess an invariant proline (corresponding to Pro62 of PAN-N) that is required to adopt a cis-conformation for coiled-coil formation [6]. Thus, the PAN-N structure suggests strongly that Rpt2, Rpt3, and Rpt5 alternate in the hetero-hexamer, which would not be the case in the (1/2/6/4/5/3) order [8].
Here, we infer the quaternary structure of the 26S proteasome AAA-ATPase hexamer given the crystallographic PAN structures, a cryo-EM map of the 26S proteasome from Drosophila melanogaster [9], and published physical protein-protein interactions. Moreover, we devise a quaternary structure model of the AAA-ATPase-CP sub-complex in the 26S proteasome. The resulting model provides some insights into the chaperone-mediated assembly of CP and RP.
Materials and Methods
Density segmentation
We segmented the density of the AAA-ATPase from the EM map by exploiting the approximate 3-fold rotational symmetry using the method described in [9]. In an area approximately corresponding to the position of the AAA-ATPase hexamer, the density was symmetrized along a 3-fold rotation axis and thresholded by a gray value such that the included volume comprised 300 kDa (protein density: 1.3 mg/ml). The rotation axis was iteratively refined to maximize the correlation of the segmented map and the original data (6-dimensional search).
Comparative modeling
We built comparative models of the N-domains of Rpt1-6 using PAN-N as a structural template (PDB code 2WG6; 12-33 % sequence identity between the Rpt subunits and PAN-N). For the AAA-domains, we used the AAA-fold of PAN as a template (3H4M; sequence identities 52-60%). The structure of the PAN AAA-ring was approximated by superposing the PAN monomers on the HsLU hexamer (1DO2), as proposed in [7]. For initial model building we chose an arbitrary AAA-ATPase order (1/2/4/3/6/5). We omitted those segments from our models that were absent from the templates: the N-termini, the linker of N-ring and AAA-ring, and the C-termini. Comparative models were built by MODELLER [10] using multiple sequence alignments from T-COFFEE [11]. A comparative model of the D. melanogaster CP was built based on the bovine CP (1IRU, 55-77% sequence identity for the α subunits and 39-68% for the β subunits).
Quaternary structure of AAA-ATPase-CP
To determine the configuration of the different AAA-ATPase-subunits and the position of the CP in the 26S EM map we (i) determined the configuration of the AAA-ATPase subunits in the hexamer that are most consistent with the compiled physical protein-protein interactions and the segmented AAA-ATPase map; (ii) calculated the placement of the CP based on the best fit to the 26S proteasome EM map; (iii) inferred the rotation of the AAA-ATPase hexamer on the CP from our compiled list of protein-protein interactions.
AAA-ATPase
We fitted the N-ring and the AAA-ring, as built in their initial arbitrary order, into the 3-fold symmetrical AAA-ATPase map in Chimera (correlation coefficient 0.71) [12]. For every subunit order, two different fits of N-ring and AAA-ring were consistent with the maximum linker length (9 residues) between N- and AAA-ring (Fig. 2 B/C).
Figure 2.
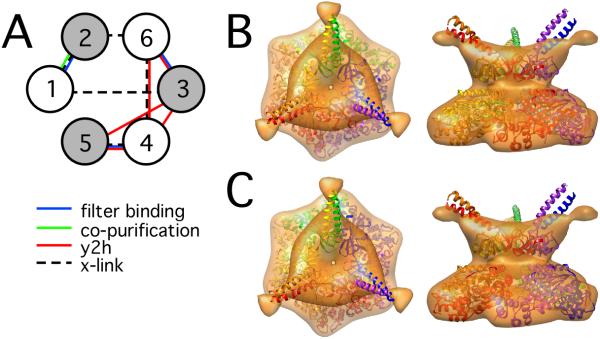
Best-scoring configurations of the AAA-ATPase subunits. A: Schematic representation of the AAA-ATPase subunit order and the contact restraints acting on them (Table 1). The gray circles represent subunits with Pro-62 in the cis-conformation. B: One of two possible quaternary structure arrangements of the N- and AAA-ring fitted into the segmented 3-fold symmetric EM map. C: The AAA-ring is rotated by 60° with respect to the N-ring in the second quaternary structure. In the two quaternary structures, a 9-residue linker spans a distance of approximately 21 Å and 26 Å, respectively, both of which are compatible with a typical Cα-Cα distance of 3.5 Å.
Subsequently, the different AAA-ATPase configurations were sampled by rotating the different Rpt subunits into all possible positions using the DOMINO optimizer for both quaternary structures [13]. Theoretically, the Rpt subunits can be positioned in 240 different configurations (120 for each quaternary structure). We only retained those 30 configurations that placed Rpt2, 3, and 5 in the cis positions of the ring (Fig. 2). The configurations were scored based on contact restraints derived from physical protein-protein interactions (Table 1), as described below. Finally, the comparative models were rebuilt for the two possible quaternary structures in the (1/2/6/3/4/5) order (Fig. 2 B/C).
Table 1.
Protein interactions in the CP-AAA-ATPase sub-complex. Protein-protein interactions were compiled from Yeast-two-hybrid screens (y2h), chemical cross-linking (x-link), protein binding to filter matrix (filter), different pulldown experiments (pulldown), and co-purification (co-puri). Since y2h screens are known to be prone to false-positive detections [17], we incorporated only those y2h interactions that have been reported in at least two independent studies. The data were obtained from 26S proteasomes of Homo sapiens (h), Saccharomyces cerevisiae (y), Sus barbatus (p), and Caenorhabditis elegans (w).
| Sub-complex | Technique | References | Species |
|---|---|---|---|
| Rpt1-Rpt2 | filter, x-link, co-puri | [8; 38; 39] | y,h |
| Rpt1-Rpt3 | x-link | [8] | h |
| Rpt1, Rpt2, Rpt3, Rpt6 | filter | [38] | h |
| Rpt2-Rpt6 | x-link | [8] | h |
| Rpt3-Rpt4 | Y2h | [40; 41; 42] | y |
| Rpt3-Rpt5 | Y2h, x-link | [8; 40; 42; 43; 44] | y,h |
| Rpt3-Rpt6 | Y2h, filter | [38; 40; 41; 43] | y,h |
| Rpt4-Rpt5 | Y2h, filter, x-link | [8; 19; 38; 43; 44] | w,y,h |
| Rpt4-Rpt6 | Y2h, x-link | [8; 41; 45] | y,h |
| Rpt4-alpha4 | Y2h | [19; 41] | y,w |
| Rpt4-alpha6 | x-link | [8] | h |
| Rpt6-alpha2 | x-link | [46] | p |
CP
We determined the position of the CP in the 26S proteasome map based on cross-correlation. Due to the pseudo 7-fold symmetry of the CP, there are 7 different similarly scoring fits of the CP in the EM map. We determined the cross-correlation coefficients of all these 7 possibilities with respect to the EM map by first rotating the model into the approximate position and then automatically refining it (rotational correlation function, RCF). The resulting RCF exhibited a significant maximum in one position (Supplementary Fig. S1) and the auto-RCF of the CP model showed a qualitatively similar shape.
AAA-ATPase-CP
We rotated the two quaternary structure of the AAA-ATPase hexamer (1/2/6/3/4/5 order) into the three different positions. The 6 conformations were scored based on the AAA-ATPase-CP contact restraints (Table 1).
Scoring of contact restraints
A given contact restraint was considered to be violated when the distance between the closest intermolecular atom pairs was larger than the sum of the atom radii plus a tolerance [14]. The tolerance value reflected the inaccuracy of the model and the resolution of the experimental technique. The model error was approximated to be 3 Å for the relative positions of the AAA-ATPase atoms. The error of the CP-AAA-ATPase interface was estimated to be higher (15 Å) due to the omitted AAA-ATPase C-termini, which bind to the CP [15], and because the positions of AAA-ATPase hexamer on the CP is likely variable (‘wobbling’) [15]. For the contact restraints from chemical cross-linking experiments, we additionally considered the length of the cross-linker arm: 16.1 Å for sEGS and 11 Å for DTBP [8]. The resulting tolerance was the square root of the sum of the squared errors of model and experiment. All calculations were performed using the Integrative Modeling Platform (IMP, http://www.salilab.org/IMP) [14].
Fitting of the coiled-coils
The coiled-coil pairs were treated as rigid bodies, connected to the remaining AAA-ATPase by flexible linkers (5 residues). The positions of the coiled-coil pairs and the flexible linker residues, as well as the ‘end-of-track bumpers’ (see below), were refined based on the correlation with the EM map, volume exclusion, and terms a molecular mechanics force field. Minimization of the score was performed using a conjugate gradients method starting from the comparative model and leaving the remaining AAA-ATPase fixed [16].
Results and Discussion
Order of the AAA-ATPase ring
In a previous analysis of the AAA-ATPase complex in the cryo-EM map of the D. melangaster 26S proteasome we assumed that the coiled-coil domains could not be resolved due to the flexible linkage of these domains [6]. Accordingly, we omitted the N-terminal coiled-coil motif from our previous AAA-ATPase model [9]. The structure of PAN is approximately 6-fold rotationally symmetric in the absence of the coiled-coils. The coiled-coil pairing leads to breaking the 6-fold symmetry, while preserving a 3-fold rotational symmetry [6]. Thus, we imposed the approximate 6-fold rotational symmetry of the hexamer for segmentation of the AAA-ATPase from the EM data [9].
On visual inspection of the original EM data we observed significant density at the sites where the coiled-coils are expected to be located in the EM density (Fig. 1 A). The coiled-coils are particularly well recognizable in the map with less density (‘without’) of the two maps lacking a variable compartment (probably Rpn10) [9]. To visualize the AAA-ATPase hexamer including the coiled-coils, we segmented the corresponding density from the 26S map using the 3-fold rotational symmetry of PAN (Materials and Methods). The resulting segmented AAA-ATPase density clearly reveals three extensions at the upper ring (Fig. 1 C).
Figure 1.

Segmentation of the AAA-ATPase from the 26S proteasome map. A. The ATPase density (orange) was segmented from the 26S proteasome map (yellow) based on the approximate 6-fold symmetry of the AAA-ring. Three distinct rod-shaped features protrude from the AAA-ATPase density (red). The displayed map is one of two 26S conformations described in [9], which lacks an additional mass (probably Rpn10). B. Segmented AAA-ATPase using 6-fold symmetry. The rod-like features are not present in the map at the chosen contour level. C. Segmented AAA-ATPase using 3-fold symmetry. At the three corners of the map rod-like features project out of the density (red).
We next attempted to fit comparative models of the AAA-ATPases into the segmented 3-fold symmetrical density (Materials and Methods). For a given subunit order, the AAA-ring can be placed in two different positions with respect to the N-ring, which are both compatible with a 9-residue linker between the two rings. We scored all possible permutations of the subunits according to physical protein-protein interactions (Table 1). Furthermore, we allowed only those configurations that place Rpt2, 3, and 5 in the cis positions of the ring (Fig. 2 A) [6]. Only one subunit order, Rpt (1/2/6/3/4/5) (enumerated clockwise when seen from the N-termini), violates the minimum of two contact restraints. Both violated contact restraints stem from yeast-two hybrid technique (Rpt3-Rpt5 and Rpt4-Rpt6), which is typically prone to false-positive detections [17]. While the subunit order is unambiguous at this stage of the analysis, two possibilities remain for the quaternary structure of N-ring and AAA-is (Fig. 2 B/C). Interestingly, omitting the positional constraint on Rpt2,3,5 does not result in any models with fewer violated contact restraints; e.g., the (1/2/6/4/5/3) topology violates three contact restraints.
In our study, we incorporated numerous additional protein-protein interactions that were reported subsequent to ref. [8], which resulted in the (1/2/6/4/5/3) topology. Importantly, we also knew the PAN-N crystal-structure, where the hexamer forms a pore of approximately 11 Å in diameter. The length of the sEGS cross-linker spacer arm (16.1 Å) used in [8] allows formation of cross-links across the pore, which was presumably not anticipated. In our model, the cross-link Rpt1-Rpt3 connects two opposite subunits spanning the entire pore. Interestingly, the intensity of the cross-linked protein pair Rpt1-Rpt3 was faint (Figure 1 b in [8]); the weak intensity could be due to the low accessibility of the cross-linked residues inside the pore. Notably, the Rpt5/Rpt1/Rpt2 order in our model is also consistent with the Hsm3/Rpn1/Rpt1/Rpt2/Rpt5 base precursor complex [18] (see below), whereas Rpt5 is not in direct contact with Rpt1 or Rpt2 in the (1/2/6/4/5/3) model.
Configuration of the CP-AAA-ATPase sub-complex
To determine the configuration of CP and AAA-ATPase in the 26S proteasome we first placed the CP into the EM density based on the maximal correlation coefficient (Materials and Methods). Subsequently, we positioned the AAA-ATPase on top of the CP (order: 1/2/6/3/4/5). The approximate 3-fold symmetry of the hexameric AAA-ATPase model and the corresponding EM map allows positioning each of the two quaternary AAA-ATPase structures (Fig. 2 B/C) in three different ways on top of the CP. Since the models of all AAA-ATPase subunits were derived from the same template and the resolution of the EM map is only approximately 20 Å, the correlation coefficient of these six models to the EM map does not exhibit significant differences. Thus, we scored the 6 AAA-ATPase-CP configurations exclusively based on the protein-protein interactions (Table 1). One configuration is consistent with a maximum of 4 out of 5 CP-ATPase contact restraints (Fig. 3); only one yeast-two hybrid interaction is violated (Rpt4-α4), which was reported as weak [19].
Figure 3.
AAA-ATPase-CP model. A: The atomic model of the AAA-ATPase-CP sub-complex fitted into the cryo-EM map of the 26S proteasome. The AAA-ATPase model is placed on the top. The six AAA-ATPase subunits and the adjacent CP α-subunits are colored as indicated in panel C, the remaining CP subunits in gold. The AAA-ATPase quaternary structure corresponds to Fig. 2C. B: Zoomed-in stereo view of the rectangular area in A. C: Schematic view of the topology of the CP-AAA-ATPase model and the CP-RP interactions (Table 1). The AAA-ATPase subunits are displayed in Arabic numbers, whereas Roman numbers indicate the CP α-subunits. The asterisks mark the AAA-ATPase subunits that have been shown to induce gate opening of the CP (Rpt2 and Rpt5).
In our model, the AAA-ATPase possesses an eccentric and slightly tilted position on top of the CP. The subunit pair Rpt3/6 is largely buried by the remaining density of the 26S proteasome corresponding to the non-ATPase subunits of the 26S proteasome. In concordance with our model, Rpt3 and Rpt6 were recently found as the only AAA-ATPases in a RP sub-complex including all non-ATPases except for Rpn1 [20]. The coiled-coils of pair Rpt1/2 are partially accessible in the 26S proteasome conformation ‘without’, whereas they are largely shielded by non-ATPases in the higher density conformation ‘with’ [9]. The coiled-coils of AAA-ATPase pair Rpt4/5, however, are exposed in both EM maps, suggesting that they mediate interactions in protein degradation. Indeed, Rpt4 was shown to be involved in the endoplasmic reticulum (ER) association of 26S proteasomes [21]. In addition, Rpt5 was reported to be central to substrate recognition, possibly even acting as an ubiquitin receptor [22], which is also compatible with an exposed positioning of Rpt5.
X-ray crystallographic analysis of the CP/11S complex suggested that the C-termini of the AAA-ATPase subunits bind at the interfaces of neighboring CP subunits [23], which was confirmed for the C-terminal peptides of PAN by cryo-EM studies [24]. Specifically, the C-termini of Rpt2 and Rpt5 are mainly responsible for gate opening [25; 26]. Our model places the C-terminus of Rpt2 between the α3 and α4 subunits, and Rpt5 between α6 and α7. Genetic studies have revealed that α3 and α7 control degradation of the CP [27], which could be explained by their interaction with Rpt2 and Rpt5 in our model.
Opening angle of the coiled-coils
It is apparent that the opening angle between the coiled-coil pairs and the pseudo-3 fold rotational symmetry axis of the AAA-ATPase hexamer in the EM map is larger than for PAN-N (Fig. 2 B,C). The PAN-N structure suggests that the opening angle is determined by two loops in the OB-folds acting as ‘end-of-track bumpers’ [6]. In particular, the sequence of the bumper limiting the outward motion is not conserved (Figure S2), which is consistent with a different opening angle of the coiled-coil domains in the Rpt hexamer compared to PAN-N.
To approximate the coiled-coil opening angles in the 26S proteasome we performed flexible fitting of the coiled-coil domains in the 26S proteasome EM map (Fig. 3 A/B). Due to the low resolution of the EM map the resulting positions are only meant to provide an approximation of the differences in coiled-coil positions in the 26S proteasome compared to PAN-N. The coiled-coil pairs are rotated by 5° (Rpt1/2), 21° (Rpt6/3), and 15° (Rpt4/5), respectively (average: 14°).
The opening angle of the coiled-coils probably changes in the functional cycle. The placement of the AAA-ATPase subunits in the EM map suggests that the coiled-coil pairs of Rpt4/5 and Rpt1/2 have substantial freedom to move, for example upon substrate recognition. In contrast, fluctuation of the Rpt3/6 coiled-coils would induce large-scale motion of the lid. Such a ‘wagging’ motion of the lid has been observed previously [28].
Implications for the assembly mechanism of the 26S proteasome
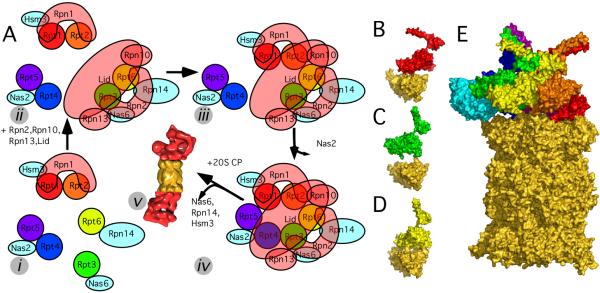
The assembly of the RP, and subsequently the assembly of the RP and CP is mediated by multiple dedicated assembly chaperones [18; 29; 30; 31; 32]. When incorporating our structural data into the recent mechanistic data the following assembly mechanism emerges (Fig. 4A). Isolation of various chaperone-bound sub-complexes indicates at least 5 different assembly steps: (i) Different base-precursor complexes form, each associated with assembly chaperones. The largest base-precursor complex consists of Rpt1/Rpt2/Rpn1/Hsm3 [30; 32], possibly also Rpt5 [18]. In our ATPase order (1/2/6/3/4/5), Rpt5 and Rpt1 physically interact in contrast to the previously suggested (1/2/6/4/5/3) order [8]. Rpt4 forms a complex with Nas2 [18], and probably also Rpt5 [20; 30; 32]. The experimental data for Rpt5 in both precursor complexes strongly supports the (5/1/2) sequence in our AAA-ATPase order. The AAA-ATPases Rpt6 and Rpt3 apparently remain unpaired and bind to Rpn14 and Nas6, respectively. It was suggested that Rpn1 and Rpn2 are centrally located in the AAA-ATPase hexamer [33], which would implicate Rpn1 forming a nucleus for the Rpt1/Rpt2/Rpn1/Hsm3 base precursor complex [30]. However, the cavity in PAN and our AAA-ATPase hexamer is much too small to accommodate either, Rpn1 or Rpn2 [6]. Thus, we suggest that the most likely horseshoe-shaped Rpn1 [34] embraces the ATPase subunits in the RP as well as the base-precursor complex, rather than forms a toroidal nucleus. (ii) When Rpt3/6 pair the RP lid as well as the base subunits Rpn1, Rpn10, and Rpn13 associate [20]. In the base, Rpn2 probably embraces Rpt3, and Rpt6, similar as Rpn1 and Rpt1/Rpt2 [18]. (iii) Nas6 and Hsm3 catalyze binding of Rpt1/Rpt2/Rpn1 to the lid/Rpn1/Rpn10/Rpn13/Rpt3/Rpt6 sub-complex [32]. (iv) The AAA-ATPase ring is completed and releases the assembly chaperone Nas2 [18; 30]. The completed RP is still associated to the assembly chaperones Nas6, Rpn14, and Hsm3. (v) The RP binds to the CP upon dissociation of Nas6, Rpn14, and Hsm3.
Figure 4.
Different assembly chaperones are required to position the AAA-ATPases on the CP. A: Model for the assembly process of the 26S proteasome. The assembly consists of at least five different steps: (i) Formation of different base-precursor complexes, each bound to a different chaperone (cyan). (ii) The lid and the remaining base subunits form a sub-complex with the AAA-ATPase pair Rpt3/Rpt6. (iii) An RP precursor lacking only Rpt4/Rpt5 forms. (iv) The Rpt4/Rpt5 dimer completes the RP and the chaperone Nas2 dissociates. (v) The three chaperones Nas6, Rpn14, and Hsm13 dissociate upon binding of RP and CP. B: Quaternary structure of Rpt1 (binds to assembly chaperone Hsm3) and the two adjacent CP alpha subunits (gold). C: Same for Rpt3 (binds to Nas6), and D: Rpt6 (Rpn14). E: Positioning Nas6 (PDB code 2DZN, [36]) on our CP-AAA-ATPase model does not lead to steric clashes with the CP.
Nas6, Rpn14, and Hsm3 bind the C-terminus of a specific AAA-ATPase: Nas6 binds to Rpt3, Hsm3 to Rpt1, and Rpn14 to Rpt6. Interestingly, the assembly chaperones likely adopt entirely different folds [31]. Why are completely different enzymes required to mediate the assembly of similar ATPase subunits with similar CP α-subunits? Based on our CP-AAA-ATPase model we propose an explanation for the requirement of 3 different types of chaperones for CP-RP assembly. In our model, the AAA-ATPase complex is positioned non-symmetrically on the CP, which seems also to be the case for the PAN-CP complex (Fig. 2 in [35]). Whereas the assembly of the 6-fold symmetrical AAA ring of PAN and the 7-fold symmetrical CP succeeds without chaperones in vitro [35], the efficient assembly of the non-symmetrical 26S AAA-ATPase hexamer and the non-symmetrical eukaryotic CP requires chaperones [18; 29; 30; 31; 32]. We hypothesize that the different chaperones are required to sterically ‘guide’ the AAA-ATPase subunits to their respective binding pockets at the CP. The chaperones would form an outer scaffold enabling the correct ‘fit’ of the ATPase ring on the CP. Since the subunits Rpt1, Rpt3, and Rpt6 adopt substantially different positions with respect to the adjacent α-subunits of the CP (Fig 4 B,C,D), structurally distinct chaperones are required. Conformational changes of the AAA-ATPase C-termini upon binding into their pockets in the CP could then trigger the chaperone release.
The putative position of Nas6 in our CP-AAA-ATPase model is consistent with the chaperone causing a unique fit of the RP on the CP: Nas6 was co-crystallized with a fragment of Rpt3 [36]. We placed Nas6 into our model by superposing the co-crystallized Rpt3 fragment to the Rpt3 subunit of our model (Fig. 4 E). In the resulting complex, Nas6 embraces Rpt3 and the adjacent CP subunits α1 and α2 without leading to major steric clashes, reminiscent of Dmp1-Dmp2 in the assembly of the CP α-ring [37]. Thus, our model indicates that physical occlusion of Nas6 and the CP subunits plays a less important role in the release of the chaperones than previously assumed [31]. We argue that the probably distinct conformations of the AAA-ATPase C-termini in the chaperone-bound and CP-bound states are mutually exclusive and hence responsible for releasing the chaperones upon binding to the CP.
Supplementary Material
Acknowledgments
FF is grateful to a long-term fellowship from the Human Frontier Science Project Organization (HFSPO). The research of KL was supported by continuous mentorship from Haim J. Wolfson as well as fellowships from the Edmond J. Safra Bioinformatics Program at Tel-Aviv University and the Clore Foundation Ph.D Scholars program and was carried out in partial fulfillment of the requirements for the Ph.D. degree at TAU. This work was supported by the DFG Cluster of Excellence “Munich-Centre for Advanced Photonics”. AS was supported by the Sandler Family Supporting Foundation, National Institutes of Health (R01 GM54762, U54 RR022220, PN2 EY016525, and R01 GM083960), National Science Foundation (IIS-0705196), Ron Conway, Mike Homer, Hewlett-Packard, NetApp, IBM, and Intel.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- [2].Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- [3].Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–68. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- [4].Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–9. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- [5].Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–71. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- [6].Djuranovic S, Hartmann MD, Habeck M, Ursinus A, Zwickl P, Martin J, Lupas AN, Zeth K. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–90. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- [7].Zhang F, Hu M, Tian G, Zhang P, Finley D, Jeffrey PD, Shi Y. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–84. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hartmann-Petersen R, Tanaka K, Hendil KB. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch Biochem Biophys. 2001;386:89–94. doi: 10.1006/abbi.2000.2178. [DOI] [PubMed] [Google Scholar]
- [9].Nickell S, Beck F, Scheres S, Korinek A, Förster F, Lasker K, Mihalache O, Sun N, Nagy I, Sali A, Plitzko JM, Carazo JM, Mann M, Baumeister W. Insights into the molecular architecture of the 26S proteasome. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0905081106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- [11].Notredame C, Suhre K. Computing multiple sequence/structure alignments with the T-coffee package. Curr Protoc Bioinformatics. 2004 doi: 10.1002/0471250953.bi0308s04. Chapter 3. Unit3 8. [DOI] [PubMed] [Google Scholar]
- [12].Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- [13].Lasker K, Topf M, Sali A, Wolfson HJ. Inferential optimization for simultaneous fitting of multiple components into a CryoEM map of their assembly. J Mol Biol. 2009;388:180–94. doi: 10.1016/j.jmb.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Alber F, Forster F, Korkin D, Topf M, Sali A. Integrating Diverse Data for Structure Determination of Macromolecular Assemblies. Annu Rev Biochem. 2008;77:443–477. doi: 10.1146/annurev.biochem.77.060407.135530. [DOI] [PubMed] [Google Scholar]
- [15].Saeki Y, Tanaka K. Unlocking the proteasome door. Mol Cell. 2007;27:865–7. doi: 10.1016/j.molcel.2007.09.001. [DOI] [PubMed] [Google Scholar]
- [16].Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fields S. High-throughput two-hybrid analysis. The promise and the peril. Febs J. 2005;272:5391–9. doi: 10.1111/j.1742-4658.2005.04973.x. [DOI] [PubMed] [Google Scholar]
- [18].Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–70. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Davy A, Bello P, Thierry-Mieg N, Vaglio P, Hitti J, Doucette-Stamm L, Thierry-Mieg D, Reboul J, Boulton S, Walhout AJ, Coux O, Vidal M. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2001;2:821–8. doi: 10.1093/embo-reports/kve184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Thompson D, Hakala K, Demartino GN. Subcomplexes of PA700, the 19S regulator of the 26S proteasome, reveal relative roles of AAA subunits in 26S proteasome assembly, activation, and ATPase activity. J Biol Chem. 2009 doi: 10.1074/jbc.M109.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lipson C, Alalouf G, Bajorek M, Rabinovich E, Atir-Lande A, Glickman M, Bar-Nun S. A proteasomal ATPase contributes to dislocation of endoplasmic reticulum-associated degradation (ERAD) substrates. J Biol Chem. 2008;283:7166–75. doi: 10.1074/jbc.M705893200. [DOI] [PubMed] [Google Scholar]
- [22].Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–7. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- [23].Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–99. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- [24].Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–8. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–44. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–22. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kohler A, Cascio P, Leggett DS, Woo KM, Goldberg AL, Finley D. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol Cell. 2001;7:1143–52. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- [28].Walz J, Erdmann A, Kania M, Typke D, Koster AJ, Baumeister W. 26S proteasome structure revealed by three-dimensional electron microscopy. J Struct Biol. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- [29].Saeki Y, Toh EA, Kudo T, Kawamura H, Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137:900–13. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- [30].Funakoshi M, Tomko RJ, Jr., Kobayashi H, Hochstrasser M. Multiple Assembly Chaperones Govern Biogenesis of the Proteasome Regulatory Particle Base. Cell. 2009;137:887–99. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, Lee BH, Zhang F, Shi Y, Gygi SP, Finley D. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–5. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kaneko T, Hamazaki J, Iemura S, Sasaki K, Furuyama K, Natsume T, Tanaka K, Murata S. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–25. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- [33].Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat Struct Mol Biol. 2008;15:573–80. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Effantin G, Rosenzweig R, Glickman MH, Steven AC. Electron microscopic evidence in support of alpha-solenoid models of proteasomal subunits Rpn1 and Rpn2. J Mol Biol. 2009;386:1204–11. doi: 10.1016/j.jmb.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–98. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- [36].Nakamura Y, Umehara T, Tanaka A, Horikoshi M, Padmanabhan B, Yokoyama S. Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem Biophys Res Commun. 2007;359:503–9. doi: 10.1016/j.bbrc.2007.05.138. [DOI] [PubMed] [Google Scholar]
- [37].Yashiroda H, Mizushima T, Okamoto K, Kameyama T, Hayashi H, Kishimoto T, Niwa S, Kasahara M, Kurimoto E, Sakata E, Takagi K, Suzuki A, Hirano Y, Murata S, Kato K, Yamane T, Tanaka K. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat Struct Mol Biol. 2008;15:228–36. doi: 10.1038/nsmb.1386. [DOI] [PubMed] [Google Scholar]
- [38].Richmond C, Gorbea C, Rechsteiner M. Specific interactions between ATPase subunits of the 26 S protease. J Biol Chem. 1997;272:13403–11. doi: 10.1074/jbc.272.20.13403. [DOI] [PubMed] [Google Scholar]
- [39].Takeuchi J, Tamura T. Recombinant ATPases of the yeast 26S proteasome activate protein degradation by the 20S proteasome. FEBS Lett. 2004;565:39–42. doi: 10.1016/j.febslet.2004.03.073. [DOI] [PubMed] [Google Scholar]
- [40].Cagney G, Uetz P, Fields S. Two-hybrid analysis of the Saccharomyces cerevisiae 26S proteasome. Physiol Genomics. 2001;7:27–34. doi: 10.1152/physiolgenomics.2001.7.1.27. [DOI] [PubMed] [Google Scholar]
- [41].Chen C, Huang C, Chen S, Liang J, Lin W, Ke G, Zhang H, Wang B, Huang J, Han Z, Ma L, Huo K, Yang X, Yang P, He F, Tao T. Subunit-subunit interactions in the human 26S proteasome. Proteomics. 2008;8:508–20. doi: 10.1002/pmic.200700588. [DOI] [PubMed] [Google Scholar]
- [42].Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- [43].Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. Embo J. 2001;20:7096–107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Russell SJ, Sathyanarayana UG, Johnston SA. Isolation and characterization of SUG2. A novel ATPase family component of the yeast 26 S proteasome. J Biol Chem. 1996;271:32810–7. doi: 10.1074/jbc.271.51.32810. [DOI] [PubMed] [Google Scholar]
- [46].Satoh K, Sasajima H, Nyoumura KI, Yokosawa H, Sawada H. Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry. 2001;40:314–9. doi: 10.1021/bi001815n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.