Abstract
We present here a new two-hybrid smart pool array (SPA) system in which, instead of individual activation domain strains, well-designed activation domain pools are screened in an array format that allows built-in replication and prey-bait deconvolution. Using this method, a Saccharomyces cerevisiae genome SPA increases yeast two-hybrid screening efficiency by an order of magnitude.
Mapping protein interactions on a genomic scale is one of the major goals of systems biology1,2. The yeast two-hybrid system has been an important platform in these efforts3. Owing to the high demand in time and resources for interactome mapping, even with the help of robotic equipment, the need for highly efficient ‘smart pooling’ designs has been well recognized4-6. Recently, we developed a new pooling-deconvolution method to pool probes or baits, which improves the accuracy, coverage and efficiency of large-scale array screening simultaneously5. This method assigns 2n bait (BD) strains into n pairs of pools, screens bait pools against prey (AD) array and deconvolutes the hits based on the pattern of positive signals in the 2n array experiments5. Bait pooling, however, does not provide as large a benefit to most investigator-initiated research programs, which often focus on screening only one or a few select baits. Here we show that the same pooling-deconvolution principle can be applied to pool prey (AD) strains, permitting efficient screening of individual baits with high accuracy and coverage. Prey-based pools are advantageous over bait-based pools because once prey pool arrays are prepared they can be maintained indefinitely and reused for new screens.
Another advantage of prey pooling is apparent upon considering the established two-hybrid selection procedures. Owing to fortuitous activating sequences, a considerable fraction of bait-BD fusions can activate the two-hybrid reporter gene (for example, the HIS3 gene herein) without the presence of any prey-AD fusion. Addition of 3-amino-triazole, an enzymatic inhibitor of His3, can compensate for auto-activation. Thus, it is possible to optimize two-hybrid selection conditions based on the auto-activation level of the individual bait (by varying 3-amino-triazole concentration in the medium). Given the small number of proteins in the genome that would bind to the GAL DNA sites (or other sequences) in the reporter construct, frequency of auto-activation by prey-AD fusions is insignificant compared to that caused by bait-BD fusions, and after prey pooling selection conditions can still be optimized for individual baits when necessary.
The SPA scheme for pooling 16 (= 24) strains using the pooling-deconvolution method is illustrated in Supplementary Figure 1 online. Briefly, 16 strains are mixed into 4 pairs of pools (pairs 0–3), with 8 (= 23) strains per pool. For example, strain 4 will be pooled into the ‘−’ pool of pairs 3 and 2, and the ‘+’ pool of pairs 1 and 0. This pooling scheme has two important properties of deconvolution and redundancy. First, deconvolution is possible because every strain is pooled into 4 different pools (one from each pair), so if one of the 16 strains is two hybrid–positive (for a given bait) then 4 of the 8 pools will yield a positive colony. Thus we can deconvolute the identity of the two-hybrid positive strain owing to its presence only in a specific combination of 4 pools and absence in the other pools.
The second important property of SPA is the built-in redundancy. Using the example in Supplementary Figure 1, each AD strain is tested four times against the bait resulting in a situation that is equivalent to four separate individual screens. This inherent replication can facilitate removal of false positives because false positives are unlikely to be observed reproducibly. Likewise, replicated screens will cover more true positives, because losing the same true positive repeatedly as a result of experimental variation is also less likely. In general, this scheme generates 2n pools from 2n strains, with a built-in ‘screen redundancy’ of n. Thus, when the number of strains is increased exponentially, for example, from 16 (= 24) to 32 (= 25), the number of pools only needs to be increased linearly, that is, from 8 (= 2 × 4) to 10 (= 2 × 5).
We constructed SPA arrays from a published yeast genome-wide two-hybrid AD array, which has been successfully used to screen hundreds of bait proteins7,8. It contains ~6,000 AD strains which are maintained in 64 96-well plates. To determine screen sensitivity on the pool arrays, we constructed three sets of smart pool arrays (SPA-4, SPA-5, SPA-6) with pool sizes of 8, 16 and 32 AD strains, resulting in screen redundancies of 4, 5 and 6, respectively. For example, to construct SPA-4, all the AD strains in the original prey array were divided into sets of 16 (= 24) strains. For each 16-strain set, we constructed 8 pools (Supplementary Fig. 1), and the resulting pool array (SPA-4) was half the size of the original prey array. Similarly, we pooled sets of 32 (= 25) and 64 (= 26)preys to create SPA-5 and SPA-6, respectively (Table 1 and Supplementary Note online). The unit of robotic pooling in use was a whole (96-well) plate (instead of single wells), allowing many (96 here) pools to be made at once (see Supplementary Table 1 online).
Table 1.
Yeast two-hybrid screen on SPA arrays
| Array size |
|||||||
|---|---|---|---|---|---|---|---|
| Pool arrays |
Encoding capacitya |
Pool size |
Screen redundancy |
Number of pool families |
Number of spots |
Number of 96-well plates |
Number of 384-spot agar plates |
| Original | 1 | 1 | 1 | 6,144 | 6,144 | 64 | 16b |
| SPA-4 | 16 | 8 | 4 | 384 | 3,072 | 32 | 8 |
| SPA-5 | 32 | 16 | 5 | 192 | 1,920 | 20 | 5 |
| SPA-6 | 64 | 32 | 6 | 96 | 1,152 | 12 | 3 |
Encoding capacity is the number of strains a pool family can cover. The original array with single AD strains can be considered as SPA-1 array with one strain per pool.
Thirty-two plates are needed in practice when using the original array (to obtain a minimal screening redundancy of 2). The number of plates needed when using SPA arrays does not change.
We next illustrate the use of SPA arrays for two-hybrid screening using the three pool sizes above. An example of screening one bait strain against pool array SPA-6, in which each spot represents a pool of 32 (= 25) strains is illustrated in Figure 1a. Screening on this three-plate array represents a complete screen of all 6,144 yeast AD strains, with a screening redundancy of six (Table 1). Compared to screening the original array with a minimum twofold redundancy, which requires 32 selection plates, using SPA-6 requires only 3 plates, thus increasing the screen efficiency by over one order of magnitude (Table 1).
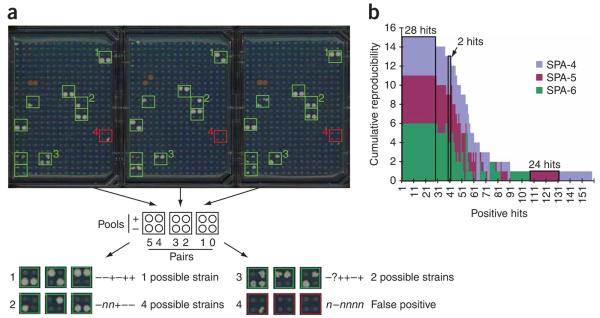
Figure 1.
Yeast genome two-hybrid screening using SPA arrays. (a) Every spot on the SPA-6 array represents a pool of 32 yeast AD strains and the original 6,144-strain yeast AD array was compressed into a SPA array with 96 12-pool sets, and each set represents 64 AD strains. The 12 spots in the three squares at the same position of the three plates belong to the same group (diagram below the plates). Green squares highlight the groups containing two-hybrid positive strains; red squares highlight a group with one false positive spot. Four examples of deconvolution are shown (examples 1–4). (b) Visualization of common positive hits across different SPA arrays. Screening 5 baits against 3 SPA arrays yields a total of 158 possible bait-prey pairs; hit detection by each of these arrays is shown. Many hits were detected on more than one SPA array, and the height of the colored areas indicates the reproducibility on the corresponding SPA array. For example, the 2 hits indicated on the graph have a reproducibility of four times on SPA-4, five times on SPA-5 and four times on SPA-6, whereas the 24 hits outlined in black were only found once on SPA-5. The method to identify common hits across different SPA arrays is described in the Supplementary Note.
We illustrate prey-bait deconvolution with SPA-6 (Fig. 1a), where a ‘pool family’ (Supplementary Note) consists of 12 (= 2 × 6) smart pools representing 64 AD strains. Because most yeast proteins bind to only 3–10 other proteins9, each set of 64 strains on SPA-6 most likely contains zero or only one two hybrid–positive strain (Supplementary Note). Therefore, the identity of the positive strain in a 64-strain set can be uniquely deconvoluted (to ‘+’ or ‘-’ profiles only) from the pattern of the corresponding 12 spots (Fig. 1a, example 1). However, when a 64-strain set contains more than one positive AD strain, there will be deconvolution ambiguity (‘?’ profiles). False positive or false negative spots can also cause ‘?’ or ‘n’ in the profile, but the profile can still be partially deconvoluted (Fig. 1a, examples 2 and 3). Profile degeneracy (‘?’ or ‘n’) is discussed further in Supplementary Note. Briefly, ‘?’ and ‘n’ will each be considered to encompass ‘+’ and ‘−’. For example, ‘+ + ? +’ will be partially deconvoluted to two possibilities (‘+ + + +’ or ‘+ + − +’); and ‘+ + n +’ yields the same two possibilities. Hits that show up only once (‘nonreproducible’ hits, such as example 4 in Fig. 1a) will be removed as false positives because false positive spots usually lack reproducibility.
We screened each of the five bait strains (Glc7, Lsm8, Nse3, Pcf11 and Pho85) against SPA-4, SPA-5 and SPA-6 to assess the performance of SPA with different pool sizes. A complete list of positive hits and their profiles is shown in Supplementary Table 2a online (see Supplementary Table 2b–d for validation of ambiguous hits).
Different SPA arrays identified comparable number of hits (Table 2). The reproducibility information can help us remove false positive signals. For example, in SPA-5 screening, although (38 out of 94 (~40%) hits were ‘non-reproducible’ (only one positive pair in the pool family), among the 56 ‘reproducible’ hits, only 4 hits were reproduced below 3 times and 9 hits below 4 times (Table 2). This strongly indicates that the ‘nonreproducible’ hits are caused by false positive signals, and screening in SPA format can remove them efficiently.
Table 2.
Performance of SPA arrays
| Reproducibility |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Pool arrays | Total | Nonreproducible | Reproducible | = 2 | = 3 | = 4 | = 5 | = 6 | Uniquely deconvoluted |
| SPA-4 | 108 | 38 | 70 | 10 | 8 | 52 | NA | NA | 48 |
| SPA-5 | 94 | 38 | 56 | 4 | 5 | 5 | 42 | NA | 40 |
| SPA-6 | 88 | 30 | 58 | 10 | 3 | 3 | 10 | 32 | 31 |
For example, for SPA-4 array, a total of 108 hits were identified, and 70 of these were reproduced at least twice among all 4 SPA pairs; 10, 8 and 52 of the reproducible hits were observed for 2, 3 and 4 times, respectively; 48 hits can each be deconvoluted to one prey. NA, not applicable.
It is important to test whether screening sensitivity is compromised when pool size is increased. We conclude that increasing pool size from 8 to 32 does not compromise the sensitivity of detecting the reproducible hits, because all three SPA arrays covered about the same number of reproducible hits as those from independent data sets, including data from our pairwise retest experiments and other independent screening projects (Supplementary Note and Supplementary Table 3 online). It is promising that although SPA-6 represents a much bigger pool size than SPA-5, the numbers of reproducible hits generated by screening SPA-5 and SPA-6 are almost the same (Table 2 and Supplementary Table 3). This trend is also illustrated in Figure 1b. It is evident that most highly reproducible hits on one SPA array can be detected on the other SPA arrays with high reproducibility also, whereas very few non-reproducible hits are common across different SPA arrays.
Finally, we compared the deconvolution performance of the SPA arrays. A large fraction of ‘reproducible’ (≥2) hits (>50%) can be unambiguously deconvoluted (Table 2). Most of the ambiguous hits can be deconvoluted fairly easily because hits with degenerate profiles can still be partially deconvoluted. Deconvolution ambiguity may be further clarified computationally with the help of bioinformatic predictions and genomic or proteomic data sets10-14, or by reciprocal (pairwise) confirmation. Ambiguity can also be resolved experimentally by testing the unresolved preys individually or, more efficiently, by using a ‘reshuffled’ pooling configuration (analogous to bait reshuffling as we previously described5). The additional deconvolution experiments to resolve ambiguity (‘ambiguity burden’) represent a very small number (<1 plate for the 5 baits) compared to that required by a duplicate screen of the original array without pooling (160 = 5 × 32 plates; Supplementary Table 4 online).
In summary, we constructed the first yeast two-hybrid SPAs of the S. cerevisiae genome and demonstrated their utility in two-hybrid screening at the genomic scale. SPA combines the advantages of both two-hybrid array screening and library screening approaches. Pooling of arrays greatly reduces array size and can increase screening efficiency by an order of magnitude. The identities of hits can be deduced without the need for extensive sequencing or secondary mating steps. The built-in redundancy in SPA facilitates efficient removal of false positive signals. Pooling does not compromise sensitivity of detecting highly reproducible hits, and once prepared, the AD strain pools can be maintained indefinitely and used repeatedly. The SPA system will greatly ease the efforts of inter-actome mapping and improve data quality, and should also be applicable to other large-scale efforts including screening small molecules that disrupt protein-protein interactions15.
Supplementary Material
ACKNOWLEDGMENTS
We thank an anonymous reviewer and H. Herschman for critical review and discussion. This research was partially supported by University of California Systemwide Biotechnology Research & Education Program Graduate Research and Education in Adaptive Bio-Technology (GREAT) Training Grant 2005-268, US National Institutes of Health National Human Genome Research Institute grant HG003729 (J.H.), and a Research Starter Grant from the PhRMA Foundation (T.H.).
Footnotes
Note: Supplementary information is available on the Nature Methods website.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Phizicky E, Bastiaens PI, Zhu H, Snyder M, Fields S. Nature. 2003;422:208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- 2.Barabasi AL, Oltvai ZN. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 3.Cusick ME, Klitgord N, Vidal M, Hill DE. Hum. Mol. Genet. 2005;14:R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 4.Zhong J, Zhang H, Stanyon CA, Tromp G, Finley RL., Jr. Genome Res. 2003;13:2691–2699. doi: 10.1101/gr.1134603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin F, et al. Nat. Methods. 2006;3:183–189. doi: 10.1038/nmeth859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thierry-Mieg N. BMC Bioinformatics. 2006;7:28. doi: 10.1186/1471-2105-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uetz P, et al. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 8.Hazbun TR, et al. Mol. Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- 9.Grigoriev A. Nucleic Acids Res. 2003;31:4157–4161. doi: 10.1093/nar/gkg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcotte EM, et al. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 11.Bader GD, Hogue CW. Nat. Biotechnol. 2002;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- 12.Schweitzer B, Predki P, Snyder M. Proteomics. 2003;3:2190–2199. doi: 10.1002/pmic.200300610. [DOI] [PubMed] [Google Scholar]
- 13.Krogan NJ, et al. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 14.Yu H, Paccanaro A, Trifonov V, Gerstein M. Bioinformatics. 2006;22:823–829. doi: 10.1093/bioinformatics/btl014. [DOI] [PubMed] [Google Scholar]
- 15.Huang J, Schreiber SL. Proc. Natl. Acad. Sci. USA. 1997;94:13396–13401. doi: 10.1073/pnas.94.25.13396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.