Abstract
The binding of a polymeric ligand to a cell surface receptor can promote its internalization. Methods to track and visualize multivalent ligands within a cell can give rise to new therapeutic strategies and illuminate signaling processes. We have used the features of the ring-opening metathesis polymerization (ROMP) to develop a general strategy for synthesizing multivalent ligands equipped with a latent fluorophore. The utility of ligands of this type is highlighted by visualizing multivalent antigen internalization in live B cells.
Advances in organic and polymer chemistry are providing access to new classes of bioactive polymers.1 With the ability to generate synthetic macromolecules that vary in structure and physical properties, the applications of polymers have expanded rapidly. Some uses include as substrates for cell growth and differentiation,2 vehicles for drug delivery,3 therapeutics,4 and molecules for studying cell signaling.5 For some biological applications, bioactive polymers must be taken up into cells. Still, little is known about the cellular internalization and trafficking of polymers. Here, we present an approach for following these processes.
The bioactive polymers that we focused upon were those generated by ruthenium carbene-initiated ring-opening metathesis polymerization (ROMP). It was shown that ROMP could be used to generate biologically-active polymers over 10 years ago.6 Since that time, ROMP has been used produce materials with a variety of biological activities.7 Continuous advances in catalyst development and polymerization techniques have provided methods to control key features of polymer structure, including their length and the functional groups they present.8 Ruthenium carbene initiators tolerate a diverse array of functional groups; therefore, they can be used synthesize highly functionalized polymers with tailored biological activities.
ROMP initially was used to generate polymers that act on the outside of the cell, but more recently, it has been employed to assemble compounds that can be internalized by cells. Our interest in polymer uptake was prompted by our studies of B cell signaling. 9 For example, polymers generated by ROMP that display antigenic epitopes can promote antibody production in vivo. The ability of these polymers to activate this process depends upon their interactions with the antigen-specific B cell receptor (BCR), a membrane-bound immunoglobulin on the surface of B cells. These polymers bind to the specific B cell receptor to activate signaling but they also promote its internalization. We therefore anticipated that, like other antigens, these polymers would be taken up by endocytosis. To investigate polymer internalization, we envisioned using antigenic polymers equipped with a group that would report directly on internalization.
Most approaches to following ligand internalization rely on appending a fluorescent dye to the molecule of interest. Uptake is detected using a discontinuous assay (typically fluorescence microscopy). Because extensive cell washing steps are required to remove background fluorescence, internalization cannot be tracked in real time. The need for multiple washing steps is exacerbated when the object of the study is a macromolecule, such as a polymer. Macromolecules are more likely to engage in non-specific interactions with cells, thereby increasing the assay background. We reasoned that polymers bearing a label, whose fluorescence is unmasked only upon endocytosis, could illuminate the processes of internalization and trafficking.
The latent fluorophore developed by Raines and coworkers can provide a sensitive measure of biomolecular entry into cells.10 The reporter dye possesses a trimethyl lock11 that masks fluorescence until it is released by cellular esterases (Figure 1). While a strategy to link a profluorophore of this type to proteins has been developed,10c the process used relies on a thiol and is not readily applicable to functionalizing compounds such as the polymers generated by ROMP. Thus, we sought to develop an alternative general strategy to attach a latent fluorophore.
Figure 1.
Upon cellular internalization of the labeled compound, esterases liberate the fluorophore.
The presence of the phenolic acetate group within the trimethyl lock dictates that the chemical reactions used to append the latent fluorophore to the polymer occur under mild conditions. We envisioned that the Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC), discovered independently by the Sharpless and Meldal groups,12 might offer an attractive solution. The reactive partners (azide and copper acetylide) are stable under many conditions yet react with each other with excellent chemoselectivity. Moreover, this reaction process has been used to modify polymers13 and to attach fluorophores to species that have undergone affinity labeling.14 To synthesize the target compounds, the best disconnection appeared to be an azide-substituted polymer and an alkyne-bearing fluorogenic label, such that the azide, not the alkyne, was appended to the polymer backbone.15 This dissection should avoid unwanted metathesis side reactions of the alkyne.16
Our strategy for generating the azide-substituted polymer relies on employing a selective end-labeling strategy.17 When ruthenium carbenes are used as initiators in ROMP, the polymer terminus can be modified selectively by terminating the polymerization reaction with a functionalized enol ether.17a–b,18 An enol ether capping agent with the requisite azide could be readily synthesized from dihydropyran (Scheme 1). Exposure to aqueous acidic media generated the incipient aldehyde, which was subjected to Wittig reaction with (methoxymethyl)-triphenylphosphonium chloride to afford enol ether 1 (3:1 mixture of E:Z isomers). When this species was treated with diphenylphosphoryl azide (DPPA)19 under Mitsunobu-like conditions20 the azide-terminated capping agent 2 was obtained.
Scheme 1.
Route to Azide-Containing Capping Agent
The new capping agent was used to append a single azide group to a polymeric antigen that can promote B cell receptor internalization. We had previously used ROMP to synthesize multivalent ligands that bear antigenic dinitrophenyl (DNP) epitopes, which bind to a cell line expressing a DNP-specific B cell receptor.9 Thus, we modified the pre-existing synthetic route to generate DNP-substituted polymers bearing a terminal azide group. A norbornene derivative that bears a succinimidyl ester was polymerized using a ruthenium initiator21 to generate polymers of defined lengths (Scheme 2). The degree of polymerization was controlled by the ratio of the ruthenium initiator to monomer (1:10 or 1:500). The reaction was terminated using an excess of the azide-containing enol ether 2. To convert the polymer into an antigen that can bind to the DNP-specific B cell receptor, we conjugated DNP-substituted lysine to the backbone. Previous reports from our laboratory indicated that substitution with 0.4 equivalents of DNP-lysine (relative to the succinimidyl ester groups) afforded polymers that could activate B cell signaling; therefore, we used reaction conditions to generate polymers with this level of backbone substitution. Glucamine was added to quench any remaining succinimidyl esters and to promote water solubility of the final product polymers.
Scheme 2.
Synthesis of DNP-Substituted Polymers by ROMP.
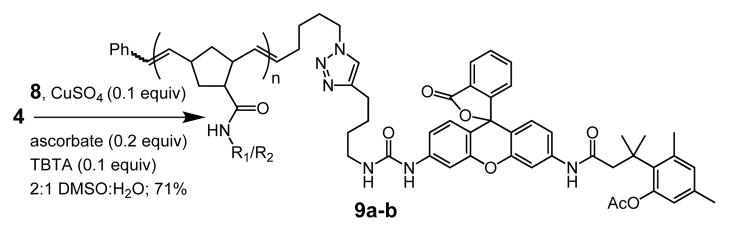
For conjugation of the latent fluorophore to the azide-terminated polymers 4a and 4b, we needed a masked dye with an alkyne handle. For this purpose, we used a rhodamine derivative bearing a urea linker.10c Previous studies had shown that a urea substituent has only a minor effect on the fluorescence intensity of the dye. The modified dye was generated by desymmetrization to afford Boc-protected rhodamine 5. The alkyne was introduced using an isocyanate generated in situ from the Curtius rearrangement of 6-heptynoic acid. The electrophilic intermediate was captured with amine 5 to afford the alkyne-terminated fluorophore 6, and the Boc protecting group was cleaved to yield the fluorescent rhodamine derivative 7. Condensation of 7 with trimethyl lock derivative 811 afforded the pro-fluorophore 9.
To attach the latent fluorophore to the polymer, we examined several conditions. The combination of CuI and CuBr in water–DMSO solvent afforded the desired triazole product but in low yield. To generate Cu(I) from Cu(II), sodium ascorbate can be used as a reducing agent.22 The Cu(I) oxidation state can be stabilized by the addition of ligands such as (benzyltriazolylmethyl)amine (TBTA), and this additive has been used to promote many bioconjugation reactions.23 We found that this reagent combination also facilitates attachment of the fluorogenic label to the polymer. Specifically, the conjugation of 4 and 7 was achieved in the presence of CuSO4, sodium ascorbate, and TBTA (Scheme 4). Importantly, no unmasking of the latent fluorophore was observed under these reaction conditions.
Scheme 4.
Profluorophore Attachment via a Cu(I)-Catalyzed Azide–Alkyne Cycloaddition.
With access to the labeled polymers, we tested whether a model esterase (pig liver esterase) could liberate the fluorophore. When polymer 9a was exposed to esterase (5 μM), the fluorophore was liberated with a kcat/KM value of 5.8 ×106 M−1s−1 (see the Supporting Information). These kinetic parameters suggest that the fluorogenic label can be used to monitor uptake processes that occur on the minute time scale. We next employed fluorescence microscopy to test whether polymer uptake could be detected in live cells. To label the DNP-specific B cell receptor without engaging multiple receptors, we used a fluorescent monovalent Cy3-conjugated anti-IgM fragment. Images collected immediately after polymer addition reveal diffuse staining of the B cell receptor, indicating that it is distributed across the cell surface (Figure 2B). No signal from the polymer was detected (Figure 2A). After 15 minutes, however, rhodamine fluorescence was apparent, indicating that the fluorogenic label serves as a sensitive reporter of polymer uptake (Figure 2C).
Figure 2.
Images of polymer-treated live A20.2J/HLTNP cells. Cells were treated for 30 min with Cy3-conjugated anti-IgM to stain the DNP-specific B cell receptor. Polymer 9b was added at 37 °C, and images were acquired without washing immediately (A,B) and after 15 minutes (C, D). Panel E depicts data from the emission of both the Cy3 and rhodamine probes. The scale bar represents 39 μm in panels A–B and 10 μm in C–F.
The observed polymeric antigen internalization is consistent with B cell receptor-mediated uptake. We had shown previously that DNP-substituted polymers promote B cell receptor internalization.9 The labeled polymers reveal that the B cell receptor and polymer 9b (M:I=500) colocalize (Figure 2E). In contrast, polymers lacking DNP groups were not taken up (see the Supporting Information). Together, the results suggest that receptor and polymer are internalized together. Most significantly, the fluorescence arising from unmasking of the fluorogenic label is present only in the interior of the cells. This result underscores the utility of the latent fluorophore for monitoring multivalent ligand uptake.
In summary, we have developed a strategy for synthesizing multivalent ligands equipped with a fluorogenic label. Our approach relies upon the attachment of a profluorophore to a polymer using the Cu(1)-catalyzed azide-alkyne cycloaddition. This approach is general; it can facilitate the conjugation of fluorophores or other prodrugs to a variety of azide-containing species. We have demonstrated its utility for generating reagents to track and visualize B cell receptor-mediated antigen uptake. This new approach provides a means to visualize the uptake and position of multivalent ligands within the cell. We anticipate that our method of fluorogenic label installation will facilitate studies of cellular internalization and trafficking of both multivalent and monovalent compounds.
Supplementary Material
Scheme 3.
Synthesis of Alkyne-Terminated Latent Fluorophore
Acknowledgments
This research was supported by the NIH (R01 AI055258). The UW-Madison Chemistry NMR facility is supported by NSF (CHE-0342998 and CHE-9629688) and the NIH (1-S10-RR13866). RTC was supported by the Chemistry–Biology Interface Training Program (T32 GM008505) The authors thank E.M. Kolonko, L.D. Lavis, and R.T. Raines for helpful discussions. TBTA was a generous gift of K.B. Sharpless. The A20.2J/HLTNP cells were a generous gift of A. Ochi.
Footnotes
Supporting Information Available: Experimental procedures and characterization data for compounds 1–4, 6, 7, 9, and 10. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hawker CJ, Wooley KI. Science. 2005;309:1200–1205. doi: 10.1126/science.1109778. [DOI] [PubMed] [Google Scholar]
- 2.(a) Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]; (b) Langer R, Tirrell DA. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]; (c) Ratner BD, Bryant SJ. Annu Rev Biomed Eng. 2004;6:41–75. doi: 10.1146/annurev.bioeng.6.040803.140027. [DOI] [PubMed] [Google Scholar]; (d) Drury JL, Mooney DJ. Biomaterials. 2003;24:4337–4351. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 3.(a) Kataoka K, Harada A, Nagasaki Y. Adv Drug Del Rev. 2001;47:113–131. doi: 10.1016/s0169-409x(00)00124-1. [DOI] [PubMed] [Google Scholar]; (b) Peppas NA, Bures P, Leobandung W, Ichikawa H. Eur J Pharm Biopharm. 2000;50:27–46. doi: 10.1016/s0939-6411(00)00090-4. [DOI] [PubMed] [Google Scholar]; (c) Christie RJ, Grainger DW. Adv Drug Del Rev. 2003;55:421–437. doi: 10.1016/s0169-409x(02)00229-6. [DOI] [PubMed] [Google Scholar]; (d) Murthy N, Xu M, Schuck S, Kunisawa J, Shastri N, Frechet JMJ. Proc Natl Acad Sci USA. 2003;100:4995–5000. doi: 10.1073/pnas.0930644100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Kiick KL. Science. 2007;317:1182–1183. doi: 10.1126/science.1145951. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Duncan R. Nat Rev Drug Discovery. 2003;2:347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]; (c) Lee CC, MacKay JA, Frechet JMJ. Nat Biotechnol. 2005;23:1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]; (d) Boas U, Heegard PMH. Chem Soc Rev. 2004;33:43–63. doi: 10.1039/b309043b. [DOI] [PubMed] [Google Scholar]; (e) Haag R, Kratz F. Angew Chem Int Ed. 2006;45:1198–1215. doi: 10.1002/anie.200502113. [DOI] [PubMed] [Google Scholar]
- 5.(a) Kiessling LL, Gestwicki JE, Strong LE. Angew Chem Int Ed. 2006;45:2342–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Borrok MJ, Kolonko EM. ACS Chem Biol. 2008;3:101–109. doi: 10.1021/cb700211s. [DOI] [PubMed] [Google Scholar]; (c) Mammen M, Choi SK, Whitesides GM. Angew Chem Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Mortell KH, Gringas M, Kiessling LL. J Am Chem Soc. 1994;116:12053–12054. [Google Scholar]
- 7.(a) Smith D, Pentzer EB, Nguyen ST. Polymer Reviews. 2007;47:419–459. [Google Scholar]; (b) Gestwicki JE, Kiessling LL. Nature. 2002;415:81–84. doi: 10.1038/415081a. [DOI] [PubMed] [Google Scholar]; (c) Lee Y, Sampson NS. Curr Opin Struct Biol. 2006;16:544–550. doi: 10.1016/j.sbi.2006.05.015. [DOI] [PubMed] [Google Scholar]; (d) Geng J, Mantovani G, Tao L, Nicolas J, Chen G, Wallis R, Mitchell DA, Johnson BRG, Evans SD, Haddleton DM. J Am Chem Soc. 2007;129:15156–15163. doi: 10.1021/ja072999x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Ladmiral V, Mantovani G, Clarkson GJ, Cauet S, Irwin JL, Haddleton DMJ. J Am Chem Soc. 2006;128:4823–4830. doi: 10.1021/ja058364k. [DOI] [PubMed] [Google Scholar]; (f) Kiessling LL, Owen RM. In: Handbook of Metathesis. Grubbs RH, editor. Vol. 3. Wiley VCH; Weinheim: 2003. pp. 180–225. [Google Scholar]; (g) Maynard HD, Okada SY, Grubbs RH. J Am Chem Soc. 2001;123:1275–1279. doi: 10.1021/ja003305m. [DOI] [PubMed] [Google Scholar]; (h) Rawal M, Gama CI, Matson JB, Hsieh-Wilson LC. J Am Chem Soc. 2008;130:2959–2961. doi: 10.1021/ja709993p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grubbs RH. Angew Chem Int Ed. 2006;45:3760–3765. doi: 10.1002/anie.200600680. [DOI] [PubMed] [Google Scholar]; (b) Schrock RR, Hoveyda AH. Angew Chem Int Ed. 2003;42:4592–4633. doi: 10.1002/anie.200300576. [DOI] [PubMed] [Google Scholar]
- 9.Puffer EB, Pontrello JK, Hollenbeck JJ, Kink JA, Kiessling LL. ACS Chem Biol. 2007;2:252–262. doi: 10.1021/cb600489g. [DOI] [PubMed] [Google Scholar]
- 10.(a) Chandran SS, Dickson KA, Raines RT. J Am Chem Soc. 2005;127:1652–1653. doi: 10.1021/ja043736v. [DOI] [PubMed] [Google Scholar]; (b) Lavis LD, Chao TY, Raines RT. ChemBioChem. 2006;7:1151–1154. doi: 10.1002/cbic.200500559. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lavis LD, Chao TY, Raines RT. ACS Chem Biol. 2006;1:252–260. doi: 10.1021/cb600132m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amsberry KL, Gerstenberger AE, Borchardt RT. Pharm Res. 1991;8:455–461. doi: 10.1023/a:1015890809507. [DOI] [PubMed] [Google Scholar]
- 12.(a) Rostovtsev VV, Green LG, Folkin VV, Sharpless KB. Angew Chem Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]; (b) Tornoe CW, Christensen C, Meldal MJ. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 13.(a) Lutz JF. Angew Chem Int Ed. 2007;46:1018–1025. doi: 10.1002/anie.200604050. [DOI] [PubMed] [Google Scholar]; (b) Fournier D, Hoogenboom R, Schubert US. Chem Soc Rev. 2007;36:1369–1380. doi: 10.1039/b700809k. [DOI] [PubMed] [Google Scholar]
- 14.Speers AE, Adam GC, Cravatt BF. J Am Chem Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 15.(a) Yang SK, Weck M. Macromolecules. 2008;41:346–351. [Google Scholar]; (b) Binder WH, Kluger C. Macromolecules. 2004;37:9329–9330. [Google Scholar]
- 16.(a) Zhang W, Moore JS. Adv Synth Catal. 2007;349:93–120. [Google Scholar]; (b) van de Weghe P, Bisseret P, Blanchard N. J Organomet Chem. 2006;691:5078–5108. [Google Scholar]; (c) Furstner A, Davis PW. Chem Commun. 2005;18:2307–2320. doi: 10.1039/b419143a. [DOI] [PubMed] [Google Scholar]; (d) Lloyd-Jones GC, Margue RG, de Vries JG. Angew Chem Int Ed. 2005;44:7442–7447. doi: 10.1002/anie.200502243. [DOI] [PubMed] [Google Scholar]
- 17.(a) Gordon EJ, Gestwicki JE, Strong LE, Kiessling LL. Chem Biol. 2000;7:9–16. doi: 10.1016/s1074-5521(00)00060-0. [DOI] [PubMed] [Google Scholar]; (b) Owen RM, Gestwicki JE, Young T, Kiessling LL. Org Lett. 2002;4:2293–2296. doi: 10.1021/ol0259239. [DOI] [PubMed] [Google Scholar]; (c) Hilf S, Berger-Nicoletti E, Grubbs RH, Kilbinger AFM. Angew Chem Int Ed. 2006;45:8045–8048. doi: 10.1002/anie.200602323. [DOI] [PubMed] [Google Scholar]; (d) Gibson VC, Okada T. Macromolecules. 2000;33:655–656. [Google Scholar]
- 18.Sanford MS, Love JA, Grubbs RH. J Am Chem Soc. 2001;123:6543–6554. doi: 10.1021/ja010624k. [DOI] [PubMed] [Google Scholar]
- 19.Lal B, Pramanik BMI, Manhas MS, Bose AK. Tetrahedron Lett. 1977;18:1977–1980. [Google Scholar]
- 20.Stork G, Niu D, Fujimoto A, Koft ER, Balkovec JM, Tata JR, Dake GR. J Am Chem Soc. 2001;123:3239–3242. doi: 10.1021/ja004325r. [DOI] [PubMed] [Google Scholar]
- 21.(a) Love JA, Morgan JP, Trnka TM, Grubbs RH. Angew Chem Int Ed. 2003;42:1743–1746. [Google Scholar]; (b) Choi TL, Grubbs RH. Angew Chem Int Ed. 2003;42:1743–1746. doi: 10.1002/anie.200250632. [DOI] [PubMed] [Google Scholar]
- 22.Chan TR, Hilgraf R, Sharpless KB, Folkin VV. Org Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 23.(a) Wang Q, Chan TR, Hilgraf R, Folkin VV, Sharpless KB, Finn MG. J Am Chem Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]; (b) Link JA, Tirrell DA. J Am Chem Soc. 2003;125:11164–11165. doi: 10.1021/ja036765z. [DOI] [PubMed] [Google Scholar]; (c) Link JA, Mock ML, Tirrell DA. Curr Opin Biotech. 2003;14:603–609. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]; (d) Speers AE, Cravatt BF. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]; (e) Sawa M, Hsu TL, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong CH. Proc Natl Acad Sci USA. 2006;103:12371–12376. doi: 10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. ACS Chem Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.