Abstract
Galactofuranose (Galf) residues are present in the cell wall glycoconjugates of numerous pathogenic microbes. UDP-galactofuranose (UDP-Galf), the biosynthetic precursor of Galf-containing glycoconjugates, is produced from UDP-galactopyranose (UDP-Galp) by the flavoenzyme UDP-galactopyranose mutase (UGM). The gene encoding UGM (glf) is essential for the viability of pathogens, including Mycobacterium tuberculosis, and this finding underscores the need to understand how UGM functions. Considerable effort has been devoted to elucidating the catalytic mechanism of UGM, but progress has been hindered by a lack of structural data for an enzyme-substrate complex. Such data could reveal not only the substrate binding interactions but how UGM can act preferentially on two very different substrates, UDP-Galp and UDP-Galf, yet avoid other structurally related UDP-sugars present in the cell. Herein, we describe the first structure of a UGM-ligand complex, which provides insight into the catalytic mechanism and the molecular basis for substrate selectivity. The structure of UGM from Klebsiella pneumoniae bound to the substrate analog UDP-glucose (UDP-Glc) was solved by X-ray crystallographic methods and refined to 2.5 Å resolution. The ligand is proximal to the cofactor, a finding that is consistent with a proposed mechanism in which the reduced flavin engages in covalent catalysis. Despite this proximity, the glucose ring of the substrate analog is positioned such that it disfavors covalent catalysis. This orientation is consistent with data indicating that UDP-Glc is not a substrate for UGM. The relative binding orientations of UDP-Galp and UDP-Glc were compared using saturation transfer difference-NMR. The results indicate that the uridine moiety occupies a similar location in both ligand complexes, and this relevant binding mode is defined by our structural data. In contrast, the orientations of the glucose and galactose sugar moieties differ. To understand the consequences of these differences, we derived a model for the productive UGM-substrate complex that highlights interactions that can contribute to catalysis and substrate discrimination.
Keywords: UDP-galactopyranose mutase, Glf, cell wall biosynthesis, mycobacteria, galactofuranose
Introduction
Galactofuranose (Galf) residues are cell wall components of numerous microorganisms, including pathogenic strains of Klebsiella pneumoniae and Escherichia coli, as well as mycobacterial species.1–6 The furanose form of galactose also is present in eukaryotic pathogens, such as Leishmania major, Aspergillus fumigatus, and Trypanosoma cruzi.7–9 The presence of Galf in many disease-causing organisms provides impetus to understand the mechanisms underlying its incorporation. The biosynthetic precursor of Galf-containing glycoconjugates is uridine 5’-diphospho-α-D-galactofuranose (UDP-Galf), which is generated by the enzyme UDP-galactopyranose mutase (UGM). The gene encoding UGM (glf) is essential for viability in mycobacteria.10 UGM is also vital in eukaryotes, as strains of Aspergillus spp. lacking the enzyme show defects in growth,11 cell wall biosynthesis,12 and virulence.13 Similarly, deletion of the gene encoding UGM in Leishmania major results in attenuated virulence.14 Because there is no human homolog of UGM,15 it has been suggested that this enzyme can serve as a target of antimicrobial agents, especially antimycobacterial agents.16–19 Intriguingly, UGM inhibitors have been synthesized that block mycobacterial growth.19 These promising developments provide incentive to extend current understanding of UGM catalysis.
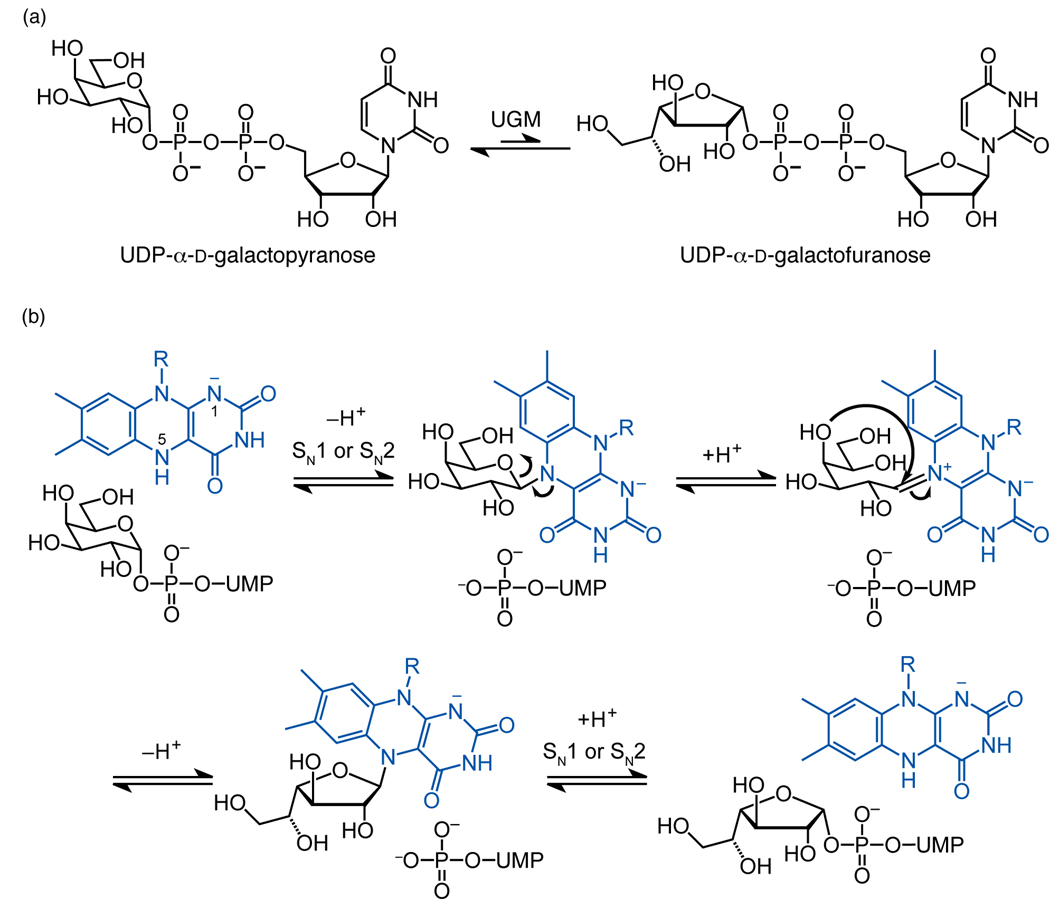
UGM is a flavoenzyme that catalyzes the interconversion of UDP-galactopyranose (UDP-Galp) and UDP-Galf (Fig. 1a).3 Mechanistic studies suggest that its noncovalently-associated flavin adenine dinucleotide (FAD) plays an important role in catalysis.20 Specifically, UGM is active only when the cofactor is reduced.20 The reduced flavin has been suggested to have several different functions, from mediating transient electron transfer (single electron transfer or hydride transfer) to serving a structural role within the protein scaffold.5,21–23 We provided evidence for the intermediacy of a flavin–galactose iminium adduct (Fig. 1b).24,25 In this mechanism, the N5 of reduced flavin acts as a nucleophile (SN1 or SN2 attack) to generate a covalent flavin–galactose adduct.24,25 A radical mechanism that also affords the iminium intermediate was subsequently put forth,26,27 but complications regarding this proposal have not yet been addressed.28 The hypothesis that a flavin-derived iminium ion mediates catalysis via a previously unknown mechanistic pathway provides incentive to determine the mode of substrate binding.
Figure 1.
Interconversion of UDP-Galp and UDP-Galf is catalyzed by UGM. (a) The reaction catalyzed by UGM. (b) Nucleophilic mechanism for catalysis by UGM; the reaction could proceed by either SN1 or SN2 as described previously.25 N1 is depicted as negatively charged as reported.22
Because a structure of a substrate or substrate analog bound to UGM has been elusive, X-ray crystallography has provided valuable but incomplete information. The first structure, of E. coli UGM, revealed that the enzyme is a dimer in which each subunit harbors a FAD cofactor.20 The cofactor sits in a cleft lined with highly conserved residues. Structures of UGM homologs from M. tuberculosis and K. pneumoniae corroborate these findings.29 Complementary results have come from studying the UGM from K. pneumoniae using site-directed mutagenesis. When the importance of highly conserved residues was evaluated,20,30 two Arg residues were found to be essential for catalysis.30 The identification of one of these, Arg174, was unexpected, because the available crystallographic data place this residue outside the putative active site. This residue is located on a mobile loop adjacent to the putative active site pointing away from the flavin.20,29 Molecular dynamics simulations suggest that this loop closes upon substate binding, thereby orienting the Arg174 side chain toward the pyrophosphoryl group of the ligand.30–32 Thus, several lines of evidence indicate that UGM undergoes a conformational change upon ligand binding, and we set out to explore this issue.
We envisioned using x-ray crystallographic analysis to address directly how UGM binds its substrate. Because UDP-glucose (UDP-Glc) differs from the substrate UDP-Galp only by an equatorial rather than axial C4-OH group, we reasoned that UDP-Glc would not undergo reaction but rather serve as an unreactive substrate analog. Indeed, we were able to crystallize the complex and determine its structure. The resulting structure of the complex defines the location of the active site, and reveals how conserved amino acids interact with the uridine diphosphoryl portion of the ligand.
Consistent with the proposal that the reaction proceeds through a flavin–galactose adduct, the sugar residue of the substrate analog is in the vicinity of the flavin. Despite its similarity to the natural substrate, however, UDP-Glc does not give rise to the spectroscopic signal indicative of the putative flavin iminium ion.25,33 This observation highlights a difference in the reactivity of UDP-Glc and the substrate UDP-Gal. It also is consistent with the orientation of UDP-Glc in the complex, which precludes bond formation with the reduced flavin. The data indicate that the equatorial C4-OH group of UDP-Glc influences the structure of the complex and thereby renders the iminium adduct inaccessible. These findings provide a structural rationale as to why UDP-Glc, which is present in significant concentrations in the cell, does not interfere with the recognition of substrate by UGM.
Results
UDP-Glc functions as a UGM ligand
The structural similarity of UDP-Glc and UDP-Galp led us to postulate that the former could illuminate how UGM binds its substrate. We assessed the ability of UDP-Glc to bind UGM, by comparing the affinities of UDP-Galp and UDP-Glc for oxidized and reduced UGM. For oxidized UGM, we utilized a binding assay in which a ligand of interest competes with a UDP-linked fluorescein probe.17 The application of this approach yielded a Kd value for the complex of UDP-Galp and oxidized UGM of 290 ± 40 µM, which is similar to that determined by using a Trp fluorescence assay (220 µM).32 This value is 5–6 fold higher than that reported for the reduced enzyme complex (52 µM27 and 66 µM32), a preference similar to that observed previously in STD-NMR experiments.31,34 The natural substrate, UDP-Galp, therefore exhibits a preference for the active, reduced enzyme. For comparison, we evaluated the selectivity of UDP-Glc. As with UDP-Galp, UDP-Glc displaces the fluorescent UDP derivative, indicating that all three UDP derivatives bind in the same active site. The Kd value for oxidized UGM binding to UDP-Glc, 550 ± 100 µM, is only two-fold weaker than that for UDP-Galp. The UDP-Glc bound to the reduced enzyme with a Kd value of 750 ± 250 µM. Thus, the oxidized UGM shows only a subtle (2-fold) penchant for binding UDP-Galp over UDP-Glc, but the reduced form exhibits a preference of over 10-fold. Despite the preference for binding UDP-Galp, the affinity data indicate that UDP-Glc can serve as a ligand for UGM.
UDP-Glc Is Not a Substrate for UGM
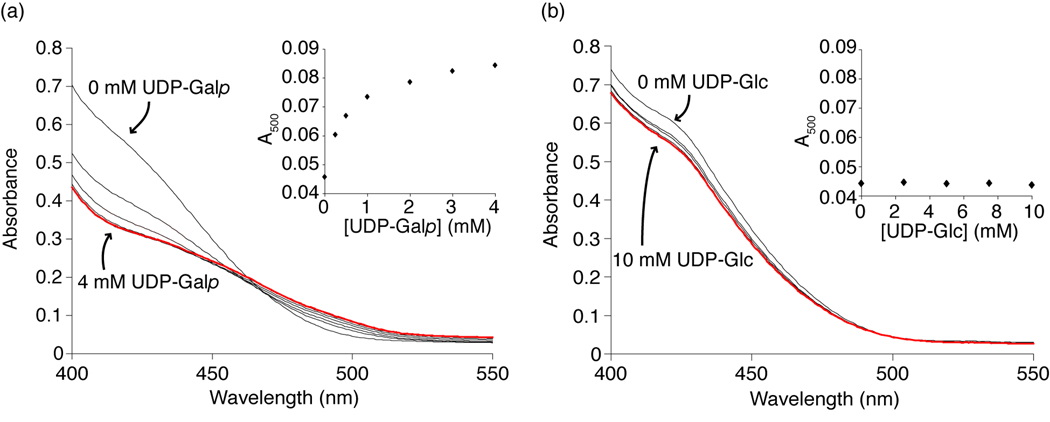
Given the similarity of UDP-Glc and UDP-Galp, we wanted to confirm that UDP-Glc was not a substrate for UGM. For structural analysis of an enzyme-ligand complex, it is desirable that a stable complex be formed. The configuration of the C4 hydroxyl group of UDP-Glc (equatorial) relative to UDP-Galp (axial) should preclude catalytic conversion of UDP-Glcp to the highly thermodynamically disfavored glucofuranose form.35 As expected, we detected no UGM-catalyzed conversion of UDP-Glcp to UDP-Glcf. It is plausible, however, that UDP-Glc could give rise to the spectroscopic signature attributed to the flavin-iminium intermediate (Fig. 1b).33 Previously, we demonstrated that when UDP-Galp and reduced UGM are mixed an increase in absorbance at 500 nm is observed.25 We therefore tested whether exposure of UGM to UDP-Glc would afford a similar result. No change was detected (Fig. 2). These data suggest that despite the structural similarity between UDP-Glc and UDP-Galp, UGM discriminates against UDP-Glc not only at the level of binding but also in catalysis. Moreover, these results indicate that UDP-Glc can serve as a stable substrate analog for structural studies of UGM.
Figure 2.
Spectrophotometric studies probing for a flavin-derived iminium ion. Spectra were recorded as 0–4 mM UDP-Galp (a) or 0–10 mM UDP-Glc (b) was titrated into a sample of reduced UGM. The red line represents the absorbance spectrum at the highest concentration of ligand. Inset shows absorbance at 500 nm, the wavelength of iminium ion absorbance. A change is detected for UDP-Galp but not UDP-Glc. Results in (a) have been published previously25 and are shown for comparison.
Structure of UGM complexes
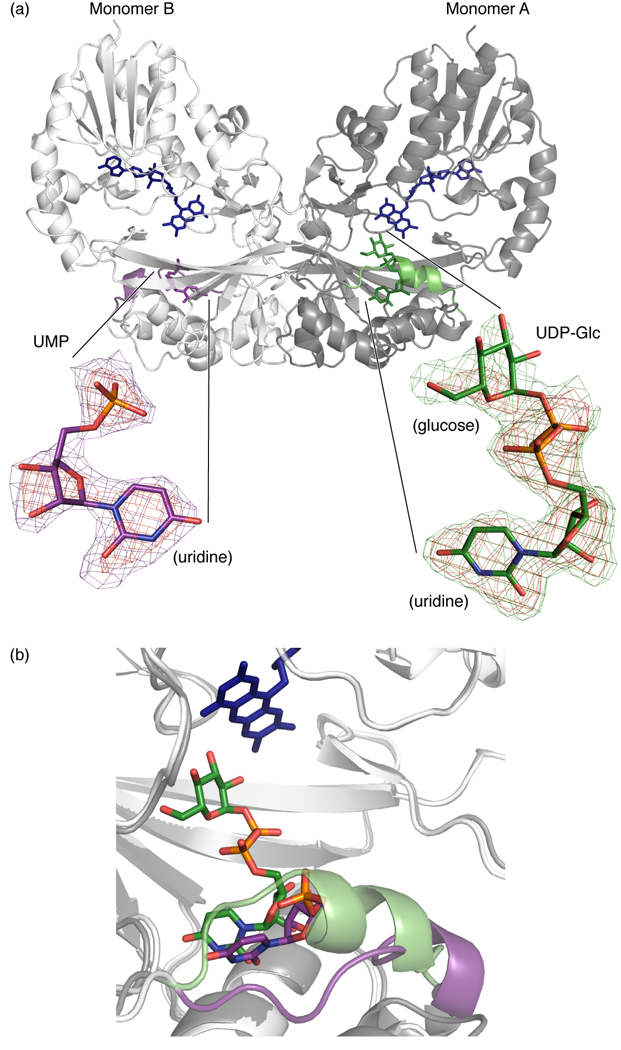
Previous attempts to obtain a structure of a complex between UGM and its substrate UDP-Galp were unsuccessful.20,29 We therefore conducted crystallization trials in the presence of the substrate analog UDP-Glc. From a solution containing 5 mM UDP-Glc, crystals were obtained. The structure of UGM bound to UDP-Glc was solved by molecular replacement and refined against 2.5 Å resolution data (Table 1). The final structure exhibits excellent geometry with 98.4% of residues in favorable Ramachandran orientations.36 The asymmetric unit contains an entire UGM dimer (Fig. 3a), but each subunit adopts a different conformation. Monomer A represents a fully liganded, closed conformation whereas monomer B represents a partially liganded, open conformation.
Table 1.
Data collection and refinement statistics.
| Data collection | |
| Resolution limits (Å) (last shell) | 50.0-2.45 (2.54-2.45) |
| Unique reflections (#) | 41,161 (4,042) |
| Completeness (%) | 98.6 (97.0) |
| Rsyma (%) | 11.0 (36.6) |
| I/σ̣I) | 14.8 (4.2) |
| Redundancy | 4.9 (4.9) |
| Wilson B-factor (Å2) | 53.9 |
| Space group | P 41 |
| Unit-cell parameters | |
| (a/b/c) (Å) | 94.0/94.0/130.3 |
| Refinement statistics | |
| Resolution range (Å) | 30.0–2.45 (2.51-2.45) |
| Rcrystb (%) / Rfreec (%) | 19.2 (25.6) / 23.8 (32.1) |
| Protein atoms | 6240 |
| Water molecules | 195 |
| Hetero atoms | 163 |
| Isotropic average temperature factors (Å2) | |
| Overall | 45.8 |
| Protein | 45.9 |
| Water | 43.4 |
| FAD (monomer A) | 39.3 |
| FAD (monomer B) | 37.6 |
| UDP-Glc | 59.7 |
| UMP | 61.4 |
| Estimated coordinate error from Luzzati plot (Å) | 0.25 |
| RMSD from ideal | |
| Bond length (Å) | 0.018 |
| Bond angles (°) | 1.7 |
| Ramachandran plot | |
| (% favored / % allowed)36 | 98.4 / 100.0 |
Rsym(I) = ΣhklΣi |Ii(hkl) – 〈I(hkl) 〉|/ ΣhklΣi Ii(hkl) where I(i) is the intensity of the ith observation of the hkl reflection and 〈I(hkl) 〉 is the mean intensity from multiple measurements of the h,k,l reflection.
Rcryst(F) = Σhkl|Fobs(hkl)–Fcalc(hkl)|/ ΣhklFobs(hkl), where Fobs(hkl) and Fcalc(hkl) are the observed and calculated structure factor amplitudes for the h,k,l reflection.
Rfree is Rcryst calculated for a randomly selected test set of reflections (5%) not included in the refinement.
Figure 3.
The structure of the UGM–UDP-Glc complex reveals the ligand orientation in the UGM active site. (a) View of the UGM dimer. Colors: FAD (blue), UDP-Glc (green), monomer A mobile loop (green), UMP (purple), monomer B mobile loop (purple). UMP inset: Omit map showing difference density (FO-FC) for UMP in monomer B. Red grid is contoured at 4σ, purple grid at 2.5σ (C purple, O red, N blue, P orange). UDP-Glc inset: Omit map showing difference density (FO-FC) for UDP-Glc in monomer A. Red grid at 4σ, green grid at 2.5σ (C green). (b) Overlay of monomer A and monomer B depicting mobile loop (residues 167–177) and ligand orientation. Closed monomer A loop and carbons are green, open monomer B loop and carbons are purple. Residues 125–166 also move significantly (up to 6 Å Cα movement) to accommodate mobile loop restructuring (not shown).
The flavin cofactor is in the active site
Despite the conformational differences between the two monomers, in both the ligand is adjacent to the flavin. Because the substrate analog is more well-defined in closed monomer A, our analysis focuses on this monomer. UDP-Glc is bound such that its glucose residue is near the isoalloxazine ring of flavin (Fig. 3b); the minimum flavin-glucose distance is only 2.9 Å. The proximity of the flavin and the sugar residue of the substrate analog is consistent with the putative mechanism involving covalent catalysis.24,25 The UDP portion of the ligand makes close contacts to the remainder of the highly conserved binding pocket (see below). In addition, the side chain of Arg174, which is pointed away from the active site in unliganded structures, is located in the active site in the complex (Fig. 4). The guanidinium moiety of Arg174 is oriented in a manner consistent with a predicted role in binding the pyrophosphoryl group of the substrate.30–32
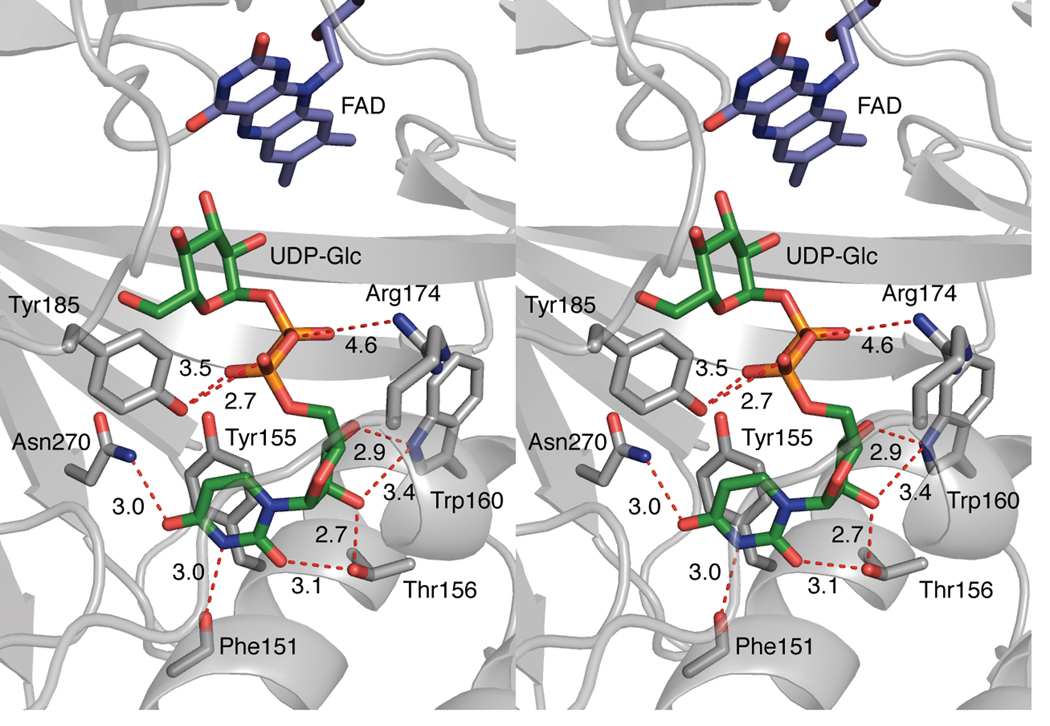
Figure 4.
Stereoview of contacts between the uridine moiety of UDP-Glc and protein atoms in monomer A. The nucleoside portion of UDP-Glc interacts with conserved amino acids (C gray, other atoms colored as described above) such as Trp160, seen through the semitransparent mobile loop. The stacking distance between Tyr155 and uracil is approximately 3.5 Å.
Monomer comparison
The differences in monomers A and B provide insight into the conformational changes that can occur upon ligand binding. A superposition of the monomers results in a Cα root mean square deviation (rmsd) of 1.45 Å. When the same operation is performed using monomer A and the unliganded K. pneumoniae UGM monomer (PDB 2bi7)29 a rmsd of 1.41 Å is obtained. In contrast, a comparison of monomer B with K. pneumoniae UGM yields a significantly smaller rmsd of 0.88 Å. The major difference between monomers A and B is in residues 125–177. When this region is excluded from the calculation (i.e., residues 2–124 and 178–384 were superimposed) a rmsd of 0.86 Å results. Together, the results indicate that monomer B adopts a conformation like that observed previously in structures of unliganded UGM homologs, whereas monomer A represents a closed form.
In the open monomer B, the active site is highly solvent-exposed, and a clear pattern of electron density was obtained for only the UMP portion of the ligand (Fig. 3a). In contrast, the entire ligand in closed monomer A is well-ordered (Fig. 3a). The positions of the uridine moieties are similar in both (Fig. 3b), but those of the α-phosphoryl groups are not—they differ by 6 Å. In the more open monomer B, the α-phosphoryl group is located toward the active site opening, while in monomer A, it is oriented toward the FAD cofactor.
Ligand binding induces conformational change
A major difference between monomers A and B is the conformation of the mobile loop.29,30 In monomer A, these residues (167–177), which are adjacent to the uridine diphosphoryl portion of the ligand, close over the active site (Fig. 3b). This conformational change involves extending a short helix composed of residues 169–171 to encompass residues 172–174. In unliganded structures,20,29 this loop adopts a variety of conformations. Only in the K. pneumoniae structure29 does this loop exhibit any secondary structure; residues 169–172 constitute a short helix. In monomer A, the loop closure and helix formation results in the movement of Arg174 to within 4.6 Å of the ligand pyrophosphoryl group. Arg174 has high B values and gives rise to relatively weak electron density, yet its ability to occupy a position near the pyrophosphoryl moiety is notable. This movement toward the ligand is consistent with previous biochemical data and with a purported role for this conserved residue in phosphoryl group binding.30–32
Orientation of the uridine nucleotide
Because complete density for UDP-Glc is observed (Fig. 3a), the monomer A structure provides insight into the contributions of highly conserved catalytic residues20,30 to substrate binding (Fig. 4). For example, the plane of the uracil moiety of UDP-Glc is parallel to that of invariant Tyr155, and the two aromatic rings are separated by approximately 3.5 Å. Their proximity and orientation indicate that this π-stacking interaction contributes to substrate binding. This observation was unexpected, as previous models and molecular dynamics simulations incorporated π-stacking interactions between uracil and invariant Trp160.20,29–32,34 Although Trp160 does not contribute to uracil binding, the indole nitrogen can form a bifurcated hydrogen bond to the C2- and C3-hydroxyl groups of the ribose ring. The uracil N3 is within hydrogen bonding distance of the backbone carbonyl of Phe151, as are the uracil O4 and the side chain carboxamide group of Asn270. The hydroxyl group of Thr156 is positioned to engage in hydrogen bonding both with the C2-OH of ribose and the O2 of uracil. Tyr185 is located in proximity of the α-phosphoryl group. Hydrophobic residues also define parts of the uridine binding pocket, and Phe152, Leu97, and Leu175 all are within van der Waals distance (3.1–3.5 Å) of the ligand.
Orientation of glucose residue
In the closed conformation, the glucose residue of UDP-Glc is adjacent to the flavin. Proximity is required for the putative covalent catalysis mechanism, in which a bond is formed between the flavin N5 and the anomeric position of the sugar (Fig. 1b). In monomer A, however, 6.0 Å separates these nucleophilic (flavin N5) and electrophilic (sugar C1) groups. In addition, the UDP leaving group21 is not oriented favorably for a substitution reaction (SN1 or SN2). The relative positions of the UDP-Glc and the flavin are consistent with the observation that no signal indicative of a flavin-derived iminium ion is detected upon the addition of UDP-Glc (Fig. 2). These differences led us to hypothesize that the orientation of the glucose moiety in the complex does not mirror that of the galactopyranose moiety.
STD-NMR reveals uridine components of UDP-Glc and UDP-Galp bind similarly
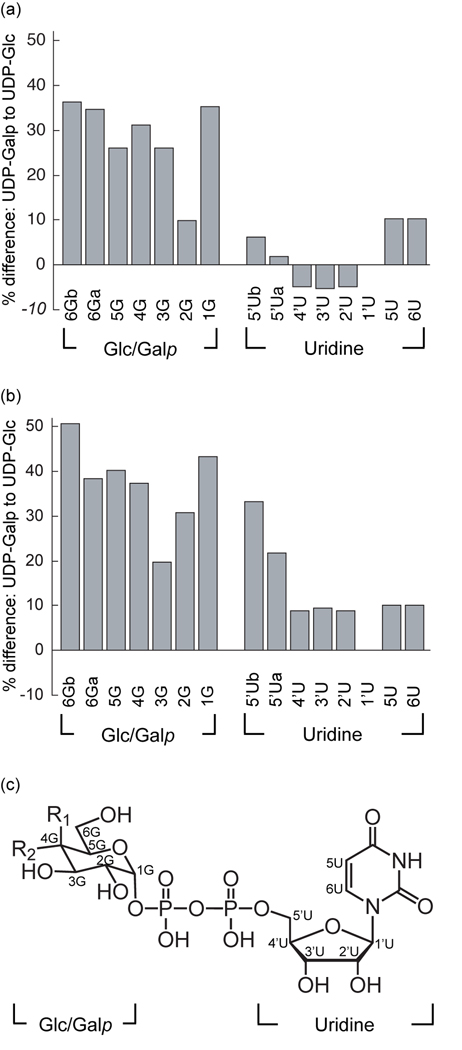
To compare the UDP-Glc and UDP-Galp binding modes, we employed saturation transfer difference-NMR spectroscopy (STD-NMR). In this method, protein resonances are saturated with a radiofrequency pulse, and magnetization is transferred to proximal ligand protons.37 The efficiency of transfer depends upon the relative proximity and orientation of the ligand and protein protons. STD values thus reflect binding interactions; therefore, a comparison of the STD values obtained for related ligands can report on their relative binding orientations. Others have used STD-NMR to investigate UGM, and these studies have indicated that UDP-Galp binds more tightly to reduced UGM than does UDP.31,34 Given this successful application of STD-NMR in monitoring ligand binding to UGM, we employed it to compare the binding of UDP-Glc and UDP-Galp, under both oxidizing (catalytically inactive) and reducing (active) conditions. When UGM is oxidized (Fig. 5a), the uridine STD signals of both UDP-Glc and UDP-Galp are similar, with an average difference (absolute value) of 6%. A much larger difference, 28%, was observed for the sugar signals from the hexose sugar moiety (Glc or Galp). These results indicate that the uridine portion of each ligand binds in a similar manner, but the interactions of the sugar moieties differ. The data from the reduced complex follow a similar trend: a 15% difference for the uridine signals compared to 37% for those of the sugar portion (Fig. 5b). The larger differences for the reduced complex are consistent with our findings that this form of UGM more effectively discriminates between UDP-Glc and the substrate UDP-Galp. We also observed that the largest differences in the uridine signal for the reduced complex are at ribose C5, the position connected to the α-phosphoryl group. As the C5 position is likely to be most strongly influenced when the orientations of the sugar moiety vary, these data further support the conclusion that the uridine groups bind similarly but that the mode of sugar binding depends upon its identity.
Figure 5.
Analysis of the binding of UDP-Galp and UDP-Glc to UGM using STD-NMR. Percent difference between UDP-Glc and UDP-Galp STD-NMR intensities in oxidized UGM (a) and reduced UGM (b) are charted by atom. All STD intensities were normalized to proton 1’U at 100%. Percent difference values represent the difference between UDP-Galp and UDP-Glc STD intensities, divided by UDP-Galp STD intensities. Results from one representative experiment are shown. U=uridine, G=glucose or galactose. (c) Structure of UDP-Glc/UDP-Galp. UDP-Glc: R1=H, R2=OH. UDP-Galp: R1=OH, R2=H. Repetition of the experiment afforded similar results (Supplementary Fig. 1).
Energy-minimized active site model for substrate binding
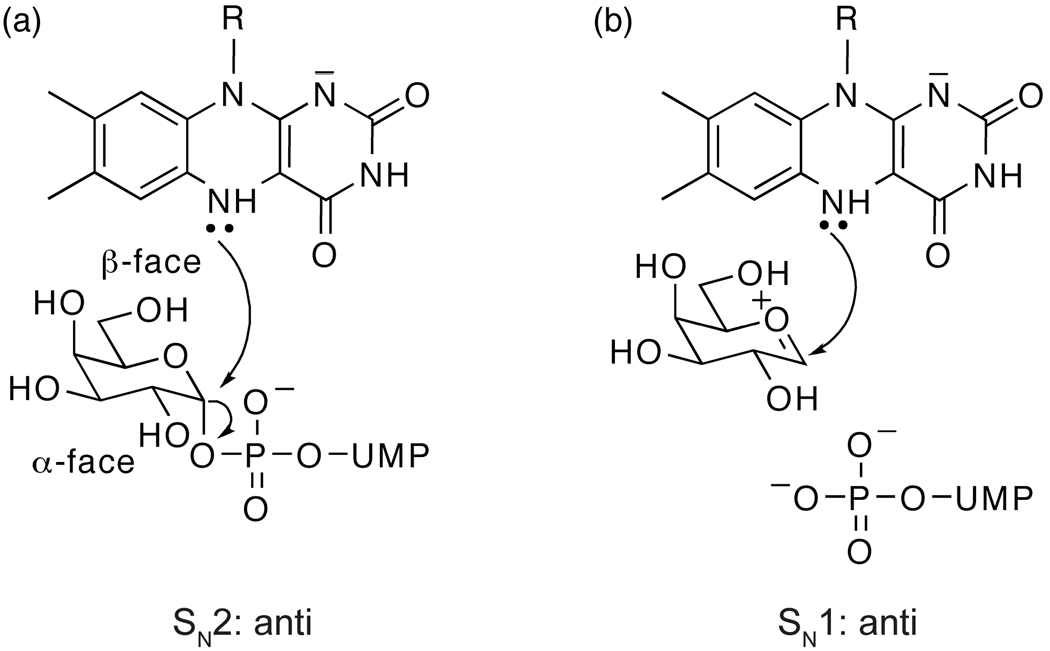
We used our x-ray structure, previous data, and the results of our comparative analysis of UDP-Glc and UDP-Galp to develop a model of the complex of UGM with its substrate UDP-Galp. In constructing such a model, we assumed that the binding mode of the UMP moiety in monomer A is preserved in the UDP-Galp complex. We then explored whether the Galp moiety could access an orientation that is consistent with our observation that a covalent flavin– galactose adduct can form under catalytic conditions. Specifically, the preferred geometry for production of this species entails an anti orientation of the flavin and pyrophosphoryl leaving group (Fig. 6).
Figure 6.
Geometries consistent with nucleophilic attack. (a) An SN2 reaction, in which the N5 of reduced flavin attacks the anomeric position from the β-face of UDP-Galp causing the UDP to leave from the α-face. (b) SN1 reaction, anti relationship. An SN1 reaction can proceed with UDP leaving from the α-face of UDP-Galp to form an oxocarbenium intermediate. The N5 of the reduced flavin can then attack from the β-face.
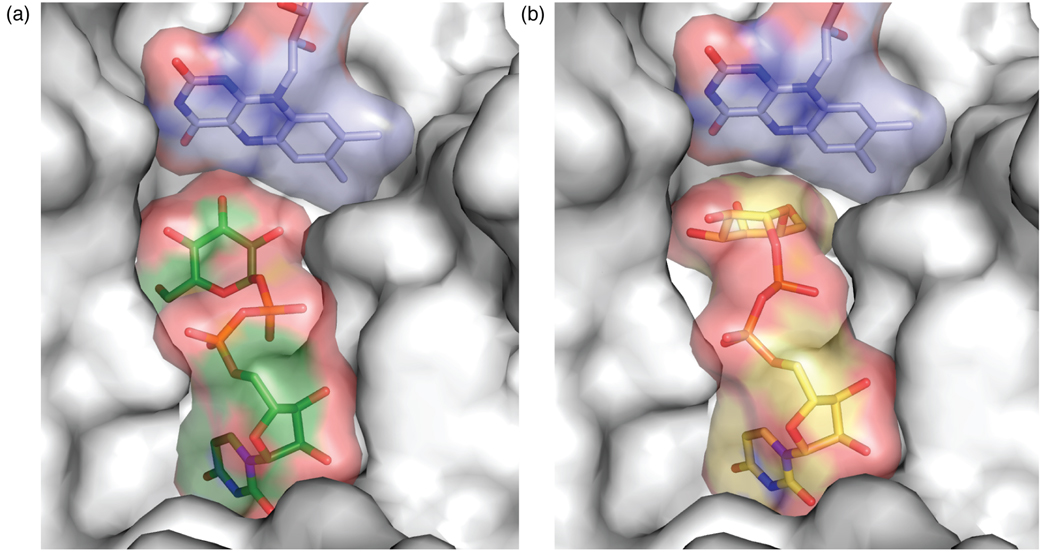
To create the model, UDP-Galp was oriented in the active site analogously to UDP-Glc (Fig. 7a). Rotatable bonds in the substrate were altered, such that the β-phosphoryl moiety could interact with Tyr314 in a manner similar to that observed for the α-phosphoryl moiety and Tyr185. In the resulting arrangement, an interaction between the β-phosphoryl group and Gln159 is maintained. The anomeric carbon–β-phosphoryl oxygen (glycosidic) bond was then positioned anti to the nucleophilic flavin N5. The complex was then subjected to energy-minimization (see Methods). In the model that results (Fig. 7b and Supplementary Fig. 2), the distance from flavin N5 to the anomeric carbon of UDP-Galp is 4.2 Å, consistent with covalent catalysis by flavin. This distance is similar to that between the N5 of flavin and anomeric carbon (C1) of the substrate, which resulted from docking studies of UDP-Galp34 and UDP-Galf.31 Still, the uridine orientation predicted by calculations is very different from that observed in our structure (UDP-Glc) and model (UDP-Galp) (see Discussion).
Figure 7.
The ligand-binding pocket of UGM. (a) Monomer A binding pocket with UDP-Glc bound. UGM is rendered as a surface to depict the binding pocket (coloring as in Fig. 3). (b) Binding pocket showing UDP-Galp model (colored as in Fig. 7a except carbons yellow). In (a) and (b), residues 163–177 and 340–384 were deleted for clarity.
The C4-OH of UDP-Galp in the minimized model points toward the backbone carbonyl of Pro59. Intriguingly, UDP-Glc would not be able to bind to UGM in this conformation. A steric clash between the equatorial C4-OH of UDP-Glc and invariant Arg280 (~1.3 Å separation) precludes the formation of a UDP-Glc complex that is poised to form a flavin-sugar bond. This steric effect is alleviated when UDP-Glc adopts the orientation observed in monomer A. In the model of the UDP-Galp complex, the β-phosphoryl group is closer to Arg280 (5.2 Å in the modeled substrate complex compared to 5.7 Å in the structure of the substrate analog). Both distances are too long for hydrogen bonding, supporting the possibility that the role of this absolutely conserved residue is not to facilitate initial substrate binding but rather to stabilize the UDP intermediate during turnover.30,31 Alternatively, Arg280 may interact with the 6-OH of Galp; the significance of this functional group to UGM catalysis has recently been investigated.38,39 In the model, the distance separating the guanidinium group of Arg280 and the 6-OH of the Gal residue is 2.9 Å, suggesting that the guanidinium side chain serves as hydrogen bond donor and the 6-OH serves as an acceptor. Either proposed role for Arg280 is consistent with its position in model. Thus, our substrate-UGM model is consistent with the findings reported herein, as well as other data.20,25,29,30,32
Discussion
The substrate-binding pocket of UGM has been identified and defined by the first X-ray crystal structure of UGM with a bound ligand. Analysis of the UGM complexes of the ligand UDP-Glc and the substrate UDP-Galp using STD-NMR and molecular modeling indicates that both bind in the same pocket. Thus, the active site can be identified directly, and it is adjacent to the flavin cofactor. This location is consistent with a role for flavin in catalysis. The uridine component of either the substrate or the substrate analog binds tightly in a pocket lined with conserved residues. Uridine binding orients the hexose (Galp or Glc) moiety toward the flavin. A key feature of ligand binding is that it can induce a conformational change in a mobile loop that closes over the active site,31,32,34 thereby nudging the sugar moiety and cofactor into close proximity.
Role of conserved residues in ligand binding
The structure of the complex of UDP-Glc and UGM reveals features that illuminate, expand upon, and refute ideas inferred from previous models. For example, our results indicate an unexpected role for conserved Trp160. Previous mutagenesis experiments indicate the activity for the Trp160Ala variant is decreased by six orders of magnitude.30 To account for this dramatic decrease, docking studies were conducted that suggest that the uracil ring of the substrate stacks upon Trp160.20,29–31,34 This orientation also was invoked in a study in which it was postulated that the substrate bridges the Trp160 and flavin sites.32 Our crystallographic structure refinement of the UDP-Glc complex indicates a very different role for Trp160. It engages in an unusual tryptophan-ribose interaction in which it interacts with the C2-OH and C3-OH of the ribose residue (Fig. 4). Additionally, we have found that the uracil ring does indeed engage in a stacking interaction, but not with Trp160; rather, it interacts with Tyr155, an absolutely conserved active site residue. Phenylalanine substitution of this residue results in a significant decrease in substrate binding affinity (Km value diminished by 15-fold).30 Our finding that Tyr155 participates in a stacking interaction provides a rationale for this decrease. It remains unclear, however, whether this decrease in affinity is due to a fundamental difference in the stacking ability of tyrosine and phenylalanine40 or a misalignment of residues due to loss of hydrogen bonding from the tyrosine phenol. Whatever the source of the affinity difference between wild-type UGM and the Tyr155Phe variant, uracil-tyrosine stacking interactions similar to those we have observed are found commonly in RNA-binding proteins.41
In addition to providing insight into the importance of Tyr155 and Trp160, our structure reveals the function of three other highly conserved, active site aromatic residues: Tyr185, Tyr314, and Tyr349. These residues have been shown to be important for catalysis,20,30 and have been implicated in hydrogen bonding20 or aromatic interactions.30 Of these five aromatic residues, only Tyr155 clearly engages in π-stacking in the UDP-Glc complex. It is possible, however, that Tyr349 also engages in a cation-π interaction (Supplementary Fig. 2).42 It could stabilize a positive charge (full or partial) that develops at the anomeric position of galactose in the transition state of a substitution reaction (either SN1 or SN2). Another potential role for Tyr349 is to use hydrogen bonding to facilitate an orientation of Galp that allows for bond formation between the reduced flavin and Galp. The remaining three conserved aromatic residues of the active site, Trp160, Tyr185, and Tyr314, utilize hydrogen bonding to interact with the ligand. For instance, Tyr185 is poised to interact with the α-phosphoryl group of UDP-Glc. The model of the substrate complex suggests Tyr314 contacts the β-phosphoryl group through a similar mode of interaction. In addition to their role in hydrogen bonding, the aromatic character of these three residues may serve to stabilize substrate binding. It is interesting to note that the carbohydrate binding sites of lectins often are lined with aromatic residues.43–45
A comparison of positions that differ between prokaryotic and eukaryotic UGMs suggests a conservation of function. For example, the aforementioned Tyr185, which is conserved in prokaryotes, corresponds to Trp in eukaryotic UGMs.30,46 The indole NH, like the phenol of Tyr185, could serve as a hydrogen bond donor to the α-phosphoryl group (Supplementary Fig. 3). Similarly, another residue that is highly conserved in prokaryotes, Thr156, is in position to engage in hydrogen bonding to both uracil (O2) and ribose (C2-OH). In eukaryotes, however, an asparagine is typically found at this position.30,46,47 Like threonine, asparagine could engage in hydrogen bonding with two sites on the ligand. Asn270 is another conserved residue in prokaryotes that differs from the corresponding tyrosine residue in eukaryotes; however, both can form a hydrogen bond with uracil (O4). Although many side chain variations can be rationalized, some differences between the conserved positions in prokaryotes versus eukaryotes are not readily understood. The prokaryotic residue Gln159, which is in position to bind the β-phosphoryl group, is commonly a valine in eukaryotes. The basis for this preference awaits a structure of a eukaryotic UGM.
Conformational changes and loop movement
Our structure provides snapshots of two distinct modes of substrate interaction. Open monomer B, with its partially-ordered ligand, appears to represent an encounter complex, in which the nucleoside is bound, but the glucose moiety is disordered. Monomer A provides an alternative view, in which the mobile loop is closed over the active site. We anticipate this conformation is closer to the catalytically active form of the enzyme. The closed and open conformations observed in one dimer of UGM are reminiscent of other crystal structures showing monomer asymmetry, some of which have been consistent with known evidence for positive cooperativity48 or extreme negative cooperativity (half-sites reactivity).49 Neither type of cooperativity has been reported for UGM. Based on inspection of crystal lattice interactions in our UGM structure, it is likely that packing interactions explain the asymmetry in this case. Monomer A participates in crystal packing that appears to stabilize the closed orientation, whereas similar interactions are lacking in open monomer B.
The differences between monomers A and B provide new information about the UGM mobile loop. Results from extensive molecular dynamics simulations have shown that loop movement in the presence of substrate allows for transient closing of the active site and reorientation of Arg174, such that it can interact with ligand phosphoryl groups.30–32 The key changes that occur in the mobile loop during this process are now apparent. The Arg174 side chain is not re-oriented by translocation of the loop alone, but rather its change in position is a consequence of the extension of the helix within the loop region. Although our results indicate Arg174 is situated in the active site, even in closed monomer A it is 4.6 Å from the nearest ligand phosphoryl oxygen. This distance is longer than might be expected; however, our STD-NMR data indicate that additional adjustments in the active site must occur when the natural substrate UDP-Galp binds. We note that Arg174 may also stabilize the UDP leaving group, as suggested above for Arg280.30,31
Two additional residues, Phe152 and Leu175, also undergo significant movements as UGM goes from the unbound to bound state (Supplementary Fig. 3). Aromatic side chains Phe152 and Tyr155 sandwich the uracil ring of the ligand. To adopt this position, Phe152 swivels approximately 2 Å from its location in the open monomer B or the unliganded Klebsiella UGM structure. During closure, Leu175 of the mobile loop translocates approximately 8 Å from its position in the open form. In the complex, it is adjacent to the bridging oxygen of the ribose moiety and Phe152. Thus, Phe152 and Leu175 form a significant portion of the uridine binding site and their movement contributes to creating shape complementarity. The significant repositioning of these residues in the bound complex underscores the difficulty of modeling the uridine binding orientation in the absence of a structure.20,29–32,34
Comparison between UDP-Glc and UDP-Galp binding to UGM
Despite the structural similarity of UDP-Glc and UDP-Galp, their interactions with UGM are quite different. Both ligands bind to the enzyme, yet UDP-Glc binds 10-fold more weakly to the catalytically active, reduced state. Moreover, its addition to the active enzyme fails to give rise to the absorbance attributed to the flavin iminium ion. The detection of this spectroscopic signature only in the presence of the substrate provides further support for a covalent flavin intermediate during catalysis. Moreover, our crystallographic data are consistent with this observed substrate specificity; the glucose moiety is improperly oriented for participation in flavin-mediated covalent catalysis. STD-NMR experiments also indicate that there are differences between the UDP-Glc and UDP-Galp complexes. Although the uridine moieties of the nucleotide-sugars bind similarly, the hexopyranose (Glc or Galp) moieties do not. Indeed, the uridine-binding pocket exhibits excellent shape complementarity (Fig. 7a). In contrast, the region near the sugar moiety is more open to solvent, and cavities between the ligand and enzyme are apparent. The excess space around the sugar residue also is consistent with the findings that UDP contributes a substantial portion of the substrate binding energy.17,27
This space around the sugar enabled the in silico reorientation of the Galp moiety in the active site to generate the minimized model (Fig. 7b). With the exception of the potential hydrogen bond between the Galp 6-OH and Arg280, the Galp moiety in the minimized model does not make tight contacts to the protein, and empty cavities between the Galp residue and protein are retained. These cavities allow space for molecular movement and are consistent with the chemical requirements for turnover. Specifically, UGM must be able to recognize both the pyranose and furanose substrates and the galactose–flavin adduct. Thus, the data suggest that the UGM active site must accommodate all of these species.
Our model provides insight into how UGM can distinguish between related nucleotide-sugars. This ability appears to be a physiological imperative. Estimates from other bacterial species place the cellular concentration of UDP-Glc in the low millimolar range, with the UDP-Galp concentration generally believed to be many fold lower.50–53 This level of competing UDP-sugar could halt UDP-Galf production were UGM unable to discriminate against related UDP-sugars. Our data show that the catalytically active form of UGM prefers to bind to UDP-Galp over UDP-Glc. Moreover, our model suggests that UGM cannot place the sugar of UDP-Glc in the same orientation as that of UDP-Galp, because the equatorial C4-OH group of UDP-Glc would undergo a steric clash with the protein. This result explains why exposure of UDP-Glc to UGM fails to give rise to the spectroscopic signal indicative of the flavin iminium ion. Our model of the complex provides a solution to the apparent paradox of how UGM can discriminate between the two highly related structures UDP-Glc and UDP-Galp, yet turn over either UDP-Galp or UDP-Galf, two entities that are structurally very different.
In addition to providing insight into UGM catalysis, our structural studies can guide the identification of UGM inhibitors. The cavities of the UGM binding pocket that allow substrate discrimination are locations to which small molecule inhibitors16–19 could bind. Knowledge of the function of conserved residues gained from this structure along with an understanding of conformational changes upon binding can facilitate the design of highly potent inhibitors. Such compounds may serve not only as valuable probes of cell wall biosynthesis in organisms like M. tuberculosis but also as therapeutic leads.
Materials and methods
Enzyme preparation
A previously described25 pGEM-T Easy (Promega, Madison, WI) construct containing the UGM gene (glf) from K. pneumoniae with two alterations: the native C-terminal arginine was changed to a glycine codon and a C-terminal hexahistidine tag was encoded. The construct was transformed into BL21(DE3) E. coli cells (Novagen, Madison, WI). Terrific Broth supplemented with 200 mg/L ampicillin was inoculated with isolated transformants and the bacteria were incubated at 37 °C for 20 hours without induction. Cells were harvested by centrifugation (10,000 × g) and resuspended in buffer containing 20 mM sodium phosphate, pH 7.4, 25 mM imidazole, and 300 mM NaCl. Cells were disrupted by treatment with lysozyme, ribonuclease A, 0.1% Triton X-100, and sonication (Branson 450 sonifier). Lysate was cleared by centrifugation (15000 × g) and filtration (0.2 µM). Cleared lysate was purified by immobilized metal-ion affinity chromatography (IMAC) using a HisTrap HP column (GE Healthcare, Piscataway, NJ) on an AKTA FPLC system (Amersham Biosciences, Piscataway, NJ). UGM was eluted with a linear gradient of 0–500 mM imidazole in 50 mM sodium phosphate buffer at pH 7.4 with 300 mM sodium chloride. Typical yields were > 30 mg/L and pooled UGM fractions were > 90% pure. Glycerol solutions (10% v/v) of UGM were vitrified in liquid nitrogen and stored at – 80 °C.
Kd determination
Fluorescence polarization assays measuring ligand displacement by a UDP-fluorescein (UDP-Fl) probe were performed as described previously.17 Purified UGM was thawed and dialyzed against 50 mM sodium phosphate pH 7. Assays were conducted in 384-well format in 30 µL final volume, with 15 nM UDP-Fl probe and 75 nM UGM. Concentrations of UDP (Sigma-Aldrich, St. Louis, MO), UDP-Galp (Calbiochem, San Diego, CA) or UDP-Glc (Calbiochem, San Diego, CA) were varied from 0.5 µM for all ligands to 10 mM (UDP), 22 mM (UDP-Galp), or 50 mM (UDP-Glc). Millipolarization values were measured using an Envision 2100 plate reader (Perkin-Elmer, Waltham, MA). Data were fit using Kaleidagraph software (Synergy Software, USA) to y = (a–d)/(1 + (x/c)−b + d) (Equation 1) where a = maximum FP signal, b = slope, c = apparent Kd value, and d = minimum FP signal. Kd values were calculated from apparent Kd values using the equation Kd = Kd(app)/(1 + [UGM]/ Kd(UDP-Fl)) (Equation 2) where Kd(UDP-Fl) is the Kd value of UDP-Fl probe binding to UGM determined for the particular enzyme preparation (90 nM). The Kd value for UDP-Glc binding to reduced UGM was determined based on a decrease in FAD fluorescence using a QuantaMaster1 photon-counting fluorimeter (Photon Technology International, NJ) as described previously.27
UV/Vis spectroscopy and HPLC assay of UDP-Glc
Ligand titration profiles were performed similarly to previous methods25 using a Cary 50 UV-Visible spectrophotometer (Varian, Palo Alto, CA). After subtracting buffer (50 mM sodium phosphate pH 7) background absorbance, the absorbance of a solution containing 250 µM UGM, 20 mM sodium dithionite (Fluka, Steinheim, Germany) and phosphate buffer was measured using the scan mode (400–550 nm) before and after addition of aliquots of 100 mM UDP-Galp or UDP-Glc to generate titration curves.
UDP-Glc was tested as a potential substrate for UGM using an HPLC-based assay.54 In this assay, substrate and product (typically UDP-Galp and UDP-Galf) are separated with an isocratic elution of 200 mM ammonium acetate pH 8 on an anion exchange column (CarboPac PA-100, Dionex, Sunnyvale, CA). UDP-Glcp was found to coelute with UDP-Galp, and no additional peak (for the “product” UDP-Glcf) is observed following incubation of reduced UGM with UDP-Glcp.
Crystallization of UGM
Prior to crystallization, IMAC-purified UGM was further purified by anion-exchange chromatography. UGM was dialyzed into 50 mM HEPES buffer at pH 7.0. Dialyzed samples were purified on a Mono Q 5/50 GL column (GE Healthcare) equilibrated with 50 mM HEPES pH 7.0. UGM was eluted with a 0–30% linear gradient of 50 mM HEPES buffer at pH 7.0, 1 M sodium chloride. Fractions containing UGM (identified by SDS-PAGE) were pooled and concentrated. Concentrated UGM was > 99% pure as assayed by SDS-PAGE. UGM was dialyzed into 20 mM HEPES buffer at pH 7.0 prior to crystallization.
All crystals were obtained using hanging-drop vapor diffusion,55 with drops consisting of 1.5 µL mother liquor and 1.5 µL UGM (5 mg/mL). Sparse matrix screening using The Cryos Suite (Qiagen) was performed in the presence of 2 mM UDP-Glc, with each precipitant diluted by 1/3 by the addition of L-cysteine to a final concentration of 7 mM. L-cysteine was previously identified as helpful for certain UGM crystallization conditions.56 Initial conditions (Cryos condition #87) were optimized to give single yellow crystals that grew from clumps of crystalline precipitate over 1–2 weeks using a mother liquor of 85 mM ammonium acetate, 42 mM tri-sodium citrate pH 5.6, 12.3% PEG 4000, 7.5% glycerol, and 15 mM L-cysteine, with the protein solution containing 5 mM UDP-Glc. Crystals were vitrified in liquid N2 after swiping through mother liquor supplemented with 0.9 M ammonium acetate and 2% glycerol. Data were collected at the Advanced Photon Source (BioCARS, Argonne, IL). Phases were determined by molecular replacement using Phaser57 with the K. pneumoniae structure (PDB code 2BI8) as a search model.29 Refinement was performed using Refmac (v5.2.0019).58 In the omit map of residue Arg174 in monomer A, electron density (3σ) can be seen to the δ-carbon (data not shown). The entire residue is modeled due to the strong biochemical evidence indicating a role in binding the pyrophosphoryl group.30 In open monomer B, electron density (3σ) is seen to the β-carbon for Arg174. Figure 3, Figure 4, Figure 6, and Figure 7 were generated in PyMOL, and superposition (Fig. 3d) was performed using the default algorithms.59
STD-NMR spectroscopy
STD-NMR was performed similarly to previous methods.34 Final concentrations were 20 µM UGM (40 µM active sites), 4 mM ligand (UDP-Galp or UDP-Glc), and 10 µM deuterated 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS) in 50 mM K2HPO4/KH2PO4 pH 7.6 in 99% D2O. For reduced samples, concentrated sodium dithionite was added to a final concentration of 20 mM to otherwise identical samples under scrubbed argon gas. STD-NMR spectra were recorded on a DMX 750 with Cryoprobe (Bruker, Billerica, MA) at 300K with 1024 scans. Continuous wave saturation of protein resonances was performed at −1.0 ppm with a saturation time of 2 seconds. Phase cycling was used to subtract saturated spectra from reference spectra. A spin lock pulse was found to provide a better baseline and was used for all spectra. No STD signal was present in experiments performed in the absence of enzyme, indicating that saturation transfer from protein to ligand is the reason for observed STD spectra. Ligand resonances were identified using previous assignments34 and available databases (MMCD: mmcd.nmrfam.wisc.edu, HMDB: www.hmdb.ca). Peak areas of STD spectra were measured using XWINNMR. STD values for UDP-Galp were overall quite similar to those seen previously34 so we referenced our spectra in the same manner, with the largest STD peak, from protons 1’U/5U, set at 100%, and all other peak areas referenced to that value. The same assumptions regarding overlap were made for UDP-Galp samples. The overlapping protons 3G and 6G(a) of UDP-Glc were assumed for simplicity to have equal STD values.
Energy minimized active site model
The UDP-Glc molecule of the oxidized UGM structure was modified to UDP-Galp by inverting the stereochemistry at C4. The β-phosphoryl and sugar moieties were rotated manually to position the carbon–β-phosphoryl oxygen bond in line with N5 of flavin. The UMP moiety was not moved. Energy minimization was performed on monomer A using Sybyl (Tripos, St. Louis, MO). Water molecules were removed, hydrogen atoms were added, and Gasteiger-Marsilli charges were calculated. Energy minimization used the Tripos force field and system defaults, utilizing 100 rounds of minimization. The UDP-Galp position was not fixed relative to the protein during minimization; for example the distance between the N5 position of flavin and the anomeric carbon (C1) was free to vary as minimization progressed.
Accession number
Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 3GF4.
Supplementary Material
Supplementary Figure SF1. Repeat of STD-NMR experiments. Experiment depicted in Fig. 2 was repeated. (a) Oxidizing conditions. (b) Reducing conditions. (c) Atom labels.
Supplementary Figure SF2. Minimized model of UDP-Galp. Contacts between substrate and specific active site residues are shown (Å). Uridine moiety contacts are equivalent to those observed for UDP-Glc and are not shown (Fig. 4).
Supplementary Figure SF3. Additional active site residues of monomer A. Binding residues that are conserved in prokaryotes but differ in eukaryotes are shown (light gray carbons), with contacts to the UDP-Glc ligand (Å). Also depicted (dark gray) are hydrophobic residues that move significantly between monomer A and B to form the uridine binding pocket. UDP-Glc and FAD are colored as in Fig. 7a.
Acknowledgments
This research was supported by the NIH (AI 063596 to LLK). TDG was supported by the NIH Biotechnology Training Program (GM08349). MJB was funded by the NIH Molecular Biosciences Training Grant (GM07215) and a Steenbock Fellowship from the UW-Madison Department of Biochemistry. Use of the Advanced Photon Source was supported by the U.S. D.O.E, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. Use of the BioCARS Sector 14 was supported by the NIH, National Center for Research Resources (RR 007707). NMR experiments were performed at the National Magnetic Resonance Facility at Madison, supported by NIH grants P41RR02301 (BRTP/ NCRR) and P41GM66326 (NIGMS). This facility is also supported by the University of Wisconsin, the NIH (RR02781, RR08438), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA. We thank Dr. Kenneth Satyshur for data collection at BioCARS Beamline 14-ID-B. We thank Dr. R.T. Raines for use of fluorimeter equipment and H. A. Steinberg for assistance with figures.
Abbreviations
- FAD
flavin adenine dinucleotide
- Galf
galactofuranose
- Galp
galactopyranose
- Glc
glucose
- Glcp
glucopyranose
- IMAC
immobilized metal-ion affinity chromatography
- PDB
Protein Data Bank
- rmsd
root mean square deviation
- STD
saturation transfer difference
- UGM
UDP-galactopyranose mutase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain
References
- 1.Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu. Rev. Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 2.Jann B, Shashkov AS, Kochanowski H, Jann K. Structure of the O16 polysaccharide from Escherichia coli O16:K1: an NMR investigation. Carbohydr. Res. 1994;264:305–311. doi: 10.1016/s0008-6215(05)80014-x. [DOI] [PubMed] [Google Scholar]
- 3.Nassau PM, Martin SL, Brown RE, Weston A, Monsey D, McNeil MR, Duncan K. Galactofuranose biosynthesis in Escherichia coli K-12: Identification and cloning of UDP-Galactopyranose mutase. J. Bacteriol. 1996;178:1047–1052. doi: 10.1128/jb.178.4.1047-1052.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Köplin R, Brisson J-R, Whitfield C. UDP-galactofuranose precursor required for formation of the lipopolysaccharide O antigen of Klebsiella pneumoniae Serotype O1 is synthesized by the product of the rfbDKPO1 gene. J. Biol. Chem. 1997;272:4121–4128. doi: 10.1074/jbc.272.7.4121. [DOI] [PubMed] [Google Scholar]
- 5.Stevenson G, Neal B, Liu D, Hobbs M, Packer NH, Batley M, Redmond JW, Lindquist L, Reeves P. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J. Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Latge JP, Kobayashi H, Debeaupuis JP, Diaquin M, Sarfati J, Wieruszeski JM, Parra E, Bouchara JP, Fournet B. Chemical and immunological characterization of the extracellular galactomannan of Aspergillus fumigatus. Infect. Immun. 1994;62:5424–5433. doi: 10.1128/iai.62.12.5424-5433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McConville MJ, Homans SW, Thomas-Oates JE, Dell A, Bacic A. Structures of the glycoinositolphospholidpids from Leishmania major. A family of novel galactofuranose-containing glycolipids. J. Biol. Chem. 1990;265:7385–7394. [PubMed] [Google Scholar]
- 9.Previato JO, Gorin PA, Mazurek M, Xavier MT, Fournet B, Wieruszeski JM, Mendonca-Previato L. Primary structure of the oligosaccharide chain of lipopeptidophosphoglycan of epimastigote forms of Trypanosoma cruzi. J. Biol. Chem. 1990;265:2518. [PubMed] [Google Scholar]
- 10.Pan F, Jackson M, Ma YF, McNeil MR. Cell wall core galactofuran synthesis is essential for growth of mycobacteria. J. Bacteriol. 2001;183:3991–3998. doi: 10.1128/JB.183.13.3991-3998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Ganiny AM, Sanders DA, Kaminskyj SG. Aspergillus nidulans UDP-galactopyranose mutase, encoded by ugmA plays key roles in colony growth, hyphal morphogenesis, and conidiation. Fungal Genet. Biol. 2008;45:1533–1542. doi: 10.1016/j.fgb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Damveld RA, Franken A, Arentshorst M, Punt PJ, Klis FM, van den Hondel CA, Ram AF. A novel screening method for cell wall mutants in Aspergillus niger identifies UDP-galactopyranose mutase as an important protein in fungal cell wall biosynthesis. Genetics. 2008;178:873–881. doi: 10.1534/genetics.107.073148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmalhorst PS, Krappmann S, Vervecken W, Rohde M, Muller M, Braus GH, Contreras R, Braun A, Bakker H, Routier FH. Contribution of galactofuranose to the virulence of the opportunistic pathogen Aspergillus fumigatus. Eukaryot. Cell. 2008;7:1268–1277. doi: 10.1128/EC.00109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleczka B, Lamerz A-C, van Zandbergen G, Wenzel A, Gerardy-Schahn R, Wiese M, Routier FH. Targeted Gene Deletion of Leishmania major UDP-galactopyranose Mutase Leads to Attenuated Virulence. J. Biol. Chem. 2007;282:10498–10505. doi: 10.1074/jbc.M700023200. [DOI] [PubMed] [Google Scholar]
- 15.Pederson LL, Turco SJ. Galactofuranose metabolism: a potential target for antimicrobial chemotherapy. Cell. Mol. Life Sci. 2003;60:259–266. doi: 10.1007/s000180300021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scherman MS, Winans KA, Stern RJ, Jones V, Bertozzi CR, McNeil MR. Drug targeting Mycobacterium tuberculosis cell wall synthesis: development of a microtiter plate-based screen for UDP-galactopyranose mutase and identification of an inhibitor from a uridine-based library. Antimicrob. Agents Chemother. 2003;47:378–382. doi: 10.1128/AAC.47.1.378-382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soltero-Higgin M, Carlson EE, Phillips JH, Kiessling LL. Identification of inhibitors for UDP-galactopyranose mutase. J. Am. Chem. Soc. 2004;126:10532–10533. doi: 10.1021/ja048017v. [DOI] [PubMed] [Google Scholar]
- 18.Carlson EE, May JF, Kiessling LL. Chemical Probes of UDP-Galactopyranose Mutase. Chem. Biol. 2006;13:825–837. doi: 10.1016/j.chembiol.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Dykhuizen EC, May JF, Tongpenyai A, Kiessling LL. Inhibitors of UDP-galactopyranose mutase thwart mycobacterial growth. J. Am. Chem. Soc. 2008;130:6706–6707. doi: 10.1021/ja8018687. [DOI] [PubMed] [Google Scholar]
- 20.Sanders DAR, Staines AG, McMahon SA, McNeil MR, Whitfield C, Naismith JH. UDP-galactopyranose mutase has a novel structure and mechanism. Nat. Struct. Biol. 2001;8:858–863. doi: 10.1038/nsb1001-858. [DOI] [PubMed] [Google Scholar]
- 21.Barlow JN, Girvin ME, Blanchard JS. Positional isotope exchange catalyzed by UDP-galactopyranose mutase. J. Am. Chem. Soc. 1999;121:6968–6969. [Google Scholar]
- 22.Fullerton SWB, Daff S, Sanders DAR, Ingledew WJ, Whitfield C, Chapman SK, Naismith JH. Potentiometric analysis of UDP-galactopyranose mutase: stabilization of the flavosemiquinone by substrate. Biochemistry. 2003;42:2104–2109. doi: 10.1021/bi027077f. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Liu H-w. Mechanistic investigation of UDP-galactopyranose mutase from Escherichia coli using 2- and 3-fluorinated UDP-galactofuranose as probes. J. Am. Chem. Soc. 2001;123:6756–6766. doi: 10.1021/ja010473l. [DOI] [PubMed] [Google Scholar]
- 24.Soltero ML, Carlson EE, Kiessling LL. Abstracts of the 29th Steenbock Symposium: Cofactors, coenzymes and catalysis. University of Wisconsin-Madison; 2003. A Proposal for the Catalytic Mechanism of UDP-Galactopyranose Mutase. [Google Scholar]
- 25.Soltero-Higgin M, Carlson EE, Gruber TD, Kiessling LL. A unique catalytic mechanism for UDP-galactopyranose mutase. Nature Struct. Mol. Biol. 2004;11:539–543. doi: 10.1038/nsmb772. [DOI] [PubMed] [Google Scholar]
- 26.Huang Z, Zhang Q, Liu H-w. Reconstitution of UDP-galactopyranose mutase with 1-deaza-FAD and 5-deaza-FAD: analysis and mechanistic implications. Bioorg. Chem. 2003;31:494–592. doi: 10.1016/j.bioorg.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Itoh K, Huang Z, Liu Hw. Synthesis and Analysis of Substrate Analogues for UDP-Galactopyranose Mutase: Implication for an Oxocarbenium Ion Intermediate in the Catalytic Mechanism. Org. Lett. 2007;9:879–882. doi: 10.1021/ol0631408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller SM. A new role for an old cofactor. Nature Struct. Mol. Biol. 2004;11:497–498. doi: 10.1038/nsmb0604-497. [DOI] [PubMed] [Google Scholar]
- 29.Beis K, Srikannathasan V, Liu H, Fullerton SWB, Bamford VA, Sanders DAR, Whitfield C, McNeil MR, Naismith JH. Crystal Structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-Galactopyranose Mutase in the Oxidised State and Klebsiella pneumoniae UDP-Galactopyranose Mutase in the (Active) Reduced State. J. Mol. Biol. 2005;348:971–982. doi: 10.1016/j.jmb.2005.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chad JM, Sarathy KP, Gruber TD, Addala E, Kiessling LL, Sanders DAR. Site-Directed Mutagenesis of UDP-Galactopyranose Mutase Reveals a Critical Role for the Active-Site, Conserved Arginine Residues. Biochemistry. 2007;46:6723–6732. doi: 10.1021/bi7002795. [DOI] [PubMed] [Google Scholar]
- 31.Yuan Y, Bleile DW, Wen X, Sanders DAR, Itoh K, Liu H-w, Pinto BM. Investigation of Binding of UDP-Galf and UDP-[3-F]Galf to UDP-galactopyranose Mutase by STD-NMR Spectroscopy, Molecular Dynamics, and CORCEMA-ST Calculations. J. Am. Chem. Soc. 2008;130:3157–3168. doi: 10.1021/ja7104152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao X, Bleile DW, Yuan Y, Chao J, Sarathy KP, Sanders DA, Pinto BM, O'Neill MA. Substrate directs enzyme dynamics by bridging distal sites: UDP-galactopyranose mutase. Proteins. 2008;74:972–979. doi: 10.1002/prot.22206. [DOI] [PubMed] [Google Scholar]
- 33.Williams RF, Bruice TC. The kinetics and mechanisms of 1,5-dihydroflavin reduction of carbonyl compounds and flavin oxidation of alcohols. 2.Ethyl pyruvate, pyruvamide, and pyruvic acid. J. Am. Chem. Soc. 1976;98:7752–7768. [Google Scholar]
- 34.Yuan Y, Wen X, Sanders DAR, Pinto BM. Exploring the Mechanism of Binding of UDP-Galactopyranose to UDP-Galactopyranose Mutase by STD-NMR Spectroscopy and Molecular Modeling. Biochemistry. 2005;44:14080–14089. doi: 10.1021/bi0513406. [DOI] [PubMed] [Google Scholar]
- 35.Angyal SJ, Pickles VA. Equilibria Between Pyranoses and Furanoses. Aust. J. Chem. 1972;25:1695–1710. [Google Scholar]
- 36.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer M, Meyer B. Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angew. Chem. Int. Ed. Engl. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 38.Eppe G, Peltier P, Daniellou R, Nugier-Chauvin C, Ferrières V, Vincent SP. Probing UDP-galactopyranose mutase binding pocket: a dramatic effect on substitution of the 6-position of UDP-galactofuranose. Bioorg. Med. Chem. Lett. 2009;19:814–816. doi: 10.1016/j.bmcl.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Errey J, Mann M, Fairhurst S, Hill L, McNeil MR, Naismith J, Percy J, Whitfield C, Field R. Sugar nucleotide recognition by Klebsiella pneumoniae UDP-d-galactopyranose mutase: Fluorinated substrates, kinetics and equilibria. Org Biomol Chem. 2009;7:1009. doi: 10.1039/b815549f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rutledge LR, Campbell-Verduyn LS, Hunter KC, Wetmore SD. Characterization of nucleobase-amino acid stacking interactions utilized by a DNA repair enzyme. J. Phys. Chem. B. 2006;110:19652–19663. doi: 10.1021/jp061939v. [DOI] [PubMed] [Google Scholar]
- 41.Ellis JJ, Broom M, Jones S. Protein-RNA interactions: Structural analysis and functional classes. Proteins. 2007;66:903–911. doi: 10.1002/prot.21211. [DOI] [PubMed] [Google Scholar]
- 42.Zacharias N, Dougherty DA. Cation-pi interactions in ligand recognition and catalysis. Trends Pharmacol. Sci. 2002;23:281–287. doi: 10.1016/s0165-6147(02)02027-8. [DOI] [PubMed] [Google Scholar]
- 43.Vyas NK, Vyas MN, Quiocho FA. Sugar and signal-transducer binding sites of the Escherichia coli galactose chemoreceptor protein. Science. 1988;242:1290–1295. doi: 10.1126/science.3057628. [DOI] [PubMed] [Google Scholar]
- 44.Hashimoto H. Recent structural studies of carbohydrate-binding modules. Cell. Mol. Life Sci. 2006;63:2954–2967. doi: 10.1007/s00018-006-6195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borrok MJ, Kiessling LL, Forest KT. Conformational changes of glucose/galactose-binding protein illuminated by open, unliganded, and ultra-high-resolution ligand-bound structures. Protein Sci. 2007;16:1032–1041. doi: 10.1110/ps.062707807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beverley SM, Owens KL, Showalter M, Griffith CL, Doering TL, Jones VC, McNeil MR. Eukaryotic UDP-Galactopyranose Mutase (GLF Gene) in Microbial and Metazoal Pathogens. Eukaryot. Cell. 2005;4:1147–1154. doi: 10.1128/EC.4.6.1147-1154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bakker H, Kleczka B, Gerardy-Schahn R, Routier F. Identification and partial characterization of two eukaryotic UDP-galactopyranose mutases. Biol. Chem. 2005;386:657–661. doi: 10.1515/BC.2005.076. [DOI] [PubMed] [Google Scholar]
- 48.Koellner G, Bzowska A, Wielgus-Kutrowska B, Luić M, Steiner T, Saenger W, Stepiński J. Open and closed conformation of the E.coli purine nucleoside phosphorylase active center and implications for the catalytic mechanism. J. Mol. Biol. 2002;315:351–371. doi: 10.1006/jmbi.2001.5211. [DOI] [PubMed] [Google Scholar]
- 49.Morris VK, Izard T. Substrate-induced asymmetry and channel closure revealed by the apoenzyme structure of Mycobacterium tuberculosis phosphopantetheine adenylyltransferase. Protein Sci. 2004;13:2547–2552. doi: 10.1110/ps.04816904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tweeddale H, Notley-McRobb L, Ferenci T. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. J. Bacteriol. 1998;180:5109–5116. doi: 10.1128/jb.180.19.5109-5116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bochner BR, Ames BN. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- 53.Ramos A, Boels IC, de Vos WM, Santos H. Relationship between glycolysis and exopolysaccharide biosynthesis in Lactococcus lactis. Appl. Environ. Microbiol. 2001;67:33–41. doi: 10.1128/AEM.67.1.33-41.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee R, Monsey D, Weston A, Duncan K, Rithner C, McNeil MR. Enzymatic synthesis of UDP-galactofuranose and an assay for UDP-galactopyranose mutase based on high-performance liquid chromatography. Anal. Biochem. 1996;242:1–7. doi: 10.1006/abio.1996.0419. [DOI] [PubMed] [Google Scholar]
- 55.McPherson A. Preparation and analysis of protein crystals. New York: John Wiley & Sons, Inc; 1982. [Google Scholar]
- 56.Sanders DAR, McMahon SA, Leonard GL, Naismith JH. Molecular placement of experimental electron density: a case study on UDP-galactopyranose mutase. Acta Crystallogr. 2001;D57:1415–1420. doi: 10.1107/s0907444901009829. [DOI] [PubMed] [Google Scholar]
- 57.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr. 2004;D60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 58.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 59.Delano WL. The PyMOL Molecular Graphics System. CA, USA: Delano Scientific, Palo Alto; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure SF1. Repeat of STD-NMR experiments. Experiment depicted in Fig. 2 was repeated. (a) Oxidizing conditions. (b) Reducing conditions. (c) Atom labels.
Supplementary Figure SF2. Minimized model of UDP-Galp. Contacts between substrate and specific active site residues are shown (Å). Uridine moiety contacts are equivalent to those observed for UDP-Glc and are not shown (Fig. 4).
Supplementary Figure SF3. Additional active site residues of monomer A. Binding residues that are conserved in prokaryotes but differ in eukaryotes are shown (light gray carbons), with contacts to the UDP-Glc ligand (Å). Also depicted (dark gray) are hydrophobic residues that move significantly between monomer A and B to form the uridine binding pocket. UDP-Glc and FAD are colored as in Fig. 7a.