Abstract
We describe the development of innovative plasmon resonance energy transfer (PRET)-based molecular imaging of biomolecules in living cells. Our strategy of in vivo PRET imaging relies on the resonant plasmonic energy transfer from a gold nanoplasmonic probe to conjugated target molecules, which creates “quantized quenching dips” within the Rayleigh scattering spectrum of the probe. The positions of these quantized quenching dips exactly match with the absorption peaks of the target molecule since we intentionally design nanoantennas (i.e., nanoplasmonic probes) to overlap the electronic dipoles of the molecule and the plasmonic resonance dipole of nanoantennas. Such the quenching dips allow quantitative and long-term dynamic imaging of the target molecule without the drawbacks of photobleaching and blinking inherent to fluorescent markers, which cannot provide chemical fingerprints. Compared with other imaging methods, our PRET spectroscopic imaging method allows us to generate nanoscale specific wavelengths of local light sources in living systems via nanoantennas and transmit back the nanospectroscopic imaging data of biochemical activities in living cells. As a first demonstration of in vivo PRET imaging, we performed a visualization of the dynamics of intracellular cytochrome c in HepG2 cells under ethanol-induced apoptosis.
Our understanding of biological systems is increasingly dependent on our ability to visualize and quantify biomolecules and biological events with high spatial and temporal resolution in the cellular context. In this regard, the development of cellular and molecular imaging techniques of living systems is of considerable interest in many areas of research, from molecular and cellular biology to medical diagnostics and molecular medicine.1–4
However, there is no existing spectroscopic imaging system for living cells. Current techniques for in vivo cellular imaging are primarily based on organic fluorophores including genetically encoded fluorophores.5–11 In spite of excellent demonstrations of fluorescent or quantum dot (QD)-based imaging in biology, both methods cannot provide chemical fingerprint information. Moreover, organic fluorophores have limitations in long-term imaging and “multiplexing” without sophisticated instrumentation and processing due to broad absorption/emission profiles, photobleaching, and blinking problems. In addition, multiple filter sets must be used for each corresponding fluorescent label in the case of using multiple fluorescent labels to observe various biomolecules.
In order to overcome these limitations of organic fluorophores, QD as fluorescent labels have been extensively developed.3,12–16 Because of the benefit of broad absorption with size-dependent narrow, symmetric photoluminescence (PL) spectra, QD of different sizes can be excited by the same laser and still be distinguished by their size-dependent PL spectra. In addition, QD resist bleaching longer than their organic counterparts. However, in general, their luminescence is subject to blinking, and they also bleach eventually. Moreover, QD-based imaging techniques still need to be improved in terms of reproducible and robust surface functionalization and flexible bioconjugation. Accumulating in vitro evidence suggests that some QD can cause cellular toxicity, which may perturb the observed cellular components severely.17,18
However, gold nanoplasmonic particles are very appealing localized biological light source for dynamic molecular and cellular imaging, because they are highly water-soluble, biocompatible, and do not suffer from photobleaching nor blinking. Cellular components targeted by these particles within cells can be imaged in optical microscopy based on the intense Rayleigh scattering of the particles19,20 or photothermal interference contrast induced by the strong optical absorption of the particles at plasma resonance.21 It is extremely desirable to acquire fingerprint (i.e., spectroscopic) information of a target molecule, but it is not possible with Förster resonance energy transfer (FRET) method.
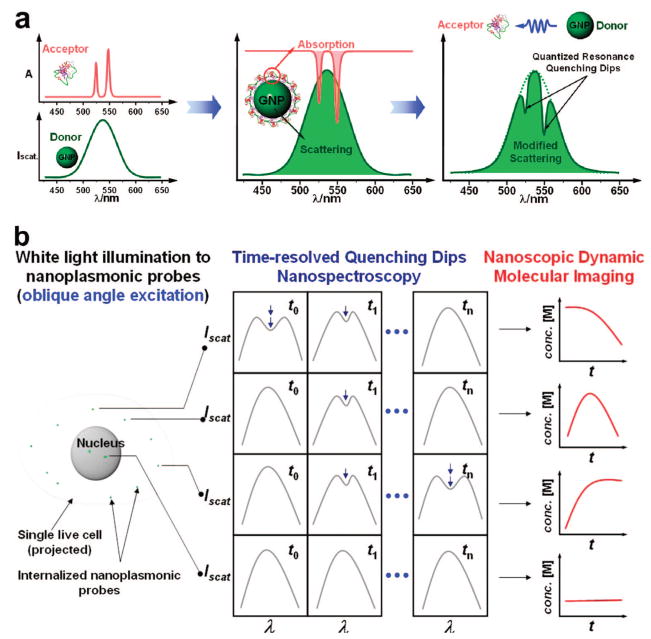
In one approach to this goal, we have recently developed plasmonic resonance energy transfer (PRET)-based nanospectroscopy22 by intentionally matching the plasmon resonance frequency of a gold nanoparticle with the frequency of the electronic transition energy of a biomolecule (Figure 1a). The underlying principle of PRET nanospectroscopy is the energy transfer between plasmon resonance of the gold nanoparticle and the envelope of molecular resonant peaks (i.e., absorption peaks) of the conjugated biomolecule that gives rise to resonant quenching dips in Rayleigh scattering spectrum of the particle. As a demonstration, we confirmed that the plasmon resonance energy of the 50 nm gold particle is transferred to conjugated cytochrome c (Cyt c), which results in the resonant spectral quenching dips in the particle Rayleigh spectrum. The positions of quenching dips exactly correspond to the absorbance peaks of cytochrome c.
Figure 1.
(a) Plasmonic resonance energy transfer (PRET)-based molecular imaging of living systems. When the frequencies of electronic transitions of a molecule overlap with the plasmon resonance frequency of gold nanoparticle upon conjugation of the particle with the molecule, this intentional spectral overlap allows the selective energy transfer and generates distinguishable spectral resonant quenching dips on the Rayleigh scattering spectrum of the particle. (b) In vivo dynamic cellular imaging of resonant biomolecules in a single live cell. In the example shown, as a white light illuminates into a live cell, gold nanoplasmonic probe can monitor the fluctuations of intracellular Cyt c in real time with nanoscopic spatial resolution by exploiting quenching dips which results from the selective energy transfer from the probe to the target Cyt c.
Here we show how to capture in vivo nanospectroscopic imaging of Cyt c in living cells based on PRET (Figure 1b). We demonstrate its utility for in vivo molecular imaging of intracellular Cyt c within HepG2 cells. Cyt c is chosen as a model protein for (i) its physiological importance. Cyt c acts as the charge transfer mediator23 and plays a crucial role in bioenergy generation, metabolism, and cell apoptosis;24 (ii) frequency matching for PRET. Cyt c has molecular resonances (optical absorption peaks) in visible range overlapping the plasmon resonance profiles (Rayleigh scattering spectra) of gold nanoparticles.
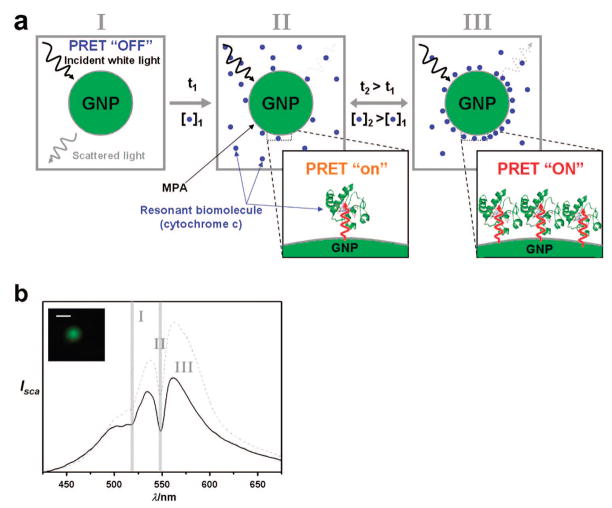
An important prerequisite for PRET-based nanospectroscopy to be applied to dynamic cellular imaging as a strong future application is to address the response time, the detection limit, and the reversibility of our system in vitro. In particular, in order to achieve the reversibility which is a very important capability of a molecular probe for dynamic imaging (Figure 2a), we selected 3-mercaptopropionic acid (MPA) as a ligand, because carboxylic acid is known to induce weak binding of Cyt c to the surface of a gold nanoparticle.25 MPA-modified, 50 nm gold nanoparticles were attached to the surface of glass slides. The scattering images and spectra of the MPA-modified probes were acquired using a dark-field microscopy system with a true-color imaging charge-coupled device (CCD) camera and a spectrometer (Figure 2b). In vitro dynamic imaging of Cyt c by both a single particle showing the plasmon resonance peak centered at 530~540 nm with green color and aggregates as judged by color and scattering profile (Figure 3a) were tested in detail.
Figure 2.
(a) Scheme of nanoscopic local and in situ imaging of resonant biomolecule using the ligand-functionalized probe. The adsorbed resonant biomolecule reversibly yields time, t (incubation time with cytochrome c (Cyt c)) and concentrations-dependent scattering spectra resulting from PRET between the probes and the resonant biomolecule conjugated on the surface of the probe. (b) Representative time-resolved Rayleigh scattering spectra of the single PRET probe in inset. The coupling of quantized electronic energy levels of molecules and plasmonic resonance dipole of nanoantennas via PRET is represented as quenching dips in nanoparticle Rayleigh scattering spectrum, and the positions (gray vertical lines) of dips match with the molecular absorption peak positions of Cyt c. The dip depth is found to be functions of incubation time (t) and concentrations of Cyt c. Bar corresponds to 2 μm. I, II, and III in figure are corresponding to the incubation time of 0, 10, and 30 min, respectively.
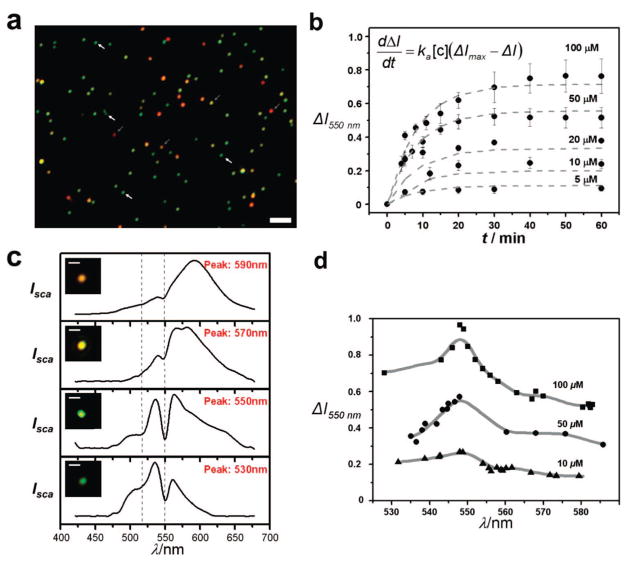
Figure 3.
In vitro PRET-based spectro-nanoscopy of Cyt c. (a). Dark-field reflectance image of carboxylic-acid functionalized 50 nm gold particles on a glass slide. The bar corresponds to 5 μm. (b) Time course of the transient quenching dip changes with varying concentrations of Cyt c. The dashed lines are theoretical fit to the first-order Langmuir equation (see inset) using the parameter (ΔImax) obtained from panel a. Note: to fit the data in panels a and b, the two lowest concentrations were excluded. (c) Representative Rayleigh scattering spectra and corresponding colors showing the effect of plasmon resonance frequency, λmax, of the probe on the resonant quenching dip depths. (d) Plot of the differential resonant quenching dip depths versus λmax with varying Cyt c concentrations.
We first investigated the sensing performance of the probes. To evaluate the sensitivity, different concentrations of Cyt c from a single stock solution were tested. Since a quenching dip at 550 nm is more dominant than that at 525 nm (Figure 2b) and no quenching dip at 525 nm was observed at Cyt c concentrations lower than 20 μM, only the quenching dip at 550 nm was analyzed. A response curve was constructed by measuring the relative dip depth, ΔI550nm= (Iscat,550nm,[cyt]=0 − Iscat,550nm,[cyt])/Iscat,550nm,[cyt]=0, after exposure to a given concentration of Cyt c between 100 nM to 100 μM (Supporting Information, Figure S2). The surface-confined thermodynamic affinity constant, Ka, can be estimated by ΔI550nm = I550nm,max (Ka [Cyt c])/(1 + Ka [Cyt c]). This curve reveals important characteristics that describe the binding of Cyt c to the carboxylic acid-terminated probe. An important binding characteristic determined by the curve is Ka of 2.5 × 10−4 M−1 for the interaction of Cyt c and the probe, which suggests the interaction can be reversible (Note that no resonance quenching dips were observed without a ligand (MPA)). As expected, the quenching dips of the probe Rayleigh scattering spectrum is found to be completely reversible irrespective of the Cyt c concentrations. By replacing the Cyt c solution with buffer (50 mM PBS), the scattering spectrum is fully recovered. Detection limit (operationally defined as 5.0% change in the differential quenching dip) is as low as around 500 nM.
Next, we evaluated the response time. A representative example of a time trace of the probe (a single gold nanoparticle) measured at 100 μM of Cyt c is shown in Figure 2b. As the incubation time increases, the scattering intensity of the dips gradually decreases, and reaches the equilibrium over 30 min. Figure 3b shows time course of the transient quenching dip changes varying concentrations of Cyt c. A significant fit is obtained when the entire time course data are applied to a first order Langmuir equation, dΔI550nm/dt = ka [Cyt c](ΔImax − I550nm), using the parameter (ΔImax) obtained from Supporting Information, Figure 2S and the reaction is described by a single rate constant (gray dot lines in Figure 2b).
Since plasmon resonance frequency, λmax, of internalized colloidal particles can be red shifted throughout the visible region of the spectrum depending on the degree of aggregation under physiological condition, it is very important to know the effect of λmax on the quenching dip depth with varying the concentrations of Cyt c. The correlation of λmax with the molecular absorption peak (550 nm) was investigated by using the probes which have plasmon resonance wavelength, λmax, in the range of 530~590 nm (Figure 3c,d). The resonant quenching dip depths display interesting behavior when scanning λmax from 530 to 590 nm. When λmax is as close as the molecular resonance, the dip depth is increased. When molecular resonance maximum (550 nm) directly overlaps with λmax, the scattering intensity at the quenching dip drops sharply up to 86% than its initial value. As λmax gradually tuned to the red from the molecular resonance, a decrease in the induced dip depth is found. Off molecular resonance (λmax > 600 nm), no resonant quenching dips (at 525 and 550 nm) were detected. This trend was maintained at three different concentrations. It should be again noted that at higher concentrations (Cyt c > 20 μM), two quenching dips are observed whereas only the quenching dip at 550 nm is detected at lower concentrations.
The successful use of the plasmonic probes for monitoring changes in Cyt c concentrations based on PRET in vitro led us to establish the utility for in vivo biological dynamic imaging. Since there are few proteins known in which their molecular absorbances overlap with those of Cyt c,26 we focused on monitoring “changes” in intracellular Cyt c concentrations. Spectral interference can be avoided if the surface of gold plasmonic particles may be tailored to interact with Cyt c through a specific ligand–receptor interaction instead of using a nonspecific ligand approach in this study.
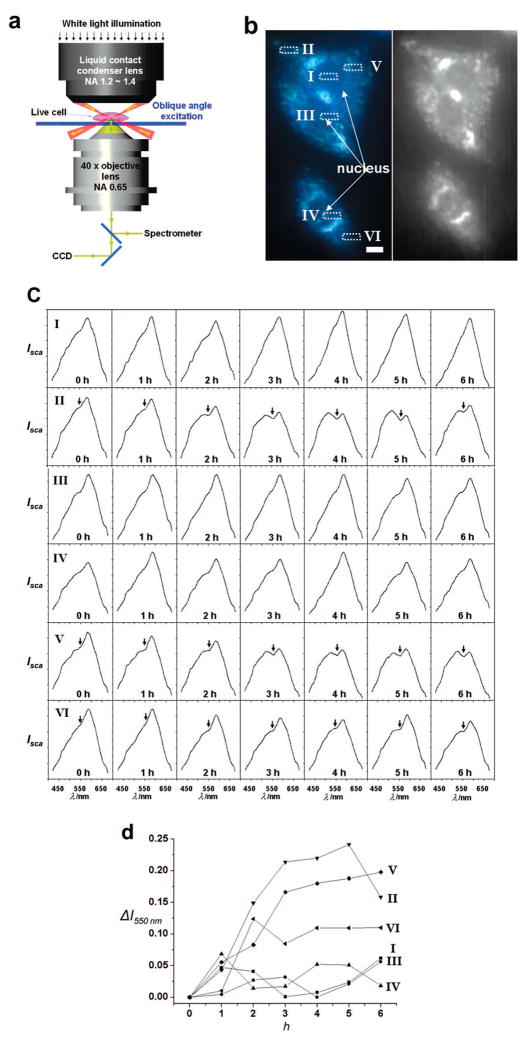
As a proof-of-concept, we applied PRET imaging to measure the changes in intracellular Cyt c in living HepG2 cells during ethanol-induced apoptosis. It is well known that Cyt c is released from mitochondria to cytoplasm as the response to proapoptotic stimuli, such as ethanol, due to increased permeability of the outer membrane of mitochondria.24,27,28 MPA-modified 50 nm gold nanoparticles were internalized into live HepG2 cells, and we monitored the spectra within HepG2 cells with 3 × 7 μm spatial resolution (Supporting Information, Figure S3). Dark-field images (Figure 4a) were taken at 1 h intervals (Figure 4a,b).
Figure 4.
In vivo dynamic imaging of Cyt c by PRET-based spectro-nanoscopy (a) The microscopy system consisting of a Carl Zeiss Axiovert 200 inverted microscope equipped with a dark-field condenser, true-color digital camera, and monochromator with a cooled spectrograph CCD camera. (b) Left: Representative dark-field image (from CCD camera) of HepG2 cells labeled with carboxylic acid-terminated probes prior to exposure with 100 mM ethanol. The bar corresponds to 10 μm. Right: Corresponding B/W dark-field scattering image from spectrometer. Scattering spectra were measured by an entrance slit in front of detector and by defining a region of interest (ROI) in the software (Supporting Information, Figure 2). (c) Imaging intracellular Cyt c dynamics: Representative scattering spectra from the numbered positions in panel a after exposure with 100 mM ethanol. Arrows in the positions II, V, and VI indicate the spectral quenching dips at 550 nm. (d) Time-course of the differential quenching dip changes at different positions compared to values at 0 h.
Compared to cells with internalized plasmonic probes, the striking differences of the scattered light intensity of the region-of interest and brightness of the dark-field image were seen in cells containing no probes. On average, maximum scattering intensities at λmax is ca. 2 times lower than those of cell with the probes (103 particles per one cell). The enhancement of scattering intensities also depends on the number of internalized particles (data not shown). Internalized probes can be distinguished by their colors as the dosage of the particles per cell is increased over 104/cell (Supporting Information, Figure S4). Representative dark-field image of HepG2 cells after internalization of the probes is shown in Figure 4b. The positions I, III, IV correspond to the nucleus, whereas the positions II, V, and VI indicate the cytoplasm.
As shown in Figure 4c, before exposure to 100 mM ethanol, only 3 positions among 69 regions of interest (4.3%) show very weak dip (maximum ΔI550nm ~ 0.05). By stark contrast, after exposure to ethanol, significant quenching dip at 550 nm at positions II, V, VI after 2 h is observed. After 6 h, 46% of total spectra show the spectral dip (minimum ΔI550nm > 0.10). More specifically, the average ΔI550nm from 3 to 6 h is ca. 0.17 in cytoplasm. On the other hand, the average ΔI550nm in the nucleus region is negligible (<0.032) as shown in positions I, III, and IV. Unlike the spectroscopic results, dark-field images of HepG2 cells with the probes did not give any significant difference during the 6 h exposure to ethanol (Supporting Information, Figure S5). We attributed the observed dip at 550 nm in the scattering spectra to the release of Cyt c from mitochondria to cytoplasm induced by ethanol. To confirm that the observed quenching dips are indeed due to increased intracellular release of Cyt c, as a control, HepG2 cells without ethanol treatment were monitored for 6 h. Similarly, the scattering spectrum and corresponding dark-field images were obtained (Supporting Information, Figures S6 and S7). In this case, before exposure to ethanol only 5.3% of total spectra obtained at 169 positions show very weak dip (maximum ΔI550nm < 0.10), which is similar to that of HepG2 cells with the probes prior to exposure. There is no noticeable variation in ΔI550nm after 6 h.
In conclusion, we have demonstrated PRET-based in vivo spectro-nanoscopy and visualized the dynamics of intracellular Cyt c within living HepG 2 cells during ethanol-induced apoptosis. The PRET between gold nanoplasmonic probe and a conjugated molecule creates the mirror image of absorption spectrum (i.e., quenching dips in Rayleigh scattering) of target molecule. By analyzing the quenching dip depth, we explored the sensitivity limits and response time of our plasmonic probe and found that even weak ligands-functionalized nanoplasmonic probe could detect reversibly low concentrations of Cyt c down to 500 nM in vitro. Having established the excellent sensing performance of our PRET imaging toward Cyt c in vitro, we successfully visualized the changes in intracellular Cyt c within living HepG 2 cells during ethanol-induced apoptosis. Our in vivo PRET imaging system has several key advantages over conventional imaging methods. PRET imaging neither blinks nor bleaches unlike fluorescence imaging and does not need to be discriminated from a strong background signal which is common problem in other plasmonic marker-based imaging. More importantly, PRET imaging provides the spectroscopic information of target molecule. In addition, since gold particles are good labels for electron and X-ray microscopy, which enable better crosschecks and permits the development of novel multimodal imaging technique. Since PRET imaging depends on molecular resonances of target molecules, it is easy to multiplex and distinguish different molecules by injecting different nanoplasmonic probes with different resonant frequencies. In the presence of other molecules in which molecular resonances overlap those of the target, it is easy to distinguish the target molecule since other molecules cannot overlap with matched frequency of specific nanoantenna. Spectral interference can be avoided by either using tailoring the surface of the probe through a specific ligand–receptor interaction or by modulating the resonant frequency of nanoantennas. We believe that in vivo PRET-based spectro-nanoscopy of living systems will have huge impacts on the fields of in vivo cellular imaging, systems biology, molecular diagnostics, drug discovery, and future quantitative biomedicine.
Supplementary Material
Acknowledgments
We acknowledge financial support by the Center for Nanostructured Materials and Technology (CNMT) under the 21st Century Frontier Research Programs of the Korea government (Code No. 08K1501-02300, 08K1501-2310, 08K1501-2320), NIH’s Nanomedicine Development Centers funding (3PN2 EY01824), and National Academies Keck Futures Initiative funding (NAKFI Nano09).
Footnotes
Supporting Information Available: Experimental details, equilibrium differential quenching dip change as a function of Cyt c concentrations, and dark-field reflectance images and scattering spectra of live HepG2 cells are available. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Miyawaki A. Dev Cell. 2003;4:295–305. doi: 10.1016/s1534-5807(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 2.Tsien RW, Tsien RY. Annu Rev Cell Biol. 1990;6:715–760. doi: 10.1146/annurev.cb.06.110190.003435. [DOI] [PubMed] [Google Scholar]
- 3.Michalet A, et al. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lang P, Yeow K, Nichols A, Scheer A. Nature Rev Drug Discovery. 2006;5:343–356. doi: 10.1038/nrd2008. [DOI] [PubMed] [Google Scholar]
- 5.Lippincott-Schwartz J, Patterson GH. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 6.Fradkov AF, et al. FEBS Lett. 2000;479:127–130. doi: 10.1016/s0014-5793(00)01895-0. [DOI] [PubMed] [Google Scholar]
- 7.Janetopoulos C, Jin T, Devreotes P. Science. 2001;291:2408–2411. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- 8.Miyawaki A, et al. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 9.Chang CJ, et al. Proc Natl Acad Sci USA. 2004;101:1129–1134. doi: 10.1073/pnas.0308079100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu J, et al. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 11.Cai L, Friedman N, Xie XS. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 12.Bruchez M. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 13.Chan WCW, Nie SM. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 14.Larson DR, et al. Science. 2000;300:1434–1436. [Google Scholar]
- 15.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 16.Courty S, et al. Nano Lett. 2006;6:1491–1495. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- 17.Hardman R. Environ Health Perspect. 2006;114:165–172. doi: 10.1289/ehp.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovric J, Cho SJ, Winnik FM, Maysinger D. Chem Biol. 2005;12:1227–1234. doi: 10.1016/j.chembiol.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Schultz S, Smith DR, Mock JJ, Schultz DA. Proc Natl Acad Sci USA. 2000;97:996–1005. doi: 10.1073/pnas.97.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X, El-Sayed IH, Qian W, El-Sayed MA. J Am Chem Soc. 2006;128:2115–2120. doi: 10.1021/ja057254a. [DOI] [PubMed] [Google Scholar]
- 21.Cognet L, et al. Proc Natl Acad Sci USA. 2003;100:11350–11355. doi: 10.1073/pnas.1534635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu GL, et al. Nat Methods. 2007;4:1015–1017. doi: 10.1038/nmeth1133. [DOI] [PubMed] [Google Scholar]
- 23.Wuttke DS, Bjerrum MJ, Winkler JR, Gray HB. Science. 1992;256:1007–1009. doi: 10.1126/science.256.5059.1007. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, et al. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 25.Bayraktar H, You CC, Rotello VM, Knapp MJ. J Am Chem Soc. 2007;129:2732–2733. doi: 10.1021/ja067497i. [DOI] [PubMed] [Google Scholar]
- 26.Kirschenbaum DM. Atlas of Protein Spectra in the Ultraviolet and Visible Regions. Plenum Press; London: 1972. [Google Scholar]
- 27.Szalai G, Krishnamurthy R, Hajnoczky G. EMBO J. 1999;18:6349–6361. doi: 10.1093/emboj/18.22.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama N, Eichhorst ST, Müller M, Krammer PH. Exp Cell Res. 2001;269:202–213. doi: 10.1006/excr.2001.5319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.