Abstract
Human high-density lipoprotein (HDL) plays a key role in the reverse cholesterol transport pathway that delivers excess cholesterol back to the liver for clearance. In vivo, HDL particles vary in size, shape and biological function. The discoidal HDL is a 140–240 kDa, disk-shaped intermediate of mature HDL. During mature spherical HDL formation, discoidal HDLs play a key role in loading cholesterol ester onto the HDL particles by activating the enzyme, lecithin:cholesterol acyltransferase (LCAT). One of the major problems for high-resolution structural studies of discoidal HDL is the difficulty in obtaining pure and, foremost, homogenous sample. We demonstrate here that the commonly used cholate dialysis method for discoidal HDL preparation usually contains 5–10% lipid-poor apoAI that significantly interferes with the high-resolution structural analysis of discoidal HDL using biophysical methods. Using an ultracentrifugation method, we quickly removed lipid-poor apoAI. We also purified discoidal reconstituted HDL (rHDL) into two pure discoidal HDL species of different sizes that are amendable for high-resolution structural studies. A small rHDL has a diameter of 7.6 nm, and a large rHDL has a diameter of 9.8 nm. We show that these two different sizes of discoidal HDL particles display different stability and phospholipid-binding activity. Interestingly, these property/functional differences are independent from the apoAI α-helical secondary structure, but are determined by the tertiary structural difference of apoAI on different discoidal rHDL particles, as evidenced by two-dimensional NMR and negative stain electron microscopy data. Our result further provides the first high-resolution NMR data, demonstrating a promise of structural determination of discoidal HDL at atomic resolution using a combination of NMR and other biophysical techniques.
Keywords: discoidal high-density lipoprotein, human apolipoprotein AI, structural determination, NMR spectroscopy, tertiary structure
Introduction
Human high-density lipoprotein (HDL) is a lipoprotein particle, transporting the excess cholesterol from peripheral tissues back to the liver for clearance.1,2 Epidemiologic data suggests that HDL is the best indicator for the risk of atherosclerosis with a high HDL level correlating with a low risk of atherosclerosis.3 HDL in vivo displays a mixture of different particles that vary in size, shape, and protein/lipid compositions, such as preβ-HDL, discoidal and spherical HDL.4 These HDL particles load different lipids including phospholipids, cholesterol, cholesterol ester (CE), and triacylglycerol (TG) during HDL assembly and exhibit different biological functions. Overall, different HDL particles function as a whole to play a key role in the reverse cholesterol transport (RCT) pathway that removes the unnecessary cholesterol from the human body.5
ApoAI is the major protein component of HDL particles. This protein is responsible for the structural integrity of HDL particles and displays different conformations on different HDL species, regulating HDL formation, maturation, and assembly.6,7 Up to 10% of circulating apoAI is lipid-free and seems to be specifically linked to the uptake of cellular phospholipids.8 In addition, lipid-free apoAI activates ABC-AI for efficient cholesterol efflux.9 Upon binding to small amounts of phospholipids, apoAI forms preβ-HDL particles.10 Experimental evidence, both in vitro and in vivo, demonstrated that preβ-HDL is the most active acceptor of free cholesterol.4,11 The addition of cholesterol and phospholipids to preβ-HDL generates larger and lipid-richer particles, triggering the assembly of HDL particles. During the initial stage of HDL assembly, discoidal HDL is formed and contains two apoAI molecules. ApoAIs on discoidal HDL activate the enzyme LCAT, which converts cholesterol into cholesterol ester.12 This results in accumulation of CE in the center of discoidal HDL and eventually forms a small mature spherical HDL, HDL3. Further maturation of spherical HDL3 depends on activity of LCAT to form a large HDL particle, HDL2. ApoAI on spherical HDL particles is able to bind to the HDL-receptor, SR-BI,13,14 and to mediate selective lipid uptake that transports the cholesterol back to the liver for clearance. This process regenerates lipid-depleted apoAI, which again scavenges peripheral cell cholesterol and phospholipids before returning to the liver. ApoAI on preβ-HDL and discoidal HDL particles also activates lipid transfer proteins (CETP and PLTP), which regulate dynamic interconversions between HDL and LDL.15,16
The discoidal HDL is a critical intermediate between lipid-poor apoAI and mature spherical HDL during HDL assembly, which plays a key role in loading cholesterol ester onto HDL. Significant attention has been focused on discoidal HDL.17 It has been demonstrated that discoidal HDL contains two apoAI molecules with a molecular weight of 140–240 kDa. Previous electron microscopy studies indicated that discoidal HDL is disk-shaped. The detailed structure of apoAI associated with discoidal HDL has been debated for nearly 3 decades and several models have been proposed.17 Among these models, the “picket fence” model suggested that the α-helices of apoAI were arranged parallel to the phospholipid bilayer, which was supported by primary sequence analysis,18 dried film infrared spectroscopy,19 and earlier computer modeling.20 However, recent experimental data using computer modeling based on the X-ray crystal structure of apoAI(44–243),21 FRET techniques,22–24 and infrared spectroscopy25 favors a “belt model,” which suggests that the α-helices of apoAI are perpendicular to the phospholipid bilayer. Recently, the Davidson and Thomas laboratories utilized high-resolution mass spectroscopy combined with cross-linking techniques to investigate apoAI structure on discoidal HDL particles.26–28 Their results support the general principle of the “belt model” and provide some details of the tertiary structure of apoAI bound to discoidal HDL. In particular, a report from Thomas' laboratory constructed a refined model of apoAI on discoidal HDL particles, suggesting that the N- and C-terminal regions of apoAI may have tertiary interaction to stabilize this lipid-bound apoAI conformation.29 Using H-D exchange mass spectroscopy, a refined molecular model of nascent discoidal HDL was reported, supporting the “double belt” model of two molecules of apoAI on discoidal HDL with residues 159–180 of each apoAI forming a protruding solvent-exposed loop. This report further showed that this solvent exposed loop, including Tyr166, a preferred target for site-specific oxidative modification within atheroma, directly interacts with and activates LCAT.30
X-ray crystallography and NMR spectroscopy are two powerful tools to study the structures of bio-molecules at atomic resolution. Both methods require pure samples of high homogeneity. Significant efforts have been made towards the crystallization of discoidal HDL particles, but were not successful, presumably due to the dynamic nature of these lipoprotein particles or the heterogeneity of the samples. Recently, crystallization of apoE bound to DPPC has been reported and the apoE molecular envelope determined to a 10 Å resolution.31 A model of apoE on DPPC particles suggests that each apoE molecule is folded into a helical hairpin with the binding region for the LDL receptor at its apex.31 NMR structural studies, on the other hand, are certainly challenging because these discoidal HDLs are large particles of protein/lipid complexes that previously far exceeded the molecular weight limit of solution NMR spectroscopy. However, solution NMR provides an ideal tool to study dynamic structures of bio-molecules and bio-complexes such as discoidal HDL particles.32,33 In addition, recently developed TROSY techniques removed the size limit of NMR, allowing for structural studies of large proteins and protein complexes with molecular weights >200,000 Da.34
In preparation of such NMR studies of discoidal HDL particles, we made a major effort to obtain ultra pure discoidal HDL samples. Our initial goal was to prepare ultra pure reconstituted HDL (rHDL) particles that are suitable for NMR structural studies. Our data indicates that rHDL particles prepared by the widely used cholate dialysis method35,36 usually contain 5–10% lipid-poor apoAI, which severely interfere with NMR spectral analysis. Using a quick ultracentrifugation method, we removed lipid-poor apoAI. We further carried out a density gradient ultracentrifugation that allowed us to purify two stable populations of discoidal rHDL particles with different sizes. One is a large rHDL particle with a diameter at 9.8 nm, and the other is a small rHDL particle with a diameter of 7.8 nm. We characterized both the discoidal HDL particles using circular dichroism spectroscopy (CD), chemical denaturation, and a phospholipid-binding assay. Our results indicated that these two discoidal HDL particles displayed different stability and phospholipids-binding activity, even though they share identical α-helix content. More importantly, NMR data indicated that apoAI protein displayed different NMR spectra on the different sized rHDL, suggesting that apoAI might adopt different tertiary conformations. Negative stain EM data also indicated a different shape of discoidal rHDL between small and large rHDL, supporting the notion that apoAI may adopt a different tertiary conformation between small and large rHDLs. In addition, our results provided the first high-resolution NMR data of discoidal HDL particles, demonstrating a promise of structural determination of discoidal HDL at atomic resolution using a combination of NMR and other biophysical techniques.
Results
Preparation of rHDL under different conditions
To carry out meaningful structural studies of discoidal HDL particles, high particle purity/homogeneity is an absolute necessity. Using the cholate dialysis method, we prepared rHDL under several different conditions. Figure 1, Panel A shows a gradient native gel of the rHDL particles prepared using different ratios of POPC:apoAI (POPC/cholate = 0.737). This figure clearly demonstrates that POPC:apoAI ratio is critical to prepare rHDL particles using cholate dialysis method. For example, at POPC:apoAI ratios <60:1, no homogenous rHDL is formed. Only a fused band representing a mixture of different particles was observed, potentially including lipid-free and lipid-poor apoAI (Panel A, Lanes 2–5). At and above a POPC:apoAI ratio of 60:1, only a single band of rHDL particles was observed, suggesting that a predominant population of rHDL particles were formed. Interestingly, the particle size of rHDL slightly increases with a larger ratio of POPC:apoAI. Based on native gel, at POPC:apoAI = 60:1 or 80:1, the particle size is ∼8.1 nm, whereas at POPC:apoAI = 100:1, the particle size is ∼9.8 nm. The results shown in Panel A are confirmed by crosslinking experiments using BS3 as the crosslinker. At POPC:apoAI ratios ≥60:1, rHDL particles predominantly contain two molecules of apoAI (Fig.1, Panel B). At POPC:apoAI ratios <60:1, particles contain a mixture of monomer, dimer, trimer, and higher oligomers (Lanes 2–5), similar to lipid-free apoAI (Lane 1b). Even in Lanes 6–8 of Panel B, very weak bands of trimer and tetramer are still visible.
Figure 1.
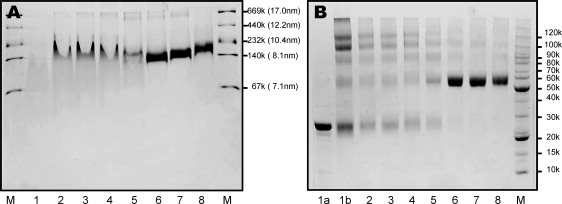
Panel A: A 6–20% gradient native PAGE gel of rHDL particles prepared using different POPC:ApoAI ratios (POPC/cholate = 0.737). Panel B: A 6–12% tricine gel of crosslinking results of rHDL particles using different POPC:ApoAI ratios. In both panels, lane 2: POPC:ApoAI = 5:1, lane 3: POPC:ApoAI = 10:1, lane 4: POPC:ApoAI = 20:1, lane 5: POPC:ApoAI = 40:1, lane 6: POPC:ApoAI = 60:1, lane 7: POPC:ApoAI = 80:1, lane 8: POPC:ApoAI = 100:1, lane M: molecular marker. In Panel A: lane 1: lipid-free apoAI. In Panel B: lane 1a: lipid-free apoAI without crosslinker, lane 1b: lipid-free apoAI with crosslinker.
We also tested the effects of different cholate concentrations on the rHDL particles at several different POPC:apoAI ratios. Our results indicates that, in contrast to the POPC:apoAI ratio, the POPC:cholate ratio has a limited effect. At all tested POPC:cholate ratios (80:60, 80:70, 80:80, 80:90, 80:100, 80:110, 80:120, and 80:140 with a fixed POPC/apoAI ratio of 80:1), the prepared rHDL run as a single band, except at POPC:cholate = 80:60 (data not shown). However, the POPC:cholate ratio seems to play a role in the purity of rHDL. For example, it seems that POPC:cholate ratios between 80:100 and 80:120 produce cleaner rHDL. Lower or higher ratios produced less pure rHDL particles. However, even at POPC:cholate ratios between 80:100 and 80:120, minor lipid-poor and higher molecular weight discoidal rHDL species can still observed in the crosslinking gel of these discoidal rHDL preparations (data not shown). Further purification of discoidal rHDL preparation using cholate dialysis method may be necessary for high-resolution structural studies of discoidal HDL particles.
To confirm if the discoidal rHDL particles need to be further purified, we carried out NMR studies. Using triple-labeled apoAI, we prepared an NMR sample of the rHDL particles generated using a ratio of POPC: apoAI:cholate = 80:1:100, and recorded [15N, 1H]-TROSY-HSQC spectra. Figure 2, Panel B shows the spectrum, displaying a well-dispersed NMR spectrum. As a comparison, Panel A shows the spectrum of Lp1AI that contains one molecule of apoAI. The NMR data indicates that apoAI in both cases display similar spectral patterns. An exception is the central region, which is more crowded in Panel B. This suggests that rHDL preparation may contain a fraction of lipid-poor Lp1AI. Indeed, new crosspeaks, as highlighted by arrows in Panel B, are not observed in Panel A. These new crosspeaks seem to be much broader than the crosspeaks from Lp1AI, indicating that they, along with the crowded crosspeaks in spectral center, may arise from rHDL particles. These results confirm that the discoidal rHDL preparation using cholate dialysis method contains a population of lipid-poor Lp1AI and further purification of this rHDL preparation is necessary for NMR studies of discoidal rHDL particles.
Figure 2.
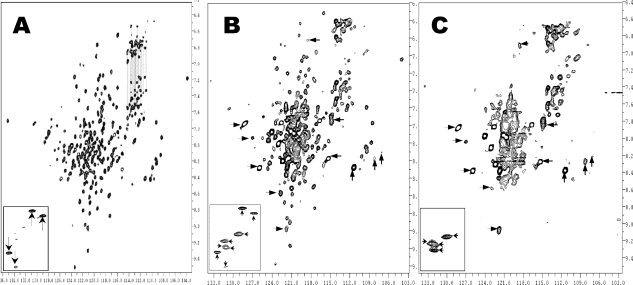
[15N, H] TROSY-HSQC spectra of Lp1AI (Panel A), rHDL sample before the first ultracentrifugation (Panel B) and rHDL sample after the first ultracentrifugation (Panel C). These spectra were recorded on a Varian INOVA 500 MHz spectrometer with a triple-resonance room temperature probe. The insets of different panels show the crosspeaks of the Trp sidechain Hɛ1/Nɛ1 atoms. The NMR samples are in 50 mM phosphate buffer at pH7.0, with 50 mM NaCl, 5 mM EDTA, 0.5 mM NaN3, and 0.05 mM DSS.
Purification of discoidal rHDL from lipid-poor apoAI
To purify discoidal rHDL particles, we utilized ultracentrifugation method with a density gradient to separate lipid-free/lipid poor apoAI from rHDL particles. Figure 3 shows the result with technical details described in the Materials and Methods. This ultracentrifugation method can be used to separate rHDL from lipid-free/lipid-poor apoAI and is also capable of preparing a large amount of pure rHDL. A total of 20 fractions were obtained, with the lightest particles on the top and the heaviest particles at the bottom. Although fractions 1–5 and 19–20 show no apoAI, fractions 6–13 display a single broad band that is above the 140 kDa and fractions 14–18 display a mixture of several different bands between 67 and 140 kDa. Because fractions 6–13 are the top fractions that are less dense than the bottom fractions and the molecular weight of these fractions is above 140 kDa, we conclude that fractions 6–13 contain purified rHDL particles. The rHDL particles contain two apoAI molecules and ∼160 phospholipids, thus are lighter than lipid-free/lipid-poor apoAI. In contrast, fractions 14–18 contain a mixture of different sizes with the smallest particles being ∼67 kDa and the largest being similar to rHDL. We suggest that the smaller particles are the lipid-poor particles, including Lp1AI. To verify this suggestion, we pooled fractions 6–13 together to make a NMR sample and recorded a [15N, 1H]-TROSY-HSQC spectrum (Fig. 2, Panel C), which displayed a completely different spectrum from that of Lp1AI (Fig. 2, Panel A). Indeed, all of the crosspeaks highlighted by arrows in Panel B are also shown in Panel C, in addition to the center crowded crosspeaks. Further, Panel C contains no crosspeaks similar to that of Lp1AI, suggesting that lipid-free/lipid-poor apoAI were completely removed by this ultracentrifugation method.
Figure 3.
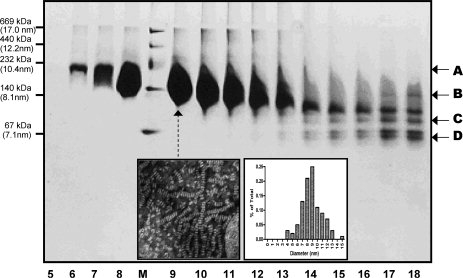
A 4–20% native gel of the fractions of the first ultracentrifugation of an rHDL preparation. A total of 20 fractions have been collected and fractions 1–4 and 19–20 contain no apoAI proteins. Fraction 1 is the top fraction and fraction 20 is the bottom fraction. M: High molecular weight marker. Arrows A and B show the broad range of the purified discoidal rHDL. Arrows C and D show several fractions of lipid-poor apoAI. Inset: Negative stain electron microscopic images of rHDL particles of Lane 9 (Left) and the size distribution determined by electron microscopy (Right). The average size of the discoidal HDL: 8.8 ± 3.7 nm.
The inset of Panel B in Figure 2 shows the crosspeaks of the Trp sidechain Hɛ1/Nɛ1 atoms of the prepared rHDL before ultracentrifugation. For comparison, the insets of Panels A and C in Figure 2 show the crosspeaks of the Trp sidechain Hɛ1/Nɛ1 atoms of Lp1AI (Panel A) and purified rHDL (Panel C). Human apoAI contains four Trp residues, thus four crosspeaks should be observed. Four well-dispersed Trp sidechain crosspeaks are observed in the spectrum of Lp1AI (Panel A, highlighted by “↓ or ↑”), whereas four less dispersed Trp Hɛ1/Nɛ1 sidechain crosspeaks are also observed in the purified rHDL (Panel C, highlighted by “←” or “→”). In contrast, Panel B shows a total of eight crosspeaks with four crosspeaks identical to the Trp sidechain crosspeaks of Lp1AI and four crosspeaks identical to the Trp sidechain crosspeaks of purified rHDL. This result confirms that the rHDL preparation using cholate dialysis method is a mixture of Lp1AI and rHDL, demonstrating that the purified rHDL particle using this ultracentrifugation method is a pure discoidal rHDL fraction.
Purification of two different sizes of discoidal rHDL particles
We noticed that the purified discoidal rHDL particles shown in Figure 3 displayed a very broad band, ranging between 140 and 232 kDa according to the molecular weight marker. Negative stain of EM showed that fraction 9 displayed discoidal shape rHDL particle with a very large size distribution (8.8 ± 3.7 nm) (Fig. 3, Inset). A native gel with a less loading clearly showed two fused bands, one is at ∼140 kDa and the other is at ∼200 kDa. A third band, which is between the two bands, is also seen which is much lighter, seems not very stable. A similar result was also reported recently, indicating three populations of the discoidal rHDL prepared using the cholate dialysis method, with a much less stable medium sized discoidal HDL population.37 To further separate these two size rHDLs, we carried out a second ultracentrifugation using a four-layer density gradient (1.15 g/mL, 1.10 g/mL, 1.08 g/mL and 1.07 g/mL). This ultracentrifugation can efficiently separate large rHDL from small rHDL. However, we have to repeat this four-layer density gradient ultracentrifugation several times, to obtain NMR quantities of purified smaller and larger particles. Figure 4, Panel A shows the result of the final spin using this ultracentrifugation, demonstrated mL that two different sizes of discoidal rHDL particles are separated. Clearly, large rHDL particle has a molecular weight ∼200 kDa as shown in Lanes 1 and 2 and small rHDL particle has a molecular weight of ∼140 kDa as shown in Lanes 7–10. Figure 4, Panel B shows the purified large (Lane 1) and small (Lane 2) rHDL particles, confirming the purity of the rHDL preparation. The question remains if these two rHDL particles display any morphology/structure/functional difference despite their different sizes.
Figure 4.
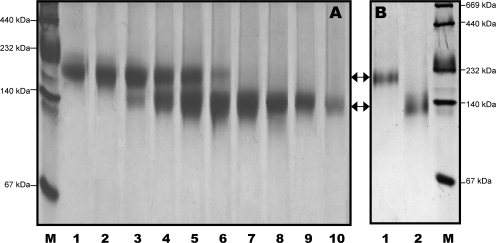
Panel A: A 4–20% native gel of the fractions of the last ultracentrifugation, showing the purification of small and large discoidal rHDL particles can be achieved using this ultracentrifugation method. Panel B: A 4–20% native gel of the purified large (Lane 1) and small (Lane 2) discoidal rHDL. M: High molecular weight marker.
Biophysical characterizations of small and large rHDL
We carried out experiments to characterize the shape and biophysical properties of both purified rHDL particles. We prepared purified rHDL using a ratio of POPC:apoAI:cholate = 80:1:108. Electron microscopy shows that both purified rHDL particles are disk-shaped (Fig. 5), with a particle size of 7.6 ± 1.6 nm for small particles and 9.8 ± 1.7 nm for large particles. In contrast to the rHDL particles shown in the inset of Figure 3, which have a diameter of 8.8 ± 3.7 nm, the size distribution of both purified small and large rHDL particles are significantly narrower (Fig. 5). This result confirms the native gel result shown in Figure 4, indicating that both purified rHDL particles are homogenous in size. To characterize apoAI protein structure and stability, we carried out CD and GdnHCl denaturation experiments. Figure 6, Panel A compares far-UV CD spectra, clearly shows no difference in terms of secondary structure of apoAI on these two rHDL particles. CD data further suggests that apoAI on both rHDL particles displays ∼68% helical content (Table I). In contrast, lipid-free apoAI displays significantly less α-helical content (∼53.4%). Figure 6, Panel B shows the GdnHCl denaturation data of these purified rHDL particles. To our surprise, a stability difference of apoAI on purified small and large rHDL is observed. The small rHDL has a [GdnHCl]1/2 at 1.90M, whereas the large rHDL has a [GdnHCl]1/2 at 2.32M. This difference indicates that apoAI protein on small rHDL is less stable than it is on large rHDL. This is confirmed by the calculated 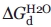 , indicating apoAI on small rHDL has a
, indicating apoAI on small rHDL has a 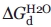 of 5.3 ± 0.2 kcal/mol and apoAI on large rHDL has a
of 5.3 ± 0.2 kcal/mol and apoAI on large rHDL has a 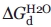 of 7.9 ± 0.7 kcal/mol (Table I). Before purification, the stability of apoAI on the mixed size of rHDL is between the stabilities of small and large rHDL (
of 7.9 ± 0.7 kcal/mol (Table I). Before purification, the stability of apoAI on the mixed size of rHDL is between the stabilities of small and large rHDL (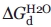 = 6.2 ± 0.9 kcal/mol and [GdnHCl]1/2 = 2.14M).
= 6.2 ± 0.9 kcal/mol and [GdnHCl]1/2 = 2.14M).
Figure 5.
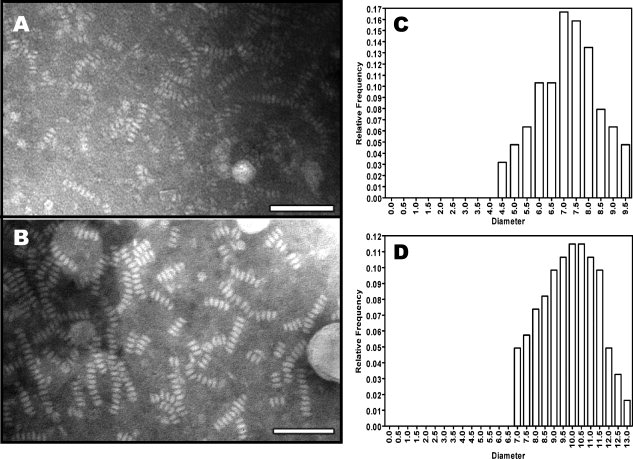
Panel A: PTA negative stain electron microscopy of purified small discoidal rHDL. Panel B: Negative stain electron microscopy of the purified large discoidal rHDL. Bar = 50 nm. Panel C: Size distribution of purified small discoidal rHDL. Panel D: Size distribution of purified large discoidal rHDL. The average diameter of small discoidal rHDL is 7.6 ± 1.6 nm and average diameter of large discoidal rHDL is: 9.8 ± 1.7 nm.
Figure 6.
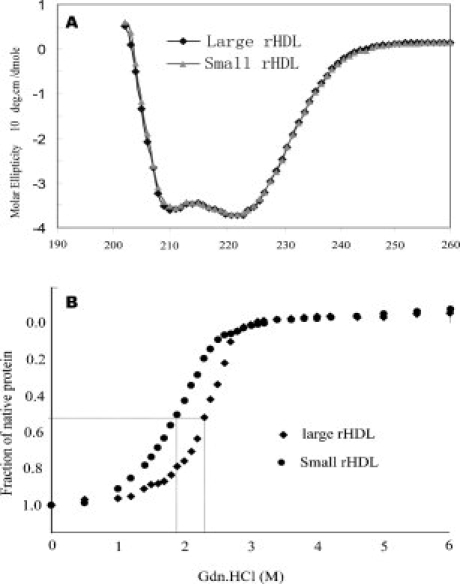
Panel A: Far-UV CD spectra of small (Black) and large discoidal rHDL (Gray) at 0.2 mg/mL in 50 mM sodium phosphate, 150 mM NaCl, pH 7.2. Panel B: GdnHCl denaturation of small (Circle) and large discoidal rHDL (diamond) under the same condition. The fraction of unfolding was plotted as a function of GndHCl concentration.
Table I.
CD and GdnHCl Denaturation Data of Protein Secondary Structure and Stability of: Lipid-Free apoAI, apoAI/smaller rHDL, apoAI/larger rHDL, and apoAI/rHDL(mixed) Discoidal Particle (80:1)
| ApoAI sample |
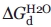 (kcal/mol) (kcal/mol) |
[GdnHCl]1/2 (M) | α-helical (%) | Number of helical residues in apoAI |
|---|---|---|---|---|
| Lipid-free apoAI | 4.3 ± 0.3 | 1.09 | 53.4 | 130 |
| ApoAI/rHDL(mixed)a | 6.2 ± 0.9 | 2.14 | 68.1 | 166 |
| ApoAI/smaller rHDLb | 5.3 ± 0.2 | 1.90 | 68.1 | 166 |
| ApoAI/larger rHDLb | 7.9 ± 0.7 | 2.32 | 68.1 | 166 |
ApoAI/rHDL(mixed) stands for discoidal rHDL particles after the first ultracentrifugation to remove the lipid-poor apoAI.
ApoAI/smaller rHDL and apoAI/larger rHDL stand for the smaller and large reconstituted discoidal rHDL particles after the second ultracentrifugation purification.
Phospholipid-binding activity of small and large rHDL
To assess lipid-binding activity, we carried out a DMPC clearance assay of both small and large rHDL. The DMPC clearance assay is widely used to characterize lipid-binding activity of exchangeable apolipoproteins. Figure 7 shows the results of DMPC clearance assay of different apoAI:DMPC ratios: an rHDL:DMPC ratio of 1:2 (Panel A) and an rHDL:DMPC ratio of 1:1 (Panel B). As a control, we also carried out DMPC clearance assays using lipid-free apoAI. Figure 7 demonstrates the difference between small and large rHDL in terms of phospholipid-binding activity, suggesting that small rHDL displays a stronger phospholipid-binding activity than that of large rHDL. Large rHDL displays a nearly saturation of phospholipid-binding activity, similar to the DMPC solution alone. Small rHDL displays an intermediate phospholipid-binding activity, which is much weaker than that of lipid-free apoAI. An increase in rHDL/DMPC ratio (1:1) enhances the clearance rates of all three apoAI states, because it takes less time to reach to the same turbidity level. However, a higher apoAI/DMPC ratio does not alter the turbidity level for both small and large rHDL/DMPC solution, but does decrease the turbidity level of lipid-free apoAI/DMPC solution, suggesting a difference in phospholipid-binding activity between lipid-free and discoidal rHDL particles.
Figure 7.
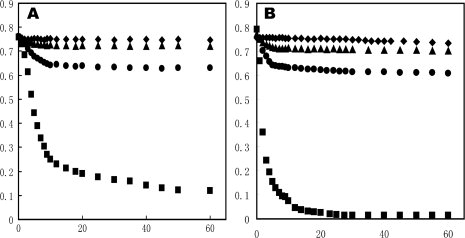
Phospholipid vesicle clearance. DMPC vesicles were incubated at 23.9°C in the absence (diamond) and presence of lipid-free apoAI (square), large rHDL (triangle) and small rHDL (Circle) in 20 mM Tris-HCl, pH 7.2, 250 mM NaCl, and 1 mM EDTA. Vesicle clearance as a function of time was followed by OD at 490 nm. Panel A: DMPC to apoAI (w/w) is 2:1 and Panel B: DMPC to apoAI (w/w) is 1:1.
Structural characterization of small and large rHDL
Although apoAI protein shows an identical secondary structure on small and large rHDLs, these rHDL particles display different stability and phospholipid-binding activity. This raises an important question: How does apoAI structure affects its stability and lipid-binding activity of rHDL? To address this question, we carried out high-resolution biophysical experiments, including solution NMR and negative stain EM, to investigate apoAI tertiary structure and morphology. Because both small and large rHDLs are apoAI/POPC complexes that are >140 kDa, we have to prepare triple-labeled apoAI with 13C, 15N, and 2H for two-dimensional [1H, 15N] TROSY-HSQC NMR spectra. Figure 8 shows two [1H, 15N] TROSY-HSQC NMR spectra overlaid on top of each other, with the black spectrum from small rHDL and blue spectra from large rHDL. It clearly indicates that apoAI protein on small and large discoidal rHDL particles displays a different HSQC spectral pattern, suggesting a different apoAI tertiary structure. This spectral difference is further highlighted by the inset of Figure 8 showing four crosspeaks of the 15Nɛ and 15NHɛ of the Trp sidechain. Indeed, blue crosspeaks (Large rHDL) displays a different pattern from those of black corsspeaks (small rHDL), suggesting that Trp sidechain of apoAI may adapt a different conformation on large rHDL from that of small rHDL.
Figure 8.
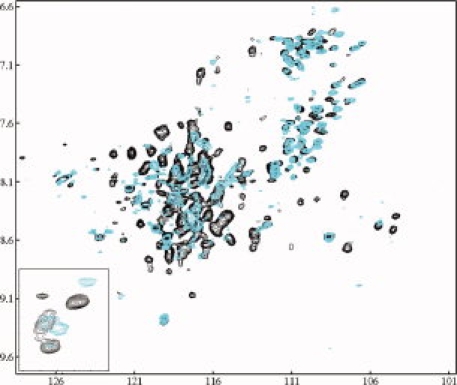
A comparison of [15N, 1H] TROSY-HSQC spectra of small rHDL (black) and large rHDL (Blue) collected on a 600 MHz NMR instrument with a cold probe at 30°C. NMR samples contain 0.2–0.5 mM triple-labeled rHDL in 50 mM phosphate buffer containing 50 mM NaCl, 0.1 mM NaN3, 1 mM EDTA, 5–7% D2O at pH 7.2. Inset: the crosspeaks of Trp sidechain Hɛ1/Nɛ1 atoms of the apoAI on small (Black) and large (Blue) discoidal rHDL particles.
Although Figure 5 shows that both purified small and large rHDLs are in discoidal shape with homogeneous sizes, the stacked nature of the rHDL particles did not allow us to analyze the detailed morphology of these rHDL particles. We further negatively stained more dilute solutions of rHDL particles with uranyl formate for detailed ultrastructural observation. Figure 9 shows the results. To demonstrate the details of the morphology and structure of these particles, 30 high-quality representative particles have been selected for either small (Panel A) or large (Panel B) rHDL, zoomed in and displayed in window size of 15 nm. In both Panels, rows 1–2 show a side view, rows 3–4 show a top view, and rows 5–6 show a tilted top view of the purified small and large rHDL. Figure 9 clearly demonstrates that both small and large rHDL particles are in disk shape with homogenous sizes. Size measurement confirms that small rHDL is 7.6 ± 1.6 nm, whereas large rHDL is 9.8 ± 1.7 nm. However, a careful examination of the morphology of rHDL particles shows significant differences in shape between small and large rHDLs. Although Rows 1–2 in Panel B show that large rHDL displays a typical discoidal shape with particle diameter is about two times larger than the particle height, the Rows 1–2 in Panel A indicate a much smaller ratio of particle diameter over particle height, suggesting a more “globular shape” for small rHDL. In addition, Rows 3–6 in Panel B show that large rHDLs display a black background in the middle of the particles, whereas Rows 3–6 in Panel A indicate that the center of small rHDL is filled with white materials that is different from what has observed in the particle center of large rHDL. We suggest that the black background is the phospholipids moiety whereas the white material is the imaging of apoAI protein. Thus, the difference observed in particles morphology between small and large rHDLs may suggest a potential conformational difference of apoAI on these different rHDL particles. Overall, apoAI on small rHDL seems to adopt a more “globular structure,” which is more compact and tighten in packing, and apoAI on large rHDL seems to display a more extended α-helical structure. This conformational difference of apoAI on small and large rHDL is supported by our NMR data shown in Figure 8.
Figure 9.
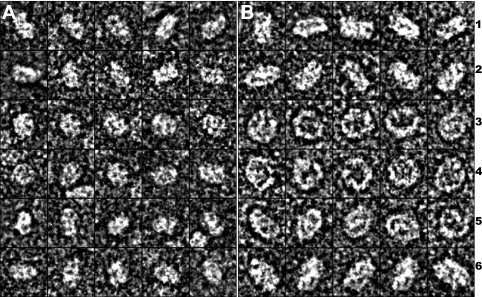
Uranyl formate negatively stained electron microscopy of purified small and large discoidal rHDLs. Panel A displays 30 representative small rHDL particle and Panel B displays 30 representative large rHDL particles. Rows 1 and 2 are the side view; Rows 3–4 are the top view; Rows 5–6 are the tilted top view of both small (A) and large (B) rHDL particles. Each box corresponds to a 15 × 15 nm area.
Discussion
The discoidal HDL particles are critical intermediates between lipid-poor apoAI and mature spherical HDL during HDL assembly. It plays a key role in loading the “bad cholesterol” into HDL particles via LCAT activity. Over the past 2 decades, significant efforts have focused on structural studies of discoidal HDL due to its important role during HDL assembly. These studies resulted in an improved knowledge of structure-function relationship of discoidal HDL.2 However, controversial results were obtained for the details of apoAI structures on discoidal HDL using different structural techniques, including FRET, EPR, and crosslinking with mass spectroscopy.29,27 Recently, a 10 Å resolution X-ray crystal structure of an apoE/DPPC particle was reported as the model structure of apoE on the DPPC particles,31 suggesting that each apoE molecule is folded into a helical hairpin with the binding region for the LDL receptor at its apex. This model provided significant structural information of apoE protein, such as global fold, however, molecular details of the apoE structure are still lacking due to a limited resolution (10 Å). In addition, an atomic resolution structure of apoAI on discoidal HDL particles is presently not available.
The detailed structure of apoAI on discoidal HDL has been debated for nearly 3 decades and several models have been derived.2 Two models, the “picket fence” and the “belt” model, are the most popular models. Recent biophysical data favors the “belt” model and several “belt” model sub-types were further proposed, including the “hairpin double belt,” the “Z double belt,” and the “double belt.”17 However, current published data does not unambiguously identify which model is the correct structure of apoAI on discoidal HDL particles. One proposal is the existence of flexible registry of two apoAI belts.23 Further, coexistence of the “belt” and the “hairpin” conformations of apoAI on the same discoidal HDL has been also proposed.24 Another report suggested, using FRET and EPR techniques, that apoAI structure on discoidal HDL is flexible and contains a flexible, less structured loop segment between residues 134–145. It is suggested that this flexible loop segment confers an intrinsic ability of apoAI to adapt its structure to changes in particle lipid content.38 This suggestion is supported by a recent publication, which indicates that a protruding solvent-exposed loop exists in the middle of apoAI on discoidal HDL, although the location of this loop (residues 159–180) is different.30 Although no conclusion about apoAI structure on discoidal HDL has been reached at this moment, a general consensus is that the apoAI structure displays significant conformational flexibility that allows this protein to adopt several different conformations. This consensus suggests that apoAI conformational dynamics is critical to its biological function and any structural studies of apoAI in different states should include studies of the conformational dynamics of this protein.
NMR techniques provide a powerful tool to study protein structure and dynamics at atomic resolution.39,40 For high-resolution NMR structural studies, an ultra pure NMR sample is required. To obtain such an ultra pure NMR sample of discoidal HDL, we first tested the effects of the ratios of POPC:apoAI:cholate on rHDL formation in terms of particle morphology, size, and purity. Our data clearly indicated that the POPC:apoAI ratio was critical to rHDL formation (Fig. 1, Panel A). Crosslinking data indicated that these rHDL particles predominantly contained two molecules of apoAI, regardless of their particle size (Fig. 1, Panel B). Our data further indicated that at a fixed POPC:apoAI ratio, cholate concentration played a role in the purity of rHDL preparation.
Although prepared rHDL seemed to be pure and homogenous as judged by native gel and crosslinking experiments, NMR characterization indicated that these rHDL preparations contained a small population of lipid-poor apoAI, Lp1AI. The [15N,1H] TROSY-HSQC spectrum of the prepared rHDL contained two sets of NMR crosspeaks, with one set of sharp crosspeaks and one set of broader corsspeaks (Fig. 2, Middle Panel). Our data indicated that the sharper NMR signals originated from Lp1AI (Fig. 2, Left Panel). The broader NMR signals were attributed to discoidal rHDL, which displayed a completely different spectral pattern (Fig. 2, Right Panel). The Insets of Figure 2 further demonstrated two sets of spectral pattern for Trp sidechain atoms, with one set of sharper spectral pattern identical to that of Lp1AI and one set of broader spectral pattern identical to discoidal rHDL. This data indicated that the rHDL preparation using cholate dialysis method was not pure and the impurity of Lp1AI significantly interfered with NMR spectral analysis, hindering a complete NMR assignment. A quick ultracentrifugation method allowed us to remove the lipid-poor apoAI impurity and obtain a large amount of pure discoidal rHDL for NMR studies (Figs. 2 and 3). The sharper NMR crosspeaks in Figure 2, Panel B were completely removed, suggesting that this ultracentrifugation method could be used to isolate pure rHDL for high-resolution NMR structural studies.
Figure 3 indicated that only 5–10% lipid-poor apoAI existed in the rHDL preparation. Interestingly, the crosspeaks from a very low concentration of lipid-poor apoAI showed a similar spectral intensity to those of the crosspeaks from a much higher concentration of discoidal rHDL. This is due to the particle size difference between lipid-poor apoAI (<67 kDa) and rHDL (140–200 kDa). A smaller particle produced sharper NMR signals, resulting in a better signal-to-noise. In contrast, larger discoidal rHDL particles produced much broader NMR signals due to a slower molecular tumbling, resulting in a much worse signal-to-noise, even using TROSY NMR experiments with a triple-labeled NMR sample.41,42 In this case, a 5–10% lipid-poor apoAI impurity may cause a serious mistake in spectral analysis of larger discoidal rHDL. This may not be a problem when the contamination comes from a protein of similar size to the protein under investigation, because a 5–10% contamination does not produce strong NMR signals to interfere with NMR spectral analysis of the protein under investigation. Thus, our data demonstrates that one has to be extremely careful in terms of NMR sample purity when working with large proteins. A small population of smaller protein contamination (5–10%) may cause a serious mistake in the NMR spectral analysis of the larger protein.
Using a second density gradient ultracentrifugation, we further separated discoidal rHDL particles into two pure stable populations. We also carried out experiments to characterize the structure, stability, and phospholipid-binding activity of the purified small and large discoidal rHDLs. As expected, both pure small and large discoidal rHDL particles had homogenous size and disk shape morphology, as demonstrated by native gel and negative stain EM (Figs. 4 and 5). The standard deviations of the EM measurements for both small and large discoidal rHDL were much smaller than the deviation of the EM measurement before the second purification, suggesting homogenous size of the purified discoidal rHDL particles. Far-UV CD spectral data indicated that apoAI protein on both small and large discoidal rHDL displayed an identical α-helical content of 68.1%, suggesting that a total of 166 residues out of 243 residues in apoAI adopt α-helical structure. In contrast, the GdnHCl denaturation data showed a different stability. The large discoidal rHDL (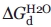 = 7.9 ± 0.7 kcal/mol) is more stable than the small discoidal rHDL (
= 7.9 ± 0.7 kcal/mol) is more stable than the small discoidal rHDL (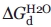 = 5.3 ± 0.2 kcal/mol). Interestingly, the discoidal rHDL obtained before the second ultracentrifugation was a mixture of both smaller and larger discoidal rHDL and displayed a stability (
= 5.3 ± 0.2 kcal/mol). Interestingly, the discoidal rHDL obtained before the second ultracentrifugation was a mixture of both smaller and larger discoidal rHDL and displayed a stability (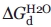 = 6.2 ± 0.2 kcal/mol) that was between small and large particles. DMPC clearance assay also indicated that small rHDL had a stronger phospholipid-binding activity than that of large rHDL, but overall both small and large rHDL displayed a much weaker phospholipid-binding activity than lipid-free apoAI. Because both small and large rHDL are enriched with phospholipid, POPC, their phospholipid binding activity should be weaker than lipid-free apoAI. Small rHDL might have less POPC than large rHDL, thus it displayed a stronger phospholipid-binding activity.
= 6.2 ± 0.2 kcal/mol) that was between small and large particles. DMPC clearance assay also indicated that small rHDL had a stronger phospholipid-binding activity than that of large rHDL, but overall both small and large rHDL displayed a much weaker phospholipid-binding activity than lipid-free apoAI. Because both small and large rHDL are enriched with phospholipid, POPC, their phospholipid binding activity should be weaker than lipid-free apoAI. Small rHDL might have less POPC than large rHDL, thus it displayed a stronger phospholipid-binding activity.
A question remains unanswered as to what is the structural reason of apoAI for its different stability and phospholipid-binding activity between small and large rHDLs. To address this question, we carried out 2D NMR 1H-15N TROSY-HSQC and high-resolution negative stain EM experiments. Our data clearly demonstrated that apoAI adopted different tertiary structures between small and large rHDLs, even though an identical α-helical content of apoAI was observed using far-UV CD spectroscopy. The HSQC NMR spectroscopy correlates apoAI backbone amide protons with 15N-labeled nitrogen and each amino acid displays one HSQC crosspeak. Thus, a crosspeak in the HSQC spectrum provides a fingerprint of an amino acid in apoAI. In addition, the spectral patterns of a HSQC spectrum provide both the secondary and tertiary structural information of apoAI. Because far-UV CD spectroscopy indicated that apoAI adapted the same secondary structure on both smaller and larger discoidal rHDL particles, the different HSQC spectral pattern of apoAI suggests that apoAI may adopt a different tertiary structure. These different apoAI tertiary structures may provide an explanation for the different stabilities between small and large rHDL particles. Furthermore, our negative stain EM data of individual rHDL particles also indicated a morphology difference between small and large rHDL. Indeed, a much smaller ratio of particle diameter over particle height and a compact shape of apoAI protein were observed for small rHDL particles. In contrast, large rHDL display a larger ratio of particle diameter over height and looser shape of apoAI protein. This EM data suggests that apoAI on small rHDL seems to adopt a more compact “globular structure,” whereas apoAI on large rHDL may display a more extended α-helical structure. Such an EM morphological difference between small and large discoidal rHDL further suggest a possible tertiary structural difference of apoAI between these two discoidal rHDL particles.
Conclusions
In summary, our high-resolution structural data indicates that apoAI protein adopts different tertiary structures on different discoidal rHDL particles, which may attribute to their different stability and lipid-binding activity. Our result further provides the first high-resolution NMR data, demonstrating a promise of structural determination of discoidal HDL at atomic resolution using a combination of NMR and other biophysical techniques. First, based on TROSY-HSQC spectrum shown in Figure 8, the backbone 1HN chemical shift dispersion of small rHDL is only 2.4 ppm (6.7–9.1 ppm), suggesting a potential intermolecular (between two ApoAI) or intramolecular (within apoAI) helix-bundle. Second, our NMR data suggests that two apoAI molecules have an identical chemical environment, because only one set of NMR signals is observed for two molecules of apoAI on discoidal rHDL. An identical chemical environment suggests that the conformation of two apoAI molecules is symmetric on rHDL, thus the double belts of α-helix structure of apoAI should be identical whether it forms an intermolecular or an intramolecular bundle. Thus, our data agrees with the “double belt model” of discoidal HDL,21 however, this NMR data argues against several different “belt” models, such as the hairpin and flexible registry,2 because two apoAI molecules are not identical in these models. A recent publication using mass spectroscopy and crosslinking experiments suggests that the C-terminal region of one apoAI forms a symmetric intermolecular helix-bundle with the N-terminal region of another apoAI (28–29),28 which is consistent with our NMR data. Thus, our preliminary NMR data provides significant important structural information of apoAI on discoidal HDL particles.
Materials and Methods
High-level expression and purification of isotopically labeled human apoAI
The human apoAI/pET30a DNA construct was generous gift from Professor Anna Jonas (University of Illinois, Urbana-Champaign). This DNA vector was modified to replace the long his-tag of the pET30a vector by a His6-Ser-Ser-tag. A factor Xa cleavage site was also introduced between the his-tag and apoAI sequence. With this DNA construct, expression was carried out using E. coli BL-21(DE3) cells. The expression of triple-labeled apoAI followed an auto-induction procedure43 with a significant modification (Sivashanmugam et al., submitted). The cells were harvested by centrifugation and the cell pellet was resuspended in binding buffer (10 mL/g cells, 20 mM Tris HCl, 500 mM NaCl, 2.5 mM imidazole, and 0.1 mM PMSF at pH 7.9) and sonicated on ice. The pellet was further resuspended in 6M urea and sonicated. All supernatant fractions were combined and applied on a His·Bind® resin column (Novagen). The column was first washed with a large amount of binding buffer, and then with washing buffer containing 20–30 mM imidazole. Purified triple-labeled apoAI was eluted from the column with a buffer containing 1.0M imidazole. The eluted protein was dialyzed against 10 mM ammonium bicarbonate to remove imidazole and then lyophilized. With this method, we routinely obtain 15 mg of triple-labeled apoAI from a 50-mL cell culture. Mass spectroscopy confirmed that the labeling efficiency was greater than 99% for 13C and 15N, and ∼90% for 2H.
Preparation of Lp1AI particle
The purified triple-labeled apoAI was dissolved in 6M GdnHCl overnight and then stepwise dialyzed against solution with reduced GdnHCl concentrations and finally against 50 mM sodium phosphate, pH 7.2, 50 mM NaCl, and 0.01% NaN3 (PBS). The protein was concentrated using centriplus YM-10 (Millipore, Bedford, MA) with MWCO 10 kDa. The Lp1AI was prepared by cosonication of POPC and apoAI with a molar ratio of 5:1 (POPC/apoAI).11 Briefly, a defined amount of POPC in chloroform was dried under nitrogen in a 12 × 75 mm glass tube and 800 μL of PBS was added and sonicated for 1 min at a constant output in a 15°C water bath, then incubated for 30 min at 37°C. The solution was sonicated again for 5 min at 95% duty cycle. ApoAI (2 mg at 1.4 mg/mL in PBS solution) was added to the lipid suspension and the protein-lipid mixture was sonicated four times with1 min each time at 90% duty cycle followed by a 1 min cooling period between the two sonications. All particles were then filtered through a 0.45 μm syringe filter. The phospholipid concentration was measured by an enzymatic colorimetric method using a phospholipid B kit from Wako Chemicals GmbH (Germany). For the NMR sample of Lp1AI, further concentration was carried out using a centricon YM-30 (Millipore, MWCO 30 kDa).
Preparation of rHDL particle
Discoidal rHDL particles were prepared using the cholate dialysis technique with varied molar ratios of POPC:apoAI:cholate.35,36 A defined amount of POPC in chloroform was added to a 15 mL glass tube, dried under nitrogen, and redissolved in PBS buffer to yield a POPC concentration of 20 mM. The mixture was vortexed thoroughly and sodium cholate in PBS was added to create solutions of varying POPC:cholate molar ratios, followed by further vortexing again for 3 min. The dispersion was then incubated for 15 min at 37°C and vortexed. This procedure was repeated several times until the solution was completely clear. ApoAI protein was then added to the clear solution with different molar ratios of POPC:apoAI:cholate. The mixture was diluted to apoAI concentration at 1 mg/mL using PBS and incubated for 1 h at 37°C. Sodium cholate was removed by extensive dialysis at 4°C.
Purification of rHDL using density gradient ultracentrifugation
The rHDL were further purified using KBr density gradient ultracentrifugation. The amount of KBr added was calculated based on www.roarbiomedical.com/cgi/descalc/calculator.pl. After adding a defined amount of KBr, the total volume of rHDL was adjusted to 2 mL (density 1.27 g/mL). The samples were transferred to 5 mL Beckman open centrifuge tubes, overlaid with 2 mL of 0.9% NaCl (density: 1.063), and centrifuged at 49,000g for 13 h at 4°C using a SW 50.1 swinging bucket rotor on a Beckman optima LE-80K ultracentrifuge. After ultracentrifugation, the sample was fractionated by pipetting 200 μL of sample from the top layer for each fraction. Thus, we obtained 20 fractions from a total of 4 mL per centrifuge tube. Aliquots (2–5 μL) of the fractions were subjected to 4–20% native gradient gel and the fractions with identical particle size were combined and exhaustively dialyzed against PBS buffer. This first ultracentrifugation separates lipid-poor apoAI (Lp1AI) from the discoidal HDL fractions.
We carried out a second ultracentrifugation using a four-layer density gradient on the discoidal HDL fractions purified with the first ultracentrifugation. After adjusting the density of rHDL sample solution by adding a defined amount of KBr to 1.25 g/mL, 2 mL samples were transferred into 12 mL Open-Top Polyallomer centrifuge tubes (Denville Scientific), overlaid gently with 2 mL of 1.15 g/mL KBr, 2 mL of 1.10 g/mL, 2 mL of 1.08 g/mL, 1 mL of 1.07 g/mL KBr PBS buffer, finally with 0.5 mL of 0.9% NaCl, and centrifuged at 39,000g for 16 h at 4°C using a SW 41 swinging bucket rotor on a Beckman Coulter optima™ LE-80K ultracentrifuge. After ultracentrifugation, the sample was fractionated from the top to bottom by pipetting 200 μL of sample solution for each fraction. Aliquots (5 μL) of the fractions were subjected to native gradient gel, and the fractions with identical particle size were combined and dialyzed against PBS buffer exhaustively for further analysis. For NMR samples, triple-labeled, purified rHDL fractions were concentrated using a centricon YM-30 (Millipore, MWCO 30 kDa).
Crosslinking experiments
Purified rHDL in PBS were incubated for 30 min with BS3, (Pierce, final concentration 0.5 mM). The reaction mixture was quenched by adding 1M Tris-HCl and 5× SDS-PAGE sample buffer and then loaded on a Tricine gel. The protein concentration was determined by a modified Lowry assay.44
CD spectroscopy and stability measurement
CD measurements were carried out on an Olis DSM 17 CD spectrophotometer (Bogart, GA) with variable temperature capability under computer control within ±0.2°C. GdnHCl denaturation was performed as described45 and monitored by CD at 222 nm in a 0.1 cm path length cuvette at 20°C. Protein concentrations were determined by the Markwell modification of the Lowry protein assay. The concentrations of denaturant were determined using the density formulas given by Pace et al.46 The α-helical content in apoAI was calculated from molar ellipticity at 222 nm.
NMR spectroscopy
The apoAI/Lp1AI NMR sample contained 50 mM phosphate buffer (pH 7.2), 50 mM NaCl, 0.01 mM NaN3, 10 mM EDTA, 5% D2O, and 0.5–1.0 mM 99% 2H/13C/15N-labeled apoAI/Lp1AI. The purified rHDL NMR sample contained 50 mM phosphate buffer, pH 7.2, 50 mM NaCl, 0.01 mM NaN3, 10 mM EDTA, 5% D2O, and 0.1–0.5 mM triple-labeled apoAI/rHDL particle. [15N, 1H] TROSY-HSQC spectra were acquired at 30°C on Varian INOVA or Bruker Avance spectrometers operating at 1H resonance frequencies of 500, 600, 800 MHz, which are equipped with pulsed-field gradient triple resonance probes (500 MHz: room temperature probe, 600/800 MHz: cold probe). Proton chemical shifts were referenced to DSS. Data were processed and analyzed on a SGI workstation using NMRPipe47 and NMRView.48
DMPC binding assay
Ten milligrams of DMPC (Avanti Polar Lipids, AL) was dissolved in a mixture of chloroform and methanol (3:1 v/v), dried using N2, and placed under vacuum for at least 10 h. One milliliters of prewarmed buffer (10 mM Tris-HCl, pH7.2, 150 mM NaCl, 0.5 mM EDTA, 23.9°C) was added to yield a final lipid concentration of 10 mg/mL. The resulting solution was vortexed several times for 30 s each. Using a 200 nm filter, unilamellar vesicles (∼200 nm in diameter) were prepared by extrusion. The protein-induced transformation of DMPC vesicles into protein/DMPC discoidal complexes was monitored as a function of time.45 Lipid-free apoAI/Lp1AI/rHDL and DMPC (1:1 = w/w) were used in this assay. ApoAI/Lp1AI/rHDL and DMPC vesicles in buffer were added into a 1 mL thermostat cuvette and mixed for 5–10 s at 23.9°C. The clearance of solution was monitored using a Perkin-Elmer spectrophotometer (model Lambda 3B) at 490 nm. All solutions were preincubated at 23.9°C before reaction.
Electron microscopy
For initial quantitative analysis of particles size, the rHDL particles were absorbed to hydrophilic, carbon, and formvar-coated grids. Samples were negatively stained for 20 s with 2% phosphotungstic acid, pH 7.0. Digital images were taken using a Philips CM-30 electron microscope equipped at 80 KeV accelerating voltage. At least 10 arbitrarily selected fields were chosen and more than 25 particles per field were measured (>250 particles per condition). Using the above method, discoidal HDL occurred as stacks of disks. This allowed a good quantitative measurement of particle sizes, but did not reveal details of morphology of these discoidal rHDL particles. To examine the morphology, further more dilute aliquots (2.5 μL) samples were adhered to carbon-coated, 400-mesh copper grids previously rendered hydrophilic by glow discharge. The grids were stained with uranyl formate (2% (w/v) for 1 min (Ted Pella, Tustin, CA). Digital images at 80,000× magnification were recorded on 4K × 4K Gatan UltraScan CCD under low electron dose conditions using a Tecnai 20 electron microscope (Philips Electron Optics/FEI, Eindhoven, The Netherlands) operating at 200 kV. Each pixel of the micrographs corresponds to 1.406 Å at the level of the specimen. Particles in micrographs were selected and boxed by the box size of 128 pixels, corresponding to 150 Å in specimen.
Acknowledgments
Part of the high-field NMR experiments described in this manuscript were performed at the W. R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the U.S. Department of Energy's Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL). PNNL is operated for the Department of Energy by Battelle. Phosphotungstic acid negative staining and imaging was carried out using the facilities of Vanderbilt University's Research Electron Microscopy Resource. The authors also thank Ms Victoria Murray for critical reading the manuscript.
Glossary
Abbreviations:
- ABCAI
ATP-binding cassette AI transporter
- ApoAI
apolipoprotein A-I
- ApoE
apolipoprotein E
- BS3
bis(sulfosuccinmidyl)suberate
- CD
circular dichroism
- CE
cholesterol ester
- CETP
cholesterol ester transfer protein
- DMPC
dimyristoylephosphatidylcholine
- DPPC
dipalmitoylphosphatidylcholine
- DSS
2,2-dimethyl-2-silapentane-5-sulfonate
- EM
electron microscopy
- EPR
electron paramagnetic resonance
- FRET
fluorescence resonance energy transfer
- GdnHCl
guanidine hydrochloride
- HDL
high-density lipoprotein
- HSQC
heteronuclear single quantum coherence
- LCAT
lecithin:cholesterol acyltransferase
- Lp1AI
reconstituted preβ-HDL that contains one ApoAI
- NMR
nuclear magnetic resonance
- PLTP
phospholipid transfer protein
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- RCT
reverse cholesterol transport
- rHDL
reconstituted discoidal HDL
- SR-BI
scavenger receptor class B, type I
- TROSY
transverse relaxation optimized spectroscopy.
References
- 1.Lewis GF, Rader DJ. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ Res. 2005;96:1221–1232. doi: 10.1161/01.RES.0000170946.56981.5c. [DOI] [PubMed] [Google Scholar]
- 2.Davidson WS, Thompson TB. The structure of apolipoprotein a-I in high-density lipoproteins. J Biol Chem. 2007;282:22249–22253. doi: 10.1074/jbc.R700014200. [DOI] [PubMed] [Google Scholar]
- 3.Asztalos BF. High-density lipoprotein metabolism and progression of atherosclerosis: new insights from the HDL Atherosclerosis Treatment Study. Curr Opin Cardiol. 2004;19:385–391. doi: 10.1097/01.hco.0000126979.41946.7e. [DOI] [PubMed] [Google Scholar]
- 4.Duriez P, Fruchart JC. High-density lipoprotein subclasses and apolipoprotein A-I. Clin Chim Acta. 1999;286:97–114. doi: 10.1016/s0009-8981(99)00096-0. [DOI] [PubMed] [Google Scholar]
- 5.Martinez LO, Jacquet S, Terce F, Collet X, Perret B, Barbaras R. New insight on the molecular mechanisms of high-density lipoprotein cellular interactions. Cell Mol Life Sci. 2004;61:2343–2360. doi: 10.1007/s00018-004-4087-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segrest JP, Li L, Anantharamaiah GM, Harvey SC, Liadaki KN, Zannis V. Structure and function of apolipoprotein A-I and high-density lipoprotein. Curr Opin Lipidol. 2000;11:105–115. doi: 10.1097/00041433-200004000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Marcel YL, Kiss RS. Structure-function relationships of apolipoprotein A-I: a flexible protein with dynamic lipid associations. Curr Opin Lipidol. 2003;14:151–157. doi: 10.1097/00041433-200304000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Brouillette CG, Anantharamaiah GM, Engler JA, Borhani DW. Structural models of human apolipoprotein A-I: a critical analysis and review. Biochim Biophys Acta. 2001;1531:4–46. doi: 10.1016/s1388-1981(01)00081-6. [DOI] [PubMed] [Google Scholar]
- 9.Rothblat GH, de la Llera-Moya M, Atger V, Kellner-Weibel G, Williams DL, Phillips MC. Cell cholesterol efflux: integration of old and new observations provides new insights. J Lipid Res. 1999;40:781–796. [PubMed] [Google Scholar]
- 10.Barrans A, Jaspard B, Barbaras R, Chap H, Perret B, Collet X. Pre-beta HDL: structure and metabolism. Biochim Biophys Acta. 1996;1300:73–85. doi: 10.1016/0005-2760(95)00236-7. [DOI] [PubMed] [Google Scholar]
- 11.Sparks DL, Frank PG, Braschi S, Neville TA, Marcel YL. Effect of apolipoprotein A-I lipidation on the formation and function of pre-beta and alpha-migrating LpA-I particles. Biochemistry. 1999;38:1727–1735. doi: 10.1021/bi981945k. [DOI] [PubMed] [Google Scholar]
- 12.Neary R, Bhatnagar D, Durrington P, Ishola M, Arrol S, Mackness M. An investigation of the role of lecithcholesterol acyltransferase and triglyceride-rich lipoproteins in the metabolism of pre-beta high density lipoproteins. Atherosclerosis. 1991;89:35–48. doi: 10.1016/0021-9150(91)90005-n. [DOI] [PubMed] [Google Scholar]
- 13.Connelly MA, Williams DL. SR-BI and HDL cholesteryl ester metabolism. Endocr Res. 2004;30:697–703. doi: 10.1081/erc-200043979. [DOI] [PubMed] [Google Scholar]
- 14.Trigatti BL. Hepatic high-density lipoprotein receptors: roles in lipoprotein metabolism and potential for therapeutic modulation. Curr Atheroscler Rep. 2005;7:344–350. doi: 10.1007/s11883-005-0045-x. [DOI] [PubMed] [Google Scholar]
- 15.de Grooth GJ, Klerkx AH, Stroes ES, Stalenhoef AF, Kastelein JJ, Kuivenhoven JA. A review of CETP and its relation to atherosclerosis. J Lipid Res. 2004;45:1967–1974. doi: 10.1194/jlr.R400007-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Huuskonen J, Olkkonen VM, Jauhiainen M, Ehnholm C. The impact of phospholipid transfer protein (PLTP) on HDL metabolism. Atherosclerosis. 2001;155:269–281. doi: 10.1016/s0021-9150(01)00447-6. [DOI] [PubMed] [Google Scholar]
- 17.Davidson WS, Silva RA. Apolipoprotein structural organization in high density lipoproteins: belts, bundles, hinges and hairpins. Curr Opin Lipidol. 2005;16:295–300. doi: 10.1097/01.mol.0000169349.38321.ad. [DOI] [PubMed] [Google Scholar]
- 18.Segrest JP, Garber DW, Brouillette CG, Harvey SC, Anantharamaiah GM. The amphipathic alpha helix: a multifunctional structural motif in plasma apolipoproteins. Adv Protein Chem. 1994;45:303–369. doi: 10.1016/s0065-3233(08)60643-9. [DOI] [PubMed] [Google Scholar]
- 19.Wald JH, Coormaghtigh E, De Meutter J, Ruysschaert JM, Jonas A. Investigation of the lipid domains and apolipoprotein orientation in reconstituted high density lipoproteins by fluorescence and IR methods. J Biol Chem. 1990;265:20044–20050. [PubMed] [Google Scholar]
- 20.Phillips JC, Wriggers W, Li Z, Jonas A, Schulten K. Predicting the structure of apolipoprotein A-I in reconstituted high-density lipoprotein disks. Biophys J. 1997;73:2337–2346. doi: 10.1016/S0006-3495(97)78264-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segrest JP, Jones MK, Klon AE, Sheldahl CJ, Hellinger M, De Loof H, Harvey SC. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J Biol Chem. 1999;274:31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- 22.Tricerri MA, Behling Agree AK, Sanchez SA, Bronski J, Jonas A. Arrangement of apolipoprotein A-I in reconstituted high-density lipoprotein disks: an alternative model based on fluorescence resonance energy transfer experiments. Biochemistry. 2001;40:5065–5074. doi: 10.1021/bi002815q. [DOI] [PubMed] [Google Scholar]
- 23.Li HH, Lyles DS, Pan W, Alexander E, Thomas MJ, Sorci-Thomas MG. ApoA-I structure on discs and spheres. Variable helix registry and conformational states. J Biol Chem. 2002;277:39093–39101. doi: 10.1074/jbc.M206770200. [DOI] [PubMed] [Google Scholar]
- 24.Panagotopulos SE, Horace EM, Maiorano JN, Davidson WS. Apolipoprotein A-I adopts a belt-like orientation in reconstituted high density lipoproteins. J Biol Chem. 2001;276:42965–42970. doi: 10.1074/jbc.M106462200. [DOI] [PubMed] [Google Scholar]
- 25.Brouillette CG, Anantharamaiah GM. Structural models of human apolipoprotein A-I. Biochim Biophys Acta. 1995;1256:103–129. doi: 10.1016/0005-2760(95)00018-8. [DOI] [PubMed] [Google Scholar]
- 26.Davidson WS, Hilliard GM. The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles: a mass specrometry study. J Biol Chem. 2003;278:27199–207. doi: 10.1074/jbc.M302764200. [DOI] [PubMed] [Google Scholar]
- 27.Silva RA, Hilliard GM, Li L, Segrest JP, Davidson WS. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 2005;44:8600–8607. doi: 10.1021/bi050421z. [DOI] [PubMed] [Google Scholar]
- 28.Bhat S, Sorci-Thomas MG, Alexander ET, Samuel MP, Thomas MJ. Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J Biol Chem. 2005;280:33015–33025. doi: 10.1074/jbc.M505081200. [DOI] [PubMed] [Google Scholar]
- 29.Bhat S, Sorci-Thomas MG, Tuladhar R, Samuel MP, Thomas MJ. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 2007;46:7811–7821. doi: 10.1021/bi700384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Z, Wagner MA, Zheng L, Parks JS, Shy JM, III, Smith JD, Gogonea V, Hazen SL. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat Struct Mol Biol. 2007;14:861–868. doi: 10.1038/nsmb1284. [DOI] [PubMed] [Google Scholar]
- 31.Peters-Libeu CA, Newhouse Y, Hatters DM, Weisgraber KH. Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. J Biol Chem. 2006;281:1073–1079. doi: 10.1074/jbc.M510851200. [DOI] [PubMed] [Google Scholar]
- 32.Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K. NMR analysis of a 900K GroEL GroES complex. Nature. 2002;418:207–211. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- 33.Tugarinov V, Muhandiram R, Ayed A, Kay LE. Four-dimensional NMR spectroscopy of a 723-residue protechemical shift assignments and secondary structure of malate synthase g. J Am Chem Soc. 2002;124:10025–10035. doi: 10.1021/ja0205636. [DOI] [PubMed] [Google Scholar]
- 34.Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 35.Matz CE, Jonas A. Micellar complexes of human apolipoprotein A-I with phosphatidylcholines and cholesterol prepared from cholate-lipid dispersions. J Biol Chem. 1982;257:4535–4540. [PubMed] [Google Scholar]
- 36.Jonas A, Matz CE. Reaction of human lecithcholesterol acyltransferase with micellar substrates is independent of the phase state of the lipid. Biochemistry. 1982;21:6867–6872. doi: 10.1021/bi00269a037. [DOI] [PubMed] [Google Scholar]
- 37.Cavigiolio G, Shao B, Geier EG, Ren G, Heinecke JW, Oda MN. The interplay between size, morphology, stability, and functionality of high-density lipoprotein subclasses. Biochemistry. 2008;47:4770–4779. doi: 10.1021/bi7023354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin DD, Budamagunta MS, Ryan RO, Voss JC, Oda MN. Apolipoprotein A-I assumes a looped belt conformation on reconstituted high-density lipoprotein. J Biol Chem. 2006;281:20418–20426. doi: 10.1074/jbc.M602077200. [DOI] [PubMed] [Google Scholar]
- 39.Mittermaier A, Kay LE. New tools provide new insights in NMR studies of protein dynamics. Science. 2006;312:224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 40.Palmer AG, III, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 41.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci USA. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tugarinov V, Hwang PM, Kay LE. Nuclear magnetic resonance spectroscopy of high-molecular-weight proteins. Annu Rev Biochem. 2004;73:107–146. doi: 10.1146/annurev.biochem.73.011303.074004. [DOI] [PubMed] [Google Scholar]
- 43.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 44.Markwell MA, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 45.Ren X, Zhao L, Sivashanmugam A, Miao Y, Korando L, Yang Z, Reardon CA, Getz GS, Brouillette CG, Jerome WG, Wang J. Engineering mouse apolipoprotein A-I into a monomeric, active protein useful for structural determination. Biochemistry. 2005;44:14907–14919. doi: 10.1021/bi0508385. [DOI] [PubMed] [Google Scholar]
- 46.Pace C. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 47.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 48.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]


