Abstract
Self-assembly of artificially designed proteins is extremely desirable for nanomaterials. Here we show a novel strategy for the creation of self-assembling proteins, named “Nanolego.” Nanolego consists of “structural elements” of a structurally stable symmetrical homo-oligomeric protein and “binding elements,” which are multiple heterointeraction proteins with relatively weak affinity. We have established two key technologies for Nanolego, a stabilization method and a method for terminating the self-assembly process. The stabilization method is mediated by disulfide bonds between Cysteine-residues incorporated into the binding elements, and the termination method uses “capping Nanolegos,” in which some of the binding elements in the Nanolego are absent for the self-assembled ends. With these technologies, we successfully constructed timing-controlled and size-regulated filament-shape complexes via Nanolego self-assembly. The Nanolego concept and these technologies should pave the way for regulated nanoarchitecture using designed proteins.
Keywords: self-assembly, nanofilament, protein design, molecular modeling, protein nanomaterial
Introduction
Self-assembly, the ability of molecules to organize themselves into high-order structures via noncovalent bonds, is a physical principle for bottom-up fabrication of nanomaterials, preparation of discrete nanostructures, and efficient nano-order patterning extending to macroscales.1,2 Because mutagenesis and fusion allow the design of recombinant proteins that can undergo self-assembly and form distinctive supramolecules, artificially designed proteins are considered to have high potentiality as nanomaterials. Padilla and coworkers3,4 described a design of proteins that can self-assemble into large symmetrical nanomaterials, which was based on the construction of a fusion protein of two homo-oligomerized symmetric proteins. Ringler and Schulz5 demonstrated the production of a quadratic 2D protein network consisting of biotinylated symmetric tetrameric aldolase as a rigid four-way connector and stiff streptavidin rods as spacers. However, control of the timing and size of the nanomaterials produced has not been achieved as desired in most of the work reported. For example, in self-assembling filaments designed according to the former strategy, the homo-oligomerized symmetric proteins undergo immediate association and expand endlessly after their expression in recombinant E. coli. In the latter report, the authors pointed out a possibility that the irregularly associated form may frequently occur because the strong binding of biotin to streptavidin (KD ≈ 10−15M) prevents self-healing.5,6 Therefore, the biotinylated position might be essential to be precisely designed to diminish irregularly connection via single streptavidin/biotin bonds.
Here we propose a novel strategy for producing self-assembling proteins, which we have named “Nanolego.” By incorporating stabilization and termination technologies into the Nanolego strategy, we were able to successfully obtain timing-controlled and size-regulated filament-shape complexes using Nanolego components.
Results
Design of self-assembling Nanolego
Figure 1(A) summarizes the construction strategy of Nanolego building blocks. We defined two types of Nanolego-configuring elements, “structural elements” and “binding elements.” The structural element (Sn) is a homo-oligomeric protein of a structurally stable symmetrical form7 used as the structural center of Nanolego. In contrast, the binding element (Ba and Bb) are a heterointeraction protein pair, which are components used for self-assembling between Nanolegos. When the fusion proteins, S-Ba or S-Bb, are expressed separately, oligomerization of the structural element results in Nanolegos including multibinding elements, (S-Ba)n or (S-Bb)n. The self-assembled polymerization of Nanolego, [(S-Ba)n·(S-Bb)n]x, is expected to occur when (S-Ba)n and (S-Bb)n are mixed.
Figure 1.
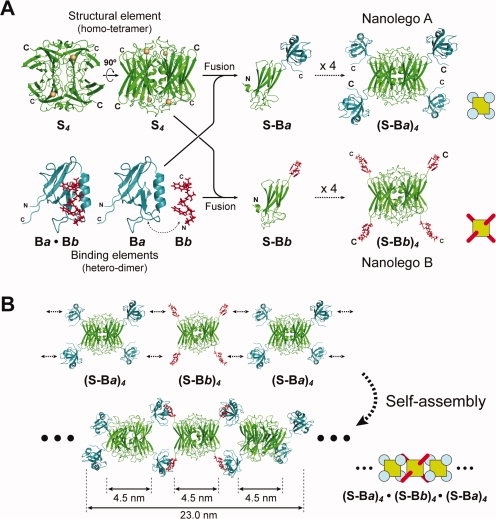
Design of self-assembling Nanolegos. A: Construction strategy of two hetero-Nanolegos, SOR-PDZ and SOR-Zpep. The basic crystal structure, indicated as structural (S) and binding elements (Ba and Bb), was composed of P. furiosus SOR and human Erbin PDZ domain and the binding peptide, respectively. For modeling of tetrameric SOR-PDZ and SOR-Zpep fusion proteins, we first constructed the model structure of monomeric SOR-PDZ and SOR-Zpep fusion proteins. The models of tetrameric fusion proteins, (S-Ba)4 and (S-Bb)4, were drawn by reconfiguration of the tetramer at the part of SOR. B: Putative self-assembling pattern of filamentous Nanolegos. These models were obtained by MD simulation based on the putative structure of the tetramer Nanolego in Figure 1(A) and reconfiguration of PDZ/Zpep-complex.
We selected a superoxide reductase (SOR) from the hyperthermostable archaeon Pyrococcus horikoshii OT38 as the structural element, and a mouse PDZ domain (PDZ) and a PDZ-binding peptide (Zpep)9,10 as the binding elements. The crystal structure of SOR is a symmetrical homotetramer cuboid in which four C-terminal ends of the proteins exist on the protein surface and are nestled at the vertexes of the cuboid. PDZ and Zpep associate reversibly with a KD of ∼10−7M. The design goal in this work was to procedure a linear filament. The fusion proteins of PDZ or Zpep at the C-terminal end of SOR (SOR-PDZ and SOR-Zpep, respectively) were designed without any spacer sequence [Fig. 1(A), and see Supporting Information Fig. S1]. The computational simulation predicted that oligomerization of the structural elements, (SOR-PDZ)4 and (SOR-Zpep)4, could occur without any physical obstacles [Fig. 1(A)] and self-assembly of Nanolegos, [(SOR-PDZ)4·(SOR-Zpep)4]n could occur by using two binding elements for each connection, which results in a linear filament [Fig. 1(B)]. We expressed both Nanolegos separately in E. coli and purified them as soluble proteins consisting of homotetramers, (SOR-PDZ)4 and (SOR-Zpep)4 (see Supporting Information Fig. S2).
To confirm the self-assembly of the SOR Nanolegos, we simply mixed equal quantities of the purified Nanolegos in solution. This led to a quick change of the bulk solution from a clear state to white turbidity [Fig. 2(A)]. The white turbidity seemed to be due to high molecular weight insoluble substances because they could be separated out by low gravity centrifugation; the SDS-PAGE analysis of the insoluble molecule revealed SOR-PDZ and SOR-Zpep with 1:1-composition [Fig. 2(B)]. We examined the substances using transmission electron microscopy [TEM, in Fig. 2(C)]; granular structures of 5–10 nm in diameter were observed before the mixing of each Nanolego mixture [Fig. 2(C), left], whereas the extended filamentous structures appeared after mixing for 2 min [Fig. 2(C), middle]. The width of these filaments was about 5 nm, which was consistent with that of the linear-structure model of self-assembled Nanolegos based on two antithetic directional extensions via every interaction with two binding elements [Fig. 1(B)]. Further, surface of the huge protein complexes, observed after 15 min from mixing of two Nanolegos [Fig. 2(C), right], also revealed several filamentous patterns where the width of these patterns also corresponded to the diameter of linear-structure model of self-assembled Nanolegos [Fig. 1(B)]. The result indicates that the SOR Nanolegos have the ability of self-assembly to build structurally ordered filamentous forms predominantly although forked filaments may be formed partly. Importantly, Nanolegos with heterobinding elements make it possible to control the self-assembled formation of nanoarchitecture through the timing of the mixing.
Figure 2.
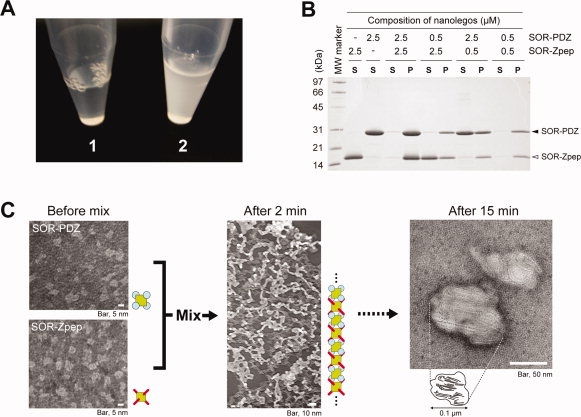
SOR Nanolegos self-assembling to the filamentous form. A: Change of state in bulk-solution by mixing purified SOR-PDZ and SOR-Zpep. The final concentration of each Nanolego was 10 μM in mixed solution. Each purified Nanolego formed a clear solution as shown in Photo 1. Photo 2 shows the solution state after 20 min from mixing these Nanolegos. B: SDS-PAGE analysis for the turbidity of SOR Nanolego mixture. The turbidity appeared in two Nanolegos-mixed solution was separated as the insoluble fraction by centrifugation at 14000g for 30 min at 4°C. The soluble and insoluble fractions of the mixture were subjected to SDS-PAGE in the lanes indicated by “S” and “P,” respectively. Each Nanolego was observed as single band with the mobility of monomer, 28.2 kDa (SOR-PDZ) and 15.7 kDa (SOR-Zpep) under the denatured condition with SDS. C: Negatively stained transmission electron micrograph of the self-assembled Nanolegos. As a rough approximation, the diameter of the filamentous form (after 2 min) was about 5 nm.
Stabilization of the assembly by disulfide bond
Next, to obtain the Nanolego filament with desired size, we tried to extend the Nanolego assembly using surface plasmon resonance (SPR); we fixed either Nanolegos on the Au surface and applied each Nanolegos step-by-step through the microfluid channel. The sensorgram increased at first extension step, although it decreased at second extension step in contrast to the first (Supporting Information Fig. S3). This is explained that the first associated Nanolegos were removed from the surface because of the association/dissociation-equilibrium state of the binding elements in solution.
Stabilization of self-assembled complexes that are formed via noncovalent bond is an important subject to maintain the assembled complexes.11 For example, in organic chemistry, Jin et al.12 reported the stable formation of self-assembled graphitic nanotube consisting of Gemini-shaped nonionic hexabezocoronene amphiphile by using olefin metathesis as a crosslinker. In this study, we tried to incorporate a cysteine (Cys) residue into each binding element of the Nanolegos by site-directed mutagenesis, where the noncovalent interaction is expected to be covalently linked by the formation of a disulfide bond between Cys-residues at each binding element.13 Considering the findings of the previous study that identified the essential amino acid residues for the interaction,14,15 we decided that there was a Cys-residue substitution at an arginine residue of PDZ and a tryptophan residue of Zpep (PDZR34C and ZpepW3C). The molecular dynamics simulation based on the crystallography of the PDZ/Zpep complex15 predicted that the Cys-residue substitution at those sites could form a disulfide bond without affecting PDZ/Zpep interaction [Fig. 3(A)]. Actually SPR analysis of the Cys-introduced PDZ/Zpep showed a distinct sensorgram from that of the wild type; the sensorgram for PDZR34C/ZpepW3C showed a specific increment at the binding phase, which was maintained even at the dissociation phase [Fig. 3(B)]. The sensorgram fitted well with the model of the two-state reactions, the noncovalent PDZ/Zpep interaction and the stabilization of PDZ/Zpep complex by the formation of a disulfide bond. The equilibrium dissociation constant of the 1st reaction (KD1), estimated by the 1:1(langmuir)-binding model, was similar to that of the wild type pair whereas the 2nd reaction, which is indicated by ka2 and kd2 values, was observed only in the mutated pair (see Supporting Information Table 2).
Figure 3.
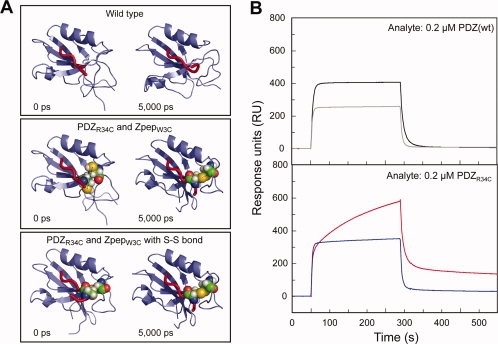
Construction of Cys-introduced Nanolegos. A: Introduction of a disulfide bond to between PDZ domain and PDZ-binding peptide. The results were MD simulation of the interaction between PDZR34C and ZpepW3C mutant. The structures on the left side are initial models before MD (0 ps), and those on the right side are the final models after MD for 5000 ps. B: Binding analyses between PDZ- and Zpep-Cys-mutant by the SPR assay. All sensorgrams indicate the response when 0.2 μM analyte was used. The kinetics values of each binding assay are shown in Supporting Information Table 2.
Size control of the self-assembled Nanolego filament using Cys-induced Nanolegos
We tried step-by-step extension of filaments from fixed Nanolegos (of either type) on the surface of matrix using Cys-induced Nanolegos. SOR-ZpepW3C were covalently fixed on the sensor chip and the purified SOR-PDZR34C was injected for 20 min and washed with running buffer for 20 min at the first extension step. Each Nanolego solution was then sequentially injected alternately by the same procedure. The sensorgram climbed at each injection step with scarcely any decrease at the washing step (see Fig. 4). Such an increment was not observed under the reduced condition using DTT-containing running buffer, or in the trial using Nanolegos with wild-type PDZ or Zpep (see Supporting Information Figs. S3 and S4). The incremental ratios from the second injection step were almost consistent with the ratio corresponding to the calculated molecular weight of two Nanolegos (SOR-PDZR34C:SOR-ZpepW3C = 1.6:1.0), which suggests that quantitative extension had been achieved and the forked filaments have not been formed extensively (Fig. 4, inset). The response unit change of the first injection step was higher than the other steps for SOR-PDZR34C injection (third, fifth, and seventh steps). The precise reason is not clear, but self-assembly activity may be altered in part of the immobilized SOR-ZpepW3C on the sensor chip, whereas unusual binding of SOR-PDZR34C may disturb further extension.
Figure 4.
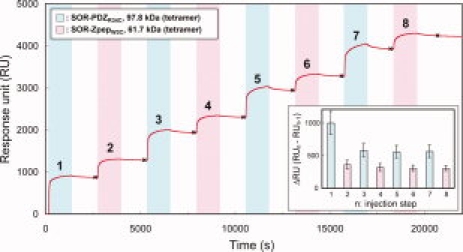
The sensorgram of 8-steps extension assembly of Cys-introduced Nanolegos. At every injection step, SOR-PDZR34C (colored sky-blue) or SOR-ZpepW3C (colored pink) was loaded for 20 min. The washing steps (not colored) were performed by passing the running buffer for 20 min. The inset graph is the summary of the increment of the SPR response for each injection of Nanolegos. The difference of RU at each injection step (ΔRU = RUn− RUn−1; n, injection step) was calculated from the RU at the data points shown as “×” in this figure. This graph presents the results of triplicate analyses, and the error bar on each column shows the standard deviation of each injection step.
Capping Nanolegos for terminating the extension of self-assembled filaments
Because extension of the Nanolego self-assembly is basically endless, it is necessary to consider how to terminate the self-assembly to regulate the filament length. We designed a new SOR Nanolego with only two of the four binding elements, which was designated as a “capping (CP) Nanolego.” Because SOR Nanolego uses two binding elements for one-step extension, incorporation of the CP-Nanolego was expected to terminate further extension. We coexpressed the SOR Nanolegos with and without ZpepW3C in recombinant E. coli, and heteromeric SOR-tetramers harboring different numbers of ZpepW3C from 0 to 4 were obtained. Finally, we purified a CP SOR Nanolego, CP-SOR-ZpepW3C, with the actual orientation of the two ZpepW3C by using two affinity columns (see Methods and Supporting Information Fig. S5). The ability of the CP Nanolego to terminate extension of the Nanolego filament was successfully monitored using a quartz crystal microbalance (QCM) as the change of the extension rate when CP Nanolegos were added to the Nanolego-mixed batch solution [Fig. 5(A)].
Figure 5.
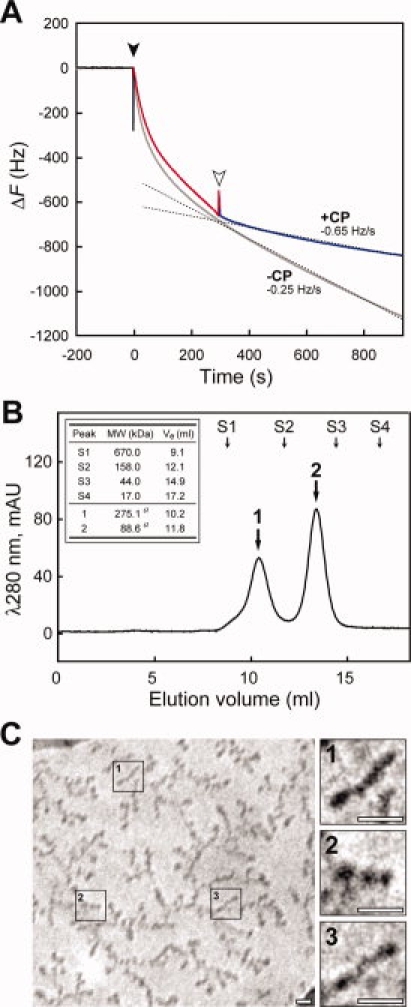
Effect of CP Nanolego on extension of Nanolego filament. A: Terminating effect of CP-SOR-ZpepW3C on filament extension observed by QCM assay for a batch solution. Black and white arrowheads indicate the injection point of SOR-PDZR34C (at 0 s) and CP-SOR-ZpepW3C (at 300 s), respectively. The main sensorgram to investigate the effect of CP-SOR-ZpepW3C is colored separately in three sections; baseline (black, −200 to –0 s), filament extension with two Nanolegos mixture (red, 0–300 s), and addition of CP-SOR-ZpepW3C (blue, 300–700 s). The gray sensorgram shows the result without the addition of CP-SOR-ZpepW3C as the control experiment. The dashed lines are the result of approximated curve fitting of the sensorgram with or without the addition of CP-SOR-ZpepW3C, and each slope of the curve as the frequency change rate is indicated in the graph with “+CP” or “−CP”, respectively. B: The result of gel-filtration chromatography of the mixture of both SOR-PDZR34C and CP-SOR-ZpepW3C in solution. The mixture of two Nanolegos was performed by the concentration ratio of SOR-PDZR34C:CP-SOR-ZpepW3C = 1:5. Peak a and b corresponded to the 3-mer Nanolego complex ([CP-SOR-ZpepW3C] × 2 + [SOR-PDZR34C] × 1) and single CP-SOR-ZpepW3C, respectively. Arrows marked with S1–4 show the eluting volumes of the molecular standard proteins. Details of elution peak a, b and the standard proteins are shown in the inset table. “Ve” indicates each elution volume. a, theoretical molecular weight. C: The results of CryoTEM observation of putative 3-mer Nanolego complexes. Photographs on the right are threefold magnifications of the three areas on the left indicated by numbers. Each white bar indicates a scale of 10 nm.
Finally, we evaluated the size-regulation of the self-assembled products with CP Nanolegos by the cryogenic TEM (Cryo-TEM) observation. When we mixed SOR-PDZR34C and CP-SOR-ZpepW3C in a batch solution with an excess molar ratio of the CP Nanolego (SOR-PDZR34C:CP-SOR-ZpepW3C = 1:5), no turbidity was observed in the mixed solution, whereas the mixture of two prototype Nanolegos at the ratio of 1:5 was turbid [see Fig. 2(B)]. The mixed solution was then subjected to gel-filtration chromatography to analyze the size of the self-assembled molecules; there were two peaks corresponding to the 3-mer Nanolego complex and single CP Nanolego, as expected [Fig. 5(B)]. SDS-PAGE analysis corresponding to the 3-mer Nanolego complex suggested that the complex consisted of two CP-SOR-ZpepW3C and a SOR-PDZR34C (see Supporting Information Fig. S6). The absence of the elution peak corresponding to the 2-mer or over 4-mer Nanolego complexes in this experiment suggested that the assembled products were stable because of the introduction of the ss-bond and that the presence of CP Nanolego successfully inhibited further association of SOR-PDZR34C. We subjected the assumed 3-mer Nanolego complex to Cryo-TEM [Fig. 5(C)]. Short rod-like structures were recognized in contrast to the monomer and the filament-like structures from the two prototype Nanolegos shown in Figure 2(C); the length of the rod shown in the magnified image in Figure 5(C) is between 13.7 and 21.2 nm, which is reasonable as the value of 23.0 nm [the length of simulated model in Fig. 1(B)] of the 3-mer Nanolego scattering in the Cryo-grid.
Discussion
Our successful demonstration of the size-regulated filamentous complex is based on the two types of Nanolego strategies. One is the concept of the structural design for the self-assembly manner of biological molecules, in which we used a symmetrical structural element and a heterobinding element. Of particular importance was the use of multipoint reversible interactions as the binding elements in the Nanolegos. These multipoint interactions are expected to reduce the twisting between Nanolegos and allow the maintaining of the linearity of the self-assembled filaments. If construction of the filaments by interaction of Nanolegos via a single binding point is attempted, a complicated design would be needed for precise and rigid spacers to connect the structural and binding elements. Another positive factor is that the binding element of a reversible weak interaction pair may also play a role in decreasing the production of the irregular assembly form via single interaction. We postulate that the equivalent state of association/dissociation eliminates the irregular assembly via single interaction, and only the proper form via two-interaction pair stably remains in the assembled product. The lock system via the disulfide bond will only be available when the proper assembly occurs; the dissociation rate of a single interaction between PDZR34C and ZpepW3C (kd1, 1.25 × 10−1 s−1) was >30 times faster than the disulfide bond formation rate (ka2, 3.92 × 10−3 s−1) whereas the dissociation rate of Nanolegos via two-interaction pairs (kd ≈ 1.6 × 10−4 s−1, data not shown) was slow enough to keep Nanolegos in the bound state until the disulfide bond formation (see Supporting Information Table 2). The other strategy leading to success was the establishment of a self-assembly control system for the Nanolegos. The disulfide linking system by spontaneous formation of a disulfide bond is very useful for stabilizing the weak multiple interactions between Nanolegos, which enabled step-by-step extension from immobilized Nanolego molecules and maintaining the size of the self-assembled complex. The CP Nanolego is also useful for terminating extension of Nanolego self-assembly, which enabled size-regulation of the self-assembled complex in solution. Although we demonstrated to produce only 3 units by CP Nanolego [Fig. 5(C)], we deem in future study that it is possible to produce 4 units over size-regulated filaments in solution by combination of multiple Nanolegos with different binding elements and CP Nanolego.
The size-regulated filamentous complex, achieved in this study, may be applicable to the quantitative accumulation of functional proteins such as fluorescent protein. Because there are free N-terminal ends on lateral face of extended SOR Nanolegos according to the simulated filament model [Fig. 1(B)], such functional proteins may be able to be incorporated quantitatively at N-terminal ends of Nanolegos, either by fusion protein technology or tagging via stable interactions, which may be useful as quantitatively amplified probes. The procedure of design and purification for CP Nanolego is also applicable for the construction of Nanolegos with two kinds of binding elements, which has the potential to arrange different Nanolegos in a fixed order. Incorporation of more than two functional proteins such as enzymes in a fixed order at N-terminal ends of Nanolegos may allow enzymatic chain reactions on a single complex.
Recent advances in genome science have led to the amassing of large amounts of information for primary and higher order protein structures and protein–protein interactions, which will increase the amount of protein information applicable for the structural and binding elements of Nanolego. It might be possible to extend the strategy to more complex structures with both further developments of our technology and the various Nanolego designed by combinations of such new elements.
Materials and Methods
Construction of Nanolegos
As the structural element of all Nanolegos, residues 1–115 of SOR of P. horikoshii OT3 (UniProt: O58810) were used. The binding element of mouse PDZ/Zpep complex was prepared from two candidate pairs as follows: residues 12–115 of tax-interacting protein-1 (TIP-1, UniProt: Q9DBG9)/β-catenin C-terminal peptide of nine residues (QLAWFDTDL, UniProt: Q0224810) and residues 1308–1402 of Erbb2 interacting protein (Erbin, UniPlot: Q80TH2)/a peptide of seven residues binding to Erbin PDZ with high affinity (TGWETWV14,15). In this study, the first candidate of PDZ/Zpep was used for the primary experiment of Nanolego self-assembly in Figure 2. To achieve structure-based selection of the amino acid residue for Cys-mutagenesis, the second candidate of PDZ/Zpep was used for the experiment in Figures 3–5. SOR and PDZ-coding double strand DNAs were prepared by PCR. On the other hand, Zpep, ZpepW3C, and poly-histidine tag (His6)-coding double strand DNAs were prepared by annealing of synthesized complementary oligo DNA sets. Cys-introduced PDZ mutant (PDZR34C)-coding DNA was constructed by overlap extension PCR with using Erbin PDZ-coding DNA as a template. To construct the fusion proteins of SOR Nanolego, the target genes were constructed by overlap extension PCR or ligation at the sticky end with T4 ligase and then cloned into the pET30a(+) expression vector (Merck KGaA, Darmstadt, Germany). To obtain the CP Nanolego, the coexpression vector harboring two genes, coding SOR-ZpepW3C without N-terminal His6, and SOR-Cohesin with C-terminal His6 was designed with pET30a(+) vector, and each gene was located at a downstream discrete T7 lac promoter region. In this case, Cohesin from CipA of Clostridium thermocellum (residues of 182–328, UniProt: Q0685116) was fused to SOR instead of ZpepW3C, SOR-CipA with N-terminal His6, to facilitate isolation of target proteins including only two binding elements by a difference of molecular size of the SOR tetramer. The sequence information of primers or oligo DNAs and physical maps of the constructed Nanolegos are shown in Supporting Information Table 1 and Figure S1, respectively. The expression vectors of all Nanolegos were transformed into E. coli strain BL21-CodonPlus (DE3)-RIL (Stratagene, La Jolla, CA).
Protein expression and purification
The proteins were produced in E. coli recombinant cells growing at 30°C in 200 mL of LB medium with 50 μg/mL kanamysin and 34 μg/mL chloramphenicol by inducing expression at an optical density of 660 nm of 0.8 with 0.1 mM isopropyl thio-β-d-galactoside and harvesting them 6 h later. The cultivated cells were collected by centrifugation (10,000g for 5 min at 4°C) and were resuspended in 60 mL of the lysis buffer (PBS, 1.0% [w/v] Triton X-100, 1.0 mM DTT). The cells were sonicated using BIORUPTOR (Cosmo Bio, Tokyo, Japan), and the lysate was clarified by centrifugation (15,000g for 30 min at 4°C). The supernatant was diluted with Buffer A (20 mM Na-phosphate buffer, 0.5 M NaCl, 20 mM imidazole, 1 mM DTT, pH 7.4) and loaded onto a Ni2+-chelate affinity column, HisTrap HP (GE Healthcare UK, Bukinghamshire, UK). Except for purification of only CP Nanolego, the proteins bound on the column were eluted using a linear gradient elution with 0–100% Buffer B (500 mM imidazole in Buffer A); the elution peak of target proteins was at 30% of Buffer B. The collected fractions included the target proteins which were concentrated with Amicon Ultra-15 (10,000 MWCO, Millipore, Billerica, MA). The proteins were further purified by size-exclusion chromatography on a HiLoad 16/60 Superdex 200pg (GE Healthcare) in Buffer C (10 mM HEPES, 0.15 M NaCl, 1 mM DTT).
Purification of CP Nanolegos
To purify CP Nanolegos which have only two binding elements, the purification from the E. coli lysate coexpressed SOR-ZpepW3C without N-terminal His6 and SOR-CipA with N-terminal His6 were consisted with two types of affinity purifications by the Ni2+-chelate affinity and the Nanolego binding element affinity. As first affinity purification, the elution strategy of boundary proteins from the HisTrap HP column was performed in incremental steps with Buffer B at various percentages against Buffer A: 5, 15, 23, 50, and 100% of Buffer B.17 The heterotetramers harboring only two binding element were collected from an eluted fraction with 23% of Buffer B. After the collected fractions were concentrated, and were further purified by size-exclusion chromatography on same condition along with the purification procedure of Nanolegos described earlier. Second, to select the actual orientation of the two binding elements for CP-Nanolego, we prepared a SOR-PDZR34C-conjugated affinity column with HiTrap NHS-activated HP, 1 mL (GE Healthcare) according to the manufacturer's protocol. The solution of gel-filtrated samples was displaced with Buffer D (10 mM HEPES, pH 7.4, 0.15 M NaCl) and was applied to the affinity column. The boundary fraction of authentic CP Nanolegos was eluted with Buffer E (10 mM HEPES, pH 7.4, 0.5 M NaCl, 10 mM DTT). The purification details for the CP Nanolegos are shown in Supporting Information Figure S5.
Model building
For modeling of tetrameric SOR-PDZ and SOR-Zpep fusion proteins, we first constructed the model structure of monomeric SOR-PDZ and SOR-Zpep fusion proteins. The SOR and PDZ, or SOR and Zpep were linearly connected by using modeling software MOE (Chemical Computing Group), and they were energy-minimized. Next, to optimize the conformation of these monomeric artificial fusion proteins, we performed simulated annealing in combination with MD simulations in an implicit water model with the generalized Born surface area method18,19 on 32 CPUs of the Riken Super Combined Cluster. During the simulated annealing process, the main-chain dihedral angles of the SOR, PDZ, and Zpep were constrained. The system was heated or cooled under alternate conditions during 500 ps as follows: heating to 50 K during 50 ps (1 K ps−1), heating from 50 to 200 K during 50–100 ps (3 K ps−1), heating from 200 to 300 K during 100–150 ps (2 K ps−1), heating from 300 to 373 K during 150–250 ps (0.73 K ps−1), and cooling from 373 to 0 K during 250–500 ps (1.492 K ps−1).
MD simulations
These MD simulations were performed by using Amber 8.019 on a personal computer (Xeon 3.2 GHz) equipped with a special-purpose computer for the MD simulations, MDGRAPE-3.20–22 The all-atom point-charge force-field ff03 was adopted,23 and the time step was set to 2.0 fs. All noncovalent bonded interactions, namely, the van der Waals and Coulomb forces and energies, were calculated directly and accurately by using MDGRAPE-3. All bond lengths were constrained to equilibrium lengths by the SHAKE method.24 All initial structures were fully solvated by the spherical water droplets of the TIP3P water molecules.25 Each system was gradually heated to 300 K during the first 50 ps with a heating rate of 6 K ps−1, and was constantly incubated at 300 K until 5000 ps. Each simulated mode on the left side was the initial model before MD (0 ps), and the one on right side was the final model after MD for 5000 ps.
TEM observation
The protein solution (0.1 μg/mL in 10 mM HEPES, 0.5M NaCl, pH 7.4) was dropped onto a carbon-coated copper grid, and drained with filter paper after allowing adsorption for 60–90 s. The samples were negatively stained with 1% uranyl acetate for 5 s, and observed with a transmission electron microscope (H7000E, Hitachi, Japan) under an acceleration voltage of 75 kV.
Surface plasmon resonance assay for the kinetic binding analysis
SPR assay was performed using a Biacore® 3000 system (Biacore AB, Uppsala, Sweden) at 25°C. The biotinylated PDZ-binding peptide was coupled to the streptavidin-coated sensor chip (Sensor Chip SA, Biacore), and a baseline increase of ∼50 resonance units (RU) was typically obtained. All measurements were carried out in HBS-EP buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.005% Tween20) as the running buffer. For the association step, the PDZ domain at five different concentrations, 0.05, 0.1, 0.2, 0.5, and 1.0 μM, was allowed to flow through at a rate of 20 μL/min for 240 s. The dissociation step was performed by sending in the running buffer at the same flow rate as the association step for 240 s. The sensorgram using each protein concentration was observed by triplicate analyses. Association (ka) and dissociation (kd) constants between the PDZ domain and the PDZ-binding peptide were obtained by the fitting of the 1:1 (Langmuir) binding model or the two-state reaction model using the BIA evaluation software version 4.1 (Biacore). The equilibrium dissociation constant (KD) is determined from the value of kd/ka.
Step-by-step extension assembly
This experiment was carried out using Biacore® 3000 (Biacore) at 25°C. SOR-ZpepW3C was immobilized to the Sensor Chip C1 (Biacore) by the procedure of amine coupling with NHS/EDC, and a baseline increase of ∼550 RU was typically obtained. All measurements were carried out in HBS-EP buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.005% Tween20) as the running buffer. In each injection step, 0.1 μM SOR-PDZR34C or SOR-ZpepW3C were allowed to flow through at a rate of 5 μL/min for 20 min. The washing steps were performed by passing the running buffer at the same flow rate as the injection step for 20 min.
Extension assay of Nanolego filaments in a batch solution
To examine the extension termination of the filaments with CP Nanolego, we assayed the association rate from immobilized ligands by the QCM technique using Affinix Q (Initium, Tokyo, Japan). SOR-ZpepW3C, 0.1 μg, was directly immobilized as the ligand on a QCM electrode. To block the activated Au surface and nonspecific binding sites, the electrode was exposed to the HBS-EP buffer with the purchased 2-methacryloyloxyethyl phosphorylcholine blocking reagent (Initium) for 2 h. As the first step in the assay, the SOR-ZpepW3C immobilized QCM electrode was soaked in 8 mL of HBS-EP buffer including 0.05 μM SOR-ZpepW3C, and we checked that the sensorgram did not change from the background level. Next, 0.05 μM SOR-PDZR34C was injected into the solution to start the filament extension. At 300 s from the addition of SOR-PDZR34C to the solution, the time course of frequency change in response to the addition of 0.1 μM CP-SOR-ZpepW3C was assessed until 1000 s. The frequency change rates for 300–1000 s with or without the addition of the CP-SOR-ZpepW3C were estimated by fitting of the approximated linear curve by the least-squares method.
Cryogenic transmission electron microscopy
A sample solution containing the Nanolego assembly (0.5 mM) was deposited onto a microgrid (STEM 150 Cu, Okenshoji, Tokyo, Japan) and the protein molecules were allowed to adsorb onto it for 1 min. Immediately after draining the excess solution with filter paper, the grid was immersed into liquid nitrogen and transferred into a cryo holder Cryotransfer System Model 626 DH (Gatan, CA) under the liquid nitrogen environment. The observation was performed using a cryo electron microscope JEM 2010 (JEOL, Japan) equipped with a cryo holder.
Acknowledgments
The authors appreciate the access to the computational resources of the RIKEN Super Combined Cluster (RSCC). They thank Hisako Imamura for technical support of the construction of Nanolego-expression vectors. They would like to thank all the members of the Laboratory for Genome Exploration Research Group (GSC) and the Genome Science Laboratory of RIKEN.
References
- 1.Whitesides GM, Boncheva M. Beyond molecules: self-assembly of mesoscopic and macroscopic components. Proc Natl Acad Sci USA. 2002;99:4769–4774. doi: 10.1073/pnas.082065899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang S. Fabrication of novel biomaterials through molecular self-assembly. Nat Biotechnol. 2003;21:1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 3.Padilla JE, Colovos C, Yeates TO. Nanohedra: using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc Natl Acad Sci USA. 2001;98:2217–2221. doi: 10.1073/pnas.041614998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeates TO, Padilla JE. Designing supramolecular protein assemblies. Curr Opin Struct Biol. 2002;12:464–470. doi: 10.1016/s0959-440x(02)00350-0. [DOI] [PubMed] [Google Scholar]
- 5.Ringler P, Schulz GE. Self-assembly of proteins into designed networks. Science. 2003;302:106–109. doi: 10.1126/science.1088074. [DOI] [PubMed] [Google Scholar]
- 6.Green NM. Avidin and streptavidin. Methods Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 7.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 8.Yeh AP, Hu Y, Jenney FE, Jr, Adams MW, Rees DC. Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states. Biochemistry. 2000;39:2499–2508. doi: 10.1021/bi992428k. [DOI] [PubMed] [Google Scholar]
- 9.Borg JP, Marchetto S, Le Bivic A, Ollendorff V, Jaulin-Bastard F, Saito H, Fournier E, Adélaïde J, Margolis B, Birnbaum D. ERBa basolateral PDZ protein that interacts with the mammalian ERBB2/HER2 receptor. Nat Cell Biol. 2000;2:407–414. doi: 10.1038/35017038. [DOI] [PubMed] [Google Scholar]
- 10.Kanamori M, Sandy P, Marzinotto S, Benetti R, Kai C, Hayashizaki Y, Schneider C, Suzuki H. The PDZ protein tax-interacting protein-1 inhibits beta-catenin transcriptional activity and growth of colorectal cancer cells. J Biol Chem. 2003;278:38758–38764. doi: 10.1074/jbc.M306324200. [DOI] [PubMed] [Google Scholar]
- 11.Lean JM. Supramolecular chemistry: concepts and perspectives. Weinheim, Germany: VCH; 1995. [Google Scholar]
- 12.Jin W, Fukushima T, Kosaka A, Niki M, Ishii N, Aida T. Controlled self-assembly triggered by olefin metathesis: cross-linked graphitic nanotubes from an amphiphilic hexa-peri-hexabenzocoronene. J Am Chem Soc. 2005;127:8284–8285. doi: 10.1021/ja051859p. [DOI] [PubMed] [Google Scholar]
- 13.Backer MV, Patel V, Jehning BT, Backer JM. Self-assembled “dock and lock” system for linking payloads to targeting proteins. Bioconjug Chem. 2006;17:912–919. doi: 10.1021/bc060037u. [DOI] [PubMed] [Google Scholar]
- 14.Laura RP, Witt AS, Held HA, Gerstner R, Deshayes K, Koehler MF, Kosik KS, Sidhu SS, Lasky LA. The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J Biol Chem. 2002;277:12906–12914. doi: 10.1074/jbc.M200818200. [DOI] [PubMed] [Google Scholar]
- 15.Skelton NJ, Koehler MF, Zobel K, Wong WL, Yeh S, Pisabarro MT, Yin JP, Lasky LA, Sidhu SS. Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain. J Biol Chem. 2003;278:7645–7654. doi: 10.1074/jbc.M209751200. [DOI] [PubMed] [Google Scholar]
- 16.Carvalho AL, Dias FM, Prates JA, Nagy T, Gilbert HJ, Davies GJ, Ferreira LM, Romão MJ, Fontes CM. Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex. Proc Natl Acad Sci USA. 2003;100:13809–13814. doi: 10.1073/pnas.1936124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howarth M, Chinnapen DJ, Gerrow K, Dorrestein PC, Grandy MR, Kelleher NL, El-Husseini A, Ting AY. A monovalent streptavidin with a single femtomolar biotin binding site. Nat Methods. 2006;3:267–273. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashford D, Case DA. Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 19.Case DA, Darden TA, Cheatham TE, III, et al. Amber 8.0. San Francisco: University of California; 2004. [Google Scholar]
- 20.Taiji M, Marumi T, Ohno Y, Futatsugi N, Suenaga A, Takada N, Konagaya A. Proceedings of Supercomputing. 2003. Protein explorer: a petaflops special-purpose computer system for molecular dynamics simulations. 2003. [Google Scholar]
- 21.Taiji M. Proceedings of Hot Chips 16. Stanford University, CA; 2004. MDGRAPE-3 chip: a 165 Gflops application specific LSI for molecular dynamics simulations. [Google Scholar]
- 22.Narumi T, Ohno Y, Okimoto N, Koishi T, Suenaga A, Futatsugi N, Yanai R, Himeno R, Fujikawa S, Ikei M, et al. Proceedings of Supercomputing. 2006. A 55 Tflops simulation of amyloid-forming peptides from yeast prion Sup35 with the special-purpose computer system MDGRAPE-3. 2006. [Google Scholar]
- 23.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 24.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 25.Jorgensen WL, Chandrasekhar J, Madura JD. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]


