Abstract
Members of the Wnt family of lipoglycoproteins initiate signaling by binding to Frizzled (Fz) receptors, and the signal is then relayed by Disheveled (Dvl). The Dvl PDZ domain is known to interact directly with a peptide derived from the KTXXXW motif of Fz7, which is conserved in all known Fz subtypes. We found that an extended region spanning the KTXXXW motif on both its N-terminal and C-terminal sides dramatically influences the affinity of peptides derived from Fz7 for Dvl PDZ. An alanine scanning study identified the specific residues external to the KTXXXW motif that are important for high-affinity binding. In a circular dichroism analysis, mutation of some of these critical residues resulted in peptide conformational changes, suggesting that the secondary structure of the peptides contributes to Fz-Dvl PDZ binding. Of the 10 known Fz subtypes, peptides derived from only Fz1, Fz2, Fz3, Fz4, and Fz7 directly bound to Dvl PDZ domain in our study. Other Fz subtypes, including some known to be involved in Wnt/β-catenin signaling (Fz5, Fz9), did not bind to Dvl, suggesting that direct interaction with Dvl PDZ does not determine the subtype-specific functionality of Fz. Molecular modeling and circular dichroism studies indicated that the Fz peptides that bind to Dvl PDZ domain form specific conformations that are different from those of nonbinding peptides.
Keywords: frizzled, dishevelled, protein-protein interaction, conformation
Introduction
The Wnt signaling pathways comprise evolutionarily conserved molecules that are important not only for their regulatory role in a wide array of developmental processes but also for their involvement in diseases, including cancer.1,2 The Wnt/β-catenin (canonical Wnt signaling) pathway entails stabilization of cytoplasmic hypophosphorylated β-catenin, which then translocates to the nucleus and associates with TCF/LEF transcription factors to upregulate gene transcription.3 Wnt is also involved in other pathways, such as control of vertebrate gastrulation through a mechanism similar to the Drosophila planar cell polarity pathway (PCP), and the stimulation of calcium flux.4 The proteins immediately downstream of Wnt in its signaling cascade are the cell surface receptors Frizzled (Fz) and low density lipoprotein-related proteins 5 and 6 (LRP5/6). Fz binds to Wnt through an N-terminal cysteine-rich domain to initiate signaling.5–8 Ten mammalian isoforms of Fz (Fz1–Fz10) are known. All Fz proteins consist of the cysteine-rich domain, seven transmembrane domains with connecting extracellular and intracellular loops, and an intracellular C-terminal tail. The Fz C-terminal region is important for Wnt signaling via Dishevelled (Dvl) proteins and for subcellular localization of Fz.9
Dvl comprises a family of three homologous proteins (Dvl1, Dvl2, and Dvl3) and is essential for all Wnt/β-catenin, PCP, and Wnt/Ca2+ signaling.10–14 Dvl proteins are composed of three major domains: DIX, PDZ, and DEP.15 The C-terminal KTXXXW (X = any amino acid) motif of Fz receptors was found to be a mediator of Wnt/β-catenin signaling in Xenopus embryos,16 demonstrating functional Fz-Dvl interaction. Although Fz1, Fz4, Fz5, and Fz9 have been shown to interact with Dvl by colocalization and coimmunoprecipitation assays,5,17–19 the only observation of their direct interaction has been between a Fz7 peptide containing the KTXXXW sequence and the PDZ domain of mouse Dvl1.20 Most known PDZ-interacting proteins bind to PDZ domain via a short peptide motif at the extreme C-terminus, and such interactions are well characterized.21 In contrast, Fz7 was identified as binding to the Dvl PDZ domain via an internal KTXXXW motif.20 PDZ-mediated interactions that occur through modes other than recognition of an extreme C-terminal motif have been identified, although they are less common.22–26 Neuronal nitric oxide synthase (nNOS) binds to the PDZ domain of syntrophin by forming a β-hairpin secondary structure, which is likely to be explained by steric hindrance caused by the amino acid loop at the end of the PDZ domain's binding groove.25 The internal PDZ ligand of the ETA endothelin receptor is also suggested to form a β-finger structure.26 There is no such specific structural information about the interactions of the Fz internal motif.
The available data on the direct interaction between Fz and Dvl, and the involvement of these two proteins in Wnt signaling pathways, suggest that this interaction may be important for Fz-mediated signal relay on Wnt stimulation. However, other data suggest that the underlying mechanism of Fz signaling via Dvl is not explained solely by their direct interaction. Although all Fz subtypes contain the KTXXXW motif, they can induce different downstream signaling pathways. For example, human Fz1, Fz5, and Fz9 and rat Fz7 induce Wnt/β-catenin signaling, whereas rat Fz2 and mouse Fz3, Fz4, and Fz6 induce Wnt/Ca2+ signaling.17,19,27,28 Fz6 has been shown to inhibit the Wnt/β-catenin signaling mediated by other Fz subtypes.29
To gain insight into the mechanism of signal transduction between Fz and Dvl, we studied the specific nature of the interaction between peptides that include the internal PDZ binding motif of Fz and the Dvl PDZ domain. We found that the Fz KTXXXW motif alone is insufficient for the high-affinity interaction between Fz and Dvl PDZ, and that other residues spanning the KTXXXW motif greatly enhance the affinity of the interaction. This extended region forms a stable secondary structure. Such interactions between the Fz peptide and Dvl PDZ domain are subtype-specific: Fz1, Fz2, Fz3, Fz4, and Fz7 interacted with Dvl, whereas other subtypes of Fz did not.
Results
Minimum sequence requirement for optimal binding of Fz7 to Dvl PDZ
To identify the minimum Fz7 sequence required for specific interaction with Dvl PDZ, we utilized peptides of varying length, all of which contained the KTXXXW motif. The interactions between these peptides and human Dvl PDZ were analyzed by Alphascreen energy transfer assay.30 The binding affinity of each peptide was determined based on its ability to compete with the binding of a biotinylated peptide and a GST-tagged PDZ domain of Dvl. Previous studies indicated that at least five amino acids C-terminal to the KTXXXW motif are essential for the full function of Xenopus Fz3,16 and a peptide [GKTLQSWRRFYH; Fz7-#1; Fig. 1(A)] that includes both the KTXXXW motif and the five amino acids (RRFYH) directly binds to mDvl1 PDZ domain.20 The affinity of this interaction is modest (Kd ∼ 10 μM) compared with that of typical reported PDZ protein domain interactions; thus, other region(s) of the Fz protein may participate in Fz-Dvl interaction and increase its affinity. In addition to the KTXXXW motif, all 10 Fz proteins possess another moderately conserved region (WIWS in Fz7) at the N-terminus adjacent to the KTXXXW motif [Fig. 1(B)]. We designed a longer Fz7 peptide of 23 amino acids [TGFWIWSGKTLQSWRRFYHRLSH; Fz7-#2; Fig. 1(A)] that included this region. We measured binding competition with self (biotin-Fz7-#2 as the probe), with Fz7-#1, and with a randomly generated nonspecific peptide of 20 amino acids. Strikingly, with an IC50 of 0.22 μM, the longer peptide Fz7-#2 showed an affinity for Dvl PDZ approximately 50 times that of the shorter Fz7-#1 peptide [Fig. 2(A)]. The IC50 of Fz7-#1 (11 μM) in this assay is consistent with previous results.20 To determine whether both regions flanking the KTXXXW motif are necessary for specific binding, we designed two additional peptides, one containing the KTXXXW motif and the N-terminal fragment of Fz7-#2 (TGFWIWSGKTLQSW: Fz7-N-fragment) and the other containing the KTXXXW motif and the C-terminal fragment of Fz7-#2 [KTLQSWRRFYHRLSH: Fz7-C-fragment; Fig. 1(A)]. The binding affinity of both fragments was much lower than that of the complete Fz7-#2 peptide [Fig. 2(B)]. Taken together, these results strongly suggest that the KTXXXW motif alone is insufficient for specific interaction between Fz7 and Dvl PDZ. Instead, a segment of the Fz7 protein that spans the KTXXXW motif on both its N- and C-terminal sides is necessary.
Figure 1.

A: Sequences of the Fz7-related peptides used in this study, aligned on the KTXXXW motif. B: Alignment of all human Fz C-terminal peptides spanning the KTXXXW motif that were used in this study.
Figure 2.
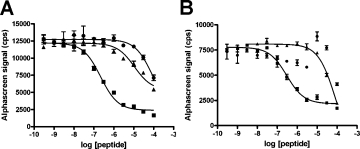
Alphascreen assays to determine the minimum sequence requirement for specific binding of Fz7 with Dvl1 PDZ. A: Comparison of Fz7-#2 peptide displacement by self (▪), by Fz7-#1 (▴), and by a nonspecific peptide (CLQEKHRILHKLLQNGNSPA, •). B: Comparison of Fz7-#2 peptide displacement by self (▪), by Fz7-N-fragment (▴), and by Fz7-C-fragment (•]). Values shown are the mean ± standard error.
Identification of Fz7 amino acids required for optimal Fz7-Dvl PDZ interaction
To investigate the contribution of specific amino acids, we conducted an alanine scanning study of Fz7-#2 peptide in which amino acids T544 to L564 were substituted sequentially. The binding affinity of each of the 21 peptides to Dvl PDZ was determined by competition with the wild-type sequence. Although some of the mutations only slightly decreased competitive affinity, others increased the IC50 by a factor as great as 40 (Table I). Significant among these were mutations of W547, K552, Q555, W557, H562, and R563. Although it eliminates the functional activity of XFz3, the mutation of T553 in the KTXXXW motif did not significantly affect the affinity and is consistent with a previous observation that substitution of the threonine in the KTXXXW motif with valine did not significantly affect binding to the PDZ domain.16 Certain nonconserved residues such as H562 also had a pronounced effect.
Table I.
The Relative Binding Affinities of Peptides Used in the Fz7-#2 Alanine Scanning Study, Shown in IC50 (μM)a
| Residue mutated | Dvl1 PDZ | Dvl3 PDZ |
|---|---|---|
| T544 | 0.5 | 0.2 |
| G545 | 0.6 | 0.2 |
| F546 | 0.7 | 0.4 |
| W547 | 1.1 | 2.3 |
| I548 | 0.4 | 0.4 |
| W549 | 0.6 | 0.9 |
| S550 | 1.1 | 0.9 |
| G551 | 0.3 | 0.5 |
| K552 | 4.9 | 6.0 |
| T553 | 0.5 | 0.6 |
| L554 | 0.7 | 1.0 |
| Q555 | 1.1 | 1.6 |
| S556 | 0.6 | 0.4 |
| W557 | 1.6 | 2.9 |
| R558 | 0.7 | 0.9 |
| R559 | 0.7 | 1.0 |
| F560 | 0.5 | 0.5 |
| Y561 | 0.7 | 0.9 |
| H562 | 7.7 | 8.6 |
| R563 | 1.3 | 1.7 |
| L564 | 0.4 | 0.8 |
| Wild-type | 0.3 | 0.2 |
The residues replaced by Ala are indicated. The IC50 value represents the 50% inhibitory concentration of each peptide required to displace the wild-type peptide bound to either Dvl1 PDZ or Dvl3 PDZ. Fz7 wild-type (Fz7#2) sequence is TGFWIWSGKTLQSWRRFYHRLSH.
Secondary structure of the PDZ-binding motif of Fz7 peptide
Given that the Dvl PDZ domain generally interacts with a sequence of only four to six amino acids in the ligand,31 it is unlikely that all residues of Fz7-#2 concurrently bind to the Dvl PDZ domain. We hypothesized that the PDZ-interacting region of Fz7 forms a secondary structure to enable optimal fitting with Dvl PDZ. To test this hypothesis, we measured the circular dichroism (CD) of our Fz7 peptides. Fz7-#2 showed negative peaks in the CD spectrum at 208 and 222 nm and a maximum positive signal at 190 nm [Fig. 3(A)], indicating the presence of a mixed α-helix/β conformation [Fig. 3(B) shows the spectra of the single-residue mutants]. The CD spectra were deconvoluted by using three different algorithms. All three analyses predicted the presence of about 20% α-helix, 30% β-sheet, and 20% β-turn structures (Supporting Information Table 2). To confirm this prediction, we calculated the molecular dynamics of Fz7-#2 peptide and its 21 mutants studied in the alanine scanning analysis. With a few exceptions, the most stable 75 conformations were very similar for all of the 22 peptides, suggesting reliability of the modeling. The modeled structure indicated the formation of a loop at the N-terminus adjacent to the KTLQSW motif involving S550, G551, and K552, followed by the KTLQSW motif with a fairly extended conformation, and a coiled region at the adjacent C-terminus [Fig. 4(A)]. Strikingly, the CD spectra of the shorter peptides Fz7-#1 and Fz7-C-fragment were typical of random coil structures [Fig. 3(A)], and that of the Fz7-N-fragment indicated the formation of a β-only structure with a single negative peak at approximately 218 nm and a positive peak at 195 nm (data not shown).
Figure 3.
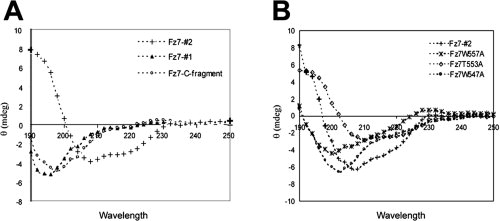
CD spectra showing secondary structure formation by Fz7 peptides. A: Different Fz7 peptides used in the study. Similar spectra were obtained at a range of concentrations (20–80 μM). B: Mutant Fz7 peptides with single residues replaced by Ala.
Table II.
The 50% Inhibitory Concentrations (IC50, μM) of Peptides Corresponding to the PDZ Binding Region of Human Fz Proteins, as Determined by Their Displacement of Biotinylated Fz7-#2 Peptide from the PDZ Domain of Human Dvl1, Dvl2, or Dvl3
| Fz subtype | Dvl1 PDZ | Dvl2 PDZ | Dvl3 PDZ |
|---|---|---|---|
| Fz1 | 2.1 | 0.6 | 1.4 |
| Fz2 | 1.3 | 0.4 | 0.7 |
| Fz3 | 1.4 | 0.6 | 1.3 |
| Fz4 | 2.2 | 0.9 | 1.8 |
| Fz5 | NS | NS | NS |
| Fz6 | NS | NS | NS |
| Fz7a | 0.2 | 0.1 | 0.3 |
| Fz8 | NS | NS | NS |
| Fz9 | NS | NS | NS |
| Fz10 | NS | NS | NS |
NS, Nonspecific binding observed only at high peptide concentrations (>100 μM).
The Fz7 peptide in this experiment is identical to that for the alanine-scanning (Table I), but measurement is performed separately in this experiment.
Figure 4.
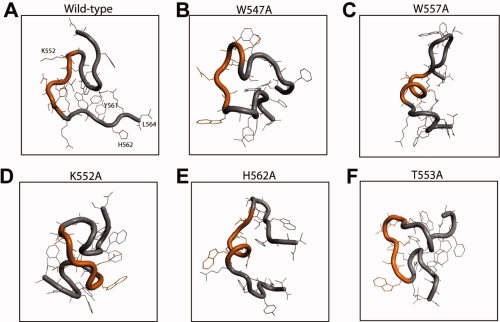
Lowest energy conformation of mutated 23 amino-acid peptides of Fz7 calculated by molecular modeling. (A) Wild-type Fz7#2, (B) W547A, (C) W557A, (D) K552A, (E) H562A, and (F) T553A mutants.
The CD analysis of single-residue mutants also indicated the involvement of Fz7 secondary structure in the interaction with Dvl PDZ [Fig. 3(B)]. The Fz7 W547A and W557A mutants, whose binding affinity to Dvl PDZ was markedly reduced, showed CD spectra very different from that of wild-type Fz: the 208 nm negative peak moved toward 200 nm, and the 222 nm negative peak decreased in intensity, indicating overall decrease in the secondary structure content of the peptide. The molecular modeling analysis of these two peptides also indicated conformational perturbation. Although wild-type Fz forms a hydrophobic cluster involving W547, the alanine residue in the W547A mutant extends outward from the cluster, thereby changing the conformation of the N-terminal region. This change apparently perturbs the conformation in which K552, T553, and L554 form a loop within the KTLQSW motif and thus is likely to affect PDZ binding [Fig. 4(B)]. The W557A mutation significantly altered the conformation of the KTLQSW motif resulting in a coiled structure [Fig. 4(C)]. In contrast, the T553A mutation barely affected the peptide's affinity and its CD spectrum was only slightly different from that of wild-type peptide [Fig. 3(B)]. In the molecular modeling, this mutation perturbs the conformation of the W557 side chain in the KTLQSW motif but not of the other regions [Fig. 4(F)].
The K552A and H562A mutants had the lowest binding affinity to Dvl PDZ. K552 lies at an optimal distance to form a hydrogen bond with Q555 in wild-type Fz [Fig. 4(A)]. When it is substituted with alanine, the conformation of the KTLQSW motif and the adjacent C-terminal region in the model is perturbed, probably by loss of this hydrogen bond. This global conformational change, together with possible accommodation changes of the K552 residue by the Dvl PDZ domain, may contribute to the reduced PDZ binding of the K552A mutant [Fig. 4(D)]. The H562A mutation dramatically changes the conformation of the KTLQSW motif, although H562 lies several residues away from this motif [Fig. 4(E)]. This effect may be understood by comparing the intramolecular interactions between the wild-type and H562A mutant peptides. H562 is preceded by the two bulky amino acids F560 and Y561. To avoid steric interactions, H562 forms a hydrogen bond with the backbone of L564, and F560 and Y561 form stacking interactions on the opposite side. W549 also forms a hydrophobic cluster with these residues, resulting in loop formation in the N-terminal region of the wild-type peptide [Fig. 4(A)]. The H562A mutation changes the orientations of these spanning amino acids, resulting in disruption of the central hydrophobic cluster. This disruption seems also to affect the N-terminal conformation by perturbing W549 and thereby affecting the global structure [Fig. 4(E)]. The H562A mutant has a coil in the KTLQSW motif, as does another weak binding mutant, W557A [Fig. 4(C)]. Taken together, these results indicate that specific binding of Fz7 to Dvl PDZ requires a specific conformation involving the KTLQSW motif and amino acids on either side of the motif.
Subtype specificity of the interaction between Fz and Dvl PDZ domains
We investigated whether Fz-Dvl interactions exhibit subtype specificity. Using the constraints revealed in the study with Fz7-#2 peptide described earlier, we generated homologous 23 amino-acid peptides corresponding to all 10 human Fz subtypes [Fig. 1(B)]. The affinity of each peptide to the PDZ domain of each human Dvl subtype was determined by displacement of biotinylated Fz7-#2 peptide from the respective PDZ domains in Alphascreen. The Fz subtypes with an affinity for PDZ afforded binding curves typical of one-site competition (examples are shown in Supporting Information Fig. 1); the IC50 values for the various Fz and Dvl subtypes are summarized in Table II. Fz1, Fz2, Fz3, Fz4, and Fz7 peptides were observed to interact specifically with Dvl PDZ. The other Fz peptides exhibited poor binding not substantially different from that of a random nonspecific peptide. To assess whether the binding affinity is different among the Dvl subtype, we determined the binding affinity of each Fz peptide against the PDZ domains of all three Dvl subtypes. The affinity of any tested Fz peptide deviated only slightly (Table II), suggesting that these sequences of Fz do not distinguish those Dvl subtypes. To investigate whether the binding and nonbinding Fz peptides differ in their specific conformations, we analyzed the peptides by molecular modeling (see Fig. 5). All modeled PDZ-binding peptides formed coils/loops at either side of a fairly extended KTXXXW motif, as does Fz7 [Figs. 4(A) and 5(A–C)]. A loop formation was commonly observed at the N-terminus that included part of the KTXXXW motif. Although this loop of PDZ-binding peptides all contained three to four residues of the KTXXXW motif and three to four residues preceding it, that of Fz5, Fz6, Fz8, and Fz9 were shorter with only two residues from the KTXXXW motif. The loop formed by Fz10 was much longer with the whole KTXXXW motif being involved. In addition, Fz6, Fz8, and Fz9 formed an additional loop within the KTXXXW region, destroying the extended conformation [Fig. 5(D–F)]. These data imply that the specific conformation of the extended KTXXXW motif with looped termini may be crucial for the specific interaction of Fz subtypes with Dvl PDZ.
Figure 5.
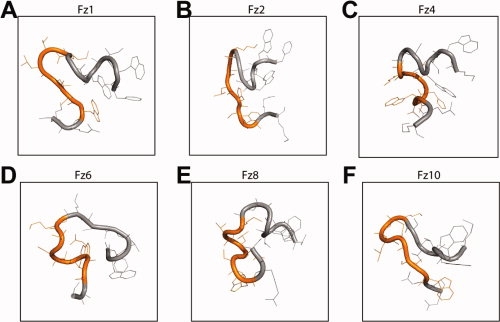
Comparison of the energy-minimized conformations of PDZ-binding and nonbinding Fz subtypes. The peptide regions shown are the 14 amino acids corresponding to the region F546 – R559 of Fz7. A–C: PDZ binding subtypes containing a looped N-terminal region followed by an extended KTXXXW motif (orange). D–F: Fz subtypes that do not bind to PDZ, containing an extended N-terminal region and/or a coiled KTXXXW motif.
Discussion
The first indication of the involvement of the C-terminal region of Fz in Wnt/β-catenin signaling was the importance of the KTXXXW motif of Fz3 in transcription of Wnt target genes in Xenopus embryos.16 The interaction partner of this motif was subsequently identified as the PDZ domain of Dvl, with a binding affinity of about 10 μM.20 Our results indicate that conserved and nonconserved amino acids other than KTXXXW in the Fz C-terminus are also essential for high-affinity interaction with Dvl PDZ.
In a previous study, the combined mutation of the three conserved residues in the Fz KTXXXW motif (Lys, Thr, and Trp) abolished the binding between mouse Dvl1 PDZ and the peptide GKTLQSWRRFYH (Fz7-#1), and single point mutations of Lys or Trp substantially reduced the binding affinity.20 Human Fz5 with a Lys-to-Ala point mutation did not coimmunoprecipitate with Dvl, whereas the wild-type Fz5 did.19 Our data confirmed that K552 and W557 of Fz7 are among the crucial residues within the longer peptide that interacts with higher affinity. However, the T553 residue did not conspicuously affect binding when changed to alanine, despite its conservation in the KTXXXW motif. The CD spectral data and molecular modeling suggest that it does not significantly affect the overall conformation of this motif except minor changes such as the direction of W557 side chain, but it may have another important role in Fz signaling function. Several studies have demonstrated that phosphorylation of a serine/threonine residue in PDZ domain ligand molecules can modulate their interaction with the PDZ domain, thereby altering their signaling function.32–35 The T553 residue of Fz may have a similar role.
Our findings indicate that other residues both N- and C-terminal to the KTXXXW motif are essential for Fz subtype-specific interaction with the Dvl PDZ domain. Binding was decreased by mutation of residues previously unknown to be crucial to Fz-Dvl PDZ interaction. Of particular interest is H562, which is not conserved among the Fz proteins but affects Fz7-Dvl PDZ binding even more than the fully conserved residues (Table I). This finding is consistent with a functional analysis showing that RRFYH (including the residue analogous to H562) C-terminal to the KTXXXW motif is required for Xenopus Fz3-mediated Wnt/β-catenin signaling.16
To evaluate the concordance between the modeled structure and the binding affinity, we have also calculated the RMSD between the wild-type and each alanine-mutated peptide (Supporting Information Table 1, column B) for the region that includes the KTXXXW motif and shows conformational similarity among the Dvl PDZ-binding peptides. Overall, we have observed a relationship between the RMSD and the Alphascreen affinity data (Supporting Information Fig. 3).
Our CD analysis revealed that the extended PDZ ligand region of Fz7 forms a stable secondary structure. Although the 23 amino-acid peptide containing regions both N- and C-terminal to the KTXXXW motif displayed an α/β mix conformation, peptides lacking either of these termini failed to induce the secondary structure and displayed much reduced binding affinity to Dvl PDZ. Secondary structure is known to play a role in the interaction of internal PDZ ligand region of nNOS, which is folded as a β-finger to bind to the syntrophin PDZ domain.25 In our molecular model of Fz7-#2 peptide, a loop is observed at the N-terminal end of the KTXXXW motif.
Subtype-specific functions of several Fz proteins have been demonstrated. For example, human Fz1, Fz5, and Fz9 and rat Fz7 induce Wnt/β-catenin signaling, whereas rat Fz2 and mouse Fz3, Fz4, and Fz6 induce Wnt/Ca2+ signaling.19,27,28,36 We analyzed the interaction between Dvl PDZ and peptides corresponding to the C-terminal region of each human Fz subtype, to identify whether Fz subtype-specific function can be correlated with Fz interaction with Dvl PDZ. Our data show that only peptides derived from Fz1, Fz2, Fz3, Fz4, and Fz7 directly bind to Dvl PDZ. Analysis of the molecular models implies that the lack of PDZ binding in Fz5, Fz6, Fz8, Fz9, and Fz10 peptides is likely to result from the difference in their conformation compared with that of PDZ-binding peptides, as described in Results in detail. The non-PDZ-binding Fz subtypes also display features seen in specific Fz7 mutants, such as changed loop formations or coils within the KTXXXW motif. It is possible that residues that are nonconserved but critical for Fz7-Dvl PDZ binding play a role in the different conformations and affinity of Fz subtypes. Although the residues in Fz1 (Asn), Fz2 (His), Fz3 (Phe), and Fz4 (His) corresponding to Fz7 Q555 are either uncharged or positively charged, those in Fz5 (Glu) and Fz8 (Glu) are negatively charged. The molecular models of these Fz subtypes indicate that these negatively charged residues result in hydrogen bonding interactions with distant amino acids, thereby making the overall conformation of Fz5 and Fz8 unlike that of Fz7. In the Fz9 and Fz10 peptides which have Q555, a different residue such as Y562 (Fz9) or S562 (Fz10) that replaces the H562 in Fz7 could be responsible for the structural change. Such multiple changes in the nonbinding Fz peptides may cooperate to change their global conformation.
The observation that some Fz subtypes (such as Fz5 and Fz9) failed to bind to Dvl PDZ via their C-terminus regardless of their involvement in Wnt/β-catenin signaling challenges the importance of their interaction with Dvl PDZ in Wnt/β-catenin signaling. Other factors such as unidentified molecules or other domains of Fz and Dvl may be involved in their interactions and function. For instance, a mutational analysis of rat Fz9 indicated that the C-terminal residues required for phosphorylation and membrane translocation of Dvl1 differ from those required for β-catenin stabilization and TCF transactivation.17 Mutations of Fz5 in the cytoplasmic regions other than the C-terminal tail have prevented coimmunoprecipitation of Fz5 with Dvl.19 We have attempted to determine the functional relevance of the Fz subtypes in Dvl binding, by using a TOPFlash luciferase reporter assay in HEK293T cells, that were titrated with cDNA encoding full-length Fz1, 4, 6, 7 and 9 in the presence or absence of cotransfection of Wnt3a cDNA. Although the cells robustly responded to cDNA encoding full-length Dvl (reporter signal increased by a factor >10; data not shown), no effect was observed in the Fz-titration. Thus, functional validation of our biochemical data may require studies in the Xenopus model16,20 or indirect analysis in other specific cells by inhibiting endogenously overexpressed Fz, as previously demonstrated in hepatocellular carcinoma.37
In summary, peptides derived from the C-terminal tail of Fz1, Fz2, Fz3, Fz4, and Fz7 interact directly and specifically with Dvl PDZ. These peptides form a secondary structure that appears to be important in the interaction. Specific point mutations in residues both N- and C-terminal to the KTXXXW motif are capable of changing the peptides' secondary structure and reducing their affinity for Dvl PDZ.
Experimental Procedures
Plasmids
The plasmids encoding the GST-tagged PDZ domains of hDvl1 and hDvl3 have been described.38 A plasmid encoding the GST-hDvl2 PDZ domain was generated by amplifying the PDZ coding region using hDvl2 cDNA as a template (5′-CCTTAAGGATCCCTCAATATCATCACAGTCAC-3′, forward primer and 5′-CCTTAACTCGAGTCTAGGGATCCCAGCACTTGGCCA-3′, reverse primer) and inserting the BamHI/EcoRI-digested PCR product into pGEX-6P2. Correct insertion was confirmed by sequencing.
Peptides
All peptides used in this study were synthesized by the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital. The peptides used for direct binding studies were biotinylated at the N-terminus, whereas those used for competition with biotinylated peptides contained no biotin.
Protein expression
Expression of the GST-PDZ domains of hDvl1 and hDvl3 has been described.38 We used a similar protocol to express hDvl2 GST-PDZ. Briefly, transfected BL21(DE3) cells were cultured in 2× Lauria Bertani medium and Dvl protein expression was induced by incubation overnight at 30°C with 0.15 mM isopropyl-l-thio-b-d-galactopyranoside. GST-tagged hDvl2 PDZ was purified from the pelleted cells by affinity chromatography in the St. Jude Protein Production Facility.
Alphascreen Assay
Alphascreen is a bead-based assay in a heterogeneous system. When a biochemical interaction brings the two beads together, a cascade of chemical reactions take place on generation of singlet oxygen, resulting in a greatly amplified signal. All binding assays in this study used the Alphascreen Glutathione-S-Transferase (GST) detection kit (PerkinElmer, Wellesley, MA), which consists of beads separately coated with anti-GST antibody and streptavidin. The optimum concentrations of biotin-tagged peptides to be used in subsequent competition assays were initially determined by titrating the biotin-tagged peptide with 100 nM hDvl3 GST-PDZ. For these dose-response assays, a solution of GST-Dvl PDZ in Alphascreen buffer (25 mM HEPES, 100 mM NaCl, 0.05% Tween 20, 1% bovine serum albumin, pH 6) was used to generate serial dilutions of each peptide in a 96-well plate. Fifteen microliters of each sample solution was transferred into a 384-well plate in triplicate and 5 μl of anti-GST acceptor beads were added to each well. After 30 min of incubation, streptavidin donor beads (5 μL per well) were added and plates were incubated for 45 min. The signal was measured with an EnVision 2103 Reader (PerkinElmer, Wellesley, MA). The binding curves were generated by GraphPad Prism software. The concentration of the biotin-Fz7-#2 peptide was 100 nM. All three GST-Dvl PDZ domains were used at 100 nM concentration. The relative affinity of different peptides for Dvl PDZ subtypes was determined by titrating the respective peptides against a solution containing a mixture of the biotin-Fz7-#2 peptide and each subtype of GST-Dvl PDZ in the Alphascreen buffer. The assay method was similar to that described earlier but had a competitor concentration range of 100 μM to 3 nM.
Circular dichroism
Circular dichroism (CD) spectra of peptides were recorded between 190 and 250 nm using an AVIV model 62 DS spectropolarimeter. All measurements were performed at 25°C in a quartz optical cell with a path length of 1 mm. Each data point in a CD spectrum was the average of five individual scans. CD measurements were performed in 10 mM phosphate buffer with 0.05% Tween 20 (pH 6), which yielded a spectrum similar to that in Alphascreen buffer but better resolved. CD analysis of Fz7-#2 was carried out at concentrations ranging from 20 to 80 μM to confirm that the observed spectral shapes were not the effect of homo-oligomerization. Fz7-N-fragment was less soluble than the other peptides and thus yielded very low signal strength.
Molecular modeling
Conformational searches were performed using the Monte-Carlo algorithm available in the MacroModel package (Schrödinger LLC, Portland, OR) with a 50 kcal/mol cutoff to generate 20,000 unique starting structures. Subsequent optimization of these structures was conducted using the OPLS2005 force field with an implicit solvation model, from which the 75 lowest energy structures were retained. The RMSD calculations were carried out using atoms of the entire peptide backbones. For most of the Fz mutants, this procedure yielded either a single set of related conformers or a small set of closely related conformers, as evidenced by the small average RMSD values shown in Supporting Information Table 1.
Acknowledgments
The authors thank Cynthia Jeffries and Andrew Lemoff for assistance in peptide purification, Dr. Richard Kriwacki for use of the CD spectrometer, Steve Otieno for technical assistance in CD measurements, the St. Jude Protein Production Facility for preparing GST-Dvl2 PDZ domain, and Sharon Naron for editorial advice.
Glossary
Abbreviations:
- CD
circular dichroism
- Dvl
Dishevelled
- Fz
Frizzled
- nNOS
neuronal nitric oxide synthase
- PDZ
PSD95/DLG/ZO1
- DIX
Dishevelled/Axin
- DEP
Dishevelled/EGL-10/Pleckstrin
- TCF
T cell factor
- LEF
Lymphoid enhancer factor
- ETA
Endothelin receptor A
- GST
Glutathione S Transferase
- RMSD
Root mean square deviation.
References
- 1.Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 2.Luo J, Chen J, Deng ZL, Luo X, Song WX, Sharff KA, Tang N, Haydon RC, Luu HH, He TC. Wnt signaling and human diseases: what are the therapeutic implications? Lab Invest. 2007;87:97–103. doi: 10.1038/labinvest.3700509. [DOI] [PubMed] [Google Scholar]
- 3.Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 4.Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of β-catenin-independent Wnt signaling. Dev Cell. 2003;5:367–377. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 5.Yang-Snyder J, Miller JR, Brown JD, Lai CJ, Moon RT. A frizzled homolog functions in a vertebrate Wnt signaling pathway. Curr Biol. 1996;6:1302–1306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- 6.Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407:535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- 7.Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 8.Wehrli M, Dougan ST, Caldwell K, O'Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S. Arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000;407:527–530. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Klein TJ, Mlodzik M. Subcellular localization of frizzled receptors, mediated by their cytoplasmic tails, regulates signaling pathway specificity. PLoS Biol. 2004;2:E158. doi: 10.1371/journal.pbio.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Axelrod JD, Miller JR, Shulman JM, Moon RT, Perrimon N. Differential recruitment of Dishevelled provides signaling specificity in the planar cell polarity and Wingless signaling pathways. Genes Dev. 1998;12:2610–2622. doi: 10.1101/gad.12.16.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boutros M, Paricio N, Strutt DI, Mlodzik M. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell. 1998;94:109–118. doi: 10.1016/s0092-8674(00)81226-x. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Yuan H, Xie W, Mao J, Caruso AM, McMahon A, Sussman DJ, Wu D. Dishevelled proteins lead to two signaling pathways. Regulation of LEF-1 and c-Jun N-terminal kinase in mammalian cells. J Biol Chem. 1999;274:129–134. doi: 10.1074/jbc.274.1.129. [DOI] [PubMed] [Google Scholar]
- 13.Capelluto DG, Kutateladze TG, Habas R, Finkielstein CV, He X, Overduin M. The DIX domain targets dishevelled to actin stress fibres and vesicular membranes. Nature. 2002;419:726–729. doi: 10.1038/nature01056. [DOI] [PubMed] [Google Scholar]
- 14.Wallingford JB, Habas R. The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development. 2005;132:4421–4436. doi: 10.1242/dev.02068. [DOI] [PubMed] [Google Scholar]
- 15.Wong HC, Mao J, Nguyen JT, Srinivas S, Zhang W, Liu B, Li L, Wu D, Zheng J. Structural basis of the recognition of the dishevelled DEP domain in the Wnt signaling pathway. Nat Struct Biol. 2000;7:1178–1184. doi: 10.1038/82047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umbhauer M, Djiane A, Goisset C, Penzo-Mendez A, Riou JF, Boucaut JC, Shi DL. The C-terminal cytoplasmic Lys-thr-X-X-X-Trp motif in frizzled receptors mediates Wnt/β-catenin signalling. EMBO J. 2000;19:4944–4954. doi: 10.1093/emboj/19.18.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karasawa T, Yokokura H, Kitajewski J, Lombroso PJ. Frizzled-9 is activated by Wnt-2 and functions in Wnt/β-catenin signaling. J Biol Chem. 2002;277:37479–37486. doi: 10.1074/jbc.M205658200. [DOI] [PubMed] [Google Scholar]
- 18.Chen W, ten Berge D, Brown J, Ahn S, Hu LA, Miller WE, Caron MG, Barak LS, Nusse R, Lefkowitz RJ. Dishevelled 2 recruits β-arrestin 2 to mediate Wnt5A-stimulated endocytosis of Frizzled 4. Science. 2003;301:1391–1394. doi: 10.1126/science.1082808. [DOI] [PubMed] [Google Scholar]
- 19.Cong F, Schweizer L, Varmus H. Wnt signals across the plasma membrane to activate the beta-catenin pathway by forming oligomers containing its receptors, Frizzled and LRP. Development. 2004;131:5103–5115. doi: 10.1242/dev.01318. [DOI] [PubMed] [Google Scholar]
- 20.Wong HC, Bourdelas A, Krauss A, Lee HJ, Shao Y, Wu D, Mlodzik M, Shi DL, Zheng J. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol Cell. 2003;12:1251–1260. doi: 10.1016/s1097-2765(03)00427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung AY, Sheng M. PDZ domains: structural modules for protein complex assembly. J Biol Chem. 2002;277:5699–5702. doi: 10.1074/jbc.R100065200. [DOI] [PubMed] [Google Scholar]
- 22.Cuppen E, Gerrits H, Pepers B, Wieringa B, Hendriks W. PDZ motifs in PTP-BL and RIL bind to internal protein segments in the LIM domain protein RIL. Mol Biol Cell. 1998;9:671–683. doi: 10.1091/mbc.9.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Huizen R, Miller K, Chen DM, Li Y, Lai ZC, Raab RW, Stark WS, Shortridge RD, Li M. Two distantly positioned PDZ domains mediate multivalent INAD-phospholipase C interactions essential for G protein-coupled signaling. EMBO J. 1998;17:2285–2297. doi: 10.1093/emboj/17.8.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu XZ, Choudhury A, Li X, Montell C. Coordination of an array of signaling proteins through homo- and heteromeric interactions between PDZ domains and target proteins. J Cell Biol. 1998;142:545–555. doi: 10.1083/jcb.142.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science. 1999;284:812–815. [PubMed] [Google Scholar]
- 26.Paasche JD, Attramadal T, Kristiansen K, Oksvold MP, Johansen HK, Huitfeldt HS, Dahl SG, Attramadal H. Subtype-specific sorting of the ETA endothelin receptor by a novel endocytic recycling signal for G protein-coupled receptors. Mol Pharmacol. 2005;67:1581–1590. doi: 10.1124/mol.104.007013. [DOI] [PubMed] [Google Scholar]
- 27.Gazit A, Yaniv A, Bafico A, Pramila T, Igarashi M, Kitajewski J, Aaronson SA. Human frizzled 1 interacts with transforming Wnts to transduce a TCF dependent transcriptional response. Oncogene. 1999;18:5959–5966. doi: 10.1038/sj.onc.1202985. [DOI] [PubMed] [Google Scholar]
- 28.Sheldahl LC, Park M, Malbon CC, Moon RT. Protein kinase C is differentially stimulated by Wnt and Frizzled homologs in a G-protein-dependent manner. Curr Biol. 1999;9:695–698. doi: 10.1016/s0960-9822(99)80310-8. [DOI] [PubMed] [Google Scholar]
- 29.Golan T, Yaniv A, Bafico A, Liu G, Gazit A. The human Frizzled 6 (HFz6) acts as a negative regulator of the canonical Wnt. β-catenin signaling cascade. J Biol Chem. 2004;279:14879–14888. doi: 10.1074/jbc.M306421200. [DOI] [PubMed] [Google Scholar]
- 30.Wilson J, Rossi CP, Carboni S, Fremaux C, Perrin D, Soto C, Kosco-Vilbois M, Scheer A. A homogeneous 384-well high-throughput binding assay for a TNF receptor using alphascreen technology. J Biomol Screen. 2003;8:522–532. doi: 10.1177/1087057103257804. [DOI] [PubMed] [Google Scholar]
- 31.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 32.Cao TT, Deacon HW, Reczek D, Bretscher A, von Zastrow M. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the β2-adrenergic receptor. Nature. 1999;401:286–290. doi: 10.1038/45816. [DOI] [PubMed] [Google Scholar]
- 33.Chung HJ, Xia J, Scannevin RH, Zhang X, Huganir RL. Phosphorylation of the AMPA receptor subunit GluR2 differentially regulates its interaction with PDZ domain-containing proteins. J Neurosci. 2000;20:7258–7267. doi: 10.1523/JNEUROSCI.20-19-07258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naren AP, Cobb B, Li C, Roy K, Nelson D, Heda GD, Liao J, Kirk KL, Sorscher EJ, Hanrahan J, Clancy JP. A macromolecular complex of beta 2 adrenergic receptor, CFTR, and ezrin/radixin/moesin-binding phosphoprotein 50 is regulated by PKA. Proc Natl Acad Sci USA. 2003;100:342–346. doi: 10.1073/pnas.0135434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung HJ, Huang YH, Lau LF, Huganir RL. Regulation of the NMDA receptor complex and trafficking by activity-dependent phosphorylation of the NR2B subunit PDZ ligand. J Neurosci. 2004;24:10248–10259. doi: 10.1523/JNEUROSCI.0546-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winn RA, Marek L, Han SY, Rodriguez K, Rodriguez N, Hammond M, Van Scoyk M, Acosta H, Mirus J, Barry N, Bren-Mattison Y, Van Raay TJ, Nemenoff RA, Heasley LE. Restoration of Wnt-7a expression reverses non-small cell lung cancer cellular transformation through frizzled-9-mediated growth inhibition and promotion of cell differentiation. J Biol Chem. 2005;280:19625–19634. doi: 10.1074/jbc.M409392200. [DOI] [PubMed] [Google Scholar]
- 37.Kim M, Lee HC, Tsedensodnom O, Hartley R, Lim YS, Yu E, Merle P, Wands JR. Functional interaction between Wnt3 and Frizzled-7 leads to activation of the Wnt/β-catenin signaling pathway in hepatocellular carcinoma cells. J Hepatol. 2008;48:780–791. doi: 10.1016/j.jhep.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujii N, You L, Xu Z, Uematsu K, Shan J, He B, Mikami I, Edmondson LR, Neale G, Zheng J, Guy RK, Jablons DM. An antagonist of dishevelled protein-protein interaction suppresses b-catenin-dependent tumor cell growth. Cancer Res. 2007;67:573–579. doi: 10.1158/0008-5472.CAN-06-2726. [DOI] [PubMed] [Google Scholar]


