Abstract
Enzymes utilize substrate binding energy both to promote ground state association and to selectively lower the energy of the reaction transition state.i The monomeric homing endonuclease I-AniI cleaves with high sequence specificity in the center of a 20 base-pair DNA target site, with the N-terminal domain of the enzyme making extensive binding interactions with the left (−) side of the target site and the similarly structured C-terminal domain interacting with the right (+) side.ii Despite the approximate two-fold symmetry of the enzyme-DNA complex, we find that there is almost complete segregation of interactions responsible for substrate binding to the (−) side of the interface and interactions responsible for transition state stabilization to the (+) side. While single base-pair substitutions throughout the entire DNA target site reduce catalytic efficiency, mutations in the (−) DNA half-site almost exclusively increase KD and KM*, and those in the (+) half-site primarily decrease kcat*. The reduction of activity produced by mutations on the (−) side, but not mutations on the (+) side, can be suppressed by tethering the substrate to the endonuclease displayed on the surface of yeast. This dramatic asymmetry in the utilization of enzyme-substrate binding energy for catalysis has direct relevance to the redesign of endonucleases to cleave genomic target sites for gene therapy and other applications. Computationally redesigned enzymes that achieve new specificities on the (−) side do so by modulating KM*, while redesigns with altered specificities on the (+) side modulate kcat*. Our results illustrate how classical enzymology and modern protein design can each inform the other.
Enzymes utilize interactions with the substrate to promote catalysis both by bringing the substrate into close proximity and proper alignment with catalytic groups on the enzyme and by selectively stabilizing the transition state for the chemical reaction.iii,iv,v Dissection of the contributions to enzyme catalysis has taken on renewed importance with the advent of computational and directed evolution approaches for engineering novel enzymatic activities for applications ranging from synthetic chemistry to therapeutics.vi,vii Reprogramming the specificity of the LAGLIDADG family of homing endonucleases for genome engineering and biotechnology purposes is one such application.viii,ix
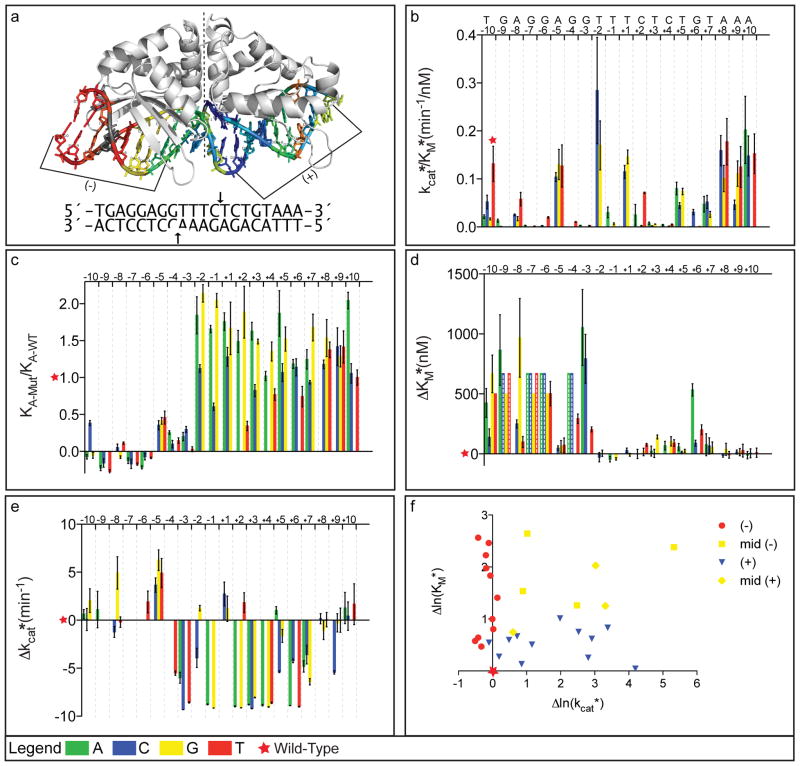
Control experiments probing the binding specificity of the I-AniI homing endonuclease, in preparation for computational redesign of specificity, revealed a striking asymmetry in the effect of base substitutions on binding affinity (Fig. 1a). DNA cleavage and DNA binding by Y2 I-AniI endonucleasex were assayed for 60 different target sites, each containing a single base-pair substitution from the wild type recognition sequence. Consistent with previous observations,xi enzyme activity assays showed that many nucleotide substitutions throughout the extended 20 base-pair recognition site abrogated or reduced cleavage, reflecting the high sequence specificity of the endonuclease (Fig. 1b). Fluorescence binding experiments showed that for mutations between −10 and −3 on the (−) side of the interface, this loss of cleavage activity is associated with a loss of binding affinity. In sharp contrast, mutations in the −2 to +10 region of the recognition site, which also eliminated or reduced cleavage, had a minimal affect on substrate binding (Fig. 1c).
Figure 1. Segregation of contributions to binding and catalysis.
The color scheme throughout the figure is A=green, C=blue, G=yellow, T=red, and error bars in all panels are standard errors from the mean (SEM).
a. Ribbon diagram of the I-AniI enzyme in complex with the wild-type target site (2QOJxi). Target site and positions of DNA cleavage are shown below: (−) side cleavage site is cut prior to (+) side site.x
b. kcat*/KM* values for the wild-type target site (red star) and each of the 60 singly-substituted target sites (vertical bars). Substitutions throughout the length of the target site abrogate enzyme activity demonstrating the high sequence specificity of the enzyme.
c. Relative binding affinities determined for each singly-substituted target site using fluorescence competition assays. Substitutions on the left side, but not the right side, significantly reduce binding affinity.
d. KM* values for each singly-substituted target site relative to the wild-type. As in c, substitutions on the left but not the right display significantly different values from wild-type.
e. kcat* values for each singly-substituted target site relative to the wild-type site. In contrast to c and d, substitutions between positions −4 and +9 have significant effects. Substitutions for which KM* was too high (> 750nM) to allow separate determination of kcat* and KM* are indicated by bars with dashed lines in d, and are left blank in e.
f. Asymmetry of the contributions to kcat* and KM*. Positions shown in red are on the left (−) side of the target site from −10 to −5 and almost exclusively contribute to KM*. Positions shown in blue are on the right (+) side of the target site from positions +3 to +7. The boundary positions, −4, −3, and +6, contribute to both kcat* and KM* and are shown in yellow. To portray the structural context of these positions, the target site in a) is colored based on the effect of the mutation on kcat*, normalized by the sum of the effects on kcat* and KM* ([|Δln(kcat*)|/|(Δln(KM*)|+ |Δln(kcat*)|)] close to 1.0, blue; close to 0.0, red; intermediate, yellow; position where KM* and kcat* could not be separately determined; grey).
To determine whether the differences between the (−) and (+) side substitutions reflected differential contributions to ground state association versus transition state stabilization, the extent of cleavage of a linear double-stranded template as a function of time was determined for all 60 singly-substituted sites under single-turnover conditions, and pseudo-Michaelis-Menten parametersxii KM* and kcat* were obtained from these data (Fig. S1–3). Comparison of the kcat*/KM* for related substrates highlights the high sequence specificity of the enzyme: for example, at position −4 the kcat*/KM* for the wild-type G:C base-pair is >2000-fold greater than for A:T and >400-fold greater than for C:G (Fig. 1b and Table S1; since specificity is determined by the differences in kcat*/KM* for different substrates,xiii these results provide perhaps the most rigorous quantification of homing endonuclease specificity to date). The contribution of target site interactions to ground state stabilization (KM*, Fig. 1d) versus transition state stabilization (kcat*, Fig. 1e) was found to be skewed: substitutions on the (−) side increased KM* significantly without reducing kcat*, while substitutions on the (+) side decreased kcat* with little effect on KM*. The overall segregation of the kinetic contributions to specificity is shown graphically in Figure 1f and in the structural schematic in Figure 1a: most single base substitutions in the target affect kcat* (blue, (+) side) or KM* (red, (−) side) but not both. The striking feature of our results is that the apparent symmetry of the binding interface is completely broken during catalysis – chemically very similar protein-DNA contacts are utilized for substrate association on the left side and selective transition state stabilization on the right side.
Influence of substrate tethering on catalysis
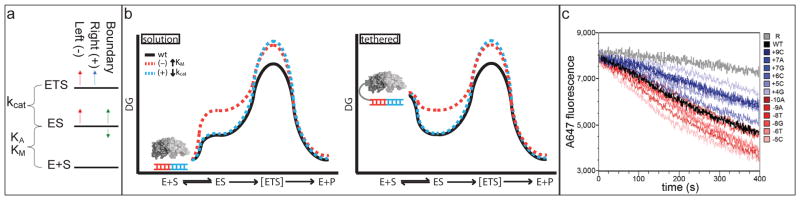
Surface display methods are widely used for the engineering of proteins with new binding specificities.xiv The sequence specificity profile obtained for singly-substituted target sites binding to I-AniI displayed on the surface of yeast closely parallels the profile observed in the solution fluorescence experiments (Fig. 1c) (JJ, BLS, and AMS, submitted). We reasoned that cleavage of mutated target sites with increased KM* should be suppressible by tethering the DNA duplex containing the target site adjacent to the displayed enzyme on the yeast surface; the increase in local substrate concentration should compensate for the decrease in ground state binding affinity (Fig. 2b). Indeed, mutations between positions −10 and −3 (red) that greatly reduced binding in solution had little effect in tethered cleavage experiments on the yeast cell surface. In contrast, substitutions on the right side of the target site (blue) that reduced cleavage in solution also reduced enzyme activity in the tethered cleavage assay, consistent with their reduction of kcat* (Fig. 2c). Substitutions that disrupt interactions involved in selective transition state stabilization cannot be overcome by increasing the local concentration of substrate.
Figure 2. Contributions to catalysis.
a. Free energy diagram showing the effect of target site substitutions on the free energies of substrate binding and transition state stabilization. The majority of substitutions on the left side increase the energy of both ES and ETS, suggesting they disrupt interactions made in both states. The majority of the positions on the right side instead raise ETS, suggesting they remove interactions present in ETS but not ES. A small subset of positions (labeled boundary) appears to selectively stabilize or destabilize ES while not affecting ETS; these substitutions may disrupt interactions present in ES but not in ETS.
b. Free energy profiles for a free (left) and tethered (right) system. Red profile: substitutions that remove interactions present in both ES and ETS; blue line, substitutions that remove interactions present in ETS but not ES. Tethering increases the free energy of free E + S to the point that the rate depends only on the free energy difference between ES and ETS. Since this free energy difference is unchanged by substitutions that remove interactions made in both ES and ETS (red profile), they do not affect the rate in the tethered case.
c. Yeast on-cell cleavage assays. Surface displayed enzyme cleaves a tethered fluorescently labeled oligo, which then diffuses away from the yeast surface resulting in loss of fluorescence. Black, wild-type target site; random DNA, grey; shades of red, left side target site substitutions; shades of blue, right side target site substitutions. Tethering suppresses decreases in cleavage rate produced by (−) side but not (+) side mutations.
Assuming the simple free energy diagram in Figure 2a, we can make inferences from the kinetic data in solution and on the yeast surface about the structures of the Michaelis and transition state complexes. Sidechain-base-pair interactions from positions −10 to −5 are present in both the Michaelis complex and the transition state (base substitutions increase KM* and KD in solution and do not affect the rate when tethered). Sequence specific base- pair interactions from +3 to +8 are formed only in the transition state (substitutions have no effect on KM* or KD, reduce kcat*, and slow the rate when tethered). A third class of interactions (at −5 and +7 for example) appear to be formed in the Michaelis complex but not the transition state (substitutions increase or decrease both kcat* and KM*/KD).
Importantly for the design calculations described in the next section, three observations suggest that the crystal structure of the complex likely resembles the transition state more than the Michaelis complex: (1) specific interactions on the (+) side of the DNA target present in the crystal structure appear to be formed in the transition state but not the Michaelis complex, (2) the third class of substitutions mentioned above that appear to stabilize only the Michaelis complex make few interactions in the crystal structure (Fig. S4), and (3) Rosetta specificity calculations based on the crystal structure correlate better with catalytic efficiency than with binding affinity (Fig. S5).
Exploitation of binding energy in engineered enzymes
Monomeric LAGLIDADG homing endonucleases, which recognize non-palindromic targets, are attractive scaffolds for genome engineering applicationsxv. An important challenge is to reprogram the substrate specificity of these enzymes towards desired target sequences.ix To redesign I-AniI specificity using Rosetta,xvi the target site in the crystal structure of the I-AniI protein-DNA complex is mutated in silico and the program searches for combinations of amino acid substitutions that allow the formation of energetically favorable interactions with the new base-pairs, but not with the wild type base-pairs.xvii Design calculations were carried out for six target site variants bearing single base pair substitutions, genes encoding the amino acid sequences of eight redesigned enzymes were constructed, and the enzymes were purified. DNA cleavage assays revealed that the designed specificity changes were for the most part achieved (Table S2). Our results demonstrate that I-AniI cleavage specificity can be reprogrammed by computational protein design, thereby providing starting points for the larger scale specificity changes required to cleave physiological target sites.
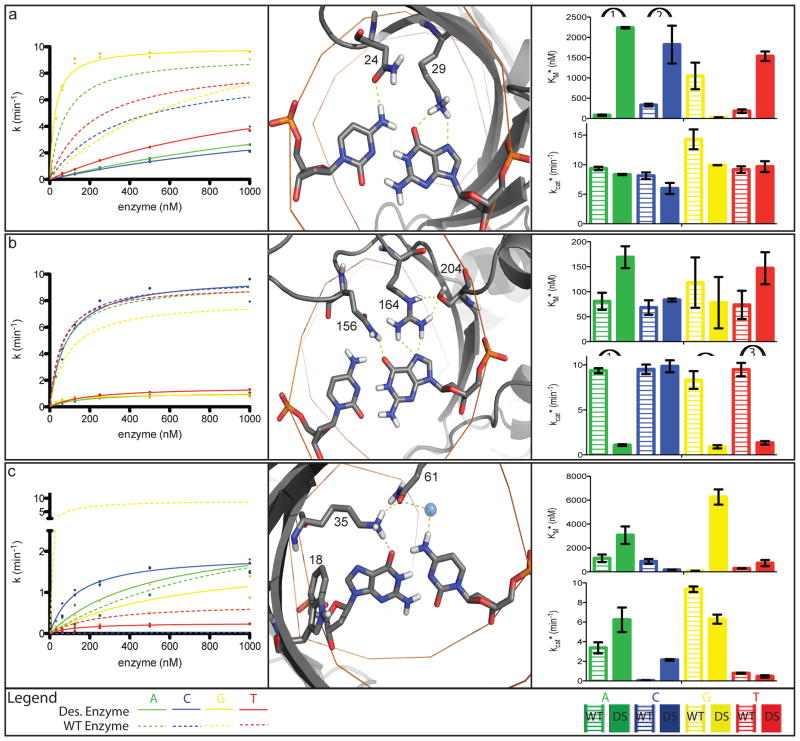
An enzyme redesigned for a new target site could achieve altered specificity either by changing kcat*, changing KM* or changing both. To determine whether the designed changes in specificity were a result of changes in KM* or kcat*, for each of eight designed endonucleases we measured the single-turnover cleavage kinetics for target substrates containing each of the four possible base-pairs at the redesign position (Table S2). A design aimed at specific recognition of a DNA target site containing base-pair −8G:C (Fig. 3a) achieved specificity exclusively by modulating KM*: the KM* decreased for the G:C, and increased for the A:T, T:A, and C:G. In contrast, a design aimed at specific recognition of +8C:G (Fig. 3b) achieved specificity entirely through kcat*: kcat* decreased for A:T, G:C, and T:A, but was unchanged for +8C:G. Both of these designed enzymes have high specificity at neighboring base-pairs, and overall specificities that are higher than the wild-type enzyme in the targeted regions (Fig. S6). A design aimed at specific recognition of the −3C:G substitution (Fig. 3c), at the boundary between KM* and kcat* influencing positions (Fig. 1), displayed changes in both kcat* and KM*, consistent with the results with the wild type enzyme at this position. These trends hold for the remaining designs as well (Table S2 and Figure S7): we find generally that the left side designs achieve specificity primarily by modulating ground state binding affinity, while the right side designs achieve specificity by modulating the free energy of the transition state.
Figure 3. Computational redesign of specificity.
The color scheme throughout the figure is A=green, C=blue, G=yellow, T=red, and error bars in right panels are standard errors from the mean (SEM).
a. Design for −8A:T to −8G:C substitution (K24N, T29K). (Middle panel) The designed residues N24 and K29 make direct hydrogen bonds to −8G and −8C respectively. (Left panel) The concentration dependence of the cleavage activity for the designed enzyme (solid lines) for different base-pairs at the −8 position differs considerably from the wild type enzyme (dashed lines). (Right panel) The kcat* values remain approximately the same for both the wild-type and designed enzymes against all target sites, but the KM* values are decreased for the target G base-pair (arrow 3) and increased significantly for the other three substitutions (arrows 1, 2, and 4).
b. Design for +8A:T to +8C:G substitution (L156Q, I164R, T204S). (Middle panel) Designed residues R164 and Q156 make direct hydrogen bonds to +8G. Designed residue S204 holds R164 in position. The kinetic traces (left panel) and bar graphs (right panel) show this design achieves altered specificity through changing kcat*. The KM* values remain approximately the same for both the wild-type and designed enzymes against all target sites, but the kcat* values are significantly decreased for all of the competitor target sites (arrows 1, 2, and 3).
c. Design for −3G:C to −3C:G substitution (Y18W, E35K, R61Q). (Middle panel) Designed residues K35 and Q61 make a direct hydrogen bond to −3G and a water-mediated hydrogen bond to −3C, respectively. Q61 and K35 also hydrogen bond with each other, and designed residue W18 further helps position K35 through packing interactions. The kinetic traces (left panel) and bar graphs (right panel) show this design achieves altered specificity through changing both kcat* and KM*. The designed enzyme has an increased kcat* (arrow 2) and decreased KM* for the −3C (arrow 1).
Functional advantage of asymmetry
Our results suggest that initial binding of I-AniI to its target site involves formation of base-specific interactions on the (−) side and lower affinity non-specific interactions on the (+) side to form the Michaelis complex (the latter are suggested by yeast display experiments which show that the enzyme binds less tightly to the (−) half-site than to the full site (JJ, BLS, and AMS, submitted)). Catalysis then requires bending of the DNA (note bend in Fig. 1a), which is stabilized at the transition state by newly formed specific interactions between the (+) side and the enzyme. Such a two-stage mechanism (see supplementary material section C) may be a general solution to the problem of specific target site recognition by enzymes that act on distorted DNA substrates. If the enzyme only bound to the distorted site, binding would require enzyme to be at the site (which may occur only once in the genome) simultaneous with fluctuation of the DNA into the distorted conformation; since both are rare events the net rate of binding, the product of two small numbers, would be very slow. If, instead, the enzyme can bind with some sequence specificity to undistorted target sites, the probability of being close enough to capture (and perhaps promote) fluctuations that distort the DNA will be very much higher. In I-AniI the total transition state binding energy appears to be roughly divided between the two steps: the N-terminal domain guides the enzyme to potential target sites which match on the (−) side, and the C-terminal domain specifically stabilizes the transition state if there is also a match on the (+) side.
Implications for enzyme engineering
There is considerable synergy between classical enzymology and modern computational design. Design should be informed by detailed analyses of the wild-type enzyme since, depending on the enzyme and substrate concentrations in the application the designed enzymes are to be used for, it may be necessary to reengineer KM*, kcat*, and/or kcat*/KM*. Conversely, computational design can provide insight into the basis for transition state stabilization. Our kinetic dissection of I-AniI cleavage activity also has implications for endonuclease re-engineering using yeast display: selection based on binding may be sub-optimal because substrate binding could be optimized at the expense of transition state stabilization, while selection for cleavage in the tethered substrate system could yield variants with decreased solution cleavage due to increased KM*. These pitfalls could potentially be overcome by selecting both for kcat* and KM*, perhaps by alternating between the two selection procedures. More generally, the union of classical enzymology with modern computational design and selection technology, as illustrated here, provides a powerful approach to revealing the mechanistic basis for, and subsequently reprogramming, sequence dependent molecular recognition.
Methods Summary
Experimental preparation and kinetic analysis
I-AniI was expressed in Escheria coli BL21(DE3) using a standard auto-induction protocol and purified over a His-trap column. Linearized plasmid substrates were prepared for each of the 60 singly-substituted target sites. Kinetic assays were carried out over a 20-fold range of enzyme (concentrations from 30 nM to 1500 nM enzyme, depending on the substrate) with 5 nM DNA substrate, and analyzed by agarose gel electrophoresis followed by integration of product and substrate band densities. The velocity versus enzyme concentration profiles were determined 2–4 independent times; reported kcat* and KM* values are the average of values determined from the independent experiments. Fluorescence competition binding assays were carried out as previously described.xviii
Computational Design
New target sequences were mapped on to the I-AniI-DNA crystal structure (2QOJxi) and the Rosetta computational design methodology was used to optimize the amino acid sequence of the protein to maximize affinity for the new site.xvi The predicted specificity of the resulting protein models for the desired target sequence was computed using Rosetta, and designs that were predicted to bind tightly and specifically were subjected to further optimization using flexible backbone protein design (supplementary methods). The tightest binding and most specific designs were again selected, and the designed amino acid substitutions were removed one at a time. If no significant loss was predicted in either specificity or binding energy, the substitution was removed from the design. The “−8G:C_A” (K24N/T29K) and “−8G:C_B” (K24N/T29Q) designs were generated instead using a genetic algorithm to simultaneously optimize binding affinity and specificity (supplementary methods). Genes encoding the designed proteins were assembled from oligonucleotides, and the designed proteins were expressed, purified, and assayed as described above.
Tethered cleavage on yeast surface
PCR-generated DNA substrates, labeled with biotin and Alexa- 647, were tethered via an antibody-streptavidin-PE bridge to the HA epitope of I-AniI expressed on the surface of S. cerevisieae in conditions which prohibit catalysis. Samples were then spiked with 10 mM MgCl2 and placed in a pre-warmed 37°C chamber and fluorescence measurements were acquired on a flow cytometer. The Alexa 647 signal from a PE-normalized population of each sample was then plotted versus time to generate the curves shown.
Methods
Protein expression and purification
Genes encoding Y2 I-AniIxix,x designs were assembled from oligonucleotidesxx, cloned into a variant of the pet15 expression vector, and sequence-verified plasmids were transformed into BL21 Star (Invitrogen). A one litre culture of auto-induction mediaxxi was inoculated with several colonies, grown at 37°C for ca. 12 hours (to approximately saturation), and expression at 18°C was continued for ca. 24 hours. Cells were harvested, resuspended in Tris 20 mM pH 7.5, 1.0 M NaCl, and 30 mM Imidazole, lysed by sonication and lysozyme. The soluble fraction was loaded onto a 1 mL HisTrap FF crude column (GE Healthcare) and I-AniI variants were purified by Imidazole gradient elution on an AKTA express (GE Healthcare). The proteins were concentrated and the buffer was exchanged to Tris 20 mM pH 7.5, 500 mM NaCl, and 50% (v/v) glycerol for storage. Purity of the proteins was assessed by SDS-PAGE gel and the concentration of samples with ca. > 95% purity was determined by measuring the absorbance at 280 nm using the calculated extinction coefficientxxii. The concentration of enzyme in the < 95% pure samples was determined by generating a standard curve of with a pure I-AniI protein, correlating protein concentration with band density (calculated with ImageJxxiii), and comparing the band density of the I-AniI protein in impure samples run on the same gel as the standard curve.
Plasmid substrate construction
All single base-pair variants from the wild-type target site in pBluescript were individually constructed by site-directed mutagenesis as describedxxiv. Sequence-verified plasmids were linearized with ScaI prior to the kinetic assays to facilitate product identification.
Endonuclease activity assays
Kinetic assays
Previous workxxv confirms that I-AniI, similar to other LAGLIDADG endonucleases, is a single-turnover enzyme, and the conditions for single-turnover kineticsxii were met in all experiments. The ionic strength of the enzyme reaction buffer was optimized for enzyme activity and stability to a final solution of 170 mM KCl, 10 mM MgCl2, and 20 mM Tris pH 9.0. Enzyme was diluted in 1.25X reaction buffer to working concentrations, serial two-fold dilutions were made, and both substrate plasmid and diluted enzyme were incubated separately at 37°C for 1 minute. The appropriate amount of plasmid (1/5 of the reaction volume) was added to each reaction for a final 1X reaction buffer and final plasmid concentration of ca. 5 nM (lowest concentration still readily visible on agarose gel). The plasmid (1/5 of reaction volume) was added to the enzyme (4/5 of reaction volume) to minimize heat loss during the transfer (found to add significant noise to the data). Reactions were halted with 200 mM EDTA, 30 % glycerol, and bromophenol blue. DNA fragments were separated on 1.2% agarose TBE gels, which were then stained in a standard ethidium bromide solution and subsequently destained in water for maximum contrast between DNA and background. All data was collected by integrating the density of the substrate (2,959 bp) and product bands (1,801 bp and 1,158 bp) using ImageJxxiii. The percent product formed is equal to the sum of the density of the two product bands divided by the total sum of the densities of the 3 bands. The progress curves fit to single exponentials for all enzyme concentrations (Fig. S1) and for all target sites except for several substitutions in the central four base-pairs between the cleavage sites on the two DNA strands (Fig. S2).
Assays for specificity positions adjacent to designed nucleotide
Two-fold serial dilutions of enzyme from 1500 nM to 11 nM were made in 1.25X reaction buffer and the enzyme was reacted with ca. 5 nM substrate (in 1X reaction buffer) for a ½ hour at 37°C. Reactions were halted and data was analyzed as described above in the “kinetic assays” section.
Fluorescence competition binding assayxviii
Unlabeled DNA oligonucleotides with each of the 60 single base-pair substitutions in the I-AniI target site (wild-type I-AniI site, 5′-TGAGGAGGTTTCTCTGTAAG-3′), a negative control sequence (5′-CTCTTCTTGCATATATCTCC-3′), an unlabeled wild-type site oligo, and a wild-type site oligonucleotide labeled with 5′ Cy3, were synthesized with six consecutive “A” flanking on each end (Integrated DNA Technology, 100-nmole scale, salt-free). Complementary oligonucleotides were ordered for all 63 sites and double stranded target DNA was preparing by annealing equal amounts of complementary strands.
His-tagged I-AniI was immobilized by incubating 200μl of 100 nM I-AniI in TBS/BSA buffer (50mM Tris-HCl (pH 7.5), 150mM NaCl, 0.2% BSA) in wells of Nickel-NTA coated HisSorb plates (Qiagen) for 2 hours at room temperature. Unbound protein was removed and the plates were washed four times with TBS/Tween-20 (50mM Tris-HCl (pH 7.5), 150mM NaCl, 0.05%Tween-20). The immobilized I-AniI in the microtiter plate was incubated for ca. four hours with both 100 nM labeled target DNA duplex and 3 μM (30-fold excess) of one unlabeled duplex per well in 200 μl of binding buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.02 mg/ml poly(dI-dC), 10mM CaCl2). The plates were washed four times with TBS (50mM Tris-HCl (pH 7.5), 150mM NaCl), and the fluorescent signal retained in each well was quantified using a SpectraMax M5/M5e micro-plate reader (Molecular Devices) (excitation: 510nm, emission: 565nm, cutoff: 550 nm). Additional negative control experiments performed in the absence of the enzyme indicated that no significant detectable fluorescent signal was retained after the protocol described above was completed. Relative binding affinities were calculated using the following equation:
where F(x), F(t), and F(n) indicate fluorescent intensities obtained from wells in which the immobilized protein was incubated with the unlabeled singly-substituted target sites, wild-type target site, and negative control sequence, respectively.
Tethered yeast display
Surface display of I-AniI on S. cerevisiae was performed using standard methodsxiv. For each sample, 5 × 106 cells were stained with biotinylated anti-HA11, followed by secondary staining with streptavidin-DNA substrate conjugates (1:3 molar ratio) on ice and in the absence of divalent cations. DNA substrates were generated by PCR using biotinylated and Alexa 647-conjugated primers complementary to the 5′ and 3′ sequences flanking the I-AniI target (or indicated target site variants). PCR products were purified by Exo1 digestion followed by size exclusion chromatography on a G-100 column (GE Healthcare) prior to conjugation with streptavidin-PE. Samples were then spiked with 10 mM MgCl2 and placed in a pre-warmed 37°C chamber and acquired at an approximate event rate of 3000/second for 400 seconds on a BD FACSAria II flow cytometer. Processing was performed using FloJo software (Treestar, Inc.). Briefly, live cells were gated by forward and side scatter properties, and doublets and clumped cells were excluded on the basis of forward scatter area versus height linearity. The Alexa 647 signal from a PE-normalized population of each sample was then plotted versus time to generate the curves shown.
Computational design methods
Single-state design (designs −9C, −8G_C, −6C, +5C, and +8C)
The computational design of homing endonuclease-DNA specificity was performed using the Rosetta design software in a manner that is specifically designed to predict new protein sequences that will bind with high affinity to novel DNA sequences.xxvi The prediction of designed proteins with novel interactions to substituted base-pairs in the I-AniI recognition sequence was performed by mutation and Monte-Carlo repacking of amino acid sidechains as described in Ashworth et al. 2006xvii. The template for the design calculations was the crystal structure of the I-AniI-DNA complex (pdb code 2QOJxi). Additionally, minor shifts of the protein backbone were modeled only in the vicinity of the designed region using a loop-rebuilding algorithmxxvii,xxviii. The specificity of each hypothetical new protein sequence for the intended new DNA recognition sequence was calculated as the Boltzmann probability of the intended complex versus a partition function consisting of each base-pair possibility at the redesigned DNA base-pair.xxix Following design, predicted protein sequences with the most favorable binding energy and highest predicted specificity were reverted position by position to the wild-type amino acid sequence to identify (and revert) designed mutations that did not significantly contribute to the energy or specificity of the designed complex.
Multi-state design (designs −8G_A and −8G_B)
Two base pair positions in the structure were computationally mutated to generate a partial match to a recognition site in the IL-2Rγ gene in a mouse model of severe combined immunodeficiency disease (SCID). Specifically, positions −9G:C and −8A:T were modeled as −9A:T and −8G:C. A multistate design calculationxxx was performed to select amino acids at positions 24Z, 26Z, 27Z, and 29Z. Three states were included in the design. The first state was the target state, which was modeled using the altered DNA structure. The second state was the original structure with the wild-type DNA sequence and served as a competitor to enforce binding specificity of the selected proteins for the altered recognition site (negative design state). The third state was the modeled structure of the best single-state design for the target state with the modified DNA sequence, and the energy associated with this state is a constant during the multi-state design procedure. It represents the best scoring protein-altered DNA complex as assessed with the Rosetta energy potential, and it is therefore impossible for the energy associated with the target state to be lower than this value. As a result, multiple calculations were performed which differed from each other only in an artificial offset applied to the third state. Progressively larger offsets bias the calculations to select sequences that achieve higher specificity for the first state over the second state at the expense of achieving Rosetta scores that are allowed to be progressively worse than the third state.
A genetic algorithm was used to evolve a population of sequences that prefer the target state to the two competitors. An initial population of 2000 sequences was generated by selecting random amino acids at the four design positions. The side chain conformations of these four residues (with the rest of the protein and DNA structure held fixed) were predicted for the first and second states using a Monte Carlo algorithm, and the Rosetta score recorded. As noted above, the energy of the third state is a constant. A ‘fitness’ score for each sequence i in the population is calculated:
Where Etarget is the energy of the target state, and the brackets denote an ensemble (Boltzmann weighted) average over the energies of the competitors. Conceptually, the fitness corresponds to the transfer free energy of the protein from the ensemble of competitors to the target state. Subsequent generations were constructed using the following procedure. First, the sequence with the best (lowest) fitness was promoted automatically. Next 1980 sequences were created by recombining two members of the population using uniform crossover of two parents chosen by tournament selection.xxxi Finally, the remaining 19 sequences were generated by mutating a single parent chosen by tournament selection with a 25% chance of randomizing each position in turn. A fitness value was calculated for each new sequence, and the population was propagated for 30 generations.
Supplementary Material
Acknowledgments
This work was supported by a NSF graduate research fellowship to S.B.T., the US National Institutes of Health (#GM084433 and #RL1CA133832), the Foundation for the National Institutes of Health through the Gates Foundation Grand Challenges in Global Health Initiative, and the Howard Hughes Medical Institute. We thank Arshiya Quadri for help with plasmid substrate preparation and Michelle Scalley-Kim for I-AniI cleavage data collected in the presence of Mn2+.
Footnotes
Supplementary information accompanies this paper
Author Contributions
S.B.T and J.J.H. performed computational design calculations and S.B.T. performed kinetic characterization of all designed and wild-type enzymes. R.T. performed the fluorescence competition binding experiment. J.J. performed the surface expressed tethered cleavage assay. J.A. and J.J.H. developed computational design procedures. S.B.T. and D.B. wrote the paper. Multiple discussions of shared data among all authors at Northwest Genome Engineering Consortium (http://research.seattlechildrens.org/centers/immunity_vaccines/ngec/) group meetings contributed to the recognition of binding/catalysis asymmetry in I-AniI Y2 and conceptual development of this manuscript.
References
- i.Jencks WP. Mechanism of enzyme action. Annu Rev Biochem. 1963;32:639–676. doi: 10.1146/annurev.bi.32.070163.003231. [DOI] [PubMed] [Google Scholar]
- ii.Bolduc JM, et al. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 2003;17:2875–2888. doi: 10.1101/gad.1109003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- iii.Wells TN, Fersht AR. Use of binding energy in catalysis measured by mutagenesis of tyrosyl-tRNA synthetase. Biochemistry. 1986;25:1881–1886. doi: 10.1021/bi00356a007. [DOI] [PubMed] [Google Scholar]
- iv.Fersht AR. Relationships between apparent binding energies measured in site-directed mutagenesis experiments and energetics of binding and catalysis. Biochemistry. 1988;27:1577–1580. doi: 10.1021/bi00405a027. [DOI] [PubMed] [Google Scholar]
- v.Benkovic SJ, Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- vi.Röthlisberger D, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- vii.Collins CH, Yokobayashi Y, Umeno D, Arnold FH. Engineering proteins that bind, move, make, and break DNA. Curr Opin Biotechnol. 2003;14:371–378. doi: 10.1016/s0958-1669(03)00091-0. [DOI] [PubMed] [Google Scholar]
- viii.Smith J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ix.Redondo P, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- x.Takeuchi R, Certo M, Caprara MG, Scharenberg AM, Stoddard BL. Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation. Nucleic Acids Res. 2008;37:877–890. doi: 10.1093/nar/gkn1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xi.Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of a homing endonuclease and its host target sequence. J Mol Biol. 2007;372:1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xii.Halford SE, Johnson NP, Grinsted J. The EcoRI restriction endonuclease with bacteriophage lambda DNA. Kinetic studies. Biochem J. 1980;191:581–592. doi: 10.1042/bj1910581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xiii.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Analysis and Protein Folding. W. H. Freeman; 1998. [Google Scholar]
- xiv.Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol. 2007;17:467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xv.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xvi.Ashworth J, et al. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xvii.Ashworth J, et al. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xviii.Zhao L, Pellenz S, Stoddard BL. Activity and specificity of the bacterial PD-(D/E)XK homing endonculease I-Ssp6803I. J Mol Biol. 2008;385:1498–1510. doi: 10.1016/j.jmb.2008.10.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xix.Bolduc JM, et al. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 2003;17:2875–2888. doi: 10.1101/gad.1109003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xx.Stemmer WPC, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- xxi.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- xxii.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xxiii.http://rsbweb.nih.gov/ij/
- xxiv.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- xxv.Geese WJ, Kwon YK, Waring RB. In vitro analysis of the relationship between endonuclease and maturase activities in the bi-functional group I intron-encoded protein, I-AniI. Eur J Biochem. 2003;270:1543–1554. doi: 10.1046/j.1432-1033.2003.03518.x. [DOI] [PubMed] [Google Scholar]
- xxvi.Havranek JJ, Duarte CM, Baker D. A simple physical model for the prediction and design of protein-DNA interactions. J Mol Biol. 2004;344:59–70. doi: 10.1016/j.jmb.2004.09.029. [DOI] [PubMed] [Google Scholar]
- xxvii.Canutescu AA, Dunbrack RL., Jr Cyclic coordinate descent: robotic algorithm for protein loop closure. Protein Sci. 2003;12:963–972. doi: 10.1110/ps.0242703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xxviii.Das R, et al. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69:118–128. doi: 10.1002/prot.21636. [DOI] [PubMed] [Google Scholar]
- xxix.Ashworth J, Baker D. Assessment of optimization of affinity and specificity at protein-DNA interfaces. Nucleic Acids Res. 2009;37:e73. doi: 10.1093/nar/gkp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xxx.Havranek JJ, Harbury PB. Automated design of specificity in molecular recognition. Nat Struct Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- xxxi.Mitchell M. An Introduction to Genetic Algorithms. MIT Press; Cambridge, MA: 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.