Abstract
Protein abundance profiling from tissue using liquid chromatograph—tandem mass spectrometry-based ‘shotgun’ proteomics and label-free relative quantitation was evaluated for the investigation of estrogen-regulated protein expression in the mouse brain and uterus. Sample preparation involved a 30-min protein extraction in 8 M aqueous urea solution, followed by disulphide reduction, thiol alkylation and trypsin digestion of the extracted proteins, and was performed on 3–4 mg of tissue in order to evaluate the suitability of this methodology to expedite the survey of cellular pathways that are affected in vivo by an experimental therapeutic intervention in an animal model. The label-free proteomic approach (spectral counting) was suitable to identify even subtle changes in cortical protein levels and revealed significant estrogen-induced upregulation of ATP synthase (both α- and β-isoforms), aspartate aminotransferase 2 and mitochondrial malate dehydrogenase without any prior subcellular fractionation of the tissue or the use of multidimensional chromatographic separation. The methodology was also suitable to observe various up- and downregulated proteins in the uterine tissue of ovariectomized mice upon treatment with 17β-estradiol. In addition to confirming a very significant decrease in the abundance of glutathione S-transferase recognized as a marker of estrogen’s impact, our studies have also revealed potential new protein markers such as desmin and lumican that are critical components of cytoskeletal arrangement and, hence, regulation of their abundance could contribute to major morphological changes in the uterus occurring upon estrogenic stimulation.
Keywords: estrogen, cortex, uterus, LC—MS/MS, label-free differential proteomics, spectral counting, extracted-ion chromatograms
INTRODUCTION
The demand for high-throughput methodologies for the analysis of differentially expressed proteins from a variety of biological matrices (cell culture, tissue, etc.) has prompted the development of rapid sample preparation and mass spectrometry-based proteomic techniques. For example, a recent study in which minimal sample processing for protein extraction was performed identified approximately 1,000 proteins from a single LC-MS/MS analysis of mouse liver tissue.1
Conventional methodologies typically used for this purpose have employed two-dimensional (2D) gel electrophoresis (GE) followed by mass spectrometric analysis. Unfortunately, a large percentage of hydrophobic/membrane proteins as well as proteins exhibiting extremely basic or acidic isoelectric point values and exceedingly large or small molecular weights are often under-represented using this technique.2 In addition, quantitation inaccuracies can occur given the high variation in protein mobility on a 2D gel as a result of common in vivo and/or ex vivo protein modifications such as phosphorylation, oxidation, or proteolytic processing. Mass spectrometry-based methods using stable-isotope labeling3 have enhanced proteomic analysis in this regard; however, certain limitations of this approach have become evident in our experience including decreased experimental throughput and sensitivity as a consequence of labeling and purification steps, limited dynamic range of quantitation, compromise of sample integrity due to side-reactions upon chemical derivatization and the strong dependency of labeling efficiency on sample composition. As a result, label-free approaches such as spectral counting have gained recent interest for abundance-based profiling and have benefited significantly from current advances in MS instrumentation regarding rapid data-dependent acquisition, ultra-high sensitivity and dynamic range. The combination of tissue proteomics that involves a 30-min protein extraction from tissue followed by LC-MS/MS-based shotgun proteomics1 with label-free relative quantitation through spectral counting4,5 or integration of extracted ion chromatograms5,6 could potentially allow for a powerful methodology to identify differentially expressed proteins from relatively small amounts of tissue. Based on the high protein coverage obtained from mouse liver tissue by a single LC—MS/MS run, Shi et al. has suggested that this approach, with its obvious advantages over 2DE-based proteomics, would be useful in toxicological or clinical studies of the liver.1
Drug discovery is an additional area where accelerated tissue proteomics would be beneficial in the process of target identification—both by aiding the elucidation of primary mechanisms of action and by surveying unanticipated “off-target” actions.7 Efforts to find new potential therapeutical value for many already established drugs8 and the growing recognition of multiple-ligand design as an emerging drug discovery paradigm,9 especially to combat neurodegenerative diseases,10 provide further impetus to introduce such methods in this field.
Estrogens have been implicated in the maintenance of a healthy brain in females via multiple, yet often poorly understood molecular mechanisms,11 and estrogen deprivation has been associated with various neurological conditions and vulnerability to neurodegenerative diseases.12 The strategy to first observe in vivo the overall effect of estrogens on tissue to gain an initial insight into their most relevant effect(s) at the protein level has been recommended previously.13 Using this “global” approach and relying on 2D-GE, image analysis and LC—MS/MS-based protein identification from the gel spots that showed differential stain intensity compared to those of untreated control animals, estrogen-regulated proteins (linked to vesicular transport and membrane trafficking, neuronal plasticity, energy production and signal transduction pathways) have been identified from the rat ventromedial nucleus of the hypothalamus recently.14 Although regulation of a few mitochondrial proteins involved in energy production has been inferred directly from the analysis of the brain tissue, half of them were matched only a single peptide to the reported protein. Also employing 2D-GE methodology, in vivo regulation of mitochondrial proteins by E2 has been explored through the isolation of the organelle from the entire forebrain region.15 ATP synthase alpha was found to be upregulated in both studies. However, upregulation was reported for NADH dehydrogenase (ubiquinone) Fe—S protein 1 (Ndufs1) upon analyzing the tissue directly,14 while downregulation of this protein was observed when the mitochonria were isolated before 2D-GE.15
The known restrictions and limited throughput of proteomics strategies based on 2D-GE2,13 have prompted us to consider a gel-free methodology for global tissue proteomics, especially when mice are preferred as experimental species or small brain areas are to be analyzed without resorting to sample pooling. Also considering requirements to increase throughput for applications in drug discovery, we have tested whether suitable protein profiles manifesting the impact of E2 could be obtained by adapting the method explored for the liver by Shi et al.1 and employed a 30-min protein extraction with aqueous urea solution followed by shotgun proteomics based on one-dimensional reversed-phase LC—MS/MS analysis for other tissues. In addition to the brain that is relatively soft compared to tissues of other organs such as the liver, our studies included the mouse uterus representing a much harder tissue. The latter organ is a well known target for estrogens functioning as primary female sex hormones.16 Therefore, any therapeutic strategy that targets estrogen-associated pathways in the central nervous system has to address a potential impact on the uterus. Our aim also was to introduce, according to the recommendation of Shi et al., and evaluate label-free differential quantitation to pursue tissue proteomics.
Experimental Methods
Chemicals and Reagents
All chemicals and reagents were purchased from Sigma Aldrich (St. Louis, MO) unless otherwise specified.
Animals
Swiss-Webster mice (30±2 g body weight) were used for all experiments conducted in accordance with the guidelines set forth in the Declaration of Helsinki and the Guiding Principles in the Care and Use of Animals (DHEW Publication, NIH 80-23) and approved by the Institutional Animal Care and Use Committee at the University of North Texas Health Science Center.
Ovariectomy of the animals was done by their supplier (Charles River Laboratories, Wilmington, MA). The animals were shipped approximately one week after ovariectomy and were allowed to adapt in the animal facility of the University of North Texas Health Science Center for approximately two weeks before starting daily injections with the vehicle (corn oil, 60 μl per injection) control or E2 (50 μg/kg body weight in corn oil vehicle) for 5 consecutive days between 10:00 am and 12:00 am. The animals were sacrificed by cervical dislocation, decapitated, and their brains were removed. An abdominal incision was then made and the uterus was removed by cutting at the junction of the uterus and vagina and at the site of the ovariectomy on each horn. Excess fat and connective tissues were removed and the organ was blotted and weighed. All tissues were stored at -80 °C until analysis.
Sample Preparation
Three to four milligrams of tissue (mouse brain from frontal cortex and uterus, respectively) were incubated in 25 μl of 8 M aqueous urea solution for 30 min, according to the procedure we adapted from Shi et al.1 For the verification of the spectral counting method under our experimental conditions, different amounts of bovine serum albumin (BSA) corresponding to 1 μg, 500 ng, 250 ng, and 125 ng were spiked into brain tissue samples from separate control animals prior to incubation in the 8 M aqueous urea solution. After centrifugation, 10 μl of the supernatant was treated with 1mM dithiothreitol and incubated at 65 °C for 30 min. Carbamidomethylation of thiol groups was performed by addition of 5 mM iodoacetamide and incubation for 30 min in the dark at room temperature. The samples were then diluted 4-fold with 50 mM ammonium bicarbonate and incubated overnight at 37 °C with 1 μg of sequencing grade trypsin (Promega, Madison, WI). The enzymatic digestion was then quenched by acidifying the sample with 1 μl of acetic acid.
For comparison with the overnight reaction with trypsin, processing of two control brain samples were finished with a microwave-assisted procedure in a CEM (Matthews, NC) Discover system. Maximum power and temperature were set to 50 W and 55 °C, respectively. After 60 min of digestion, the solution was allowed to cool and 1 μl of acetic acid was added.
LC-MS/MS Analyses
Online reversed-phase LC-MS/MS analysis of mouse brain protein digests was performed using a hybrid linear ion trap (LTQ) — Fourier transform ion cyclotron resonance (FTICR, 7 Tesla) mass spectrometer (LTQ-FT, Thermo, San Jose, CA) equipped with a nanoelectrospray ionization source and operated with the Xcalibur (version 2.2) data acquisition software. Protein digests were loaded onto a PepMap C18 capillary trap (LCPackings, Bannockburn, IL) and desalted with 3% acetonitrile, 1% acetic acid for 5 min prior to injection onto a 75 μm i.d. × 10 cm PicoFrit C18 analytical column (New Objective, Woburn, MA). Following peptide desalting and injection onto the analytical column, a linear gradient provided by a NanoLC-2D (Eksigent) was carried out to 40% acetonitrile in 60-120 min at 250 nL/min. A 30 min gradient was utilized for BSA relative quantitation experiments as well as for the experiments to evaluate biological and analytical variability. ESI spray voltage and capillary temperature during the gradient run were maintained at 2.0 kV and 250 °C, respectively. The data-dependent mode of acquisition was utilized in which an accurate m/z survey scan is performed in the FTICR cell followed by parallel MS/MS linear ion trap analysis of the top five most intense precursor ions. FTICR full-scan mass spectra were acquired at 100,000 mass resolving power (m/z 400) from m/z 350 to 1500 using the automatic gain control mode of ion trapping. CID in the linear ion trap was performed using a 3.0-u isolation width and 35% normalized collision energy with helium as the target gas. The precursor ion that had been selected for CID was dynamically excluded from further MS/MS analysis for 60 s.
Database Searching
MS/MS data generated by data dependent acquisition via the LTQ-FT were extracted by BioWorks version 3.3 and searched against a composite IPI mouse (version 3.35, number of entries is 51490 × 2) protein sequence database containing both forward and randomized sequences using the Mascot v 2.2 (Matrix Science, Boston, MA) search algorithm. Mascot was searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 15.0 ppm assuming the digestion enzyme trypsin with the possibility of one missed cleavage. Carbamidomethylation of cysteine was specified as a fixed modification while oxidation of methionine, N-terminal protein acetylation, N-terminal peptide and lysine carbamoylation, and phosphorylation of serine and threonine were specified as variable modifications. In general, probability-based MOWSE scores corresponding to a significance threshold of P<0.05 were considered for peptide identification in addition to manual validation of the MS/MS data.
Data Compilation and Relative Quantitation
The software program Scaffold (version Scaffold 2.0, Proteome Software Inc., Portland, OR) was employed to validate MS/MS-based peptide and protein identifications. Initial peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm.17 Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm.18 These identification criteria typically established a <0.01% false discovery rate based on a decoy database search strategy. Scaffold allows for simultaneous comparison of multiple proteomic data sets in which the list of identified proteins can be sorted by various parameters. The spectral counting technique for relative protein quantitation utilizes the total number of MS/MS spectra identified for a particular protein as a measure of protein abundance and consequently this parameter was used to classify the mouse brain protein identifications within Scaffold. The method for relative quantitation followed published protocols5,6 in which the change in abundance was determined by the ratio as follows:
| (1) |
where nE2-treated and ncontrol are the total number of identified MS/MS spectra (normalized spectral count from Scaffold) for a particular protein in the E2-treated and control group, respectively. A 50% peptide probability was used for the initial list of high-confidence identifications (99% protein confidence, 95% peptide confidence and containing 2 unique peptides) in order to include peptides with lower Mascot scores that represent true positive identifications and would improve the overall spectral counting sensitivity. Ratios for proteins showing zero spectral counts in either control or E2-treated groups were tabulated as UC (unique in control) or UE (unique in E2-treated). A G statistic test (likelihood ratio test for independence) was then utilized to determine statistical significance for each protein ratio:5,19
| (2) |
where G is the G test statistic; ccontrol is (ncontrol + 1); cE2-treated is [(nE2-treated + 1)]; and tct is (ccontrol + cE2-treated)/2. The G statistic value is approximately characterized by a χ2 distribution with one degree of freedom, allowing P-value calculations for each ratio value. Ratios were considered significant at P<0.05 unless validated by integration of corresponding peptide extracted ion chromatograms or Western blotting in which the criterion P<0.05 was utilized.
Relative quantitation based on extracted ion chromatograms, which was utilized to validate spectral counting results for selected peptides and proteins, followed a similar routine as the spectral counting method. However, rather than total number of identified spectra as a measure of protein abundance, the mass spectrometric response value (based on ion current) for individual peptides from a given protein was utilized. Extracted ion chromatograms (XICs) of peptides were generated automatically via the Sieve™ program (version 1.1.0; Thermo) using its default parameters for chromatographic alignment and data framing typical for data acquired on the LTQ-FT instrument.
Functional Classification
Cytoscape 2.6.0 equipped with the BiNGO 2.0 plug-in (http://www.psb.ugent.be/cbd/papers/BiNGO/) was used to determine over- or under-represented gene ontology (GO) categories from the mouse brain and uterus proteome data sets corresponding to proteins identified at 99% protein confidence, 95% peptide confidence, and contained at least 2 unique peptides. Parameters selected in the analysis include hypergeometric statistical test, cellular component GO terms, and correction for multiple term testing by Benjamini and Hochberg false discovery rate corrections.20 GO annotations from GOA Mouse (version 38.0, available at http://www.ebi.ac.uk/GOA/) that contained 173476 total associations and 32068 total distinct proteins were used to create a reference set for the IPI mouse v3.35 protein database. Ingenuity Pathway Analysis 5.0 (Ingenuity Systems, Redwood City, CA) was also utilized to determine over-represented biological processes from the same proteome data sets. Over-represented molecular functions and biological processes were accepted at the significance level of P<0.05 (right-tailed Fisher’s exact test).
Results and Discussion
Method Development
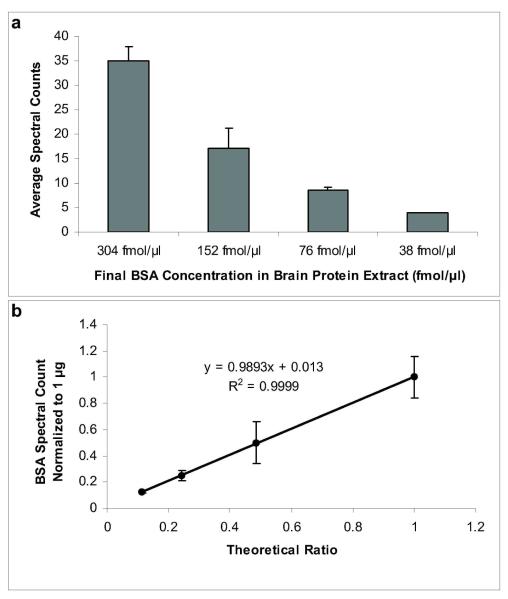
We have adapted an abundance-based protein-profiling assay based on a method previously reported by Shi et al.1 that involves minimal sample preparation for protein extraction (incubation of 2 mg tissue in 8M urea for 30 min) and combined it with label-free MS-based relative quantitation4,5 (Figure 1). Approximately 3-4 mg of tissue is used for the procedure, and the throughput of the method allows for a proteomic survey of two samples (e.g., control versus treatment) within several hours, excluding the overnight proteolytic digestion done simultaneously for all samples. Correlations between spectral counts and protein abundance levels have been demonstrated convincingly by Liu et al.4 To verify the applicability of the method for quantitative survey under our experimental conditions, mouse brain tissue was prepared and spiked with different amounts of bovine serum albumin (BSA) prior to incubation in 8M urea, protein digestion with trypsin and subsequent LC-MS/MS analysis. The spectral counting approach permitted the identification of ≥2-fold changes in protein abundance within the concentration range of BSA used for assay verification (Figure 2a). Similarly to an earlier report that introduced the methodology,4 high correlation (r2=0.9999) was obtained between the experimentally derived relative abundance ratios, using the largest BSA concentration (304 fmol/μl) as a reference point, and the theoretical ratios calculated based on the known BSA quantities spiked into the brain samples (Figure 2b).
Figure 1.
Schematic representation of the experimental design employed for rapid differential protein expression profiling from tissue.
Figure 2.
Method verification for relative protein quantitation using rapid tissue proteomics. a) The total identified MS/MS spectra for BSA decreased proportionally with the decrease in amount of BSA spiked into the mouse brain tissue. b), High correlation was observed with the normalized BSA spectral count (using the highest concentration of 304 fmol/μl as a reference point) and the theoretical ratios calculated from the known BSA quantities. Error bars represent + SD.
Comparison of protein extracts obtained from the frontal cortex of three different control animals and duplicate injections of each biological replicate were performed to evaluate potential biological and analytical variability. Several proteins derived from blood were identified, but were not included in subsequent protein expression analyses, since variation in contamination or biologically significant changes in abundance of these proteins is difficult to determine. Additionally, differentially expressed proteins containing peptide sequences identical with other isoforms were reported with multiple accession numbers; however, these particular proteins were not reported if grouping ambiguity occurred (as indicated by the Scaffold program).
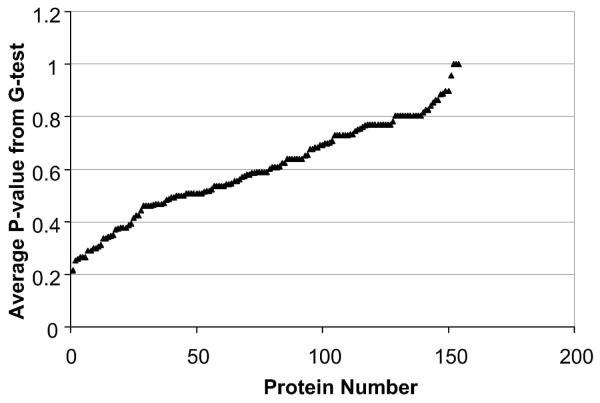
The analytical replicate analysis showed no statistically significant changes in spectral counts for proteins identified at >99% probability (containing at least 2 peptides) and based on a statistical G-test (P<0.05). The same statistical test was then utilized for comparison of biological replicates in order to determine the potential impact of biological and protein extraction variability on the quantitation sensitivity of the label-free methodology. The median P-value obtained from all proteins identified at >99% probability (containing at least 2 unique peptides) was 0.59 ± 0.18 (± SD) with the lowest P-value equal to 0.22 suggesting P<0.05 is an adequate criterion for the identification of statistically significant changes in protein expression using this approach and tissue type (Figure 3).
Figure 3.
Proteins identified (≥ 99% confidence and containing two unique peptides) after 60-min LC-MS/MS analysis of tryptic digests derived from mouse brain frontal cortex, compared across three different control animals. The median P-value was 0.59 + 0.18 (+ s.d.) with the lowest P-value equal to 0.22 suggesting the conventional statistical cutoff of P<0.05 is adequate to account for potential analytical and biological variability associated with the technique.
In a pilot feasibility study to further accelerate sample processing, we have evaluated microwave-assisted tryptic digestion.21 The latter procedure, requiring about an hour to complete for the protein extract from brain tissue, afforded protein coverage similar to that of the overnight incubation in a water bath (Supplementary Tables 1 and 2). Microwave-assisted and conventional digestion (overnight in shaking water bath) afforded 2642 (782 identified) and 2019 (671 identified) MS/MS spectra, respectively, after one-dimensional reversed-phase LC separation from dupplicate samples. Altogether, 112 proteins were identified after both microwave-assisted and conventional digestion, while 16 and 20 proteins were only fund in tissue processed with and without employing microwaves, respectively. However, subsequent experiments were performed using the conventional digestion procedure to match with conditions employed by others in previous studies aimed at finding estrogen-regulated proteins in the brain proteome or its mitochondrial subproteme.14,15
A Rapid Survey of Proteins Regulated by 17β-Estradiol in the Female Mouse Brain
We conducted our study in an animal model for menopausal depressive-like symptoms (manifested by, e.g., the prolongation of immobility time in a forced swim test and can be induced by ovariectomy in female mice) that are effectively relieved by E2 treatment.22 We confirmed by the forced swim test that E2-treatment, indeed, mitigated the depression-like state caused by ovariectomy (Supplementary Figure 1a online).
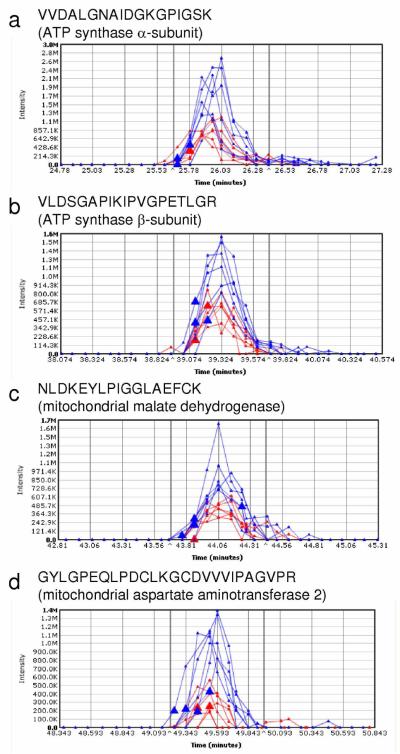
Our label-free proteomic survey using the spectral counting-based approach has been suitable to identify even subtle (as low as a 1.4-fold) change in cortical protein expression in the ovariectomized mouse upon treatment with an estrogen (Table 1). However, the number of proteins identified with high confidence from the mouse cortex after 30-min protein extraction with 8 M aqueous urea solution followed by shotgun proteomics employing reversed-phase LC—MS/MS analysis without any prior sample fractionation (Supplementary Table 1 online) fell short of that reported for mouse liver (>1,000) using the procedure we adapted from Shi et al.1 Apparently, the brain tissue would need either a more efficient protein extraction procedure than the one adapted for our survey, or would require sample fractionation to reduce complexity of the extracted protein mixture before LC—MS/MS analysis. Nevertheless, several differentially expressed proteins were identified and validated via integration of XICs by SIEVE™ (Figure 4). Specifically, XICs of the tryptic peptides VVDALGNAIDGKGPIGSK (matched to the ATP synthase α-subunit), VLDSGAPIKIPVGPETLGR (matched to the ATP synthase β-subunit), NLDKEYLPIGGLAEFCK (matched to the mitochondrial malate dehydrogenase) and DKEYLPIGGLAEFCK (matched to the mitochondrial aspartate aminotransferase 2) showed increased expression of the proteins in the cortex of E2-treated mice (blue traces) consistently and in a statistically significant extent (P<0.05) compared to the ovariectomized control animals (red traces). An increased abundance of the ATP synthase β-subunit in the E2-treated animals was also confirmed by Western blot analysis (Supplementary Figure 2a online).
Table 1.
Examples of differential protein expression in mouse brain after E2 treatment identified by spectral counting and validated by integration of extracted ion chromatograms
| Protein Name | IPI accession number |
Number of unique peptides |
Sequence coverage (%) |
Average normalized spectral count ± standard deviation, Controla |
Average normalized spectral count ± standard deviation, E2- treateda |
P-value from G testb |
Fold change |
|---|---|---|---|---|---|---|---|
| ATP synthase, alpha | IPI00130280 | 18 | 39 | 13 ± 4 | 21 ± 1 | 0.007 | 1.6 |
| ATP synthase, beta | IPI00468481 | 22 | 57 | 39 ± 5 | 53 ± 1 | 0.008 | 1.4b |
| Aspartate aminotransferase 2 |
IPI00117312 | 11 | 31 | 7 ± 2 | 14 ± 5 | 0.01 | 2.0 |
| Malate dehydrogenase, mitochondrial |
IPI00323592 | 14 | 50 | 15 ± 4 | 23 ± 3 | 0.03 | 1.5 |
N=3 (brain tissue from three different animals).
Spectral counts from each animal in control and E2-treated groups were subjected to a summation-based G statistical test..
Validated by Western blot analysis (Supplementary Figure 2a online).
Figure 4.
Representative extracted ion chromatograms (XIC) for peptides derived from differentially expressed proteins identified by initial spectral counting-based screening in the cortices of ovariectomized mice (Table 1). The blue and red traces represent XICs for peptides of a particular protein from E2-treated and control, respectively: a) VVDALGNAIDGKGPIGSK from ATP synthase alpha (ratio E2/control=2.2, P-value=3.1 × 10-4); b) VLDSGAPIKIPVGPETLGR from ATP synthase beta (ratio E2/control=1.8, P-value=2.7 × 10-3); c) GYLGPEQLPDCLKGCDVVVIPAGVPR from mitochondrial malate dehydrogenase (ratio E2/control=3.5, P-value=1.6 × 10-4); d) DKEYLPIGGLAEFCK from mitochondrial aspartate aminotransferase 2 (ratio E2/control=2.0, P-value=1.0 × 10-3).
Interestingly, the differentially expressed brain proteins upon estrogen treatment we found in the present study (Table 1 and Fig. 4) were resident in or associated with the mitochondrial inner membrane, which confirmed the validity of mitochondria-focused studies regarding estrogen’s actions on the brain.15,23 The mitochondrion is an intracellular compartment that serves as a major energy source within the cell by ATP production through the oxidation of cellular nutrients.24 Two subunits of Complex V (ATP synthase) of the electron transport chain revealed higher abundance in the frontal cortex after E2 treatment when compared to control animals. This protein complex represents a component of the oxidative phosphorylation pathway in which ATP is synthesized from ADP, a process that is coupled to proton translocation across the mitochondrial membrane through channels within the complex. The accumulation of protons occurs partly in response to the conversion of NAD+ to NADH, a cofactor utilized in the mitochondrial electron transport chain for subsequent ATP production. NADH and NAD+ cannot permeate through the inner mitochondrial membrane and, consequently, biochemical mechanisms such as the malate-aspartate shuttle exist to translocate electrons across this membrane to provide an electron source for the oxidative phosphorylation pathway.24 This particular shuttle system contains both cytosolic and mitochondrial forms of aspartate aminotransferase and malate dehydrogenase responsible for the interconversion of various small molecules such as malate and oxaloacetate. The biochemical reactions associated with the malate-aspartate shuttle are coupled to the production of NADH and NAD+ in the mitochondrial matrix and cytosol, respectively. NADH produced in the mitochondrial matrix through the malate-aspartate shuttle is an efficient mechanism for ATP production in terms of the enhanced quantity of ATP generated from the electron transport system as compared to other pathways that produce FADH2 for this purpose.24 An increased abundance of Complex V components and key proteins involved in the malate-aspartate shuttle after E2 treatment suggests a potentially novel mechanism for estrogen-mediated effects in the brain through the enhancement of pathways associated with mitochondrial-mediated cellular bioenergetics.
A recent study also identified differentially expressed proteins from rat brain mitochondria isolated after E2 treatment using 2D-GE followed by MS analysis,15 and several proteins were common to our spectral counting-based survey including ATP synthase α and β subunits, aspartate aminotransferase (glutamate oxaloacetate transaminase) 2, and malate dehydrogenase. ATP synthase α and β subunits and aspartate aminotransferase showed a similar increase in expression after E2 treatment; however, a decrease in malate dehydrogenase expression was reported from the 2D-GE analysis. Discrepancies such as these are not surprising given the high variation in protein mobility and subsequent heterogeneous representation on a 2D gel due to various in vivo and/or ex vivo protein modifications. The benefit of the employed rapid proteomics approach is apparent in this regard, since minimal purification and separation steps are introduced in the analysis limiting, thus, experimental bias for proteome constituents that exhibit certain physicochemical properties. Although our G-test did not reach statistical significance at the measured low spectral counts (3 ± 1) for the specific protein probably due to the small number animals per group (N=3), we also detected NADH dehydrogenase (ubiquinone) Fe—S protein 1 (Ndufs1) exclusively in the cortex of E2-treated animals, which was in agreement with a recent study that indicated an upregulation from direct tissue analysis14 and opposed to the one that implicated downregulation of this protein from isolated brain mitochondria.15 Additional experiments and, perhaps, a targeted LC—MS/MS approach25 are necessary to verify the in vivo effect of E2 on Ndufs1 upon using the tissue extraction method adapted for our present study,1 which will be addressed (together with a comparative evaluation involving the isolation of mitochondria) by a separate investigation.
A Rapid Survey of Proteins Regulated by 17β-Estradiol in the Mouse Uterus
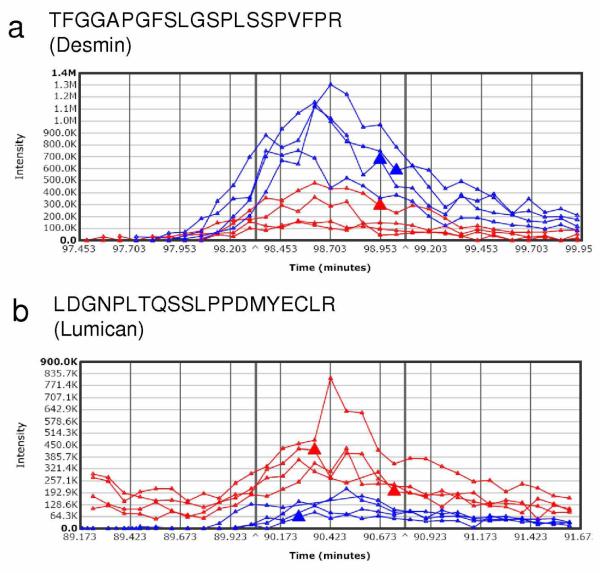
Not surprisingly, the administration of E2 to overiectomized mice resulted in a profound effect on the uterus. In addition to the apparent increase of wet weight (Supplementary Figure 1b online), the uteri of the control group appeared to have little vascularization, whereas the uteri of the E2 group showed high vascularization. Our rapid tissue proteomics-based approach also revealed significant changes, as opposed to the subtle differences in cortical protein levels, in uterine protein expression upon E2 treatment. Out of approximately 118 proteins identified at 99% confidence and containing at least 2 unique peptides, 11 and 17 were shown to be up- and down-regulated, respectively (Table 2). Despite the limited protein coverage by the adapted methodology compared to that of the mouse liver proteome, the apparent responsiveness of the uterine proteome to E2 treatment has permitted the identification of numerous differentially expressed proteins. Several of the estrogen-regulated proteins have been previously reported including the down-regulation of glutathione S-transferase (GST) by estrogen in the uterus, which has been used as a validation assay prior to cDNA microarray analysis probing the effects of E2 in the tissue.26 We have performed validation of our spectral-counting data for E2-induced changes in the expression of ATP synthase alpha subunit (Atp5a1) by Western blotting (Supplementary Figure 2b online), while results for desmin (Des) and lumican (Lmna) were validated by integration of XICs using the Sieve™ software, as shown in Figure 5. Although detailed mechanisms associated with the uterotrophic actions of E2 are unclear, we have revealed potential new estrogen-sensitive protein markers Des (upnregulated) and Lmna (downregulated) that are critical components of cytoskeletal arrangement and, therefore, variation in their abundance could contribute to major morphological changes in the uterus after E2 treatment.
Table 2.
Differential protein expression in mouse uterus after E2 treatment identified by spectral counting
| Protein Name | IPI accession number |
Number of unique peptides |
Sequence coverage (%) |
Average normalized spectral count ± standard deviation,a Control |
Average normalized spectral count ± standard deviation,a E2- treated |
P-value from G testb |
Fold change |
|---|---|---|---|---|---|---|---|
| Eif4a1 Eukaryotic initiation factor 4A-I |
IPI00118676 IPI00761992 |
3 | 13 | 0 | 3 ± 1 | 0.0005 | UEc |
| Krt19 Keratin, type I cytoskeletal 19 |
IPI00112947 | 10 | 36 | 1 ± 1 | 16 ± 3 | 3E-15 | >10 |
| Eef2 Elongation factor 2 |
IPI00466069 | 4 | 8.4 | <1 | 5 ± 1 | 7E-05 | >10 |
| Flna Isoform 1 of Filamin-A |
IPI00131138 IPI00830793 |
22 | 15 | 3 ± 1 | 30 ± 8 | 4E-24 | 10 |
| Rplp1 60S acidic ribosomal protein P1 |
IPI00113377 | 3 | 80 | 2 ± 1 | 10 ± 1 | 7E-06 | 5.0 |
| Hspa5 78 kDa glucose- regulated protein precursor |
IPI00319992 | 7 | 17 | 2 ± 1 | 9 ± 1 | 7E-06 | 4.5 |
| Cnn1 Isoform Alpha of Calponin-1 |
IPI00116645 IPI00228258 |
3 | 13 | 1d | 4 ± 1 | 0.003 | 4.0 |
| Atp5a1 ATP synthase subunit alpha, mitochondrial precursor |
IPI00130280 IPI00857439 |
3 | 10 | 1d | 4 ± 1 | 0.01 | 4.0e |
| Eef1a1 Elongation factor 1-alpha 1 |
IPI00307837 | 4 | 18 | 3 ± 2 | 9 ± 1 | 0.0003 | 3.0 |
| Des Desminf | IPI00130102 | 15 | 45 | 25 ± 10 | 54 ± 7 | 2E-11 | 2.2 |
| Hist1h2bf Histone H2B | IPI00114642 IPI00134534 IPI00227930 IPI00265768 IPI00282266 IPI00282269 IPI00348270 IPI00461514 IPI00554853 IPI00648991 IPI00761713 |
2 | 20 | 20 ± 4 | 31 ± 2 | 0.002 | 1.6 |
| Myl6 Isoform Smooth muscle of Myosin light polypeptide 6 |
IPI00354819 IPI00409817 |
7 | 59 | 36 ± 7 | 22 ± 4 | 0.0002 | 0.6 |
| Tln1 Talin-1 | IPI00465786 | 10 | 8.9 | 10 ± 4 | 4 ± 2 | 0.006 | 0.4 |
| Arhgdia Rho GDP- dissociation inhibitor 1 |
IPI00322312 | 3 | 31 | 11 ± 2 | 4 ± 1 | 0.0003 | 0.4 |
| Lum Lumican precursor | IPI00313900 | 5 | 25 | 20 ± 2 | 7 ± 1 | 6E-07 | 0.4 |
| Calm3;Calm2;Calm1 Calmodulin |
IPI00761696 | 3 | 31 | 14 ± 2 | 5 ± 1 | 1E-05 | 0.4 |
| Vcl Vinculin | IPI00405227 | 11 | 19 | 23 ± 5 | 7 ± 1 | 7E-09 | 0.3 |
| Park7 Protein DJ-1 | IPI00117264 | 4 | 37 | 7 ± 2 | 2 ± 1 | 0.002 | 0.3 |
| Tpi1 Triosephosphate isomerase |
IPI00467833 | 4 | 29 | 12 ± 1 | 4 ± 1 | 2E-05 | 0.3 |
| Ptrf Polymerase I and transcript release factor |
IPI00117689 | 4 | 15 | 14 ± 3 | 4 ± 1 | 1E-06 | 0.3 |
| Hnrnpa2b1 Isoform 3 of heterogeneous nuclear ribonucleoproteins A2/B1 |
IPI00405058 IPI00622847 IPI00828488 IPI00853914 |
3 | 19 | 5 ± 2 | 1 ± 1 | 0.0004 | 0.2 |
| Ogn Mimecan precursor |
IPI00120848 | 3 | 12 | 16 ± 3 | 3 ± 1 | 8E-10 | 0.2 |
| Anxa6 annexin A6 | IPI00310240 IPI00554894 IPI00649152 |
6 | 16 | 7 ± 1 | 1 ± 1 | 0.0001 | 0.2 |
| Pebp1 Phosphatidylethanol- amine-binding protein 1 |
IPI00137730 | 5 | 58 | 10 ± 2 | 1 ± 1 | 5E-08 | 0.1 |
| Gstm1 Glutathione S- transferase Mu 1 |
IPI00230212 IPI00649135 |
3 | 20 | 7 ± 2 | 0 | 4E-08 | UCg |
| Prelp Prolargin precursor |
IPI00122293 | 2 | 6.9 | 5 ± 2 | 0 | 2E-06 | UCg |
| Prdx6 Peroxiredoxin-6 | IPI00555059 IPI00758024 |
3 | 26 | 4 ± 1 | 0 | 3E-05 | UCg |
| Fabp4 Fatty acid- binding protein, adipocyte |
IPI00116705 | 2 | 20 | 3 ± 2 | 0 | 0.0002 | UCg |
N=4 (uterine tissue from four different animals)
Spectral counts from each animal in control and E2-treated groups were subjected to a summation-based G statistical test (P<0.05).
Unique in E2-treated animals.
Standard deviation <0.5
Validated by Western blotting (Supplementary Figure 2b online).
Protein grouping ambiguity reported by Scaffold was resolved by XIC quantitation of sequence-specific peptide
Unique in control (vehicle-treated) animals.
Figure 5.
Representative extracted ion chromatograms (XIC) for peptides derived from differentially expressed proteins identified by initial spectral counting-based screening in the uterus of ovariectomized mice. The blue and red traces represent XICs for peptides of a particular protein from E2-treated and control, respectively: a) TFGGAPGFSLGSPLSSPVFPR from desmin (ratio E2/control=4.1, P-value=3.8 × 10-3); b) LDGNPLTQSSLPPDMYECLR from lumican (ratio E2/control=0.3, P-value=2.5 × 10-3).
Bioinformatics Analysis
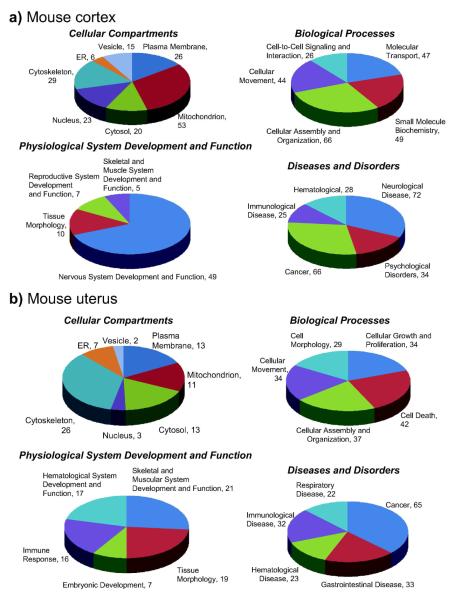
Upon gene ontology (GO) and pathway analyses (Ingenuity) of the mouse brain frontal cortex and uterine proteome data sets from ovariectomized female animals (Figure 6), reasonable representations of various cellular compartments, biological processes, physiological system development and function, and diseases and disorders were obtained. The extraction and analysis procedures were sufficient for the characterization of intracellular and membrane protein components which included greater than 80% and 20% representation of the GO-annotated proteins, respectively. Specifically, identified proteins assigned to cellular compartments distributed among mitochondrial (31%), cytoskeletal (17%), plasma membrane (15%), nuclear (13%), cytosolic (12%), vesicular (9%) and endoplasmic reticulum (ER, 3%) species in the mouse cortical extract, and covered 232 biological processes corresponding to cellular assembly and organization, small molecule biochemistry, molecular transport, cellular movement and cell-to-cell signaling and interaction. Approximately 70% of the proteins assigned according to physiological system development and function were associated with nervous system development and function. Regarding the 225 diseases and disorders, almost half of them represented neurological diseases and psychological disorders, while association with was also considerable (30% of the total). In the mouse uterus extract, identified proteins assigned to cellular compartments distributed among cytoskeletal (35%), plasma membrane (17%), cytosolic (17%), mitochondrial (15%), ER (9%), nuclear (4%) and vesicular species (3%). Biological processes (176 total) represented cell death, cellular assembly and organization, cellular growth and proliferation, cellular movement and cell morphology. Skeletal and muscular system development and function, tissue morphology, hematological system development and function, immune response and embryonic development were implicated with, and cancer was the most abundant disease and disorder (37%) linked to the proteins identified from extract of the mouse uterus.
Figure 6.
Gene Ontology (GO) and Ingenuity Pathway analyses. a) mouse brain frontal cortex; b) uterus. When proteome data sets from ovariectomized female animals were analyzed (≥ 99% confidence containing 2 unique peptides), adequate representations of various cellular compartments, biological processes, physiological system development and function pathways, and diseases and disorders were obtained. The extraction and analysis procedures were sufficient for the characterization of intracellular and membrane protein components which included greater than 80% and 20% representation of the GO-annotated proteins, respectively.
Conclusions
A label-free proteomic approach has been suitable to identify even subtle changes in the expression of several cortical proteins in the ovariectomized mouse upon treatment by an estrogen and has confirmed that the mitochondria are targets for the hormone’s action in the brain. The method has also revealed significant estrogen-induced changes in protein levels of the uterine tissue. Nevertheless, adaptation of the accelerated screening methodology that has been reported to yield high coverage for the mouse liver proteome1 and, thus, we adapted to perform our study afforded global proteomic analysis of cortical and uterine proteins in a significantly reduced depth than expected and, therefore, may need improvement in the extraction procedure for the latter tissues. However, to put our results in perspectives, exploration of the rat brain proteome directly from whole brain tissue by 2D-GE had started with the identification of 210 proteins27 (a number comparable to what we covered by our rapid proteomics survey in the mouse cortex), which implicated the applicability of the methodology only to abundant brain proteins. Additional developments, including the use of large-gel formats to improve electrophoretic separation28 and attention to pre-electrophoretic enrichment of low-abundance gene products,29,30 has maintained the viability of this methodology despite its recognized shortcomings upon analyses of complex protein mixtures2,13 and, unfortunately, at the cost of decreasing throughput. The premise of the presented label-free approach to explore differential protein expressions should also be viewed in a similar context. Our future development effort will, therefore, address improvement of methodology regarding both protein extraction from brain tissue and optimization of separation to increase protein coverage without significantly compromising throughput. While the accelerated gel-free methodology offers less experimental bias for proteome constituents that exhibit certain physicochemical properties (Figure 5) compared to methods based on 2D-GE, preference to assay simplicity and speed over maximizing protein coverage that requires meticulous sample fractionation and multidimensional analytical separation results in apparent limitations. Rapid screening differential proteomics by label-free spectral counting may only reveal changes in abundant proteins and, in case of protein-grouping ambiguities, require additional validation by, e.g., integration of XICs for specific peptides. Taken together, our study has demonstrated the utility of a mass spectrometry-based label-free approach to screen tissue for protein biomarkers, which may be explioted in numerous applications ranging from drug discovery and development, toxicology, and characterization of pathological specimens.
Supplementary Material
Acknowledgment
This research was supported by the National Institutes of Health (grant numbers NS044765, AG025384 and AG027956). Laszlo Prokai is the Robert A. Welch Professor of the University of North Texas Health Science Center (endowment number BK-0031). The authors thank Darius V. Bonds and Jingwei Fu for their excellent technical assistance and Thermo Electron for the evaluation copy of the SIEVE™ software.
Abbreviations
- 2DE-GE
two dimensional gel electrophoresis
- BSA
bovine serum albumin
- Des
desmin
- E2
17β-estradiol
- FTICR
Fourier transform ion cyclotron resonance
- GO
gene ontology
- LC
liquid chromatography
- Lmna
lumican
- LTQ
linear quadrupole ion trap
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- XIC
extracted ion chromatogram
Footnotes
Supporting Information Available: A representative list of proteins identified from mouse cortex extract by conventional (overnight) versus microwave-assisted (60 min) tryptic digestion, immobility times in forced swim test before tissue harvesting, uterine wet weights, Western blot analysis of ATP synthase β-subunit in cortical protein extracts and ATP synthase α-subunit in uterine tissue extracts of control versus E2-treated ovariectomized mice. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Shi R, Kumar C, Zougman A, Zhang Y, Podtelejnikov A, Cox J, Wisniewski JR, Mann M. Analysis of the mouse liver proteome using advanced mass spectrometry. J. Proteome Res. 2007;6:2963–2972. doi: 10.1021/pr0605668. [DOI] [PubMed] [Google Scholar]
- 2.Wu CC, Yates JR., 3rd. The application of mass spectrometry to membrane proteomics. Nat. Biotechnol. 2003;21:262–267. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Sun L, Yu Y, Xue Y, Yang P. Amino acid-coded tagging approaches in quantitative proteomics. Expert Rev. Proteomics. 2007;4:25–37. doi: 10.1586/14789450.4.1.25. [DOI] [PubMed] [Google Scholar]
- 4.Liu H, Sadygov RG, Yates JR., 3rd. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 5.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Higgs RE, Knierman MD, Gelfanova V, Butler JP, Hale JE. Comprehensive label-free method for the relative quantification of proteins from biological samples. J. Proteome Res. 2005;4:1442–1450. doi: 10.1021/pr050109b. [DOI] [PubMed] [Google Scholar]
- 7.Sleno L, Emili A. Proteomic methods for drug target discovery. Curr. Opin. Chem Biol. 2008;12:46–54. doi: 10.1016/j.cbpa.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 8.Cavalla D. Therapeutic switching: A new strategic approach to enhance R and D productivity. IDrugs. 2005;8:914–918. [PubMed] [Google Scholar]
- 9.Morphy R, Rankovic Z. Designed multiple ligands. An emerging drug discovery paradigm. J. Med. Chem. 2005;48:6523–6543. doi: 10.1021/jm058225d. [DOI] [PubMed] [Google Scholar]
- 10.Cavalli A, Bolognesi ML, Minarini A, Rosini M, Tumiatti V, Recanatini M, Melchiorre C. Multi-target-directed ligands to combat neurodegenerative diseases. J. Med. Chem. 2008;51:347–372. doi: 10.1021/jm7009364. [DOI] [PubMed] [Google Scholar]
- 11.Behl C. Oestrogen as a neuroprotective hormone. Nat. Rev. Neurosci. 2002;3:433–442. doi: 10.1038/nrn846. [DOI] [PubMed] [Google Scholar]
- 12.McEwen B. Invited review: Estrogens effects on the brain: multiple sites and molecular mechanisms. J. Appl. Physiol. 2001;91:2785–2801. doi: 10.1152/jappl.2001.91.6.2785. [DOI] [PubMed] [Google Scholar]
- 13.Pastorelli R, Carpi D, Airoldi L, Chiabrando C, Bagnati R, Fanelli R, Moverare S, Ohlsson C. Proteome analysis for the identification of in vitro estrogen-regulated proteins in bone. Proteomics. 2005;5:4936–4945. doi: 10.1002/pmic.200401325. [DOI] [PubMed] [Google Scholar]
- 14.Mo B, Callegari E, Telefont M, Renner KJ. Estrogen regulation of proteins in the rat ventrometrial nucleus of the hypothalamus. J. Proteome Res. 2008;7:5040–5048. doi: 10.1021/pr8005974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilsen J, Irwin RW, Gallaher TK, Brinton RD. Estradiol in vivo regulation of brain mitochondrial proteome. J. Neurosci. 2007;27:14069–14077. doi: 10.1523/JNEUROSCI.4391-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganong WF. Review of Medical Physiology. McGraw-Hill; New York: 2005. pp. 441–442. [Google Scholar]
- 17.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 18.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 19.Sokal RR, Rohlf FJ. Biometry: The Principles and Practice of Statistics in Biological Research. W.H. Freeman and Company; New York, NY: 1995. pp. 729–731. [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 21.Lill JR, Ingle ES, Liu PS, Pham V, Sandoval WN. Microwave-assisted proteomics. Mass Spectrom. Rev. 2007;26:657–671. doi: 10.1002/mas.20140. [DOI] [PubMed] [Google Scholar]
- 22.Bekku N, Yoshimura H. Animal model of menopausal depressive-like state in female mice: prolongation of immobility time in the forced swimming test following ovariectomy. Psychopharmacology. 2005;183:200–207. doi: 10.1007/s00213-005-0179-0. [DOI] [PubMed] [Google Scholar]
- 23.Simpkins JW, Wang J, Wang X, Perez E, Prokai L, Dykens JA. Mitochondria play a central role in estrogen-induced neuroprotection. Curr. Drug Targets CNS Neurol. Disord. 2005;4:69–83. doi: 10.2174/1568007053005073. [DOI] [PubMed] [Google Scholar]
- 24.Lodish H, et al. Molecular Cell Biology. 6th Ed W.H. Freeman and Company; New York, NY: 2008. pp. 479–510. [Google Scholar]
- 25.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong SH, Nah HY, Lee JY, Gye MC, Kim CH, Kim MK. Analysis of estrogen-regulated genes in mouse uterus using cDNA microarray and laser capture microdissection. J. Endocrinol. 2004;181:157–167. doi: 10.1677/joe.0.1810157. [DOI] [PubMed] [Google Scholar]
- 27.Fountoulakis M, Schuller E, Hardmeier R, Berndt P, Lubec G. Rat brain proteins: Two-dimensional protein database and variations in the expression level. Electrophoresis. 1999;20:3572–3579. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3572::AID-ELPS3572>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 28.Klose J, Nock C, Herrmann M, Stuhler K, Marcus K, Bluggel M, Krause E, Schalkwyk LC, Rastan S, Brown SDM, Bussow K, Himmelbauer H, Lehrach H. Genetic analysis of the mouse brain proteome. Nature Genet. 2002;30:385–393. doi: 10.1038/ng861. [DOI] [PubMed] [Google Scholar]
- 29.Krapfenbauer K, Fountoulakis M, Lubec G. A rat brain protein expression map including cytosolic and enriched mitochondrial and microsomal fractions. Electrophoresis. 2003;24:1847–1870. doi: 10.1002/elps.200305401. [DOI] [PubMed] [Google Scholar]
- 30.Xixi E, Dimitraki P, Vougas K, Kossida S, Lubec G, Fountoulakis M. Proteomic analysis of the mouse brain following protein enrichment by preparative electrophoresis. Electrophoresis. 2006;27:1424–1431. doi: 10.1002/elps.200500562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.