Abstract
p63 and p73, members of the p53 family, have been shown to be functionally distinct from p53. Vitamin D receptor (VDR) is a ligand (vitamin D3)-dependent transcription factor, which is shown to play a major role in calcium homeostasis and keratinocyte differentiation. Vitamin D and its analogues in combination with DNA-damaging agents are extensively used for cancer chemotherapy. In this report, we examined whether p53 affects p63-mediated induction of VDR and studied the effect of DNA damage on VDR induction in p53 null cell lines. Our results demonstrate that p53 itself does not induce VDR expression, nor does it affect p63-mediated VDR induction in the cell lines tested in this study. Furthermore, we observed p53-independent activation of VDR upon DNA damage and associated the induction of VDR to p73. We have demonstrated that ectopic expression of various p73 isoforms can induce VDR expression. Inhibition of p73 in cells treated with DNA-damaging agents exhibited decreased VDR expression. Finally, we show that upon DNA damage, induction of VDR sensitizes the cells to vitamin D treatment. In conclusion, our results indicate that VDR is regulated by p63 and p73 and that the induction of VDR expression upon DNA damage is p73-dependent.
p53, the most frequently altered gene in human cancers, controls multiple signaling pathways by regulating the genes involved in cell cycle arrest, apoptosis, DNA repair, cellular senescence, and inhibition of angiogenesis (1). Stabilization of p53 protein occurs in response to various stress stimuli including DNA damage, viral infection, or oncogenic activation (1, 2). Although the functional significance of p53 and its interacting proteins have been studied extensively, it was not until 1997 that the functional homologues of p53, namely p63 and p73, were discovered. Despite being structurally similar to p53, p63 and p73 were shown to be functionally more diverse and distinct from p53 (3, 4). p63 knock-out mice were born with severe developmental defects, which included lack of skin and various epithelial tissues, whereas p73 knock-out mice were born with neuronal defects as well as changes in sexual behavior (5, 6).
Functional diversities associated with p63 and p73 are partly due to their ability to generate multiple transcripts. Differential promoter usage by both p63 and p73 results in the generation of either transactivation domain containing isoforms (TAp63 and TAp73) or NH2-terminally truncated isoforms (ΔNp63 and ΔNp73) (7). Additionally, multiple carboxyl termini variants, α, β, and γ of p63 and α, β, γ, ε, and δ of p73, are generated due to the differential splicing of COOH terminus. Due to their structural similarity with p53, TA isoforms of both p63 and p73 have been shown to activate various p53-responsive genes and promote cell cycle arrest and apoptosis (8, 9). On the contrary, elevated levels of ΔNp63 and ΔNp73 isoforms have been reported in several human cancers, and these isoforms have also been associated with the induction of various genes involved in promoting cell proliferation (10–14). Additionally, ΔN isoforms of p63 and p73 have also been shown to act in a dominant negative manner and suppress the TAp63 and TAp73 isoforms and p53-mediated antiproliferative functions (15).
Several reports have implicated that, upon DNA damage, both TAp63 and TAp73 isoforms can lead to cell cycle arrest and apoptosis (16). DNA damage-induced expression of TAp63α promotes apoptosis in hepatocarcinoma cell lines (17). TAp73 variants were also shown to be induced by a subset of DNA-damaging agents in various cancer cell lines (18). In addition, TAp73α and TAp73β play a pivotal role in p53-independent chemosensitivity (19). In particular, transcriptional activity of p73α is required for E2F1-mediated apoptosis of T-cells (20). Although the precise role of p63 and p73 in p53-dependent apoptosis is still unclear, a significant down-regulation of p53-mediated apoptosis upon γ-irradiation in p63- and p73-ablated mouse embryo fibroblasts suggests that both p63 and p73 are required for the biological activity of p53 (21).
1α,25-Dihydroxyvitamin D3, the active form of secosteroid hormone vitamin D, exerts its biological actions through vitamin D receptor (VDR).2 VDR is a ligand-dependent transcription factor, which belongs to the nuclear receptor family of proteins (22). Activation of VDR by the active form of vitamin D results in heterodimerization of VDR with the retinoic acid receptor and subsequent transcriptional regulation of its responsive genes (22). Vitamin D receptor primarily regulates the genes involved in calcium homeostasis and bone formation (23). Apart from exerting its classical functions, vitamin D receptor has been shown to exert antiproliferative actions in several different cancer cell lines (24, 25). Recently, VDR has been implicated to play a broader role in the differentiation of epithelial keratinocytes and immunomodulatory activities (26, 27). Although extensive work has been performed on the biological function of vitamin D and its analogs, little is known about the transcriptional regulation of VDR. In this report, we studied the transcriptional regulation of VDR by p53 family members in both stressed and unstressed conditions. Our results demonstrate the differential regulation of VDR by p63 and p73 as well as the p53-independent activation of VDR upon DNA damage. Examination of the mechanism of VDR induction upon DNA damage demonstrated the requirement of p73 for DNA damage-induced expression of VDR.
EXPERIMENTAL PROCEDURES
Cell Lines and Plasmids
Human non-small lung carcinoma cell line H1299 and human osteosarcoma cell line SaoS2 were obtained from ATCC and were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine calf serum, 250 units of penicillin, and 250 μg of streptomycin. Expression plasmids encoding TAp63γ, TAp73β, ΔNp73β, and TAp73α were constructed in cytomegalovirus-driven pcDNA3.1A vector as described earlier (28). VDR full-length and minimal promoter luciferase constructs were constructed as described previously (29). p53 expression plasmid was a generous gift from Dr. Steven Berberich (Wright State University). VDRE-luciferase (VDRE-Luc) reporter was a generous gift from Dr. Alberto Munoz (University of Madrid). Etoposide, doxorubicin, and 1α,25-dihydroxyvitamin D3 were purchased from Sigma. For constructing VDR short hairpin RNA expression vector, shVDR target sequence was selected using algorithms obtained from Dharmacon, Inc. (Chicago, IL) and Genscript Corp. (Piscataway, NJ). Subsequently, the target sequence to VDR was cloned into the PstI site in the pSilencer 3.1 vector obtained from Ambion Inc. (Austin, TX). Target sequence used for shVDR was as follows: shVDR-sense, 5′-GATCCGCAGCGCATTGCCATATTCTGCAGATATGGCAATGATGCGCTGCTGTTTTTTGGAAA-3′; shVDR-antisense, 5′-AGCTTTTCCAAAAAACAGCAGCGCATCATTGCCATATCTGCAGAATATGGCAATGATGCGCTGCG-3′.
Transfections
Cells were seeded onto 6-well plates prior to the day of transfection. At around 80% confluence, transient transfections were performed using Lipofectamine2000 reagent (Invitrogen) with the desired plasmids in serum-free Dulbecco's modified Eagle's medium. For all of the studies, a total of 3 μg of expression plasmids, encoding TAp63γ, TAp73β, ΔNp73β, p53, and TAp73α, were used. For p53 dose-dependent studies, 1, 2, and 3 μg of p53 plasmid was used. After a 5-h incubation, the medium was replaced with Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine calf serum and 1% PS.
p73 Knock-down by siRNA
Cells were seeded in 6-well plates prior to the day of transfection. At around 30- 40% confluence, cells were subjected to calcium phosphate-mediated transfection. Briefly, 1 h before transfection, medium was changed, and the cells were kept in an incubator with 3% CO2. For each transfection reaction, siRNA (Qiagen) was mixed with CaCl2 (125 mm final concentration) and 2× BBS (50 mm BES, 280 mm NaCl, 1.5 mm Na2HPO4). The transfection mixture was incubated for 3 min at room temperature and then added to the respective wells. As control, an AllStars negative control nonsilencing RNA (Qiagen) was used. Six hours post-transfection, medium was changed, and cells were transferred to an incubator with 5% CO2. At 24 h, another round of transfection was performed as described above. After an additional 48 h, total RNA was harvested using the RNAeasy kit as per the manufacturer's protocol (Qiagen, Valencia, CA). Sequences used for p73 siRNA were as follows: sense, r(CGGGAUGCUCAACAACCAU)dTdT; antisense, r(AUGGUUGUUGAGCAUCCC G)dGdG.
Reverse Transcription-PCR
1 μg of total RNA was used to synthesize cDNA by using the TaqMan reverse transcription kit (Applied Biosystems, Foster City, CA). Quantitative real time PCR analysis was performed in a 96-well microtiter plate format on an ABI Prism7900HT sequence detection system using TaqMan Universal master mix and Assays on Demand specific for VDR (Hs_0017213_m1), p21 (Hs_00355782_m1), p73 (Hs_00232088_m1), and CYP27B1 ((Hs_00168017_m1) (PerkinElmer Life Sciences). Relative mRNA quantitation was performed by using the comparative ΔΔCt method as described earlier (28, 29).
Protein and Immunoblot Studies
At 24 h after transfection or treatments, total protein was extracted from cells using radioimmune precipitation buffer (0.5% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS, phosphate-buffered saline, pH 7.4). Protein extracts were run on 10% SDS-PAGE and transferred onto polyvinylidene difluoride membrane and blocked with 5% blocking milk solution. Membranes were subsequently immunoblotted with antibodies to detect specific proteins. Monoclonal anti-VDR, rabbit polyclonal anti-p21, rabbit polyclonal anti-p53 FL-193, monoclonal anti-p63 4A4 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and monoclonal anti-β-actin (Sigma) antibodies were used to detect VDR, p21, p53, p63, and β-actin expression, respectively. Monoclonal Ab-6 (Calbiochem) was used to detect p53. Appropriate horseradish peroxidase-conjugated antibodies (Promega, Madison, WI) were used as secondary antibodies, and the Supersignal West-pico Chemiluminescent Substrate kit (Pierce) was used to detect the chemiluminescence signal.
Immunoprecipitation for Endogenous p73α
After 24 h of doxorubicin and etoposide treatments, cells were harvested for total protein using radioimmune precipitation buffer, and 1 mg of total protein was used for immunoprecipitation. Briefly, total protein was precleared with 20 μl of rec-protein G-Sepharose beads (Invitrogen) for 1 h at 4 °C. After preclearing, beads were removed, and total protein was incubated with rotation overnight at 4 °C with 1 μg of monoclonal anti-p73 antibody (Ab-4 Lab Vision Corp., Fremont, CA). The next day, immunoprecipitated samples were incubated with rec-protein G-Sepharose beads for 1 h, followed by four washes with radioimmune precipitation buffer to remove the unbound proteins. Immunoprecipitated samples with beads were run on 10% SDS gel and immunoblotted with rabbit polyclonal anti-p73 antibody (Bethyl Laboratories Inc., Montgomery, TX).
Transactivation Assays
To measure the VDR-Luc and VDR(min)-Luc reporter activities, cells were seeded in 6-well plates at 2.25 × 105 cells/well. At 24 h after seeding, cells were transfected with 100 ng of reporter constructs along with the desired plasmids as indicated. After 24 h of post transfection, cells were washed once with phosphate-buffered saline and harvested in passive lysis buffer (Promega, Madison, WI). Dual luciferase assays were performed to detect both firefly and Renilla luciferase activity using the Dual-Luciferase Reporter 1000 Assay System as per the manufacturer's protocol (Promega, Madison, WI). The relative luciferase activity was measured by calculating the ratio of Renilla luciferase activity to firefly luciferase activity. To measure the vitamin D3-mediated transcriptional activity of VDR, H1299 cells were seeded in 12-well plates at 6 × 104 cells/well and transfected with 50 ng of VDRE-Luc reporter along with the desired plasmids as indicated. The next day, cells were treated with 8 μm etoposide for 12 h, and then medium was replaced with fresh medium containing 1α,25-dihydroxyvitamin D3 (VD3). After 24 h of post VD3 treatments, cells were subjected to a dual luciferase assay as mentioned earlier.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation analysis was performed by using a chromatin immunoprecipitation kit (Upstate Cell Signaling Solutions, Waltham, MA) according to manufacturer's protocol. Briefly, H1299 cells were seeded onto three 15-cm dishes, and after 24 h, cells were fixed on a plate by using formaldehyde for 10 min. After fixation, cells were homogenized, and nuclei were subjected to sonication to shear the DNA. Fragmented chromatin was precleared using protein G beads for 1.5 h. Precleared chromatin was subjected to overnight immunoprecipitation with Mouse monoclonal anti-p73 Ab-4 (Lab Vision Corp., Fremont, CA) or normal rabbit IgG antibodies (Santa Cruz Biotechnology). Part of the precleared chromatin that was not subjected to immunoprecipitation was used as input DNA. After immunoprecipitation, both input DNA and immunoprecipitated chromatin were reverse cross-linked, and DNA was eluted. The eluted DNA was subjected to PCR amplification using GoTaq Green PCR master mix as per the manufacturer's protocol (Promega, Madison, WI) using primers specific for VDR (forward, 5′-CCCAAGCTTGATGATTATAGGTGCGGATACCCG-3′; reverse, 5′-CGGGGTACCCAGTAACAGGTTGCGACGGAG-3′) and p21 (forward, 5′-GGTACCGGCACTCTTGTTCCCCCAGGCTG-3′; reverse, 5′-CTCGAGACCATCCCCTTCCTCACCTGAAAA-3′). PCR conditions used for both VDR and p21 were as follows. A total of 40 cycles were performed, each consisting of 30 s at 94 °C, 30 s at 60 °C, and 45 s at 68 °C.
RESULTS
p53 Does Not Induce VDR Expression or Affect p63-mediated Induction of VDR
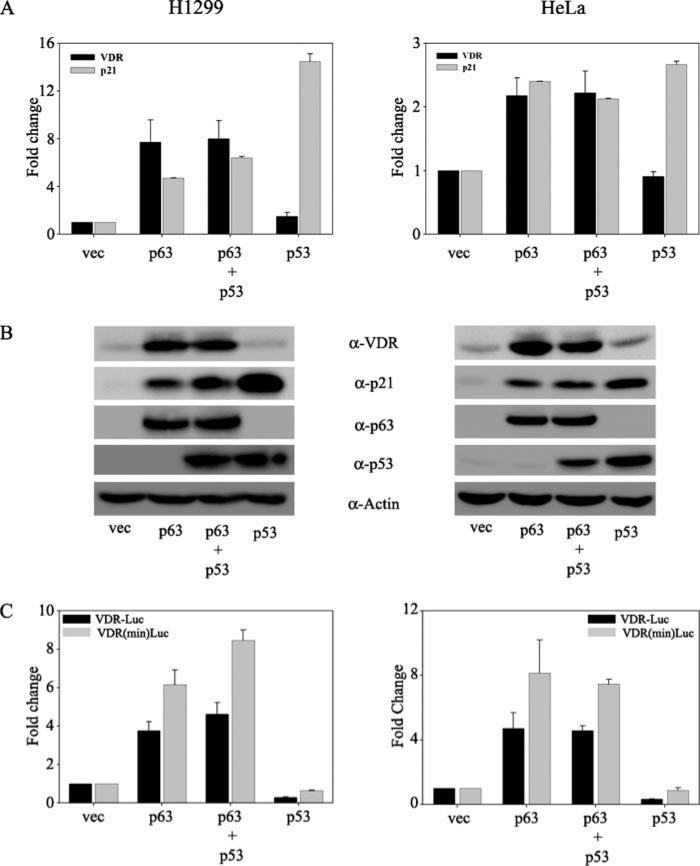
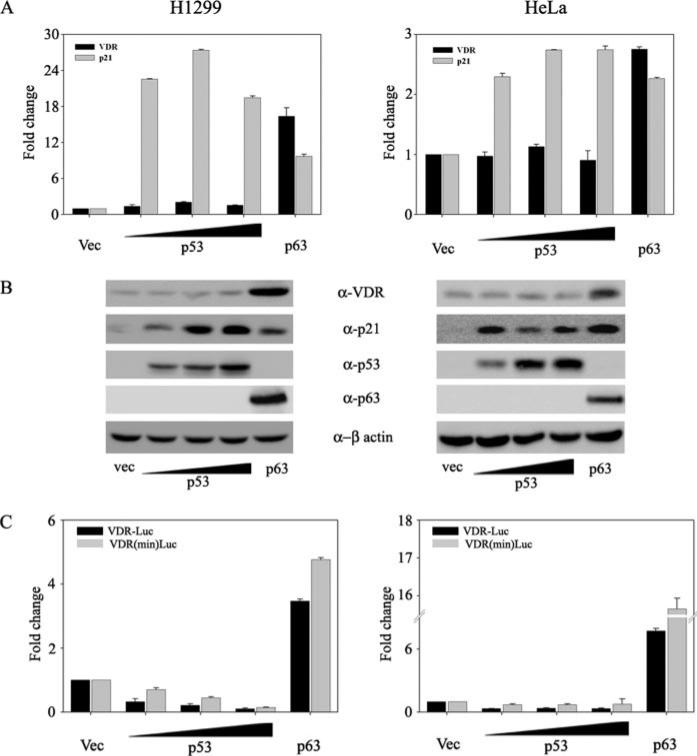
Our laboratory has previously demonstrated that p63 but not p53 induces VDR expression (29). However, a recent report has demonstrated that p53 can induce VDR expression (30). Toward understanding this discrepancy, we wanted to further understand the role of p53 family members in VDR induction. Given the significant homology between p53 and TAp63 isoforms, we asked whether p53 had any effect on p63-mediated VDR induction. To address this, we overexpressed p53 and p63 alone or together in p53–/– H1299 and HeLa cell lines (Fig. 1). We demonstrated that although TAp63γ by itself, as expected, induces VDR expression, co-transfection of p53 along with p63 did not significantly affect the ability of p63 to induce VDR expression (Fig. 1, A and B). Regulation of p21 was used as a positive control for the transactivation of both p53 and p63. Similar results were obtained when we tested the ability of p63 alone or along with p53 to regulate the activity of either the full-length VDR reporter or the minimal VDR reporter, previously shown to be activated by p63 (29). Although p63 significantly up-regulated the activity of both of these reporters, p53 did not affect p63-mediated transactivation of either of these VDR reporters (Fig. 1C). Consistent with our previous report, p53 alone was once again unable to induce expression of VDR or affect the VDR reporter activity in both of the cell lines tested (Fig. 1). In order to address whether the effect of p53 on VDR was dose-dependent, we further tested the effect of increasing concentrations of p53 on endogenous VDR expression (Fig. 2). We confirmed that p53 at all doses tested did not result in induction of VDR at either the mRNA or protein level in both of the cell lines tested (Fig. 2, A and B). As expected, p21, a downstream target of p53, was significantly induced by p53 and served as a positive control. In addition, p53 at any dose tested also did not significantly change full-length VDR or the minimal VDR reporter activity (Fig. 2C).
FIGURE 1. p53 does not affect the p63-mediated induction of VDR.
H1299 and HeLa cells were transfected with empty vector (vec), p63, p53, or p63 and p53 together as indicated. A, at 24 h post-transfection, total RNA was harvested and subjected to TaqMan reverse transcription-PCR. Glyceraldehyde-3-phosphate dehydrogenase transcript levels were used as normalization for the VDR mRNA levels. The y axis represents -fold change in VDR and p21 transcript levels relative to empty vector-transfected cells. B, whole cell extracts of transfected cells were subjected to immunoblot analysis using anti-VDR, anti-p21, anti-p53, and anti-p63 antibodies. Immunoblot analysis for β-actin served as the loading control. C, H1299 and HeLa cells were co-transfected with either full-length or the minimal VDR promoter construct along with either empty vector or expression plasmids encoding p63 alone or with p53 or p53 alone as indicated. A constant amount of expression plasmid encoding Renilla luciferase was included in all transfections to normalize for transfection efficiency. At 24 h post-transfection, cells were harvested in passive lysis buffer (PLB) and subjected to dual luciferase assays as per the manufacturer's protocol. RLU/R-Luc ratios were calculated to normalize for transfection efficiency. The y axis represents -fold change relative to empty vector control.
FIGURE 2. p53 does not induce VDR expression.
H1299 and HeLa cells were transfected with either empty vector (Vec) or increasing doses of expression plasmids encoding p53 or p63 alone as indicated. A, at 24 h post-transfection, total RNA was harvested and subjected to TaqMan reverse transcriptase PCR. The y axis represents -fold change in VDR and p21 mRNA levels relative to empty vector-transfected cells. B, whole cell lysates were subjected to immunoblot analysis using anti-VDR, anti-p21, anti-p53, anti-β-actin, and anti-p63 antibodies. C, H1299 and HeLa cells were co-transfected with either full-length VDR promoter construct or the minimal VDR promoter construct with increasing concentrations of expression plasmids encoding p53 or p63 alone as indicated. At 24 h post-transfection, cells were harvested in PLB and subjected to dual luciferase assays as per the protocol. RLU/R-Luc ratios were calculated to normalize for transfection efficiency. The y axis represents -fold change relative to empty vector control.
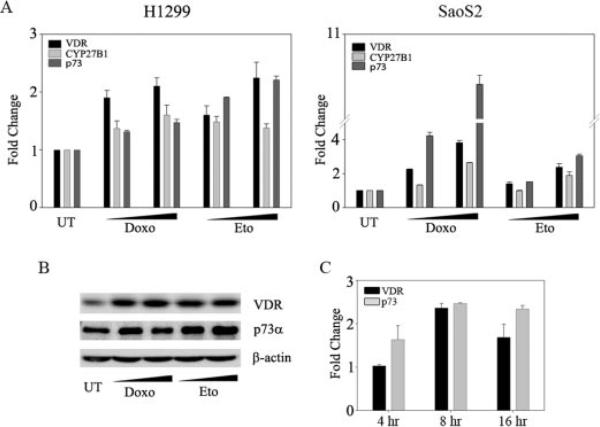
DNA Damage-induced Expression of VDR Is Independent of p53 and Correlates with p73 Expression Levels
To test whether p53 is required for DNA damage-induced VDR expression, as demonstrated earlier, we examined the effects of two DNA-damaging agents, doxorubicin and etoposide, on two p53 null cells, H1299 and SaoS2 (Fig. 3). Both DNA-damaging agents led to an induction of VDR compared with untreated cells, demonstrating that DNA damage alone can lead to the transcriptional activation of VDR (Fig. 3A). To assess whether increase in VDR upon DNA damage leads to a concomitant increase in the transcriptional activity of VDR, we examined the transcript levels of 25-hydroxyvitamin D3 1α-hydroxylase (CYP27B1), a downstream target of VDR. An increase in CYP27B1 transcript levels was also observed in the doxorubicin- and etoposide-treated cells in both the H1299 and SaoS2 cell lines (Fig. 3A). Since previous reports have demonstrated that both of these cell lines have endogenous p73, and p73 is induced upon DNA damage in various cancer cell lines (31–33), we assessed if there was a correlation between p73 and VDR expression levels. To test this, we treated both H1299 and SaoS2 cell lines with doxorubicin or etoposide and monitored transcript levels of both VDR and p73 (Fig. 3A). We demonstrated that both p73 and VDR are induced upon DNA damage. Consistent with the simultaneous increase in the transcript levels of both VDR and p73 upon DNA damage, we also observed an increase in the protein levels of both VDR and p73 upon treatment with DNA-damaging agents (Fig. 3B). Similar results were obtained in another p53 null HeLa cell line (data not shown). In order to assess whether p73 precedes the VDR induction upon DNA damage, we monitored the p73 and VDR transcript levels at different time points following the etoposide treatment. Consistent with a previous report, we observed a significant increase in p73 transcript levels at the 4 h time point following DNA damage (31). However, induction in VDR transcript levels was only observed at the 8 h time point post-treatment (Fig. 3C). Together, these results suggest that VDR induction upon DNA damage may possibly occur via p73.
FIGURE 3. DNA damage-induced expression of VDR is independent of p53 and is coupled to p73 expression.
H1299 and SaoS2 cells were either untreated (UT) or treated with doxorubicin (Doxo; 0.2 and 0.4 μm) or etoposide (Eto; 6 and 8 μm for H1299 and 2 and 4 μm for SaoS2). A, 24 h post-treatment, cells were harvested, and TaqMan real time PCR was performed to determine the transient expression levels of VDR, CYP27B1, and p73. The y axis indicates change in mRNA levels compared with untreated cells. B, 24 h post-treatment, H1299 cells were harvested and subjected to Western blot analysis to detect the protein levels of VDR and p73 using gene-specific antibodies, respectively. For the detection of p73 protein levels, whole cell lysates were first immunoprecipitated with monoclonal anti-p73 antibody and immunoblotted with polyclonal anti-p73 antibody. C, H1299 cells were treated with 8 μm etoposide, and total RNA was extracted at indicated time points post-treatment. TaqMan-based reverse transcription-PCR was performed to detect the transcript levels of p73 and VDR. The y axis represents the -fold change in VDR and p73 transcript levels relative to untreated cells at the respective time points.
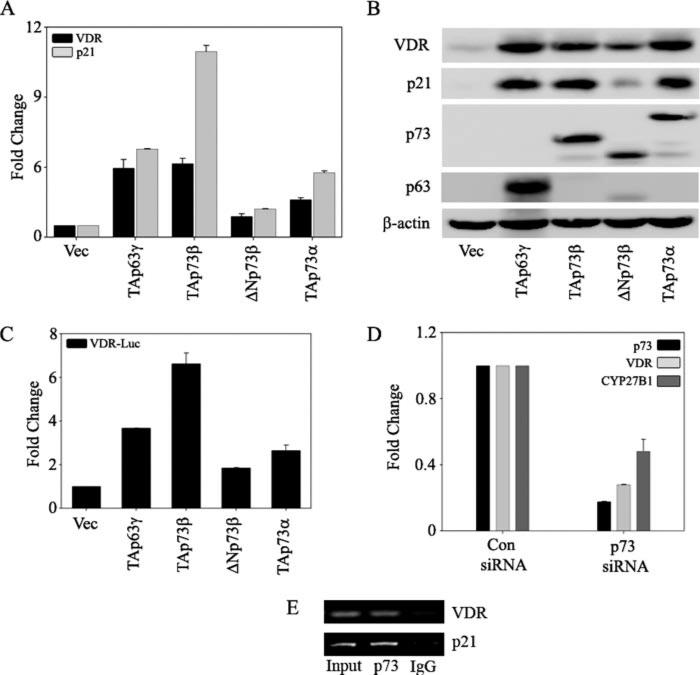
p73 Is Required for DNA Damage-induced Expression of VDR
To examine whether p73 is required for the induction of VDR upon DNA damage, we first tested whether VDR is transcriptionally activated by the p73 isoforms in a p53 null background. Ectopic expression of TAp73β and TAp73α in H1299 cells led to a significant increase in VDR and p21 expression at both mRNA and protein levels. In contrast, ΔNp73β led to a modest increase in both VDR and p21 expression levels when compared with the other TAp73 isoforms. TAp63γ was used as a positive control for both VDR and p21 induction (Fig. 4, A and B). We next tested the ability of different p73 isoforms to regulate the activity of the full-length VDR reporter, previously shown to be activated by p63. An increase in VDR reporter activity was observed with both TAp73β and TAp73α isoforms (Fig. 4C). Silencing of p73 expression in cells led to a concomitant decrease in the basal levels of both VDR and CYP27B1, a downstream target of VDR (Fig. 4D). Finally, chromatin immunoprecipitation analysis with intact H1299 cells demonstrated that endogenous p73 protein binds to the endogenous VDR promoter (Fig. 4E). Binding of p73 to the p21 promoter, a known target of p73, was used as a positive control. Together, these results demonstrate that VDR is transcriptionally activated by multiple p73 isoforms and that VDR is a direct target of p73.
FIGURE 4. p73 is essential for endogenous VDR expression.
H1299 cells were transfected with control vector, TAp63γ, TAp73β, ΔNp73β, or TAp73α as indicated. A, at 24 h post-transfection, total RNA was extracted to perform TaqMan reverse transcription-PCR to quantitate the transcript levels of VDR and p21 as indicated. The y axis represents the -fold change in mRNA levels of VDR and p21 compared with empty vector-transfected cells. B, at 24 h post-transfection, total protein was extracted to detect the endogenous VDR and p21 protein levels by using anti-VDR and anti-p21 antibodies, respectively. Ectopic expression of TAp63γ, TAp73β, ΔNp73β, and TAp73α was confirmed by using antibodies specific for p63 and p73. C, H1299 cells were co-transfected with full-length VDR promoter reporter along with the indicated plasmids. At 24 h post-transfection, cells were harvested in PLB and subjected to dual luciferase assays. RLU/R-Luc ratios were calculated to normalize for transfection efficiency. The y axis represents -fold change relative to empty vector control. D, H1299 cells were transfected with either control (Con) siRNA or p73 siRNA by using the calcium phosphate method as described under “Experimental Procedures.” At 48 h post-transfection, RNA was extracted and subjected to TaqMan real time PCR to detect p73, VDR, and CYP27B1. E, chromatin immunoprecipitation analysis was performed on H1299 cells as described under “Experimental Procedures.” Formaldehyde-cross-linked chromatin was immunoprecipitated with anti-p73 or normal IgG antibodies as indicated. Eluted DNA was PCR-amplified with primers specific for VDR and p21. p21 promoter amplification was used as a positive control for p73.
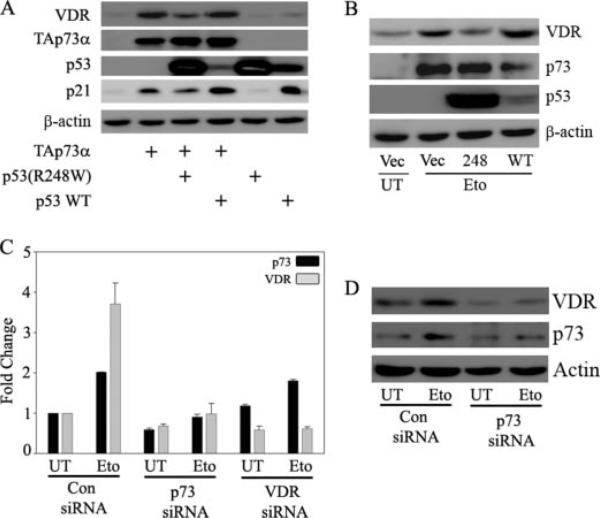
To demonstrate that the transcriptional activity of p73 is required for VDR induction upon DNA damage, we employed a dominant negative p53(R248W) mutant shown to inhibit the transcriptional activity of p73 (34). We first tested whether p53 (R248W) mutant can inhibit the ectopic p73-mediated induction of VDR protein. We observed that p53 (R248W) mutant significantly affected TAp73α-mediated induction of VDR protein levels (Fig. 5A). However, wild type p53 did not affect TAp73α-mediated induction of VDR. This is consistent with previous observations that wild type p53 cannot inhibit p73-mediated transcriptional activity (34). Next, we studied the effect of this p53 mutant on DNA damage-induced VDR induction to address whether blocking the endogenous p73α transcriptional activity affects the VDR induction seen upon DNA damage. We observed that inhibition of endogenous TAp73 transcriptional activity by p53 (R248W) mutant but not wild type p53 indeed significantly down-regulated the etoposide-induced VDR protein expression levels (Fig. 5B). To unequivocally demonstrate that p73 is required for VDR expression, we further examined whether silencing p73 expression affects VDR induction upon DNA damage. As shown in Fig. 5C, etoposide treatment alone can lead to a significant increase in expression of both p73 and VDR in cells transfected in control siRNA. VDR induction was inhibited in cells expressing siRNA against p73 in the presence or absence of etoposide treatment. Although siRNA against VDR abolishes VDR expression in the presence or absence of etoposide, it does not affect etoposide-mediated p73 induction. Similarly, in the presence of p73 siRNA, VDR protein levels were inhibited upon DNA damage (Fig. 5D). These results confirm that VDR induction upon DNA damage requires p73 induction, which occurs upstream of VDR. These findings clearly demonstrate that induction of VDR upon DNA damage in p53 null cells is occurring via induction of endogenous p73.
FIGURE 5. p73 is required for DNA damage-induced expression of VDR.
A, H1299 cells were transfected with control vector or expression plasmids encoding TAp73α, p53 wild type (WT), and p53 (R248W) mutant alone or in combination as indicated. At 24 h post-transfection, total protein was extracted for immunoblot analysis to detect the endogenous VDR protein levels. B, H1299 cells were transfected with empty vector or p53 (R248W) or p53 wild type as indicated. The next day, cells were either untreated (UT) or treated with 8 μm etoposide (Eto) as indicated. Whole cell lysate was prepared and endogenous VDR, and p73 protein levels were detected using anti-VDR and anti-p73 antibodies, respectively. Overexpression of wild type p53 and mutant p53 (R248W) was confirmed by using p53-specific antibody, and detection of β-actin was used to confirm equal amounts of protein loading. C and D, H1299 cells were transfected with control siRNA, VDR siRNA, or p73 siRNA as described under “Experimental Procedures,” and 24 h post-transfection, cells were trypsinized and replated onto 6-well plates. After cells adhered properly in wells, cells were treated with 8 μm etoposide or left untreated as indicated. At 12 h post-treatment, cells were harvested for either RNA or whole cell extract. TaqMan real time PCR was performed to detect the mRNA levels of p73 and VDR. Immunoblot analysis was performed to detect the endogenous VDR and p73 protein levels using anti-VDR and anti-p73 antibodies, respectively.
DNA Damage-induced VDR Expression Sensitizes the Cells to Vitamin D Treatment
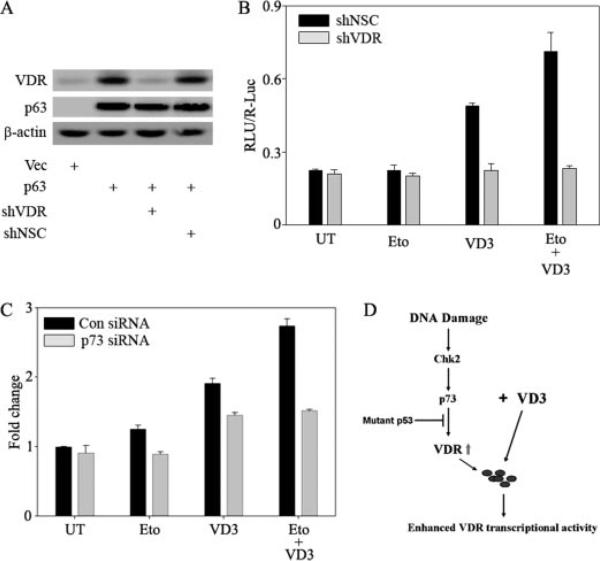
Since we have shown that VDR induction occurs upon DNA damage, and VD3 and its analogues are already used extensively for cancer chemotherapy, we wanted to determine whether VDR induction upon DNA damage sensitizes cells to the effects of VD3. To address this, we tested the VD3-mediated transcriptional activity using a reporter containing VDREs upstream of the luciferase gene in the presence or absence of VDR. Silencing of VDR was achieved by constructing a short hairpin RNA specific to VDR, and as a negative control, we used a nonsilencing control short hairpin RNA. To confirm that the shVDR can specifically silence VDR, we transfected H1299 cells with expression vector encoding TAp63γ along with short hairpin RNA to VDR or nonsilencing control. As shown in Fig. 6A, shVDR but not shNSC completely abolished the increase in VDR levels induced by TAp63γ, confirming the specificity of the shVDR. The effects of DNA damage and/or VD3 treatment on a VDRE-Luc reporter were studied in cells expressing shVDR or shNSC. Although etoposide alone did not lead to a significant increase in VDRE-Luc reporter activity, VD3 treatment led to a significant increase in the reporter activity, which was further increased in the presence of etoposide treatment (Fig. 6B). This suggested that although etoposide leads to increased VDR induction, as shown in Fig. 3, the increase in VDRE-Luc reporter activation is dependent on activation of VDR by its ligand. To confirm this, we showed that silencing VDR abolishes VDRE-Luc reporter activity in the presence or absence of etoposide and VD3. Furthermore, to confirm that p73-mediated induction of VDR is essential for the etoposide-mediated sensitization of VD3, we monitored the VD3-induced VDRE-Luc activity in the presence and absence of etoposide in cells transfected with p73 siRNA or control siRNA. We observed a significant decrease in VDRE-Luc reporter activity in cells treated with VD3 alone or in combination with etoposide treatment in cells in which p73 was silenced compared with cells transfected with control siRNA (Fig. 6C). Together, our results suggest that upon DNA damage, induction of endogenous p73 upon DNA damage leads to VDR induction, which can sensitize the cells to VD3.
FIGURE 6. DNA damage-induced VDR expression sensitizes the cells to vitamin D treatment.
A, H1299 cells were transfected with control vector or p63 alone or along with shVDR or shNSC as indicated. After 24 h of post-transfection, cells were harvested for whole cell lysates and subjected to immunoblot analysis to detect VDR, p63, and β-actin using gene-specific antibodies. B, H1299 cells were plated on 12-well plates and co-transfected with VDRE-Luc reporter along with either shNSC or shVDR as indicated. The next day, cells were either treated with 8 μm etoposide (Eto) or left untreated (UT) as indicated. 12 h of post-treatment, cells were treated with 100 nm VD3 as indicated. At 24 h after VD3 treatment, cells were harvested in PLB buffer and subjected to a dual luciferase assay. The y axis represents RLU/R-Luc relative luciferase units normalized for transfection efficiency. C, H1299 cells were transfected with control siRNA or p73 siRNA as described earlier. Immediately after two rounds of transfections with siRNA, cells were replated onto 24-well plates and transfected with VDRE-Luc reporter. The next day, cells were either treated with 8 μm etoposide or left untreated, as indicated. Subsequently, cells were treated with 100 nm VD3 as indicated. At 24 h post-VD3 treatment, cells were harvested in PLB buffer and subjected to a dual luciferase assay as mentioned earlier. D, model illustrating that DNA damage induces p73, causing downstream VDR induction, which subsequently sensitizes cells to VD3 treatment.
DISCUSSION
Identification of new transcriptional networks of p53 family members has given significant insight into the functional diversities of each member (35). However, understanding the functional cross-talk between each p53 family member based on its regulation of common and unique target genes will be crucial in deciphering their respective roles in tumorigenesis. In this report, we demonstrate that p53 can neither induce VDR expression by itself nor affect the p63-mediated up-regulation of VDR expression (Figs. 1 and 2). This therefore rules out the possibility of direct or indirect regulation of VDR by p53 in the cell lines tested in this study. Our results are contradictory to another report wherein p53 was shown to induce VDR and VDR was shown to be induced upon DNA damage in p53-containing cells. However, in depth analysis of VDR expression in multiple cancer cell lines under genotoxic stress has further ruled out the involvement of p53 in induction of VDR under physiological conditions. Induction of VDR mRNA and protein expression levels upon DNA damage was observed in HCT116 (p53–/–) as well as in SaoS2 (p53–/–) and in U2OS (p53+/+) cells (data not shown), suggesting a possible p53-independent VDR induction upon DNA damage. In addition, these results suggest transcriptional activation of VDR rather than protein stabilization. Human cancers display genetic alterations of key components involved in the DNA damage response signaling pathways, which lead to metastasis and invasiveness (36). p73 has been shown to be induced upon DNA damage in several different cancer cell lines and shown to be critical for chemosensitivity of these cells (10, 32). Interestingly, we observed a strong correlation between p73 and VDR levels upon DNA damage in these cell lines (Fig. 3). Ectopic expression of multiple p73 isoforms resulted in transcriptional activation of VDR, and silencing p73 expression led to reduction in endogenous VDR levels, suggesting a possible role of p73 in DNA damage-induced VDR expression (Fig. 4).
Recently, a subset of tumor-derived mutants of p53 has been shown to inhibit p73 transcriptional activity through direct physical interaction (37, 38). Ectopic expression of p53 (R248W) mutant resulted in down-regulation of VDR expression levels upon DNA damage, thus indicating the requirement of p73 for DNA damage-mediated VDR induction (Fig. 5). In support of this observation, with immunoprecipitation analysis, we did not observe any sort of interaction between VDR and p53 (R248W) mutant; however, as reported earlier, we observed an interaction between p73 and p53 (R248W) mutant (data not shown). Interestingly, lack of p73-mediated growth suppression has been reported in cells harboring p53 mutations (39), and knockdown of p53 mutants resulted in enhanced growth suppression various cancer cell lines (40, 41). Inhibition of DNA damage-induced p73 expression levels resulted in a concomitant decrease in VDR expression levels (Fig. 5). Furthermore, consistent with our chromatin immunoprecipitation analysis (Fig. 4E), inhibition of endogenous p73 resulted in down-regulation of basal VDR expression levels and its transcriptional activity, implying a direct transcriptional activation of VDR by p73 both at basal and stressed conditions. Our results demonstrate that DNA damage-induced VDR expression occurs via p73, which may be inhibited by tumor-derived p53 mutants (Fig. 5). Thus, researchers should consider the status of both p73 and mutant p53 when VDR-mediated growth inhibitions studies are conducted in human cancer cell lines.
It is well documented that VD3 and its analogues exert their antiproliferative activities through VDR and are extensively used for cancer chemotherapy (42–47). However, the sensitivity of VD3 analogues entirely depends on the endogenous levels of VDR (48); thus, elevation in VDR levels will further enhance the VD3-mediated antiproliferative activities. Results from our studies showed a significant increase in vitamin D-mediated VDR transcriptional activity with etoposide treatment (Fig. 6). Additionally, we have demonstrated that transcriptional activation of VDR by p73 upon DNA damage is essential for the DNA damage-mediated sensitization of vitamin D. In conclusion, we demonstrated the differential regulation of VDR by p53 family members and describe a novel p73-dependent and p53-independent activation of VDR upon DNA damage. p73-mediated induction of VDR levels upon DNA damage result in sensitization to VD3. This observation will aid in combinatorial use of DNA-damaging agents with vitamin D analogues, which will aid in better therapeutic strategies for cancer chemotherapy.
Acknowledgments
We thank Dr. Patrick Dennis and Dr. Carol Mercer for assistance with shVDR construct design and laboratory members Joshua DeWeese and Shama Khokhar for technical assistance and helpful discussions. This work was supported by NCI, National Institutes of Health, Grant CA118315-2 (to M. K.).
Footnotes
The abbreviations used are: VDR, vitamin D receptor; VD3, 1α,25-dihydroxyvitamin D3; CYP27B1, 25-hydroxyvitamin D3 1α-hydroxylase; VDRE-Luc, vitamin D-responsive elements upstream of luciferase; shVDR, short hairpin VDR; siRNA, small interfering RNA; BES, 2-[bis(2-hydroxyethyl)amino]ethanesulfonic acid; PLB, passive lysis buffer; shNCS, short hairpin non-silencing control.
REFERENCES
- 1.Giaccia AJ, Kastan MB. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 2.Schuler M, Green DR. Biochem. Soc. Trans. 2001;29:684–688. doi: 10.1042/0300-5127:0290684. [DOI] [PubMed] [Google Scholar]
- 3.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 4.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 5.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 6.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, McKeon F, Caput D. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 7.Scoumanne A, Harms KL, Chen X. Cancer Biol. Ther. 2005;4:1178–1185. doi: 10.4161/cbt.4.11.2254. [DOI] [PubMed] [Google Scholar]
- 8.Dietz S, Rother K, Bamberger C, Schmale H, Mossner J, Engeland K. FEBS Lett. 2002;525:93–99. doi: 10.1016/s0014-5793(02)03093-4. [DOI] [PubMed] [Google Scholar]
- 9.Harms K, Nozell S, Chen X. Cell Mol. Life Sci. 2004;61:822–842. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang KH, Lee JH, Kim KC, Ham SW, Kim MY, Choi KH. FEBS Lett. 2002;522:161–167. doi: 10.1016/s0014-5793(02)02921-6. [DOI] [PubMed] [Google Scholar]
- 11.Wu G, Osada M, Guo Z, Fomenkov A, Begum S, Zhao M, Upadhyay S, Xing M, Wu F, Moon C, Westra WH, Koch WM, Mantovani R, Califano JA, Ratovitski E, Sidransky D, Trink B. Cancer Res. 2005;65:758–766. [PubMed] [Google Scholar]
- 12.Senoo M, Matsumura Y, Habu S. Oncogene. 2002;21:2455–2465. doi: 10.1038/sj.onc.1205330. [DOI] [PubMed] [Google Scholar]
- 13.Sniezek JC, Matheny KE, Westfall MD, Pietenpol JA. Laryngoscope. 2004;114:2063–2072. doi: 10.1097/01.mlg.0000149437.35855.4b. [DOI] [PubMed] [Google Scholar]
- 14.Casciano I, Mazzocco K, Boni L, Pagnan G, Banelli B, Allemanni G, Ponzoni M, Tonini GP, Romani M. Cell Death Differ. 2002;9:246–251. doi: 10.1038/sj.cdd.4400993. [DOI] [PubMed] [Google Scholar]
- 15.Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. Mol. Cell Biol. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moll UM, Slade N. Mol. Cancer Res. 2004;2:371–386. [PubMed] [Google Scholar]
- 17.Gressner O, Schilling T, Lorenz K, Schulze Schleithoff E, Koch A, Schulze-Bergkamen H, Maria Lena A, Candi E, Terrinoni A, Valeria Catani M, Oren M, Melino G, Krammer PH, Stremmel W, Muller M. EMBO J. 2005;24:2458–2471. doi: 10.1038/sj.emboj.7600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozaki T, Nakagawara A. Cancer Sci. 2005;96:729–737. doi: 10.1111/j.1349-7006.2005.00116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 20.Lissy NA, Davis PK, Irwin M, Kaelin WG, Dowdy SF. Nature. 2000;407:642–645. doi: 10.1038/35036608. [DOI] [PubMed] [Google Scholar]
- 21.Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 22.Nezbedova P, Brtko J. Endocr. Regul. 2004;38:29–38. [PubMed] [Google Scholar]
- 23.Demay MB. Ann. N. Y. Acad. Sci. 2006;1068:204–213. doi: 10.1196/annals.1346.026. [DOI] [PubMed] [Google Scholar]
- 24.Campbell MJ, Gombart AF, Kwok SH, Park S, Koeffler HP. Oncogene. 2000;19:5091–5097. doi: 10.1038/sj.onc.1203888. [DOI] [PubMed] [Google Scholar]
- 25.Banerjee P, Chatterjee M. Mol. Cell. Biochem. 2003;253:247–254. doi: 10.1023/a:1026072118217. [DOI] [PubMed] [Google Scholar]
- 26.Hawker NP, Pennypacker SD, Chang SM, Bikle DD. J. Invest Dermatol. 2007;127:874–880. doi: 10.1038/sj.jid.5700624. [DOI] [PubMed] [Google Scholar]
- 27.Gombart AF, Luong QT, Koeffler HP. Anticancer Res. 2006;26:2531–2542. [PubMed] [Google Scholar]
- 28.Caserta TM, Kommagani R, Yuan Z, Robbins DJ, Mercer CA, Kadakia MP. Mol. Cancer Res. 2006;4:759–768. doi: 10.1158/1541-7786.MCR-05-0149. [DOI] [PubMed] [Google Scholar]
- 29.Kommagani R, Caserta TM, Kadakia MP. Oncogene. 2006;25:3745–3751. doi: 10.1038/sj.onc.1209412. [DOI] [PubMed] [Google Scholar]
- 30.Maruyama R, Aoki F, Toyota M, Sasaki Y, Akashi H, Mita H, Suzuki H, Akino K, Ohe-Toyota M, Maruyama Y, Tatsumi H, Imai K, Shinomura Y, Tokino T. Cancer Res. 2006;66:4574–4583. doi: 10.1158/0008-5472.CAN-05-2562. [DOI] [PubMed] [Google Scholar]
- 31.Urist M, Tanaka T, Poyurovsky MV, Prives C. Genes Dev. 2004;18:3041–3054. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Zheng Y, Zhu J, Jiang J, Wang J. Oncogene. 2001;20:769–774. doi: 10.1038/sj.onc.1204149. [DOI] [PubMed] [Google Scholar]
- 33.Costanzo A, Merlo P, Pediconi N, Fulco M, Sartorelli V, Cole PA, Fontemaggi G, Fanciulli M, Schiltz L, Blandino G, Balsano C, Levrero M. Mol. Cell. 2002;9:175–186. doi: 10.1016/s1097-2765(02)00431-8. [DOI] [PubMed] [Google Scholar]
- 34.Di Como CJ, Gaiddon C, Prives C. Mol. Cell Biol. 1999;19:1438–1449. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deyoung MP, Ellisen LW. Oncogene. 2007;26:5169–5183. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- 36.Lengauer C, Kinzler KW, Vogelstein B. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Prives C. Oncogene. 2007;26:2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- 38.Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. Mol. Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prabhu NS, Somasundaram K, Satyamoorthy K, Herlyn M, El-Deiry WS. Int. J. Oncol. 1998;13:5–9. doi: 10.3892/ijo.13.1.5. [DOI] [PubMed] [Google Scholar]
- 40.Vikhanskaya F, Lee MK, Mazzoletti M, Broggini M, Sabapathy K. Nucleic Acids Res. 2007;35:2093–2104. doi: 10.1093/nar/gkm099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scian MJ, Stagliano KE, Ellis MA, Hassan S, Bowman M, Miles MF, Deb SP, Deb S. Cancer Res. 2004;64:7447–7454. doi: 10.1158/0008-5472.CAN-04-1568. [DOI] [PubMed] [Google Scholar]
- 42.Rashid SF, Mountford JC, Gombart AF, Campbell MJ. Steroids. 2001;66:433–440. doi: 10.1016/s0039-128x(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 43.Banwell CM, Singh R, Stewart PM, Uskokovic MR, Campbell MJ. Recent Res. Cancer Res. 2003;164:83–98. doi: 10.1007/978-3-642-55580-0_5. [DOI] [PubMed] [Google Scholar]
- 44.Campbell MJ, Adorini L. Expert Opin. Ther. Targets. 2006;10:735–748. doi: 10.1517/14728222.10.5.735. [DOI] [PubMed] [Google Scholar]
- 45.Ordonez-Moran P, Larriba MJ, Pendas-Franco N, Aguilera O, Gonzalez-Sancho JM, Munoz A. Front. Biosci. 2005;10:2723–2749. doi: 10.2741/1731. [DOI] [PubMed] [Google Scholar]
- 46.Hershberger PA, McGuire TF, Yu WD, Zuhowski EG, Schellens JH, Egorin MJ, Trump DL, Johnson CS. Mol. Cancer Ther. 2002;1:821–829. [PubMed] [Google Scholar]
- 47.Torres R, Calle C, Aller P, Mata F. Mol. Cell Biochem. 2000;208:157–162. doi: 10.1023/a:1007089632152. [DOI] [PubMed] [Google Scholar]
- 48.Palmer HG, Gonzalez-Sancho JM, Espada J, Berciano MT, Puig I, Baulida J, Quintanilla M, Cano A, de Herreros AG, Lafarga M, Munoz A. J. Cell Biol. 2001;154:369–387. doi: 10.1083/jcb.200102028. [DOI] [PMC free article] [PubMed] [Google Scholar]