Abstract
The stringent response is important for bacterial survival under stressful conditions, such as amino acid starvation, and is characterized by the accumulation of ppGpp and pppGpp. ObgE (CgtA, YhbZ) is an essential conserved GTPase in Escherichia coli and several observations have implicated the protein in the control of the stringent response. However, consequences of the protein on specific responses to amino acid starvation have not been noted. We show that ObgE binds to ppGpp with biologically relevant affinity in vitro, implicating ppGpp as an in vivo ligand of ObgE. ObgE mutants increase the ratio of pppGpp to ppGpp within the cell during the stringent response. These changes are correlated with a delayed inhibition of DNA replication by the stringent response, delayed resumption of DNA replication after release, as well as a decreased survival to the response to amino acid deprivation. With this data, we place ObgE as an active effector of the response to amino acid starvation in vivo. Our data correlate the pppGpp/ppGpp ratio with DNA replication control under bacterial starvation conditions suggesting a possible role for the relative balance of these two nucleotides.
Keywords: YhbZ, ppGpp, pppGpp, stringent, replication
INTRODUCTION
Bacterial responses to stressful conditions play crucial roles in cell survival. Understanding how cells respond to these conditions can help to ultimately control their persistence and pathogenicity (Jain et al., 2006). Among the best-studied stress responses is the stringent response to amino acid starvation (Jain et al., 2006, Potrykus & Cashel, 2008). This response is characterized by the accumulation of ppGpp and pppGpp [reviewed in (Cashel, 1996)].
Under conditions of amino acid starvation, the presence of uncharged tRNAs signals to RelA to synthesize pppGpp and ppGpp from GTP or GDP, respectively, and ATP [reviewed in (Potrykus & Cashel, 2008)]. These reactions occur with similar speeds (Cochran & Byrne, 1974, Cashel, 1975), but because of the excess of GTP in Escherichia coli, RelA produces primarily pppGpp, which is then converted to ppGpp. Because of the abundance of ppGpp over pppGpp in the stringent response [see (Potrykus & Cashel, 2008)], ppGpp is thought to be the biologically relevant nucleotide, although the effects of each nucleotide are often not distinguished. An enzyme named Gpp (or GppA) is one factor that hydrolyzes pppGpp to ppGpp (Somerville & Ahmed, 1979). However, there remains at least one unidentified pppGppase in Escherichia coli (Somerville & Ahmed, 1979). The most marked outcome of the general rise in pppGpp and ppGpp, collectively known as “(p)ppGpp”, in the stringent response is an arrest of cell growth, manifest by the cessation of DNA replication and cell division, a reduced cell size, decrease in stable RNA synthesis and an increase in amino acid biosynthesis genes [(Schreiber et al., 1991, Schreiber et al., 1995, Barker et al., 2001, Cashel, 1996, Potrykus & Cashel, 2008) and references therein]. For cells to resume growth, it is necessary for them to decrease levels of ppGpp and/or pppGpp in the cell. This is accomplished by the ppGpp hydrolase, SpoT (Cashel, 1996). SpoT also can act as a (p)ppGpp synthetase and under other stressful conditions, SpoT may substitute for RelA to synthesize (p)ppGpp (Battesti & Bouveret, 2006, Seyfzadeh et al., 1993, Xiao et al., 1991, Vinella et al., 2005, Spira et al., 1995).
Several observations have indicated a connection between ObgE (Obg in E. coli) and the stringent response. Obg is a small protein conserved from bacteria to humans that is essential in all species tested to date, suggesting an important role for this enzyme within the cell (Obg has also been named “CgtA” for “conserved GTPase” but we conform to genetic nomenclature rules that give precedent to names that occur first in the literature). ObgE has been shown to have very weak GTPase activity (Tan et al., 2002) and a variety of cellular functions that have yet to be entirely defined. ObgE is required for chromosome segregation (Foti et al., 2007, Kobayashi et al., 2001), cell division (Foti et al., 2007, Foti et al., 2005, Kobayashi et al., 2001), recovery from DNA replication inhibition (Foti et al., 2005), and has effects on the levels of mature ribosomes in Escherichia coli (Sato et al., 2005). ObgE has also been shown to interact with ribosomal complexes (Wout et al., 2004, Jiang et al., 2007) and in Bacillus subtilis appears to play an important role in the general stress response (Scott & Haldenwang, 1999, Kuo et al., 2008), although it is not clear whether this role will be conserved in Escherichia coli (Scott & Haldenwang, 1999). Two crystal structures of Obg have been solved, one from Thermus thermophilus that clearly shows the three domains of this protein (Kukimoto-Niino et al., 2004), and a second from Bacillus subtilis (Buglino et al., 2002) with the C-terminal domain deleted. In the Bacillus subtilis structure ppGpp was found in half of the GTPase active sites (Buglino et al., 2002). The accidental crystallization of Obg with ppGpp may indicate a high affinity for ppGpp, or may have been a consequence of overexpression of Obg in exhausted media (Buglino et al., 2002). Initial studies in Bacillus subtilis suggested that Obg did not have an especially high affinity for ppGpp (Buglino et al., 2002). However, affinity constants have never been measured for Obg and ppGpp, and the relevance of this binding has not been confirmed.
Deficiencies of Obg/CgtA in Vibrio cholerae and Escherichia coli under certain conditions appeared to cause ppGpp accumulation constitutively and, in Vibrio cholerae, expression of a subset of genes known to be induced in the stringent response (Raskin et al., 2007, Jiang et al., 2007). Jiang et al. also showed that when (p)ppGpp accumulates, ObgE bound less to ribosomal complexes (Jiang et al., 2007). Finally, in both Escherichia coli and Vibrio cholerae, Obg/CgtA interacts with SpoT, the ppGpp synthetase and hydrolase (Wout et al., 2004, Raskin et al., 2007). However, the biological relevance of this interaction remains unknown and there are no reports of Obg/CgtA alteration of SpoT activity. These studies indicate a connection between ObgE and the stringent response and that the stringent response has an effect on ObgE localization (Jiang et. al., 2007). To date, ObgE has not been shown to have an effect on the cell’s response to amino acid starvation, which is the subject of our current study.
We investigate here the influence that ObgE activity has on the stringent response in Escherichia coli and show that it is an in vivo effector of the response to amino acid starvation. We show that ObgE binds to ppGpp in Escherichia coli with biologically relevant affinity, suggesting that this molecule is an in vivo ligand of ObgE, competitive with other nucleotides. We also show that ObgE changes the ratio of ppGpp to pppGpp in the cell during a response to amino acid starvation. ObgE subsequently affects cell survival and the control of DNA replication during the stringent response. By correlating the ratio of pppGpp/ppGpp to DNA replication inhibition, our data provide potential downstream effects of changes in the relative levels of two signaling molecules often thought of as one.
Results
ObgE binds to ppGpp with a similar affinity as GDP
Given the potential connections between ppGpp and ObgE, we first investigated the direct binding of ppGpp to ObgE to determine if the strength of the interaction was sufficient to yield biological effects.
Binding of Obg to ppGpp was measured using inhibition experiments. ObgE and its homologs from a variety of species exhibit very slow hydrolysis of GTP under standard multiple turnover conditions [all around 1–2 phosphates produced/enzyme/hour](Wout et al., 2004, Tan et al., 2002, Welsh et al., 1994, Lin et al., 1999). Because of Obg’s unusually slow hydrolysis rate, multiple turnover conditions could not be accurately utilized for inhibition experiments. Instead, single turnover experiments using excess ObgE over substrate were performed. Hydrolysis rates of GTP at varying concentrations of ObgE were determined similar to a technique used by Peluso et. al. in 2001 (Peluso et al., 2001). This technique allowed the determination of the half saturation constant, K1/2, of the GTPase activity of ObgE. [For enzymes with a rate-limiting chemical step, K1/2 is equivalent to the dissociation constant, Kd.] Utilizing inhibition assays, the Ki’s of ObgE binding to GDP and ppGpp could then be determined.
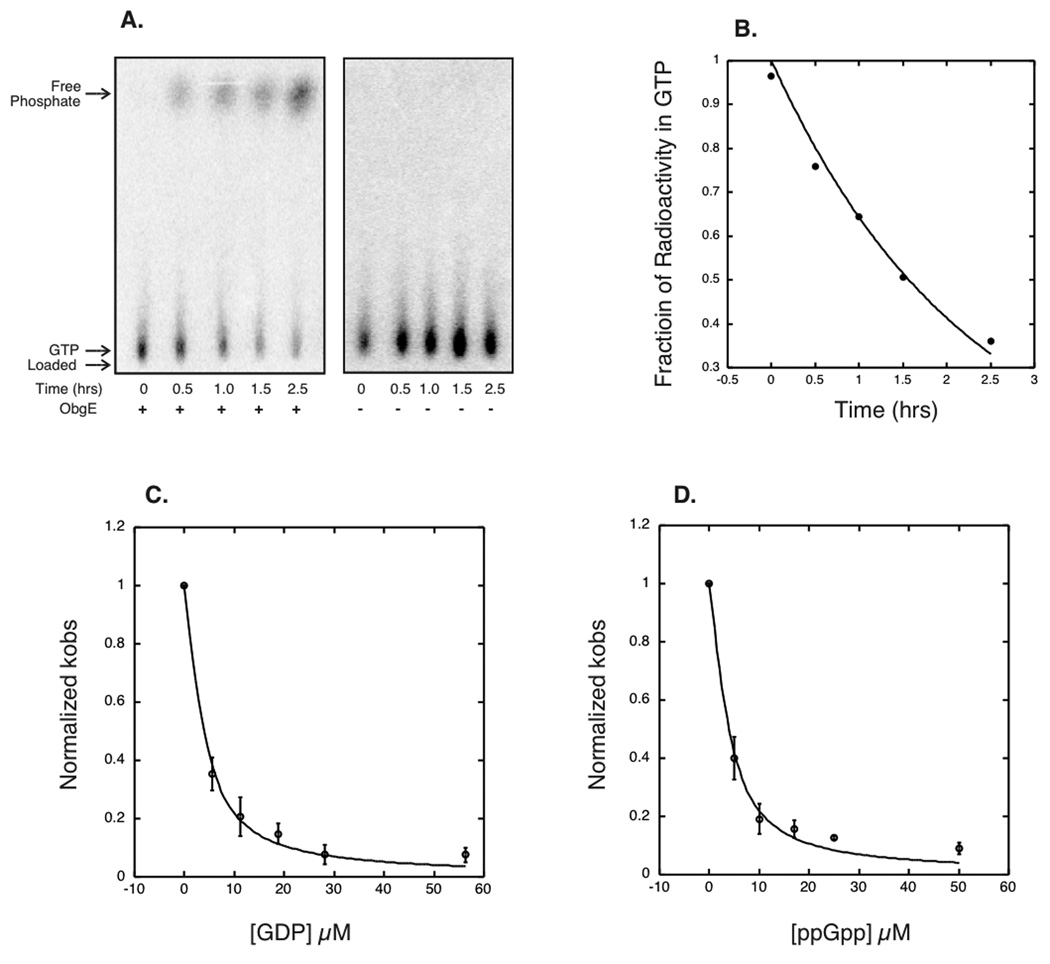
Single turnover GTPase assays were performed with ObgE in excess of substrate, γGTP32. GTP hydrolysis was followed over a time course of 2.5 hours by separating γGTP32 from P32 on thin layer chromatography (TLC). A representative experiment is shown in Figure 1A. The fraction of radioactivity remaining in the GTP form was graphed as shown in Figure 1B, and the kobs for this reaction was fit the graph (See Material and Methods and Supplementary Materials). By varying concentrations of ObgE, the K1/2 was determined to be 14 µM ± 6 µM. From this information we were also able to measure the maximal rate constant, kmax, for ObgE as 0.9 ± 0.2 products/enzyme/hour (See Materials and Methods and Supplementary Materials).
Figure 1.
ObgE GTPase and inhibition experiments. A) Representative thin layer chromatography plates for GTPase experiments showing the uninhibited ObgE GTPase (left panel), and buffer control (right panel). B) A representative fit to GTPase data. C) The average of three GDP inhibition experiments, with the standard deviation indicated as error bars. The curves were originally plotted individually to attain an average and standard deviation for the values of Ki observed. D) The average of three ppGpp inhibition experiments fit as with GDP.
Inhibition experiments were then performed by holding ObgE and substrate concentrations constant at 3.5 µM ObgE and 0.0032 µM of γGTP32 respectively and then adding GDP or ppGpp as inhibitors. These curves were then fit to determine the apparent Ki for GDP and ppGpp (see Materials and Methods and Supplementary Information). GDP and ppGpp appeared to bind to ObgE with exactly the same affinity, with apparent Ki’s of 1.6 ± 0.4 µM and 1.6 ± 0.5 µM, respectively (See Figure 1C and D). The in vivo concentrations of GTP, GDP and ppGpp during the Escherichia coli life cycle all well exceed the K1/2 and Ki for ObgE binding (ppGpp can reach concentrations in the hundreds of micromolar) (Buckstein et al., 2008). The fact that ObgE was shown to bind to ppGpp with affinity in the physiological range argues that ppGpp is a ligand of ObgE in vivo.
ObgE affects the ratio of pppGpp to ppGpp in response to amino acid deprivation
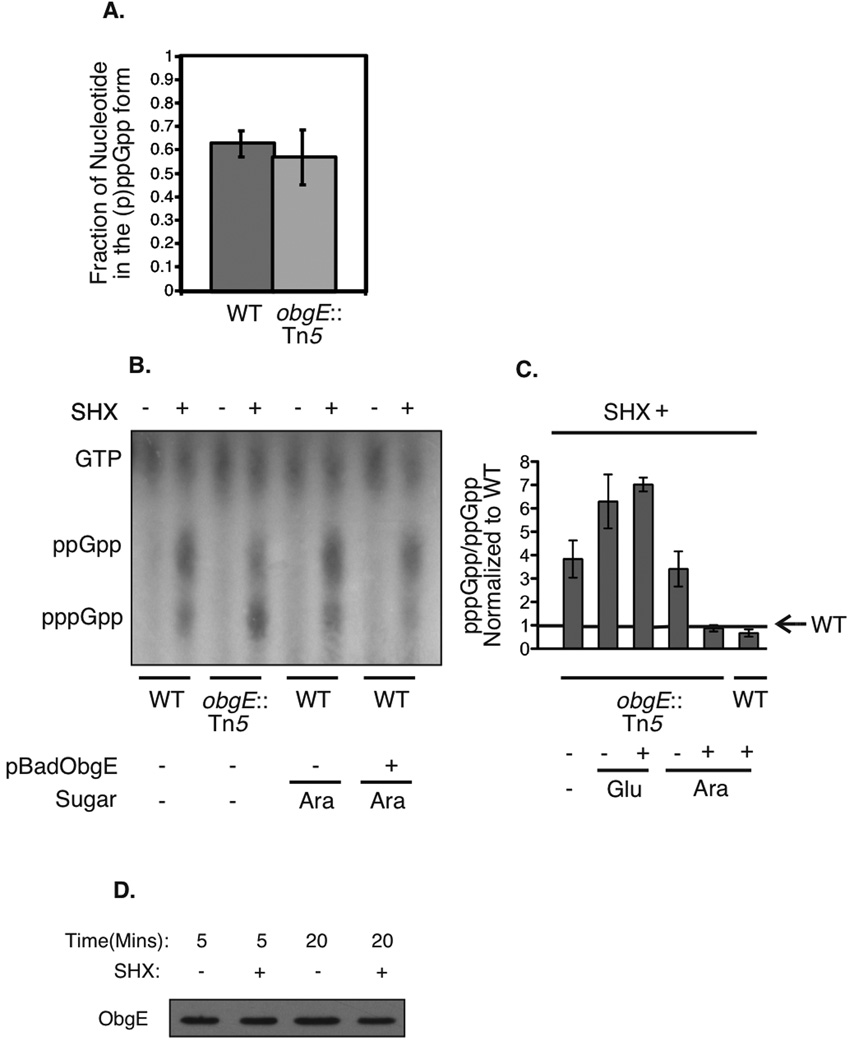
To determine if ObgE affects the levels of (p)ppGpp in vivo during the stringent response, wild-type cells were compared to those carrying a C-terminal insertion of the transposon Tn5 in ObgE. This strain contains the deletion of 9 amino acids and the addition of 68 at the C-terminal of the protein (Foti et al., 2005). We chose this mutant because it had strong defects in response to replication inhibition, but is otherwise viable (Foti et al., 2005). Both wild type and obgE::Tn5 strains were grown in LB supplemented with 32P-labelled orthophosphate. The stringent response was induced by the addition of serine hydroxamate (SHX), an inhibitor of serine tRNA synthetase (Tosa & Pizer, 1971), for twenty minutes. This treatment is known to increase the levels of ppGpp and pppGpp within the cell. The cells were lysed and relative levels of radiolabelled ppGpp, pppGpp, and GTP were detected by TLC analysis. All experiments were performed in triplicate.
To our surprise, although total levels of ppGpp plus pppGpp were not detectably different (Figure 2A), obgE::Tn5 cells had a substantially increased ratio of pppGpp/ppGpp (Figure 2B and C). This effect could be completely suppressed by over-expression of wild type ObgE from an arabinose-induced plasmid promoter (Figure 2C). This suppression supports the interpretation that any change to the pppGpp/ppGpp ratio observed in obgE::Tn5 mutants in response to amino acid starvation is due to a defect of the ObgE protein itself. Cells containing the obgE::Tn5 mutation and the ObgE+ plasmid were indistinguishable from obgE::Tn5 single mutants when the promoter was repressed by growth in glucose (Figure 2C), suggesting that the effects are sensitive to the concentration of ObgE in the cell. These results indicate that ObgE may play a role, directly or indirectly, in setting the relative levels of pppGpp to ppGpp during the stringent response in Escherichia coli.
Figure 2.
Levels of (p)ppGpp in cells treated with serine hydroxamate. A) Fraction of total guanine nucleotides in the (p)ppGpp form for wild-type and obgE::Tn5 mutant cells, averaged from three independent experiments. Error bars represent the standard deviations of the means. B) Representative thin layer chromatography plate for in vivo ppGpp and pppGpp detection, with and without serine hydroxamate treatment. The samples shown (from left to right) are: wild type (MG1655), obgE::Tn5 (STL7742), wild type (MG1655) grown in the presence of arabinose, wild-type expressing pBADObgE+ (STL7679) grown in the presence of arabinose. C) Cumulative data averaged over three experiments each showing the ratio of pppGpp/ppGpp following serine hydroxamate treatment, normalized to wild type (MG1655). Error bars represent standard deviations of the means. The samples shown (from left to right) are: obgE::Tn5 (STL7742), obgE::Tn5 (STL7742) grown in the presence of glucose, pBADObgE+ in an obgE::Tn5 background (STL13897) grown in the presence of glucose (plasmid expression off), obgE::Tn5 (STL7742) grown in the presence of arabinose, pBADObgE+ in an obgE::Tn5 background (STL13897) grown in the presence of arabinose (plasmid expression on), and pBADObgE+ in a wild type background (STL7679) grown in arabinose (plasmid expression on). D) Levels of ObgE protein before and after serine hydroxamate treatment. Samples were taken at time-points following the addition of serine hydroxamate and ObgE was detected via western blot using polyclonal anti-ObgE antibody.
ObgE controls cell survival in response to amino acid deprivation
To see if ObgE influences the response to amino acid starvation, we performed Live/Dead fluorescence staining to determine the amount of survivors within the population for wild type cells and obgE::Tn5 cells. With this procedure, only dead cells with damaged membranes stain are permeable to staining with propidium iodide (and primarily exhibit red fluorescence) whereas live cells stain with Syto 9 (green fluorescence). Cells that stained strongly with both dyes were considered “injured” and included in a separate category; our experience suggests that these cells are on the path to death. Whereas wild type cells have about 98% survival rate after treatment with SHX for 90 minutes (total cells minus the dead and injured cells), obgE::Tn5 cells drop from a 94% viability (untreated) to 79% (SHX treated) (see Table I). Mutants in relA had a slight decrease in viability and relA obgE::Tn5 double mutants had a loss of viability enhanced relative to the obgE::Tn5 single mutant. The decrease in viability of obgE::Tn5 cells suggests that they are unequipped to process amino acid starvation correctly, and more so in the absence of (p)ppGpp synthesis by RelA.
Table 1.
Live/Dead staining, septation, and cell length following SHX treatment.
| Sample | SHX | Dead (%) | Injured (%) | Septated (%) | Mean Length (µM) |
|---|---|---|---|---|---|
| wild type | − | 0.13 | 0.74 | 31 | 3.8 |
| wild type | + | 0.29 | 2.1 | 4 | 3.0 |
| obgE::Tn5 | − | 2.8 | 2.9 | 32 | 4.6 |
| obgE::Tn5 | + | 13 | 8.2 | 2 | 3.4 |
| relA251 | − | 0.6 | 2.7 | n.d. | n.d. |
| relA251 | + | 3.6 | 4.8 | n.d. | n.d. |
| obgE::Tn5 relA | − | 1.8 | 6.2 | n.d. | n.d. |
| obgE::Tn5 relA | + | 18 | 10 | n.d. | n.d. |
SHX, (+) serine hydroxamate treatment for 90 min; (−) no treatment.
ObgE protein levels are not largely altered with the initiation of the stringent response
To see if ObgE protein levels were drastically altered during its activity in the stringent response, we used Western blotting to detect ObgE protein levels in wild type cells with and without serine hydroxamate treatment. A Western blot through 20 minutes of treatment (the point at which ObgE influences the ppGpp/pppGpp ratio), shown in Figure 2D, illustrates the lack of major changes in total ObgE protein levels observed. This indicates that ObgE protein levels do not grossly change upon induction of the stringent response. We then altered the protocol to include infrared imaging quantification of our Western blots by lysing the cells and performing quantitative Western blotting on cleared cell lysates, normalized to total protein levels, and using a fluorescent secondary antibody. We found that after 20 minutes of exposure to serine hydroxamate wild type cells contained 90 ± 30% of ObgE protein present in untreated cells.
These results are consistent with previous experiments showing only mild changes (twofold) of ObgE RNA levels in wild type cells during the stringent response to amino acid starvation, although levels of ObgE may be grossly altered in (p)ppGpp null strains (Traxler et al., 2008). These data are in agreement, suggesting that ObgE’s functions in the stringent response is not accompanied by large changes in ObgE protein level.
ObgE does not affect the cell’s ability to halt growth in response to amino acid deprivation
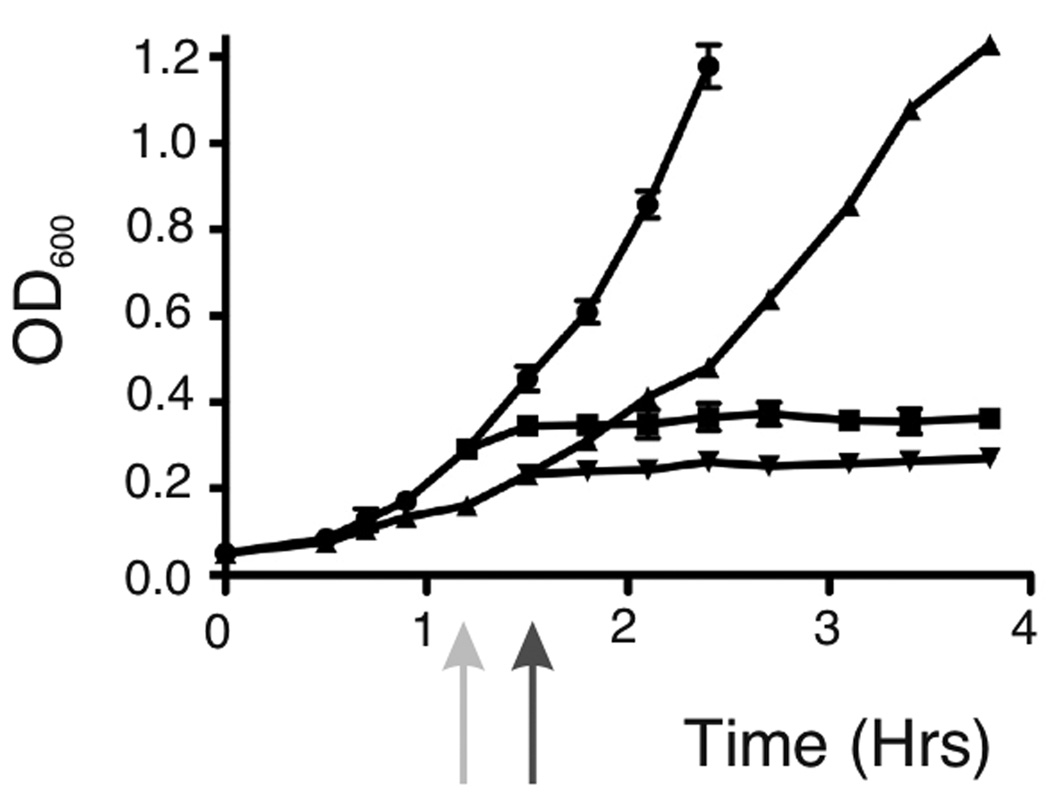
The poor survival of obgE::Tn5 cells in response to starvation may be consequence of an inability to arrest growth and division in response to increased levels of (p)ppGpp. To test this hypothesis, we looked at the obgE::Tn5 strain’s ability to halt growth as measured by OD600, its ability to form division septa in response to amino acid starvation, and cell length. We found that like wild type cells, obgE::Tn5 cells immediately halted growth as measured by OD600 upon the addition of SHX to cultures. As seen in Figure 3, both wild type cells and obgE::Tn5 cells, upon SHX induction, no longer increased OD600. In contrast, both strains continued increasing OD600 values with time when left untreated, with obgE::Tn5 cells growing slightly slower than wild type, as has been seen before. We found that obgE::Tn5 cells were able to inhibit septum formation in response to SHX treatment as has been previously reported for wild type strains (Ferullo & Lovett, 2008). As seen in Table 1, both wild type and obgE::Tn5 strains growing in early log phase exhibited signs of septation in about 30% of their population. In response to SHX treatment, septating cells in both populations dropped to around 3%. Cell lengths for obgE::Tn5 were also mildly reduced in response to SHX as has been seen before for stringent wild type cells (see Table 1), also suggesting a down-regulation of growth. Taken together, these data indicate that obgE::Tn5 mutant strains are able to halt growth in response to amino acid starvation and that their lack of survival under these conditions is not correlated with an inability to initiate these responses.
Figure 3.
Growth curves. Wild type cells and obgE::Tn5 cells were grown with aeration in LB at 37°C. The light gray arrow indicates where SHX was added to wild type cells, and the dark gray arrow indicates where SHX was added to obgE::Tn5 cells. Strains are shown as follows: untreated wild type cells (circles), SHX-treated wild type cells (squares), untreated obgE::Tn5 cells (triangles pointing up), and SHX-treated obgE::Tn5 cells (triangles pointing down).
ObgE affects DNA replication control by the stringent response
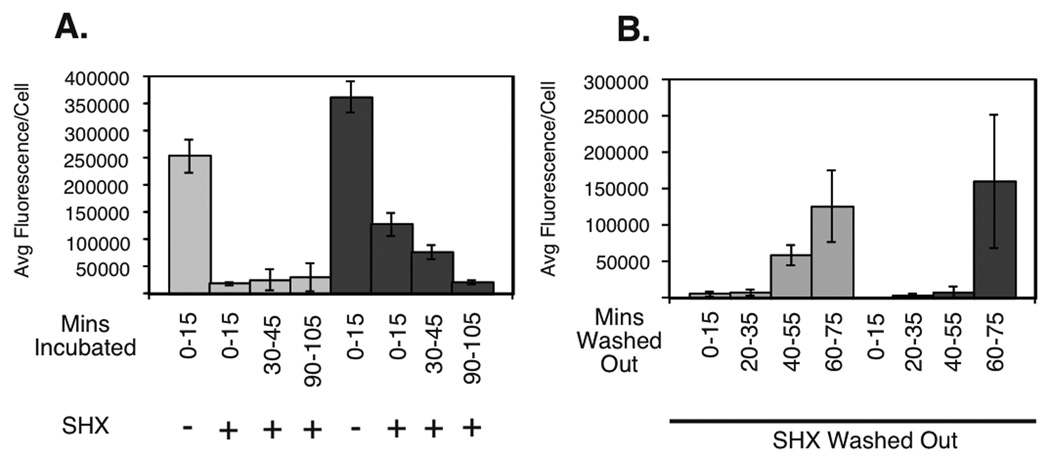
Our previous study showed that after induction of the stringent response, E. coli cells complete ongoing rounds of replication and arrest cell cycle at the level of replication initiation (Ferullo & Lovett, 2008). To determine if ObgE has an effect on DNA replication, newly replicated DNA was followed by labeling with EdU (5-ethyl-2‘deoxyuridine) conjugated to fluorescent azides. This assay was previously optimized for use in Escherichia coli by our lab for this purpose (Ferullo et al., 2009). In this manner, the amount of newly replicated DNA can be correlated to the amount of fluorescence per cell. Untreated and SHX treated cells were pulsed for 15 minutes at various time-points with EdU before fixation, labelled with fluor, and imaged via microscopy. As seen before (Schreiber et al., 1995, Ferullo & Lovett, 2008, Ferullo et al., 2009), in the presence of SHX, wild type cells halted their replication. We found that within 15 minutes of SHX treatment, replicated DNA labeling was virtually undetectable. At this same time-point, however, we found that obgE::Tn5 cells continued to incorporate label, indicating ongoing replication (See Figure 4A and Supplementary Figure S1). This data suggest that ObgE is required for the timely inhibition of DNA replication by amino acid starvation. However, by 90–105 minutes the obgE::Tn5 strains exhibited no detectable replication. This delay in obgE mutants was reproducible for multiple isolates.
Figure 4.
Arrest and restart of DNA replication in response to SHX treatment and release. (A) EdU-Click fluorescence following SHX treatment for wild-type cells (light grey) and obgE::Tn5 mutant cells (dark grey). B) EdU-Click fluorescence following SHX removal for wild-type cells (light grey) and obgE::Tn5 mutant cells (dark grey).
Interestingly, we found that upon release of these cells from serine hydroxamate wild type cells regained ability to incorporate label faster than did obgE::Tn5 cells (See Figure 4B and Supplementary Figure S1). By 55 minutes, wild type cells had significantly brighter newly replicated DNA labeling than obgE::Tn5 cells. These data indicate that ObgE also impacts the release of the stringent response and the resumption of DNA replication. By 60–75 minutes obgE::Tn5 cells did reach a wild type level of DNA replication and were apparently able to recover from replication inhibition. Together these data indicate that ObgE is either involved in the timing of onset and termination of stringent response DNA replication inhibition or that DNA damage persists that delays completion of replication and its resumption after release.
obgE::Tn5 cells have constitutively high DNA content and exhibit a partial SOS response
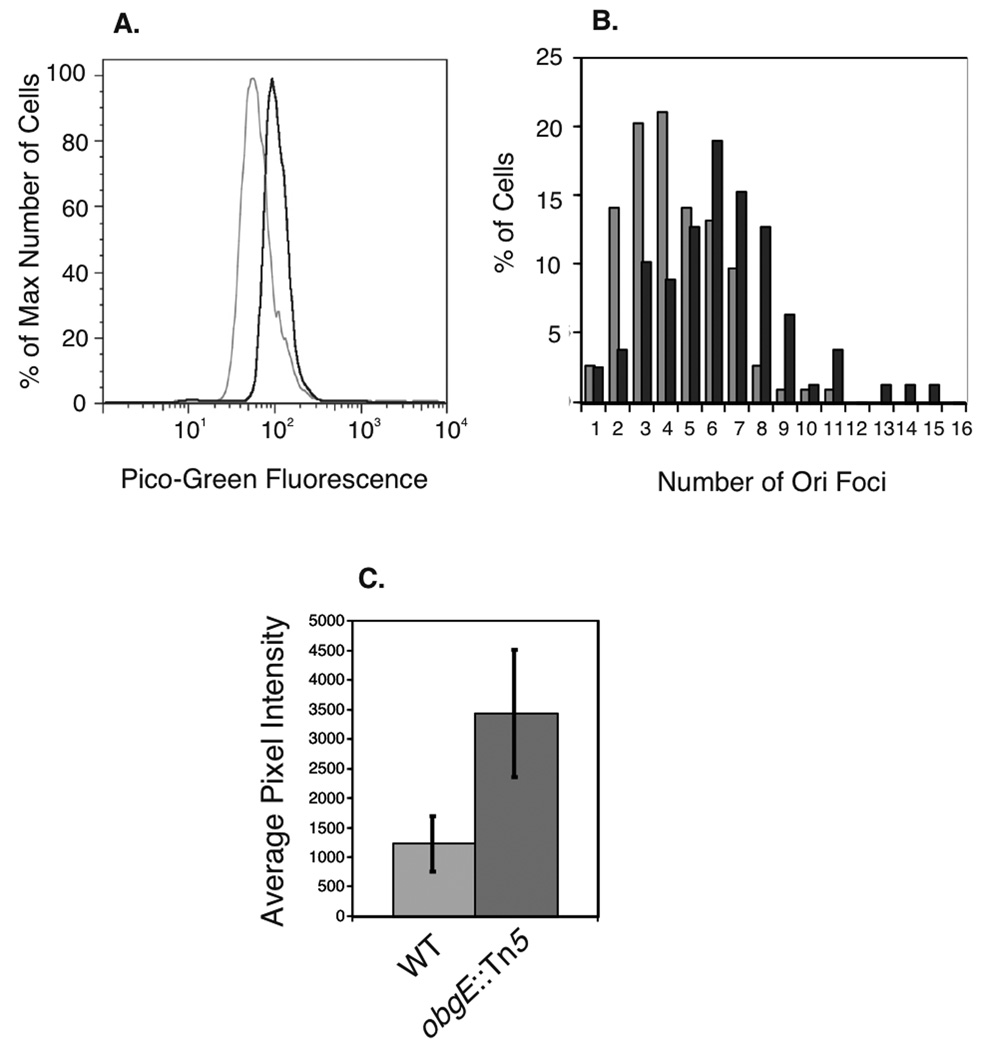
obgE::Tn5 cells have previously been shown to have longer cell lengths, which we also note here, and higher copy number of the origins of replication (oriC) in minimal media as compared to wild type cells as shown by flow cytometry (Foti et al., 2005). To confirm that obgE::Tn5, like other ObgE mutants (Kobayashi et al., 2001, Foti et al., 2007), had constitutively higher DNA content in rich medium, we measured DNA content of obgE::Tn5 grown in LB by staining cells with PicoGreen fluorescence detected by flow cytometry. As predicted, obgE::Tn5 cells exhibited constitutively higher DNA content than wild types cells as indicated by an increase in fluorescence (see Figure 5A). To confirm this, we used obgE::Tn5 strains containing the parS DNA sequences near the origin of replication as a binding site for GFP-ParB expressed from a plasmid and visualized oriC regions via microscopy. When compared to wild type, obgE::Tn5 cells showed more oriC foci (see Figure 5B). On average, wild type cells showed 4.4 ori foci, and obgE::Tn5 showed 6.3 ori foci. Run-out flow cytometry of cells with rifampicin and cephalexin confirmed higher initiation capacity of obgE::Tn5 (data not shown). Put together, this data indicate that ObgE regulates total DNA content within Escherichia coli cells, and that the obgE::Tn5 mutant cells have higher DNA content than do wild type cells.
Figure 5.
DNA content and SulA expression measurements. A) Representative flow cytometry data showing DNA content from 30,000 wild type and obgE::Tn5 cells labeled with PicoGreen fluorescence. Wild type cells are shown in light grey and obgE::Tn5 cells are shown in dark grey. B) Distribution of numbers of oriC foci per cell using a ParS/GFP-ParB system for obgE::Tn5 strains (dark grey) and wild type strains (light grey). C) SOS response induction measured by a SulA::mCherry reporter plasmid, averaged over 100 wild type and obgE::Tn5 cells.
One explanation of the replication phenotype of obgE mutants is that there may be constitutively high levels of DNA damage. To test this hypothesis, we used strains containing a reporter of the SOS response to DNA damage. This reporter consists of a plasmid containing the promoter for the SulA gene containing a tight LexA binding site highly regulated by the SOS response. When this plasmid is transformed into both wild type and obgE::Tn5 cells and analyzed for mCherry fluorescence, the obgE::Tn5 cells showed significantly higher SulA expression than did wild type (Figure 5C), indicating that obgE::Tn5 cells constitutively exhibit a partial SOS response.
Discussion
ObgE binds ppGpp similarly to GDP
We show here that ObgE binds to ppGpp with similar affinity as it binds to GDP. This data correlates with previously published biochemical data for ObgE. The single turnover kmax observed here for GTP hydrolysis of 0.9 products/enzyme/hour is very similar to the reported multiple turnover kmax of 1.0–1.2 products/enzyme/hour (Wout et al., 2004, Tan et al., 2002). The K1/2 observed here as 14 ± 6 µM matches well with the reported Kd of binding to GTP of 8 µM (Wout et al., 2004), and the reported multiple turnover Km of 18 µM (Tan et al., 2002). The Ki of ObgE binding to GDP measured here of 1.6 ± 0.4 µM is very similar to the Kd of binding of Bacillus subtilis (Welsh et al., 1994), and Salmonella typhimurium (Lamb et al., 2007) Obg to GDP of 1.7 µM and 1.4 µM, respectively.
The result that GDP and ppGpp bind with the same affinity is consistent with the Bacillus subtilis structure of Obg bound to ppGpp (Buglino et al., 2002). In the structure, the two phosphates attached to the 3’ carbon of ppGpp appear to make no contacts with the protein, but hang free (Buglino et al., 2002). Obg appears to bind to the parts of ppGpp that it has in common with GDP. This would predict the similar binding affinities to GDP and ppGpp that were observed for ObgE in this study and as suggested (Buglino et al., 2002). In this study, ppGpp showed purely inhibitory action on GTPase activity, in contrast to previous data suggesting that it could contain both inhibitory and stimulatory actions on GTPase activity of Obg (Buglino et al., 2002). The in vivo concentrations of all GDP, GTP and ppGpp during the Escherichia coli life cycle well exceed the Kd and Ki for ObgE binding (Buckstein et al., 2008) (ppGpp can reach concentrations in the hundreds of micromolar). The fact that ObgE was shown to bind to ppGpp with affinity in the physiological range argues that ppGpp can be a ligand of ObgE in vivo.
The similar affinities of ObgE for ppGpp and GDP reinforces the fact that ObgE has a fairly open active site, allowing for the possibility that ObgE may share the binding of a nucleotide such as ppGpp with another protein, such as SpoT, RelA, or Gpp. As these are all effectors of pppGpp and ppGpp levels in the cell, it may be possible that ObgE works together with these other proteins as well to produce the effects on ppGpp/pppGpp ratios observed here. ObgE’s ability to physically interact with SpoT (Wout et al., 2004, Raskin et al., 2007) makes it a likely candidate; however SpoT’s pyrophosphohydrolase activity (release of the 3’ pyrophosphate) is a different chemistry than the pppGppase activity hypothesized to be responsible for the change in ppGpp/pppGpp ratio (5’ gamma-phosphate release). If SpoT is involved in the observed ObgE phenotypes, then it must play a more complicated role than originally predicted.
The similar binding affinity of ObgE to ppGpp and GDP also re-enforces the idea that enzymes generally labeled as GTPases could potentially have nucleotide sensing capacities within the cell with less commonly tested nucleotides, such as pppGpp and ppGpp. In fact, to date, a small handful of GTPases in Escherichia coli have also been shown to interact with ppGpp. Interestingly, aside from Gpp, translation factors IF2, EF-G, and EF-Tu all have the ability to directly hydrolyze pppGpp to ppGpp in vitro (Cashel, 1996). IF2 binds ppGpp with similar affinity as it binds to GTP, and has been proposed to be a metabolic sensor within the cell (Milon et al., 2006), a role often suggested for Obg. EF-Tu slows translation with a rise in ppGpp levels, causing an increase in translation fidelity during the stringent response (Dix & Thompson, 1986). Mutants in EF-G in Salmonella typhimurium also alter basal levels of ppGpp in vivo (Macvanin et al., 2000), and in Escherichia coli EF-G controls ppGpp levels under heat shock conditions (Pao et al., 1981). These proteins share several features with ObgE: they are all GTPases that associate with the ribosome and can interact with ppGpp. In addition, some of them have also been shown to have downstream roles in the stringent response. Future work exploring the connections between these GTPases may prove exciting.
Obg may directly hydrolyze pppGpp or regulate this hydrolysis
In this study we observed that ObgE changes the ratio of ppGpp to pppGpp in response to serine hydroxamate. Currently, our understanding of the importance of pppGpp is far from complete. Much of the literature describing the effects of these nucleotides in vivo groups them both together as (p)ppGpp, and discuss their effect as one. (Distinguishing between their effects in vitro has been thwarted by the fact that only ppGpp is commercially available). The current model of the stringent response is based on the fact that RelA can synthesize ppGpp and pppGpp with a similar speed (Cashel, 1975, Cochran & Byrne, 1974). Given the immense excess of GTP in Escherichia coli over GDP at any given time it has been assumed that primarily pppGpp is made, which then is quickly converted to ppGpp within the cell, leading to the observed excess of ppGpp over pppGpp during the stringent response. We show here a correlation between a change in this nucleotide ratio and a deficiency in survival to amino acid starvation and DNA replication control in ObgE mutants. Although ObgE may be acting with other proteins to produce these effects, the simplest explanation of our observations is that ObgE is a pppGpp hydrolase and that this hydrolysis accompanies its roles in promoting fork stability and cell survival during amino acid starvation. The ability of ObgE to bind ppGpp with identical efficiency to GDP suggests that it will likely bind pppGpp and may potentially hydrolyze this molecule as it does GTP. This raises the possibility that pppGpp may itself have an active role in stringent signaling, and that the act of hydrolysis of pppGpp to ppGpp may itself be a signal in this pathway.
Does Obg/CgtA directly regulate (p)ppGpp levels?
We observed higher pppGpp levels relative to ppGpp in the obgE mutant and a suppression of this effect by expression of ObgE+ from a plasmid, although neither condition changed the overall levels of (p)ppGpp. It has been argued previously that ObgE mutants have constitutively high (p)ppGpp levels through its ability to regulate SpoT hydrolysis (Jiang et al., 2007, Raskin et al., 2007). This provided an attractive explanation for the essential nature of Obg/CgtA, to remove accumulated (p)ppGpp, and its ability to interact with SpoT. However, recent work has shown that ObgE remains essential for cell proliferation in even in relA or relA spoT strains that cannot synthesize (p)ppGpp (Shah et al., 2008, Jiang et al., 2007). Moreover, Obg/CgtA has not been shown to affect SpoT activity, in vivo or in vitro. It may be worthwhile to consider other explanations. Since (p)ppGpp levels rise naturally as cells begin to enter stationary phase and as a result of stress, the high basal levels of (p)ppGpp reported previously for ObgE/CgtA mutants could be an indirect effect of growth problems in these mutants. That is, the higher levels of ppGpp in these mutants could result from increased RelA-dependent synthesis rather than decreased SpoT hydrolysis of ppGpp.
We do not see an elevation of constitutive levels of (p)ppGpp in the obgE::Tn5 mutant, as shown here, nor in ObgE-depleted cells (data not shown) in conflict with previous reports (Jiang et al., 2007, Raskin et al., 2007). A number of experimental differences including plasmid vs. chromosomal alleles, temperature, and growth medium make comparison difficult. It is possible that the differences between the two experiments with E. coli protein are due to the site of ObgE mutation used in these two studies (i.e. the N-terminal (Jiang et al., 2007) or the C-terminal of ObgE (this work)). Recent work in Bacillus subtilis suggests that the N-terminal domain is involved in Obg’s growth phenotype whereas its C-terminal domain is involved with stress responses (Kuo et al., 2008). Although the C-terminal Tn5 mutant could either increase or decrease overall (p)ppGpp levels through hypothetical misregulation of SpoT, we note that the observed phenotypes of the mutant with respect to replication --slow to arrest and slow to resume-- is consistent with neither scenario. This suggests a role of Obg in the output of the response, over and above any effects on (p)ppGpp levels.
In addition, other observations raise doubts whether depletion of Obg/CgtA elicits a full-blown stringent response. ObgE deficient cells in a variety of conditions and in a range of organisms have been seen to have significantly longer cells with more DNA content than wild type cells (Kobayashi et al., 2001, Morimoto et al., 2002, Czyz et al., 2001, Sikora-Borgula et al., 2002, Slominska et al., 2002, Foti et al., 2005, Datta et al., 2004). In our hands, depletion of ObgE does not affect cell growth and ability to initiate replication; cells form long polyploid filaments almost immediately upon depletion. This contrasts to stringent cells, either induced by amino acid starvation or by excess pppGpp production, that arrest replication and cease cell growth, producing short cells with a reduced, integer number of chromosomes. We conclude that if a rise in constitutive (p)ppGpp levels occurs in Obg/CgtA mutants, it does not lead to a canonical, full-blown stringent response. If ObgE depletion does cause accumulation of ppGpp, then ObgE must also be required for arrest of replication and cell growth seen during the stringent response, since this fails to occur in ObgE-depleted cells. Therefore, if Obg does indeed regulate (p)ppGpp levels, its involvement must be more complicated than previously appreciated. Future experiments to address these issues should prove informative.
A role on the output of the stringent response
The effect of ObgE on DNA replication during the stringent response fits well with the known role of ObgE to promote survival following exposure to DNA replication inhibitors (Foti et al., 2005). Replication in the presence of low levels of replication inhibitors and replication during amino acid starvation may both produce replication pauses or gaps. Our previous studies showed that ObgE had strongly synergistic effect with double strand break repair factors (RecA, RecBCD), both for normal viability and in the survival of mild replication inhibition, leading us to conclude that replication forks are more vulnerable to breakage in the absence of ObgE (Foti et al., 2005). This could provide an explanation for replication defects of obgE::Tn5 seen during the stringent response: ObgE mutants have more difficulty completing replication during starvation and experience damage in the process. Interestingly, E. coli temperature-sensitive cells mutated in the N-terminal domain of ObgE have been reported to replicate DNA less efficiently than wild type cells as measured by thymidine incorporation (Sikora et al., 2006). Some of may be due to problems in initiation of replication since the temperature-sensitivity of this strain can be reduced by expression of the replication initiator protein, DnaA, and DnaA levels are reduced in this mutant at nonpermissive temperature. However, in other studies, neither the temperature-sensitive allele nor depletion of obgE led to defects in replication initiation as detected by flow cytometry (Foti et al., 2007, Kobayashi et al., 2001) or by visualization of oriC foci (Foti et al., 2007). ObgE-depleted cells continue to replicate to reach a median ploidy of 15–20 N after 3 hours of depletion; it is possible that this high DNA content may down-regulate DnaA. The hypothesis that ObgE mutants have difficulty completing replication is also consistent with our observation of an elevated constitutive SOS response in the obgE::Tn5 strain and may explain the enhanced lethality during starvation in relA strains that cannot induce a full stringent response.
In summary, we have shown here that ObgE alters the ppGpp/pppGpp ratio of Escherichia coli cells starved for amino acids. These data, together with an altered biological stringent response places ObgE protein as an effector of the response to amino acid starvation. Correlation of the ppGpp/pppGpp ratio to a delayed change in DNA replication also further supports a connection between between two functions of ObgE often considered to be disparate, namely its involvement in DNA replication control and its connection to ppGpp.
Materials and Methods
Strains and growth conditions
Except for protein purification, the strains used in this study are derived from wild-type E. coli K-12 MG1655 (Table I.) All cells were grown at 37°C on Luria-Bertani (LB) medium (1% Bacto tryptone, 0.5% yeast extract, 0.5% sodium chloride, and for plates, 1.5% agar. Antibiotics were used with the following concentrations: kanamycin (Km), 30 µg/ml ; amplicillin (Ap), 100 µg/ml; tetracycline (Tc), 15 µg/ml and chloramphenicol (Cm) 30 µg/ml. Arabinose and Glucose, when applicable, were used at a concentration of 0.2%. Strains isogenic with MG1655 were constructed by P1 vira transduction. Plasmids were isolated and purified from storage strains via miniprep (Qiagen and Sigma-Aldrich) and transformed by electroporation (Dower et al., 1988).
Purification of ObgE
ObgE was purified as described (Kobayashi et al., 2001) from STL 8248 with the following modifications: Cells were grown shaking at 37°C to OD600 ~0.6 at which point isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM. Cells continued to be incubated shaking at 37°C for 2.5–3.5 hours. Cells were then spun down, resuspended in 1/100 the volume of Tris-sucrose buffer (50 mM Tris-HCl pH 7.5, 10% sucrose), and frozen at −80°C. To lyse the cells, DTT was added to 1 mM, NaCl was added to 100 mM and the cells were incubated on ice for 15 min. Lysozyme in Tris-sucrose was added to a final concentration of 0.2 mg/mL and the mixture was incubated on ice for 45 min, heat shocked by placing into a 37°C water bath for two minutes, then left on ice for 2 minutes. This process was then repeated 4–5 times and the lysed cells were then spun for 35 minutes at 67,100 × g. To every 5 mL of cleared lysate 3.198 mL of saturated ammonium sulfate was added slowly with stirring. The precipitate was recovered by centrifugation at 13,250 g for 15 minutes and resuspended in TEGED100 buffer (20 mM Tris 7.5, 100 µM EDTA, 1 mM DTT, 10% glycerol, and 100 mM NaCl) and dialyzed overnight into 1 L of TEGED100 buffer. Insoluble particles were removed by centrifugation and the solution was loaded onto a 5 mL HiTrap Blue HP column (GE Healthcare), and step eluted from TEGED100 buffer to TEGED1400 buffer (20 mM Tris 7.5, 100 µM EDTA, 1 mM DTT, 10% glycerol, and 1400 mM NaCl). Fractions containing ObgE were collected and diluted with three volumes of TEGED100 buffer. The entire mixture was loaded onto a 5mL Q-column (GE Healthcare) and ramp-eluted from TEGED100 buffer-TEGED1000 buffer (20 mM Tris 7.5, 100 µM EDTA, 1 mM DTT, 10% glycerol, and 1000 mM NaCl). Fractions containing ObgE were pooled, dialyzed into 1x Reaction Buffer (50 mM Tris 7.5, 50 mM NaCl, 1 mM DTT), concentrated in a 10,000 MWCO PES spin column (Sartorius, Vivaspin 6). Protein was either used immediately (for K1/2 curves), or an equal volume of glycerol was added and the protein was stored at −20°C (for inhibition curves).
ObgE GTPase assays and inhibition experiments
Inhibition experiments were performed similar to experiments performed by Peluso et al. in 2001 (Peluso et al., 2001). Samples consisted of: 9.5 µL of 35.8mM Tris, 35.8mM NaCl, 1.8mM DTT, 5.3mM MgCl2, 2%glycerol, 3.5µM ObgE, and .0032µM of γ32P-GTP (EasyTides Perkin Elmer) and GDP (Sigma) or ppGpp (Trilink Technologies) with concentrations varying from 0 to 56 µM. Experiments were initiated by the addition of γ32P-GTP and incubated at 37°C. 1µL aliquots were removed at various time-points, quenched with an equal volume of 40µM cold EDTA and stored on ice. [GDP] and [ppGpp] stock solutions were diluted with water and neutralized with 50 mM Tris Base. 1µL of each reaction was spotted on a polyethyleneimine (PEI) plate (Sigma-Aldrich). Plates were developed in 1L beakers covered with parafilm containing a mobile phase of 1 M formic acid and 0.5 M LiCl. The plates were subsequently dried, exposed to a phosphoimager screen, and quantitated using Quantity One software (Bio-Rad). None of the images used included saturated pixels. (See supplementary data for K1/2 calculations and for curve-fitting information).
In vivo pppGpp and ppGpp measurements
In vivo measurements of ppGpp and pppGpp were performed as described previously in (Metzger et al., 1989) with the following modifications. Cultures were grown overnight in LB medium at 37°C and supplemented with the appropriate antibiotics and sugars (Sigma). In the morning, cells were diluted to reach early log phase following 2.25 hours of growth in LB and appropriate sugar, 0.2% glucose or 0.2%, for those experiments involving pBAD33 ObgE expression. Cells were grown for 15 minutes, at which point 32P-orthophosphate (Perkin Elmer) was added to a final concentration of 120 µCi/mL, incubated for 2 hours, followed by treatment with serine hydroxamate (Sigma-Aldrich) at 1.2 mg/mL. 60 µL samples prior to addition of the drug or after 20 minute treatment were added to 50 µL of 13N formic acid (Sigma), frozen and thawed twice on dry ice with ethanol and the samples were subjected to centrifugation to remove debris. 6 µL of sample was loaded onto polyethyleneimine (PEI) plates (Sigma-Aldrich). Chromatography was performed in 1L beakers covered with parafilm containing a mobile phase of 1.5 M KH2PO4 (Fisher Scientific). The plates were subsequently dried, exposed to a phosphoimager screen (most exposures took two or more days), and quantified using Quantity One software (Bio-Rad), none of the images used included saturated pixels. For some experiments samples were stored in formic acid at −20°C overnight before loading onto PEI plates. Unlabeled GTP and ppGpp were spotted on the plates as markers and visualized by UV light-induced fluorescence.
ObgE protein levels following serine hydroxamate treatment
Culture of wild type (MG1655) were grown to early log phase, split and 1 mg/mL serine hydroxamate (Sigma) was added to one culture. Samples were taken for 20 minutes following the addition of serine hydroxamate and the density of the culture was determined by OD600. Cells were collected from 1 mL of cells by microcentrifugation and the cell pellet was resuspended in (OD600 × 100) µL of 2×FSB (4% sodium dodecyl sulfate (SDS), 200 mM DTT, 120 mM Tris pH 6.8, 0.002% bromphenol blue, 10% glycerol). Boiled samples were subjected to SDS-polyacrylamide gel electrophoresis. Proteins were transferred onto a PVDF membrane and blotted for ObgE by using polyclonal anti-ObgE primary antibody (Foti et al., 2007). Horseradish peroxidase-linked anti-rabbit Ig, from donkey (GE Healthcare/Amersham Biosciences) was used as a secondary antibody. Western blots were visualized with SuperSignal West Pico Chemiluminescent Substrate (Pierce). For quantitative Western blots, the protocol was slightly altered: following cell collection, cell pellets were resuspended in 100 µL of Tris-Sucrose Buffer and lysed as indicated under the ObgE purification procedures. The lysates were cleared by centrifucation and normalized to total protein level as measured by the Bradford assay (Bradford, 1976) with the addition of Blank Lysis buffer (50 mM Tris-HCl pH 7.5, 10% sucrose, 1 mM DTT, 100mMNacL) to relevant samples. Samples were then combined with an equal volume of 2xFSB and Western blots were performed as indicated above, except using IRDye 800CW Goat Anti-Rabbit IgG secondary antibodies (LI-COR Biosciences). Experiments were performed in triplicate and quantified using an Odyssey machine (LI-COR Biosciences). Reported are the mean and standard deviation of fluorescent intensity normalized to untreated cells originating from the same isolate.
Live/Dead analysis following serine hydroxamate treatment
After 2 hr of growth in fresh medium, cells were treated with and without 1 mg/ml serine hydroxamate (Sigma-Aldrich) for an additional 90 minutes. 1 ml cells were resupended in 50 µl Phosphate-Buffered Saline (PBS)and stained with the LIVE/DEAD BacLight Bacterial Viability Kit (Invitrogen) according to the manufacturer's instructions. Data was acquired for 30,000 cells by using a Becton Dickinson FACSCalibur flow cytometer and analysis was done using the FlowJo 6.4.1 software (Tree Star).
Growth Curves
Two isolates each of MG1655 and STL 7742 were grown overnight at 37°C. in LB and diluted to an OD600 of 0.05. OD600 was then measured at various timepoints following dilution. When each culture reached an OD600 of 0.2–0.3, it was split with half of the cultures treatement with 1 mg/ml serine hydroxamate (Sigma-Aldrich). Shown are the average and standard deviation of the OD600 between the two isolates of each strain for each time point.
EdU-Click labeling of replicated DNA
Newly replicated DNA was detected using l EdU-Click labeling (Ferullo et al., 2009) using a commercially available Alexa fluor 488 kit “Click-IT Edu” (Invitrogen). All washes performed in PBS were performed twice, except the wash immediately following incubation in 0.5% Triton-X based buffer in PBS, using 1.5 mL of PBS for each wash. The final wash utilized only 1mL of PBS for each sample. A GFP filter with exposure times of 2.0 seconds was employed to measure cell fluorescence. All data was collected on an Olympus BX51 microscope equipped with an 100x objective and a Retiga Exi (Qimaging Inc.) camera with image analysis using Volocity imaging software (Perkin-Elmer Improvision). Figures of fluorescence shown all have a black point of 456 and a white point of 1500 intensity so that the same contrast may be shown. To quantitate the average fluorescence per cell per field of view, cells were identified as objects in phase by using −1 standard deviation of the mean intensity as the upper limit of intensity allowed. Any objects identified that were touching the edge of the field of view or smaller than 1 µm2 were not considered. Fluorescent objects were detected using a threshold pixel intensity minimum of 675 and maximum of 3000. Fluorescence was summed over the field of view and divided by the number of cells manually counted in that field of view for at least 100 cells and three fields of view for each sample; the standard deviation between fields is reported. DNA replication was measured using this assay for wild type cells (MG1655) and obgE::Tn5 cells (STL7742) at various time-points for cultures at OD600 0.1–0.2 followed by 1 mg/mL serine hydroxamate for 90 minutes. Serine hydroxamate was removed by microcentrifugation of cells, removal of the supernatant and 5-fold dilution in fresh medium to resume growth.
Septation measurements
Visualization of septating cells was aided by staining inner cell membranes with FM 4–64 (N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide (Fishov & Woldringh, 1999). Cells were stained and analyzed as described (Ferullo et al., 2009). Septating cells were counted and lengths were measured using Volocity imaging software (Perkin-Elmer Improvision).
SOS induction in obgE mutant cells using sulA::mCherry plasmid reporter
MG1655 cells and STL7742 cells were transformed with a SulA::mCherry reporter plasmid, derived from a SulA::GFP plasmid (McCool et al., 2004) by replacement with mCherry (Steven Sandler, unpublished results). The cells were grown in LB supplemented with 100 µg/mL of ampicillin to log phase (OD600= 0.3–0.4). Cells were visualized on 2% agarose-padded slides and a coverslip was added for analysis under the microscope. All data was collected on an Olympus BX51 microscope equipped with an 100x objective and Retiga Exi (Qimaging Inc.) camera. All analysis was performed using Volocity imaging software (Perkin-Elmer Improvision). To quantitate the average fluorescence per cell, cells were identified as objects as described above. The average pixel intensity for mCherry fluorescence was calculated for 100 cells and averages and standard deviations are reporter.
oriC analysis
STL 12758 and STL 14030 were grown in LB Ap medium to OD600= 0.3–0.4. GFP-labeled origins of replication within the living cells were immediately visualized using 2% agarose pads on an Olympus BX51 microscope equipped with a RGB liquid crystal color filter and a Qimaging Retiga EXi camera. Foci were counted visually using Volocity imaging software (Perkin-Elmer Improvision) with a blackpoint intensity of 456 and a whitepoint intensity of 900. 114 cells were counted for two individual isolates of STL 12758 and 79 cells were counted for two individual isolates of STL14030.
DNA content analysis using flow cytometry
DNA content per cell was determined as previously reported (Ferullo & Lovett, 2008), with the following modifications. Data was acquired for at 30,000 cells using a FACS Aria Flow Cytometer using DIVA 6.1.1 as its operating system (BD Biosciences). Data was further analyzed using FloJo 6.4.7 software (Tree Star). For clarity, a lower fluorescence gate was added in the analysis of 101 fluorescence intensity as unlabeled cell controls ran below this intensity.
Table 2.
Strain and plasmids used in this study.
| A. Strains | Relevant Genotype | Source or Derivation |
|---|---|---|
| CAG12072 | sfsB203::Tn10 | Singer et al., 1989 |
| MG1655 | K-12 wild-type rph-1 | Blattner et al., 1997 |
| STL7679 | [pSTL 346] | Cmr transformation into MG1655 |
| STL7742 | obgE::Tn5 | Foti et al., 2005 |
| STL8248 | F- ompT hsdS(rB-mB-) dcm- Tetr gal λ (DE3) [pJT130] |
Tan et al., 2002 |
| STL8783 | relA251::kan | Metzger et al., 1989 |
| STL8789 | obgE::Tn5 sfsB203::Tn10 | Kmr tranductant P1 STL7742 X STL519 |
| STL8813 |
obgE::Tn5 relA251::kan sfsB203::Tn10 |
Tcr transductant P1 STL8789 X STL8783 |
| STL12512 | [pMK8] | Apr transformation into MG1655 |
| STL12758 | pstA::parS | Ferullo & Lovett,2008 |
| STL12760 | gadB::parS | Ferullo & Lovett, 2008 |
| STL13897 | obgE::Tn5 [pSTL 346] | Cmr transformation into STL 7742 |
| STL14013 | obgE::Tn5 [pMK8] | Apr transformation into STL7742 |
| STL14030 |
obgE::Tn5 pstA::parS sfsB203::Tn10 |
Tcr transductant P1 STL8789 X STL12758 |
| STL14033 |
obgE::Tn5 gadB::parS sfsB203::Tn10 |
Tcr transductant P1 STL8789 X STL12760 |
| B. Plasmids | ||
|---|---|---|
| pBAD33 | cat araC | Schreiber et al., 1991 |
| pJT130 | pET11a obgE | Tan et al., 2002 |
| pMK8 |
sulA::mCherry in a TGV light plasmid (from strain SS2514) |
S. Sandler |
| pSTL346 | cat araC obgE+ in pBAD33 | Foti et al., 2005 |
Strains isogenic with MG1655, except STL8248, used for ObgE protein purfication
Acknowledgements
We would like to thank Noreen Francis for help with flow cytometry and Jeffery Agar and Lizbeth Hedstrom for their insight on this project. We would also like to thank the Bardwell lab and Sandler lab for their generosity in providing strains and plasmids.
Bibliography
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- Battesti A, Bouveret E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol. 2006;62:1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buglino J, Shen V, Hakimian P, Lima CD. Structural and biochemical analysis of the Obg GTP binding protein. Structure. 2002;10:1581–1592. doi: 10.1016/s0969-2126(02)00882-1. [DOI] [PubMed] [Google Scholar]
- Cashel M. Regulation of bacterial ppGpp and pppGpp. Annu Rev Microbiol. 1975;29:301–318. doi: 10.1146/annurev.mi.29.100175.001505. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VH, Vinella D. The stringent response. In: Neidhardt RCFC III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Resnikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: cellular and molecular biology. ASM Press: Washington, D.C.; 1996. pp. 1458–1496. [Google Scholar]
- Cochran JW, Byrne RW. Isolation and properties of a ribosome-bound factor required for ppGpp and ppGpp synthesis in Escherichia coli. J Biol Chem. 1974;249:353–360. [PubMed] [Google Scholar]
- Czyz A, Zielke R, Konopa G, Wegrzyn G. A Vibrio harveyi insertional mutant in the cgtA (obg, yhbZ) gene, whose homologues are present in diverse organisms ranging from bacteria to humans and are essential genes in many bacterial species. Microbiology. 2001;147:183–191. doi: 10.1099/00221287-147-1-183. [DOI] [PubMed] [Google Scholar]
- Datta K, Skidmore JM, Pu K, Maddock JR. The Caulobacter crescentus GTPase CgtAC is required for progression through the cell cycle and for maintaining 50S ribosomal subunit levels. Mol Microbiol. 2004;54:1379–1392. doi: 10.1111/j.1365-2958.2004.04354.x. [DOI] [PubMed] [Google Scholar]
- Dix DB, Thompson RC. Elongation factor Tu.guanosine 3'-diphosphate 5'-diphosphate complex increases the fidelity of proofreading in protein biosynthesis: mechanism for reducing translational errors introduced by amino acid starvation. Proc Natl Acad Sci U S A. 1986;83:2027–2031. doi: 10.1073/pnas.83.7.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferullo DJ, Cooper DL, Moore HR, Lovett ST. Cell cycle synchronization of E. coli using the stringent response, with fluorescence labeling assays for DNA content and replication. Methods. 2009 doi: 10.1016/j.ymeth.2009.02.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferullo DJ, Lovett ST. The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet. 2008;4:e1000300. doi: 10.1371/journal.pgen.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishov I, Woldringh CL. Visualization of membrane domains in Escherichia coli. Mol Microbiol. 1999;32:1166–1172. doi: 10.1046/j.1365-2958.1999.01425.x. [DOI] [PubMed] [Google Scholar]
- Foti JJ, Persky NS, Ferullo DJ, Lovett ST. Chromosome segregation control by Escherichia coli ObgE GTPase. Mol Microbiol. 2007;65:569–581. doi: 10.1111/j.1365-2958.2007.05811.x. [DOI] [PubMed] [Google Scholar]
- Foti JJ, Schienda J, Sutera VA, Jr, Lovett ST. A bacterial G protein-mediated response to replication arrest. Mol Cell. 2005;17:549–560. doi: 10.1016/j.molcel.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Jain V, Kumar M, Chatterji D. ppGpp: stringent response and survival. J Microbiol. 2006;44:1–10. [PubMed] [Google Scholar]
- Jiang M, Sullivan SM, Wout PK, Maddock JR. G-protein control of the ribosome-associated stress response protein SpoT. J Bacteriol. 2007;189:6140–6147. doi: 10.1128/JB.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi G, Moriya S, Wada C. Deficiency of essential GTP-binding protein ObgE in Escherichia coli inhibits chromosome partition. Mol. Microbiol. 2001;41:1037–1051. doi: 10.1046/j.1365-2958.2001.02574.x. [DOI] [PubMed] [Google Scholar]
- Kukimoto-Niino M, Murayama K, Inoue M, Terada T, Tame JR, Kuramitsu S, Shirouzu M, Yokoyama S. Crystal structure of the GTP-binding protein Obg from Thermus thermophilus HB8. J Mol Biol. 2004;337:761–770. doi: 10.1016/j.jmb.2004.01.047. [DOI] [PubMed] [Google Scholar]
- Kuo S, Demeler B, Haldenwang WG. The growth-promoting and stress response activities of the Bacillus subtilis GTP binding protein Obg are separable by mutation. J Bacteriol. 2008;190:6625–6635. doi: 10.1128/JB.00799-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb HK, Thompson P, Elliott C, Charles IG, Richards J, Lockyer M, Watkins N, Nichols C, Stammers DK, Bagshaw CR, Cooper A, Hawkins AR. Functional analysis of the GTPases EngA and YhbZ encoded by Salmonella typhimurium. Protein Sci. 2007;16:2391–2402. doi: 10.1110/ps.072900907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Covalle KL, Maddock JR. The Caulobacter crescentus CgtA protein displays unusual guanine nucleotide binding and exchange properties. J. Bacteriol. 1999;181:5825–5832. doi: 10.1128/jb.181.18.5825-5832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macvanin M, Johanson U, Ehrenberg M, Hughes D. Fusidic acid-resistant EF-G perturbs the accumulation of ppGpp. Mol Microbiol. 2000;37:98–107. doi: 10.1046/j.1365-2958.2000.01967.x. [DOI] [PubMed] [Google Scholar]
- McCool JD, Long E, Petrosino JF, Sandler HA, Rosenberg SM, Sandler SJ. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol Microbiol. 2004;53:1343–1357. doi: 10.1111/j.1365-2958.2004.04225.x. [DOI] [PubMed] [Google Scholar]
- Metzger S, Schreiber G, Aizenman E, Cashel M, Glaser G. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J Biol Chem. 1989;264:21146–21152. [PubMed] [Google Scholar]
- Milon P, Tischenko E, Tomsic J, Caserta E, Folkers G, La Teana A, Rodnina MV, Pon CL, Boelens R, Gualerzi CO. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc Natl Acad Sci U S A. 2006;103:13962–13967. doi: 10.1073/pnas.0606384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto T, Loh PC, Hirai T, Asai K, Kobayashi K, Moriya S, Ogasawara N. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology. 2002;148:3539–3552. doi: 10.1099/00221287-148-11-3539. [DOI] [PubMed] [Google Scholar]
- Pao CC, Fleckenstein J, Dyess BT. Role of peptide chain elongation factor G in guanosine 5'-diphosphate 3'-diphosphate synthesis. J Bacteriol. 1981;145:429–433. doi: 10.1128/jb.145.1.429-433.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso P, Shan SO, Nock S, Herschlag D, Walter P. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry. 2001;40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: Still Magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Raskin DM, Judson N, Mekalanos JJ. Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc Natl Acad Sci U S A. 2007;104:4636–4641. doi: 10.1073/pnas.0611650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Kobayashi G, Hayashi H, Yoshida H, Wada A, Maeda M, Hiraga S, Takeyasu K, Wada C. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells. 2005;10:393–408. doi: 10.1111/j.1365-2443.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- Schreiber G, Metzger S, Aizenman E, Roza S, Cashel M, Glaser G. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991;266:3760–3767. [PubMed] [Google Scholar]
- Schreiber G, Ron EZ, Glaser G. ppGpp-mediated regulation of DNA replication and cell division in Escherichia coli. Curr Microbiol. 1995;30:27–32. doi: 10.1007/BF00294520. [DOI] [PubMed] [Google Scholar]
- Scott JM, Haldenwang WG. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor sigma(B) J. Bacteriol. 1999;181:4653–4660. doi: 10.1128/jb.181.15.4653-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfzadeh M, Keener J, Nomura M. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc Natl Acad Sci U S A. 1993;90:11004–11008. doi: 10.1073/pnas.90.23.11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Das B, Bhadra RK. Functional analysis of the essential GTP-binding protein coding gene cgtA of Vibrio cholerae. J Bacteriol. 2008 doi: 10.1128/JB.02021-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora AE, Zielke R, Wegrzyn A, Wegrzyn G. DNA replication defect in the Escherichia coli cgtA(ts) mutant arising from reduced DnaA levels. Arch Microbiol. 2006;185:340–347. doi: 10.1007/s00203-006-0099-3. [DOI] [PubMed] [Google Scholar]
- Sikora-Borgula A, Slominska M, Trzonkowski P, Zielke R, Mysliwski A, Wegrzyn G, Czyz A. A role for the common GTP-binding protein in coupling of chromosome replication to cell growth and cell division. Biochem. Biophys. Res. Commun. 2002;292:333–338. doi: 10.1006/bbrc.2002.6671. [DOI] [PubMed] [Google Scholar]
- Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominska M, Konopa G, Wegrzyn G, Czyz A. Impaired chromosome partitioning and synchronization of DNA replication initiation in an insertional mutant in the Vibrio harveyi cgtA gene coding for a common GTP-binding protein. Biochem. J. 2002;362:579–584. doi: 10.1042/0264-6021:3620579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Ahmed A. Mutants of Escherichia coli defective in the degradation of guanosine 5'-triphosphate, 3'-diphosphate (pppGpp) Mol Gen Genet. 1979;169:315–323. doi: 10.1007/BF00382277. [DOI] [PubMed] [Google Scholar]
- Spira B, Silberstein N, Yagil E. Guanosine 3',5'-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J Bacteriol. 1995;177:4053–4058. doi: 10.1128/jb.177.14.4053-4058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Jakob U, Bardwell JC. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 2002;184:2692–2698. doi: 10.1128/JB.184.10.2692-2698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosa T, Pizer LI. Biochemical bases for the antimetabolite action of L-serine hydroxamate. J Bacteriol. 1971;106:972–982. doi: 10.1128/jb.106.3.972-982.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D, Albrecht C, Cashel M, D'Ari R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol. 2005;56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- Welsh KM, Trach KA, Folger C, Hoch JA. Biochemical characterization of the essential GTP-binding protein Obg of Bacillus subtilis. J. Bacteriol. 1994;176:7161–7168. doi: 10.1128/jb.176.23.7161-7168.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wout P, Pu K, Sullivan SM, Reese V, Zhou S, Lin B, Maddock JR. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J Bacteriol. 2004;186:5249–5257. doi: 10.1128/JB.186.16.5249-5257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]