Abstract
A key challenge to investigations into the functional roles of glycosaminoglycans (GAGs) in biological systems is the difficulty in achieving sensitive, stable and reproducible mass spectrometric analysis. GAGs are linear carbohydrates with domains that vary in the extent of sulfation, acetylation and uronic acid epimerization. It is of particular importance to determine spatial and temporal variations of GAG domain structures in biological tissues. In order to analyze GAGs from tissue, it is useful to couple mass spectrometry with an on-line separation system. The purposes of the separation system are both to remove components that inhibit GAG ionization and to enable the analysis of very complex mixtures. This contribution presents amide-silica hydrophilic interaction chromatography (HILIC) in a chip-based format for LC/MS of heparin, heparan sulfate and chondroitin/dermatan sulfate GAGs. The chip interface yields robust performance in the negative ion mode that is essential for GAGs and other acidic glycan classes while the built-in trapping cartridge reduces background from the biological tissue matrix. The HILIC chromatographic separation is based on a combination of the glycan chain lengths and the numbers of hydrophobic acetate groups and acidic sulfate groups. In summary, chip based amide-HILIC LC/MS is an enabling technology for GAG glycomics profiling.
Introduction
Progress in the understanding of glycosaminoglycan (GAG) biochemistry has been limited by the challenging chemical and physical properties of this glycoconjugate glycan class. As a result, techniques for GAG analysis lag behind those available for proteins and other carbohydrate classes. GAGs are linear biopolymers that may be sulfated during the biosynthetic process. Biosynthesis of GAG chains is a regulated process that gives rise to proteoglycans with cell and tissue specific structures [1–3]. Nascent heparin/HS chains are modified by a series of biosynthetic enzymes to form domains with high levels of N-sulfation, O-sulfation and glucuronyl-C5-epimerization (NS domains) interspersed with those with a high degree of N-acetylation and low sulfation (NA domains), reviewed in [4, 5]. Nascent CS/DS chains may be acted upon by a specific epimerase to form domains with high IdoAα3GalNAc4Sβ4 content interspersed by domains containing GlcAβ3GalNAc4S and varying amounts of GlcAβ3GalNAc6S, reviewed in [6]. These modification reactions are controlled by a complex interplay of factors including biosynthetic enzyme expression levels and substrate concentration during transport through the Golgi apparatus. Assays of GAGs from biological sources reflect a distribution of structures rather than a single template controlled product [4].
Heparin/HS and CS/DS GAGs participate in a large number of homeostatic, developmental, and signaling processes by interacting with many families of growth factors and growth factor receptors [7, 8]. Such interactions modulate cellular responses to growth factor stimuli at cell surfaces [9–11]. They also serve to control transport of growth factors through extracellular matrices and enable growth factor concentration gradients [12]. Although the number of proteins modified with GAG chains is relatively small, the ranges of their functions are elaborated by variation in their GAG structures. The levels of HS biosynthetic enzymes [4] and the structures of the mature chains vary according to organ [3] and developmental state. A given proteoglycan may function differently depending on the structures of its GAG chains specific to cellular and extracellular environments. The process of tumor metastasis involves remodeling of the extracellular matrix and coordinated changes in surface adhesive properties of cancer cells [13]. Changes to expression of HSPGs associated with cancer offer potential therapeutic targets [14].
An effective analytical platform must enable direct determination of the biosynthetically heterogeneous distributions of GAG domains from tissue. Much GAG structural work has relied on chemical or enzymatic depolymerization to disaccharides followed by chromatographic [15, 16], gel electrophoretic [17, 18], capillary electrophoretic [19, 20] or mass spectrometric [21, 22] analysis of the composition of the products. While these are useful measurements, it is essential to broaden them to include the analysis of extended GAG oligosaccharides.
GAG chains may be partially depolymerized by treatment with specific bacterial polysaccharide lyase enzymes [23]. Mass spectrometry enables analysis of mixtures of partially depolymerized GAGs without the need for complete purification of each component. Nonetheless, mass spectrometric analysis of complex mixtures is most effective when combined with a separation step. High performance size exclusion chromatography provides very robust performance for LC/MS of GAG mixtures [24–26]. Despite its reliability, SEC resolution is low. Furthermore, separation entails use of relatively high flow rates and thus dilutes the analyte concentration. As a result, it is not appropriate for the analysis of sample quantities below the 100 pmol level. Graphitized carbon chromatography (GCC) has been used for LC/MS of native [27–29] and permethylated [30] glycans employing acidic mobile phase systems and positive ion LC/MS. Separation of acidic glycans over GCC requires relatively high pH ammonium bicarbonate mobile phases and has been used for negative ion LC/MS of released glycoprotein oligosaccharides [31] and GAGs [32, 33]. While the high pH facilitates ionization of acidic oligosaccharides, it degrades the silica surfaces present in the spray tips typically used for sub-microliter flow rate chromatographic interfaces. Reversed phase ion pairing chromatography has provided successful results for analysis of GAG classes [34–37]. This technique involves the inclusion of a mono- di-, tri, or tetraalkylamine in the mobile phase at concentrations of 5–30 mM in the presence of an organic acid. Although the technique may be used for analysis of complex GAG mixtures, the relatively high concentration, the potential for amine oxidation by-products, and limited volatility of alkyl amines all contribute to contamination of the mass spectrometer ion source. In addition, mobile phase conditions must be tailored to the compound class. In addition, it is desirable to use stable isotope tags to enable quantitation of GAG mixtures [38].
Amide-silica stationary phase has been used for hydrophilic interaction chromatography (HILIC) positive ion LC/MS of N-glycans [39]. Very similar conditions have been reported for chromatographic separation of native and reductively aminated CS/DS oligosaccharides [40–44]. Capillary amide-HILIC LC/MS has been employed in the analysis of CS/DS chains extracted from connective tissue [45]. This work demonstrated that deuterated and non-deuterated glycomics tags are not separated.
For analysis of GAGs and other acidic glycan classes using amide-HILIC LC/MS, it is necessary to use the negative ion mode. The primary limiting factor to the application of amide-HILIC LC/MS to the generation of glycomics profiling data on GAGs and other native glycan classes is the stability of the spray interface. Our group has found conventional silica sprayers to be problematic due to clogging in the negative mode, requiring time consuming optimization. As a result, time consumed in optimization of the spray conditions has been the limiting factor for GAG glycomics profiling. Removal of contaminating proteins, lipids, nucleic acids and acidic non-GAG carbohydrates must be achieved in a manner that facilitates analysis of sub-microgram quantities of GAGs. Trapping cartridges enable success in analysis of such samples by focusing the analyte while allowing contaminants to flow through with a minimum interaction with the stationary phase. Use of a chip-based GCC system with built-in electrospray nozzle has been reported for positive ion LC/MS of human milk and mucin oligosaccharide alditols [29]. This chip system included a trapping cartridge and valve that is controlled by the chromatograph software.
This work reports the use of a novel chip-based amide-HILIC system for negative ion LC/MS of CS/DS, heparin, and HS GAG classes that removes the difficulties with the spray interface. GAGs are the most acidic biomolecules; this property dictates the use of negative ion mass spectrometry for LC detection. There is a very limited track record in the use of negative ion mode for LC/MS in general and for analysis of GAGs in particular. The chip-based system enables both robust positioning of the spray needle and the analysis of GAGs isolated from complex biological and chemical matrices. The amide-silica is packed into 75 micron internal diameter channels. The use of capillary chromatography at such low internal diameters is greatly facilitated by the integrated spray nozzle and robotic positioning relative to the spray orifice. This integrated system eliminates the need to optimize connections between chromatograph, injector, trapping column, analytical column and spray needle. The amide-HILIC separation mechanism is robust and the same conditions can be used for GAGs and other glycan classes. We demonstrate robust and reproducible analyses of complex oligosaccharide mixtures derived from partially depolymerized GAGs.
Materials and methods
Bovine kidney HS (10 µg, Sigma Chemical, St. Louis, MO) was partially digested using a mixture of heparin lyases I, II, and III (Ibex Technologies, Inc., Montreal, Canada). Digestions were carried out in 100 mM NaCl, 20 mM tris, 1 mM Ca(OAc)2 using 1 mIU of each enzyme. Digestion progress was monitored at 232 nm and the reaction was stopped by boiling when the absorbance reached a value 30% of that observed for a complete digestion. Samples were then lyophilized to dryness, reconstituted in 10 µL of 30% methanol, and injected onto a Superdex Peptide 3.2/30 column (Amersham Biosciences, Piscataway, NJ) with an isocratic mobile phase of 50 mM ammonium formate in 10% acetonitrile. The eluting oligosaccharides were collected and serially dried in a centrifugal evaporator three times with addition of 800 µL of distilled water.
Porcine intestinal mucosa heparin (100 mg, sodium salt, 182 USP/mg, Sigma) was digested with heparin lyase I (4 × 150 mIU, added at 6 hr intervals) in ammonium acetate (1 mL, 100 mM with 0.1 mg/mL bovine serum albumin) at 37 °C. The digestion products were fractionated by preparative SEC (Biorad Bio-Gel P-10, fine) using ammonium bicarbonate mobile phase (200 mM) at a flow rate of 40 µL/min [46]. Collected fractions were desalted by dialysis against pure water.
Chip-based amide-HILIC LC/MS. Amide-80 stationary phase (Tosoh Bioscience, LLC. Montgomeryville, PA, 5 µm particle size, 80 Å pore size) was packed into 75 µm × 150 mm chips by Agilent Technologies Corp., Santa Clara CA. Mobile phase A = 10% acetonitrile, 50 mM formic acid pH 4.4, B = 95% acetonitrile, 5% solvent A. The chip is made of laminated polyimide films with laser ablated channels, ports, and frit structures [47, 48]. A face sealed rotary valve permits switching between sample loading and separation modes with minimal dead volume. The device incorporates a laser ablated nano-electrospray emitter. The chip was interfaced with an Agilent 1200 series nanoflow HPLC system with microfluidic connections between trapping and analytical columns [48]. A separate microscale pump allows mobile phase to be pumped through the trapping column during sample loading while flow is maintained separately through the analytical column. Samples were injected through the trapping column at 4 µL/min and washed with 100% B for 10 min (see figure legends for volumes). The valve was turned to place the trapping column in line with the analytical column at a flow rate of 400 nL/min. The mobile phase was decreased from 95% B to 30% B over the course of 30 min. Negative ion electrospray voltage was set to 1500 V. Mass spectra were acquired using an Agilent 6300 Chip quadrupole ion trap (QIT) mass spectrometry system or an Agilent 6520 Chip-quadrupole time-of-flight (QTOF) mass spectrometry system. Spray conditions with the 6520 system were found to be consistent over the course of a three month period.
Results
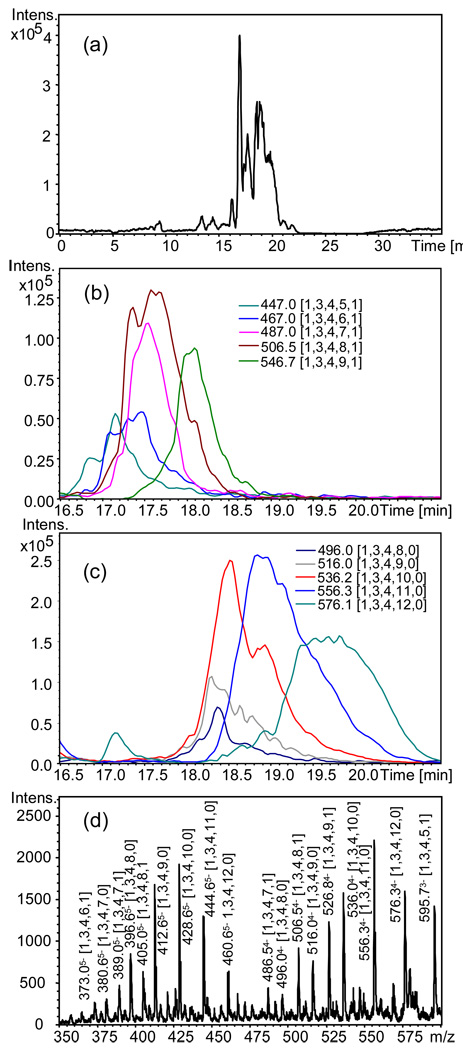
Heparin oligosaccharides contain 2–3 sulfate groups per disaccharide unit, rendering them the most challenging GAG class to analyze. Heparin was partially depolymerized using heparin lyase I and the resulting oligosaccharides were separated into size fractions. Each oligomer chain length contains a distribution with respect to the number of sulfate and acetate groups. Each oligosaccharide composition was observed with 2–3 charge states. The degree of ammonium adduction increased as charge state decreased. Under these conditions, mass spectral complexity becomes too high for direct analysis of heparin size fractions using a trapped ion instrument due to space charge limitations. Figure 1a shows the base peak chromatogram (BPC) for a heparin dp8 fraction acquired using chip-based amide-HILIC LC/QIT MS. Two series of oligosaccharides were observed, one unacetylated and the other monoacetylated. Both were observed with varying numbers of sulfate groups. Extracted ion chromatograms (EICs) for monoacetylated dp8 4- charge state are shown in Figure 1b. Oligosaccharide compositions are given as [ΔHexA, HexA, GlcN, SO3, Ac] in all figures. As a group, the monoacetylated dp8 eluted earlier than their unacetylated counterparts, Figure 1c. As the number of sulfate groups increases, the peak width appears to increase. This phenomenon will be described in greater detail using a high resolution mass analyzer, below. Figure 1d shows the mass spectrum summed over 15–21 min. The dp8 oligosaccharides are observed with 5- (m/z 370–465) and 4- (m/z 480–600) charge states. The pattern of ions differing by sulfate and acetate groups is readily apparent despite the relatively low resolution of the QIT analyzer.
Figure 1.
Chip-based amide HILIC LC/QIT-MS of heparin dp6, 0.15 µg injected; (a) BPC m/z 300–600; EICs for 4- charge state ions for (b) monoacetylated dp6 and (c) unacetylated dp6; (d) mass spectrum summed over 15–21 min. Oligosaccharide compositions are given as [ΔHexA, HexA, GlcN, SO3, Ac].
In order to fully elucidate the chromatographic and mass spectrometric behavior of heparin oligosaccharides using amide-HILIC LC/MS, a high resolution QTOF instrument was employed. Figure 2a shows the BPC obtained from a heparin dp6, dp8, dp10 mixture acquired using a QTOF instrument equipped with a chip-based chromatography interface. The elution ranges for dp6, dp8 and dp10 are shown on the BPC. Summed mass spectra for these three ranges are shown in (b–d). For dp6 (b), 4-, 3- and 2- charge states were observed. A series of dp6 oligosaccharides are detected, with compositions ranges of [1,2,3,SO3,Ac] where SO3 = 6–9 and Ac = 0,1. The extent of ammonium adduction increases with decreasing charge state. A complete table of mass assignments is given in Supplemental Table 1.
Figure 2.
Chip-based amide HILIC LC/QTOF-MS of a heparin dp6, dp8, and dp10 mixture, 0.1 µg injected; (a) BPC m/z 400–1000; summed mass spectra for (b) dp6, summed over 15.0–18.0 min; (c) dp8, 18.0–20.1 min; (d) dp10 20.1–22.5 min.
As shown in (c), dp8 oligosaccharides are observed with charge states of 5-, 4- and 3-. Complete mass assignments are given in Supplemental Table 2, showing that observed compositions include [1,3,4,SO3,Ac] where SO3 = 7–12 and Ac = 0,1. Again, the abundances of ammonium adducts increase with decreasing charge states. Note that acetylated dp10 oligosaccharides are also detected in this elution time range.
For heparin dp10 (d), the heavily ammonium adducted 3- charge state is considerably more abundant than the 4- or 5- charge states. Complete mass assignments are given in Supplemental Table 3. The dp10 oligosaccharides (d) are observed with compositions [1,4,5,SO3, Ac] where SO3 = 10–15 and Ac = 0–2. All 3- charge state dp10 ions were detected with ammonium adducts. For the mass spectra shown in (b–d) the accuracies of the mass assignments for relatively abundant ions (>100 counts) were generally in the ±2–8 ppm range. The error values increased in absolute value for relatively low abundance ions as signal strength decreased.
Figure 3 shows EICs for heparin dp6 oligosaccharide compositions [1,2,3,8,0] (a–c) and [1,2,3,9,0] (d–f). The [1,2,3,8,0] 4- ion (a) was observed as two chromatographic peaks, centered at 17.2 and 17.8 min. Only one peak (at 17.2 min), however, was observed for the 3- (b) and 2- (c) charge states. A single peak at 17.8 min was detected for composition [1,2,3,9,0] for all three charge states (d–f). It therefore appears that [1,2,3,9,0] 4- undergoes some in-source loss of SO3 to form an ion isobaric with [1,2,3,8,0] 4-. Loss of SO3 is not observed for the 3- and 2- charge states of [1,2,3,9,0]. They do not occur for any charge states of dp6 compositions containing fewer than 9 sulfate groups. For dp8, SO3 loss occurs for [1,3,4,11,0] 5- and [1,3,4,12,0] 5-. The 3- and 2- charge states do not undergo such losses. Other dp8 compositions do not undergo such losses for any charge state. For dp10, SO3 losses are observed for [1,4,5,14,0] 6- and [1,4,5,15,0] 6- but not for 5-, 4-, or 3- charge states. Other dp10 compositions do not undergo such losses for any charge state. Therefore, loss of SO3 appears to be favored particularly for the highest observed negative charge state for the oligosaccharide compositions with the most sulfate groups per disaccharide. Therefore, it is appropriate to select a lower charge state for the purpose of generating EICs for quantification.
Figure 3.
Extracted ion chromatograms from Figure 2 for dp6 compositions (a) [1,2,3,8,0] 4- m/z 411.7; (b) [1,2,3,8,0] 3- m/z 549.3; (c) [1,2,3,8,0] 2- m/z 824.5 and (d) [1,2,3,9,0] 4- m/z 431.7; (e) [1,2,3,9,0] 3- m/z 576.0; (f) [1,2,3,9,0] 2- 873.0. Oligosaccharide compositions are given as [ΔHexA, HexA, GlcN, SO3, Ac].
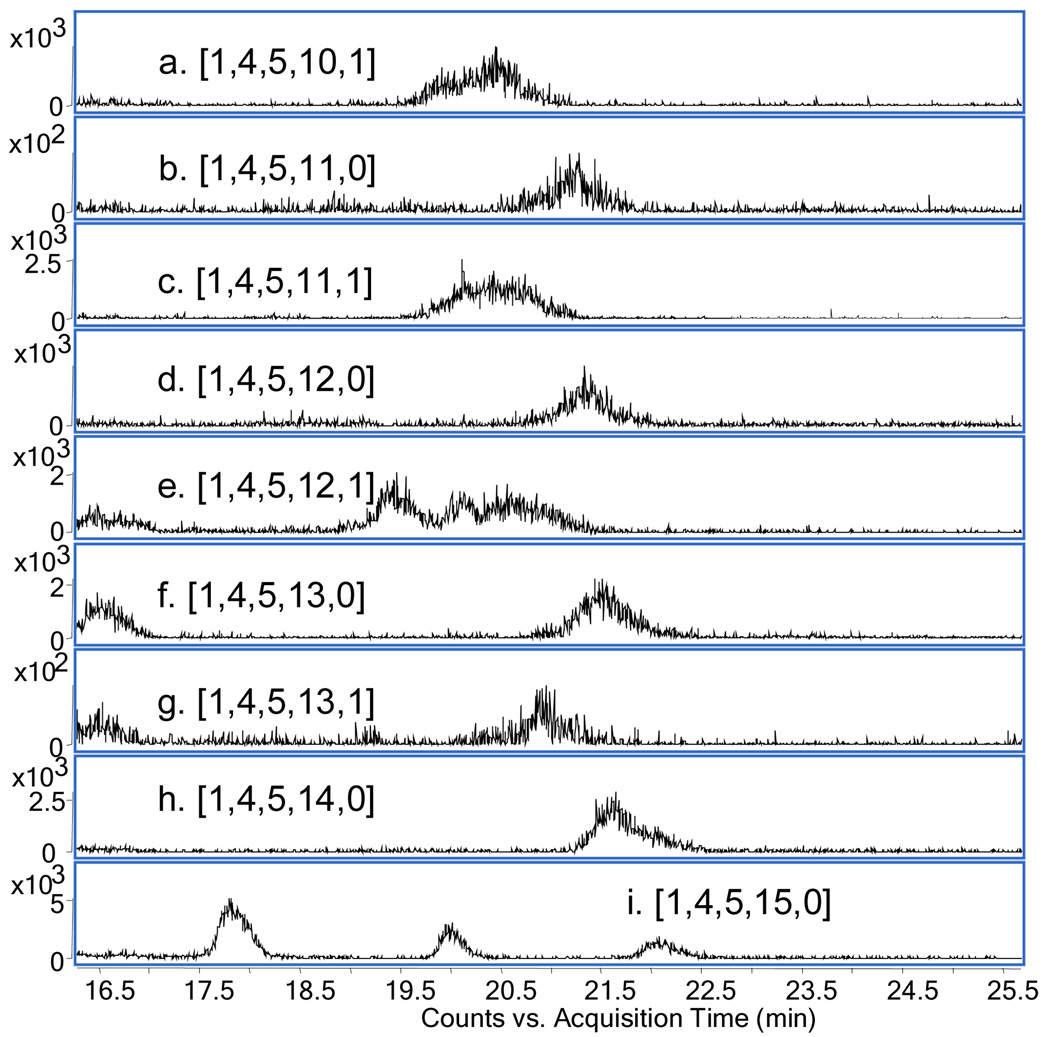
Figure 4 shows a series of EICs for the 5- charge state of a series of dp10 compositions. The acetylated compositions (a, c, e, g) were observed to elute with broader peaks than the unacetylated compositions (b, d, f, h, i). Note that for (i) m/z 576.0 is isobaric for [1,2,3,9,0] 3-, [1,3,4,12,0] 4- and [1,4,5,15,0]5-, thus three peaks were observed. Summed mass spectra (Supplemental Figure 1) for the peaks corresponding to acetylated compositions showed the presence of m/z 504.4 corresponding to [1,4,5,10,1] 5- and 504.2, corresponding to ([1,4,5,10,1] – 1Da) 5-. Similar pairs of ions separated by 1 Da were observed for [1,4,5,11,1] and [1,4,5,12,1] but not for [1,4,5,13,1]. Such losses were also observed for acetylated dp6 and dp8 ions (data not shown but were not observed for unacetylated ions. These observations are consistent with glycosylamine formation for the reducing terminal GlcN residue for acetylated compositions. Glycosylamines are typically produced by prolonged incubation of saccharides with saturating solutions of ammonium bicarbonate [49, 50]. The preparative SEC fractionation was accomplished in the presence of ammonium bicarbonate. The results are consistent with the conclusion that acetylated heparin oligosaccharides are more susceptible to glycosylamine formation during fractionation. Glycosylamine formation has been observed in studies of heparin-antithrombin III binding using amide-HILIC LC/MS [51]. Glycosylamine formation was not observed for GAG preparations not fractionated using large format SEC. It is expected that use of non-ammonium containing salts with the large format SEC will eliminate glycosylamine formation.
Figure 4.
Extracted ion chromatograms Figure 2 for dp10 compositions (a) [1,4,5,10,1] 5- m/z 504.4; (b) [1,4,5,11,0] 5- m/z 512.0; (c) [1,4,5,11,1] 5- m/z 520.4; (d) [1,4,5,12,0] 5- m/z 528.0; (e) [1,4,5,12,1] 5- m/z 536.4; (f) 1,4,5,13,0] 5- m/z 544.0; (g) [1,4,5,13,1] 5- m/z 552.4; (h) [1,4,5,14,0] 5- m/z 560.0; (i) [1,4,5,15,0] 5- m/z 576.0. Oligosaccharide compositions are given as [ΔHexA, HexA, GlcN, SO3, Ac].
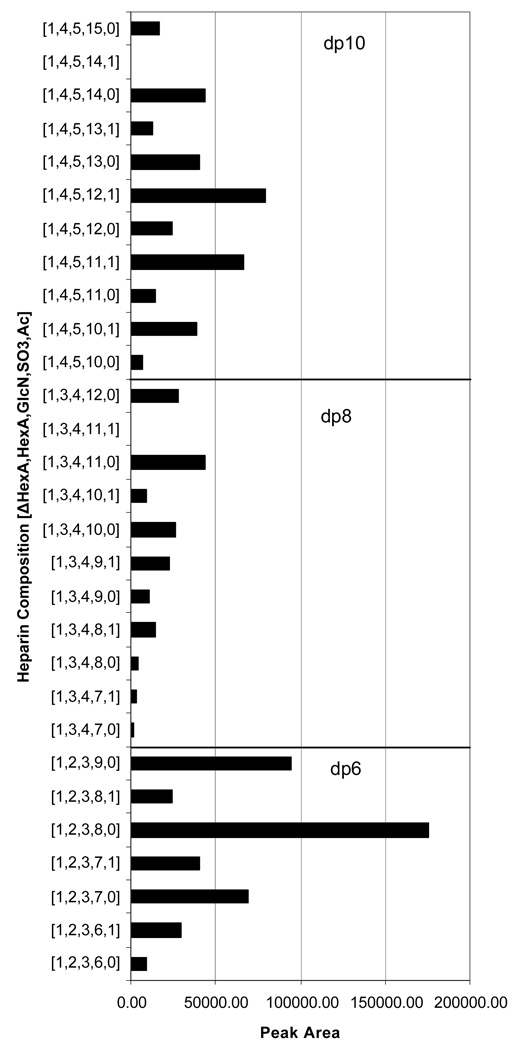
Figure 5 shows the distribution of dp6, dp8 and dp10 oligosaccharide compositions, summing unmodified and glycosylamine forms, obtained from peak areas extracted from the LC/MS data shown in Figure 2. For dp6, the 3- charge state was used and the overall distribution is very similar to that obtained using packed capillary amide-HILIC LC/MS using a different mass spectrometer and using both 2- and 3- charge states [51]. For dp8, the 4- charge state and for dp10 the 4- and 5- charge states were used. The highest charge state for each composition was avoided because of sulfate losses. The lowest charge states were observed with multiple ammonium adducts and overlapping isotopic clusters, preventing reliable extraction of ion chromatograms. The middle range of charge states thus provided the most reliable peak areas. These data demonstrate the potential for using chip-based amide-HILIC LC/MS for comparing distributions of heparin oligosaccharides among different samples.
Figure 5.
Plot of peak area for heparin dp6, dp8 and dp10 oligosaccharide compositions reconstructed from the LC/MS data shown in Figure 2. Peak areas were summed for dp63-, dp8 4- and dp10 4- and 5- charge states. Oligosaccharide compositions are given as [ΔHexA, HexA, GlcN, SO3, Ac].
HS is a GAG that is particularly disperse, resulting from the large number of combinations of enzymatic modifications that occur to chains following polymerization. The structure of HS varies according to tissue location, developmental and disease state [52, 53]. Structural information on HS from biological tissue is typically limited to disaccharide compositional analysis of exhaustively depolymerized samples. The LC/MS compositional profiles for oligosaccharides derived from partially depolymerized HS enables generation of information on the domain structure of the HS chain. This information is particularly relevant because HS-protein interactions require oligosaccharides ranging from dp6–12 for protein binding.
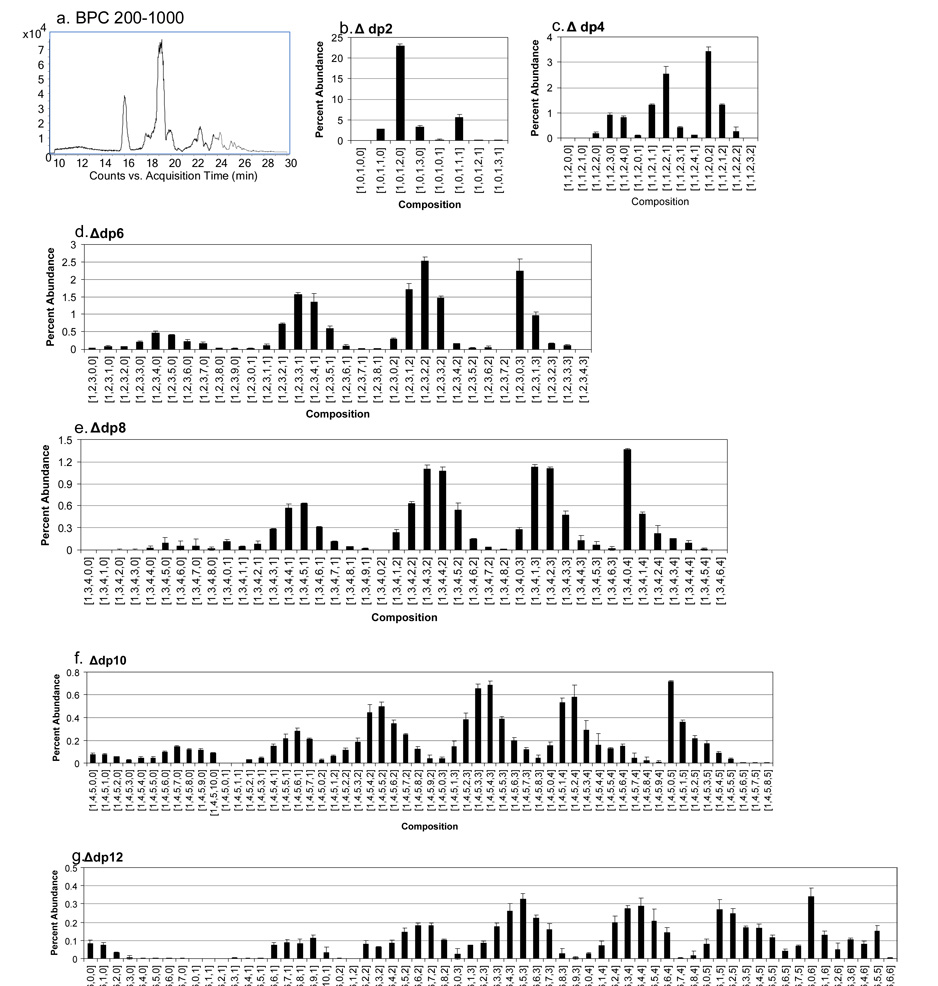
To demonstrate the effectiveness of chip-based amide-HILIC LC/MS for this purpose, bovine kidney HS was depolymerized using a mixture of heparin lyase enzymes to 30% completion. The unfractionated mixture was loaded onto an amide-HILIC chip and subjected to a gradient from high to low percent acetonitrile with a formate modifier with detection using a QTOF instrument. The BPC (Figure 6a) shows the elution range for oligosaccharides derived from HS corresponding to dp2-dp12. The relative abundances for Δ-unsaturated HS oligosaccharides from dp2-dp12 are shown in Figure 6b–g. Representative mass assignments and retention times are shown in Supplemental Table 4. The data were acquired in triplicate and error bars are shown for all HS oligosaccharide compositional abundances. For these data, ion chromatograms for all charge states observed were combined to produce results in bar graph form.
Figure 6.
Chip-based amide-HILIC LC/QTOF-MS of a 30% lyase digestion of bovine kidney HS, 0.5 µg injected; (a) BPC, m/z 200–1000; HS oligosaccharide relative abundances (b) Δdp2, (c) Δdp4, (d) Δdp6, (e) Δdp8, (f) Δdp10, (g) Δdp12. Oligosaccharide compositions are given as [ΔHexA, HexA, GlcN, SO3, Ac].
The results (a) show the presence of both sulfated and unsulfated Δdp2, with [1,0,1,2,0] the most abundant composition, as expected for HS with abundant NA domains. Oligosaccharides corresponding to Δdp4 were detected with 0, 1 and 2 acetate groups (c) with [1,1,2,0,2] the most abundant composition. This composition corresponds to an NA domain, and more highly sulfated variants are relatively low in abundances. The unsulfated Δdp4 correspond to NS domains, and [1,1,2,3,0] and [1,1,2,4,0] are equally abundant. The [1,1,2,2,1] is the most abundant mono-acetylated composition. The Δdp6 oligosaccharides (d) also show abundant NA domain compositions in which all three glucosamine residues are acetylated. The unsulfated [1,2,3,0,3] is the most abundant of these with sulfated variants decreasing in abundance. The pattern of sulfation for NA domains in which all glucosamine residues are acetylated is similar for Δdp8 (e), Δdp10 (f) and Δdp12 (g). For all oligosaccharide lengths, compositions with zero acetate groups are comparatively low in abundances. Oligosaccharides with both N-acetylated and N-sulfated glucosamine residues, as indicated by their compositions, contain a distribution of sulfated forms. The number of sulfate groups for the most abundant compositions decreases as the number of acetate groups increases.
The high quality of these data is characterized by both the small error bars and the overall consistent pattern of the HS oligosaccharide compositional abundances. The results demonstrate an approach that will be applicable for the profiling of HS structure as a function of biological variables. It is likely that amide-HILIC LC/MS will be widely applicable, especially when used with a well-engineered chromatographic interface, such as demonstrated here.
Discussion
Chip-based amide-HILIC enables LC/MS of highly complex heparin and heparan sulfate GAG oligosaccharide mixtures. Heparin/HS oligosaccharides released from tissue sources or cell culture contain a distribution of chain lengths, sulfation and acetylation patterns. The chains may be partially depolymerized by partial digestion with polysaccharide lyases, generating mixtures of oligosaccharides. Such mixtures are too complex for analysis by direct infusion.
The GAGs are the most acidic biomolecules and may be analyzed directly using negative ion mode ESI MS. Difficulties with achieving a stable electrospray in the negative mode increase as the flow rate decreases. Low flow rates are necessary in order to reduce the chromatographic scale so as to achieve the high sensitivities necessary for the analysis of glycans released from small quantities of tissue. The chip format integrates the electrospray nozzle, analytical column, trapping column and associated valves. Thus, difficulties with connections between individual components are eliminated. Robotic positioning of the spray nozzle with respect to the MS source orifice eliminates need for manual manipulation. Experience in the authors’ laboratory has shown that the most time consuming steps for the setup of a system for amide-HILIC LC/MS in the negative ion mode using conventional sprayers is the achievement of a stable electrospray. A stable spray using the described chip-based spray device is possible with no day-to-day optimization over a three month period. Thus, effort can be shifted to analyzing the glycomics profiling data set rather than achieving stable spray. The fact that the fluidic connections are made in the chip device and the spray geometry is robotically controlled enables a much needed shift of effort to the derivation of biochemical meaning of the mass spectral information.
The ultimate limitations on sensitivity of an LC/MS system for glycans arises from the presence of contaminating factors from the biological matrix or chemical workup that suppress ionization of the glycans eluting from the column or interfere with glycan signals. For any sample derived from tissue, to the degree the LC/MS step tolerates and removes such contaminants, time consuming workup steps can be eliminated. Adsorption of GAG oligosaccharides to amide-HILIC is strong enough to allow use of a short trapping column. Such a trapping column allows washing of the injected glycan mixture at increased flow rate, flushing out weakly retained contaminant compounds. In the absence of a trapping column, such contaminants would bleed abundant ions throughout the mass chromatogram that would suppress glycan signals. The trapping step facilitates analysis of the same mixtures by removing contaminants.
Amide-HILIC LC/MS enables analysis of partially depolymerized GAG oligosaccharide mixtures too complex for direct infusion MS. An ion trap instrument suffices for the analysis of relatively complex mixtures of CS/DS oligosaccharides from connective tissue. It is possible to analyze complex mixtures derived from partial digestion of HS using amide-HILIC ion trap LC/MS. Increased resolution is necessary for analysis of larger GAG oligosaccharides, in particular those derived from heparin. The described analytical system will provide a robust and sensitive LC/MS platform for partially depolymerized GAG classes that should be useful for many academic and industrial groups interested in fundamental studies of GAG biochemistry and in engineering drug and therapeutic products based on their structures. Heparins pose the greatest analytical challenge among the GAG classes, but even these can be analyzed successfully with this system.
Supplementary Material
Acknowledgments
This work was funded by NIH/NCRR P41RR10888, NIH/NHLBI R01HL74197, and an Agilent Technologies Foundation Research Project Gift. HILIC chips were provided by Carsten Kraiczek and Rudi Grimm of Agilent Technologies.
Abbreviations
- Ac
acetate
- BPC
base peak chromatogram
- CS
chondroitin sulfate
- Dp
degree of polymerization
- DS
dermatan sulfate
- ESI
electrospray ionization
- GAG
glycosaminoglycan
- GlcN
N-acetylglucosamine
- HexA
hexuronic acid
- HILIC
hydrophilic interaction chromatography
- HS
heparan sulfate
- ppm
parts per million
- QIT
quadrupole ion trap
- QTOF
quadrupole time-of-flight
- RT
retention time
References
- 1.Lindahl U, Kusche-Gullberg M, Kjellen L. Regulated diversity of heparan sulfate. J Biol Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 2.Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maccarana M, Sakura Y, Tawada A, Yoshida K, Lindahl U. Domain structure of heparan sulfates from bovine organs. J Biol Chem. 1996;271:17804–17810. doi: 10.1074/jbc.271.30.17804. [DOI] [PubMed] [Google Scholar]
- 4.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 5.Bulow HE, Hobert O. The Molecular Diversity of Glycosaminoglycans Shapes Animal Development. Annu Rev Cell Dev Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- 6.Silbert JE, Sugumaran G. Biosynthesis of chondroitin/dermatan sulfate. IUBMB Life. 2002;54:177–186. doi: 10.1080/15216540214923. [DOI] [PubMed] [Google Scholar]
- 7.Conrad HE. Heparin Binding Proteins. New York: Academic Press; 1998. [Google Scholar]
- 8.Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 9.Nugent MA, Iozzo RV. Fibroblast growth factor-2. Int J Biochem Cell Biol. 2000;32:115–120. doi: 10.1016/s1357-2725(99)00123-5. [DOI] [PubMed] [Google Scholar]
- 10.Segev A, Nili N, Strauss BH. The role of perlecan in arterial injury and angiogenesis. Cardiovasc Res. 2004;63:603–610. doi: 10.1016/j.cardiores.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 11.Fannon M, Forsten KE, Nugent MA. Potentiation and inhibition of bFGF binding by heparin: a model for regulation of cellular response. Biochemistry. 2000;39:1434–1445. doi: 10.1021/bi991895z. [DOI] [PubMed] [Google Scholar]
- 12.Dowd CJ, Cooney CL, Nugent MA. Heparan sulfate mediates bFGF transport through basement membrane by diffusion with rapid reversible binding. J Biol Chem. 1999;274:5236–5244. doi: 10.1074/jbc.274.8.5236. [DOI] [PubMed] [Google Scholar]
- 13.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 14.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 15.Toyoda H, Motoki K, Tanikawa M, Shinomiya K, et al. Determination of human urinary hyaluronic acid, chondroitin sulphate and dermatan sulphate as their unsaturated disaccharides by high-performance liquid chromatography. J Chromatogr. 1991;565:141–148. doi: 10.1016/0378-4347(91)80378-p. [DOI] [PubMed] [Google Scholar]
- 16.Toyoda H, Kinoshita-Toyoda A, Fox B, Selleck SB. Structural analysis of glycosaminoglycans in animals bearing mutations in sugarless, sulfateless, and tout-velu. Drosophila homologues of vertebrate genes encoding glycosaminoglycan biosynthetic enzymes. J Biol Chem. 2000;275:21856–21861. doi: 10.1074/jbc.M003540200. [DOI] [PubMed] [Google Scholar]
- 17.Calabro A, Benavides M, Tammi M, Hascall VC, Midura RJ. Microanalysis of enzyme digests of hyaluronan and chondroitin/dermatan sulfate by fluorophore-assisted carbohydrate electrophoresis (FACE) Glycobiology. 2000;10:273–281. doi: 10.1093/glycob/10.3.273. [DOI] [PubMed] [Google Scholar]
- 18.Calabro A, Midura R, Wang A, West L, et al. Fluorophore-assisted carbohydrate electrophoresis (FACE) of glycosaminoglycans. Osteoarthritis Cartilage. 2001;9 Suppl A:S16–S22. doi: 10.1053/joca.2001.0439. [DOI] [PubMed] [Google Scholar]
- 19.Militsopoulou M, Lamari FN, Hjerpe A, Karamanos NK. Determination of twelve heparin- and heparan sulfate-derived disaccharides as 2-aminoacridone derivatives by capillary zone electrophoresis using ultraviolet and laser-induced fluorescence detection. Electrophoresis. 2002;23:1104–1109. doi: 10.1002/1522-2683(200204)23:7/8<1104::AID-ELPS1104>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Lamari FN, Kuhn R, Karamanos NK. Derivatization of carbohydrates for chromatographic, electrophoretic and mass spectrometric structure analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;793:15–36. doi: 10.1016/s1570-0232(03)00362-3. [DOI] [PubMed] [Google Scholar]
- 21.Saad OM, Leary JA. Compositional analysis and quantification of heparin and heparan sulfate by electrospray ionization ion trap mass spectrometry. Anal Chem. 2003;75:2985–2995. doi: 10.1021/ac0340455. [DOI] [PubMed] [Google Scholar]
- 22.Desaire H, Sirich TL, Leary JA. Evidence of block and randomly sequenced chondroitin polysaccharides: sequential enzymatic digestion and quantification using ion trap tandem mass spectrometry. Anal Chem. 2001;73:3513–3520. doi: 10.1021/ac010385j. [DOI] [PubMed] [Google Scholar]
- 23.Ernst S, Langer R, Cooney CL, Sasisekharan R. Enzymatic degradation of glycosaminoglycans. Crit Rev Biochem Mol Biol. 1995;30:387–444. doi: 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- 24.Zaia J, Costello CE. Compositional analysis of glycosaminoglycans by electrospray mass spectrometry. Anal Chem. 2001;73:233–239. doi: 10.1021/ac000777a. [DOI] [PubMed] [Google Scholar]
- 25.Hitchcock AM, Costello CE, Zaia J. Glycoform quantification of chondroitin/dermatan sulfate using an LC/MS/MS platform. Biochemistry. 2006;45:2350–2361. doi: 10.1021/bi052100t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henriksen J, Ringborg LH, Roepstorrf P. On-line size-exclusion chromatography/mass spectrometry of low molecular mass heparin. J Mass Spectrom. 2004;39:1305–1312. doi: 10.1002/jms.723. [DOI] [PubMed] [Google Scholar]
- 27.Kawasaki N, Ohta M, Hyuga S, Hashimoto O, Hayakawa T. Analysis of Carbohydrate Heterogeneity in a Glycoprotein Using Liquid Chromatography/Mass Spectrometry and Liquid Chromatography with Tandem Mass Spectrometry. Anal Biochem. 1999;269:297. doi: 10.1006/abio.1999.4026. [DOI] [PubMed] [Google Scholar]
- 28.Kawasaki N, Ohta M, Hyuga S, Hyuga M, Hayakawa T. Application of Liquid Chromatography/Mass Spectrometry and Liquid Chromatography with Tandem Mass Spectrometry to the Analysis of the Site-Specific Carbohydrate Heterogeneity in Erythropoietin. Anal Biochem. 2000;285:82. doi: 10.1006/abio.2000.4739. [DOI] [PubMed] [Google Scholar]
- 29.Niñonuevo M, An H, Yin H, Killeen K, et al. Nanoliquid chromatography-mass spectrometry of oligosaccharides employing graphitized carbon chromatography on microchip with a high-accuracy mass analyzer. Electrophoresis. 2005;26:3641–3649. doi: 10.1002/elps.200500246. [DOI] [PubMed] [Google Scholar]
- 30.Costello CE, Miller MJC, Cipollo JC. A Glycomics Platform for the Analysis of Permethylated Oligosaccharide Alditols. 2007 doi: 10.1016/j.jasms.2007.07.016. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlsson NG, Wilson NL, Wirth HJ, Dawes P, et al. Negative ion graphitised carbon nano-liquid chromatography/mass spectrometry increases sensitivity for glycoprotein oligosaccharide analysis. Rapid Commun Mass Spectrom. 2004;18:2282–2292. doi: 10.1002/rcm.1626. [DOI] [PubMed] [Google Scholar]
- 32.Karlsson NG, Schulz BL, Packer NH, Whitelock JM. Use of graphitised carbon negative ion LC-MS to analyse enzymatically digested glycosaminoglycans. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;824:139–147. doi: 10.1016/j.jchromb.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 33.Estrella RP, Whitelock JM, Packer NH, Karlsson NG. Graphitized Carbon LC-MS Characterization of the Chondroitin Sulfate Oligosaccharides of Aggrecan. Anal Chem. 2007;79:3597–3606. doi: 10.1021/ac0622227. [DOI] [PubMed] [Google Scholar]
- 34.Kuberan B, Lech M, Zhang L, Wu ZL, et al. Analysis of Heparan Sulfate Oligosaccharides with Ion Pair-Reverse Phase Capillary High Performance Liquid Chromatography-Microelectrospray Ionization Time-of-Flight Mass Spectrometry. J Am Chem Soc. 2002;124:8707–8718. doi: 10.1021/ja0178867. [DOI] [PubMed] [Google Scholar]
- 35.Thanawiroon C, Rice KG, Toida T, Linhardt RJ. LC/MS sequencing approach for highly sulfated heparin-derived oligosaccharides. J Biol Chem. 2004;279:2608–2615. doi: 10.1074/jbc.M304772200. [DOI] [PubMed] [Google Scholar]
- 36.Vongchan P, Warda M, Toyoda H, Toida T, et al. Structural characterization of human liver heparan sulfate. Biochimica et biophysica acta. 2005;1721:1–8. doi: 10.1016/j.bbagen.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Henriksen J, Roepstorff P, Ringborg LH. Ion-pairing reversed-phased chromatography/mass spectrometry of heparin. Carbohydr Res. 2006;341:382–387. doi: 10.1016/j.carres.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 38.Bowman M, Zaia J. Novel Tags for the Stable Isotopic Labeling of Carbohydrates and Quantitative Analysis by Mass Spectrometry. Anal Chem. 2007;76:5777–5784. doi: 10.1021/ac070581b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wuhrer M, Koeleman CAM, Deelder AM, Hokke CN. Normal-phase nanoscale liquid chromatography - Mass spectrometry of underivatized oligosaccharides at low-femtomole sensitivity. Anal Chem. 2004;76:833–838. doi: 10.1021/ac034936c. [DOI] [PubMed] [Google Scholar]
- 40.Takagaki K, Munakata H, Majima M, Endo M. Enzymatic reconstruction of a hybrid glycosaminoglycan containing 6- sulfated, 4-sulfated, and unsulfated N-acetylgalactosamine. Biochem Biophys Res Commun. 1999;258:741–744. doi: 10.1006/bbrc.1999.0697. [DOI] [PubMed] [Google Scholar]
- 41.Takagaki K, Munakata H, Kakizaki I, Majima M, Endo M. Enzymatic reconstruction of dermatan sulfate. Biochem Biophys Res Commun. 2000;270:588–593. doi: 10.1006/bbrc.2000.2452. [DOI] [PubMed] [Google Scholar]
- 42.Takagaki K, Munakata H, Majima M, Kakizaki I, Endo M. Chimeric glycosaminoglycan oligosaccharides synthesized by enzymatic reconstruction and their use in substrate specificity determination of Streptococcus hyaluronidase. J Biochem (Tokyo) 2000;127:695–702. doi: 10.1093/oxfordjournals.jbchem.a022659. [DOI] [PubMed] [Google Scholar]
- 43.Takagaki K, Ishido K. Synthesis of chondroitin sulfate oligosaccharides using enzymatic reconstruction. Trends in Glycoscience and Glycotechnology. 2000;12:295–306. [Google Scholar]
- 44.Takagaki K, Munakata H, Kakizaki I, Iwafune M, et al. Domain structure of chondroitin sulfate E octasaccharides binding to type V collagen. J Biol Chem. 2002;277:8882–8889. doi: 10.1074/jbc.M106479200. [DOI] [PubMed] [Google Scholar]
- 45.Hitchcock A, Costello C, Zaia J. Comparative Glycomics of Connective Tissue Glycosaminoglycans. 2007 doi: 10.1002/pmic.200700787. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziegler A, Zaia J. Separation of heparin oligosaccharides by size-exclusion chromatography. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;837:76–86. doi: 10.1016/j.jchromb.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 47.Yin H, Killeen K, Brennen R, Sobek D, et al. Microfluidic chip for peptide analysis with an integrated HPLC column, sample enrichment column, and nanoelectrospray tip. Anal Chem. 2005;77:527–533. doi: 10.1021/ac049068d. [DOI] [PubMed] [Google Scholar]
- 48.Yin H, Kileen K. The fundamental aspects and applications of Agilent HPLC-Chip. J Sep Sci. 2007;30:1427–1434. doi: 10.1002/jssc.200600454. [DOI] [PubMed] [Google Scholar]
- 49.Likhosherstov LM, Novikova OS, Derevitskaja VA, Kochetkov NK. A new simple synthesis of amino sugar [beta]--glycosylamines. Carbohydr Res. 1986;146:C1–C5. [Google Scholar]
- 50.Manger ID, Rademacher TW, Dwek RA. 1-N-glycyl beta-oligosaccharide derivatives as stable intermediates for the formation of glycoconjugate probes. Biochemistry. 1992;31:10724–10732. doi: 10.1021/bi00159a012. [DOI] [PubMed] [Google Scholar]
- 51.Naimy H, Leymarie N, Bowman M, Zaia J. Characterization of heparin oligosaccharides binding specifically to antithrombin III using mass spectrometry. Biochemistry. 2008;47:3155–3161. doi: 10.1021/bi702043e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ori A, Wilkinson MC, Fernig DG. The heparanome and regulation of cell function: structures, functions and challenges. Front Biosci. 2008;13:4309–4338. doi: 10.2741/3007. [DOI] [PubMed] [Google Scholar]
- 53.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.