Abstract
Abstract We revisit an application developed originally using abductive Inductive Logic Programming (ILP) for modeling inhibition in metabolic networks. The example data was derived from studies of the effects of toxins on rats using Nuclear Magnetic Resonance (NMR) time-trace analysis of their biofluids together with background knowledge representing a subset of the Kyoto Encyclopedia of Genes and Genomes (KEGG). We now apply two Probabilistic ILP (PILP) approaches - abductive Stochastic Logic Programs (SLPs) and PRogramming In Statistical modeling (PRISM) to the application. Both approaches support abductive learning and probability predictions. Abductive SLPs are a PILP framework that provides possible worlds semantics to SLPs through abduction. Instead of learning logic models from non-probabilistic examples as done in ILP, the PILP approach applied in this paper is based on a general technique for introducing probability labels within a standard scientific experimental setting involving control and treated data. Our results demonstrate that the PILP approach provides a way of learning probabilistic logic models from probabilistic examples, and the PILP models learned from probabilistic examples lead to a significant decrease in error accompanied by improved insight from the learned results compared with the PILP models learned from non-probabilistic examples.
1 Introduction
There is currently considerable interest within Artificial Intelligence in Probabilistic Logic Learning (De Raedt et al., 2008) and the closely allied area of Statistical Relational Learning (Getoor and Taskar, 2007). These research fields are concerned with the integration of probabilistic reasoning with first order logic representations and machine learning. This integration is needed in order to face the challenge of real-world data mining problems in which the data consists of sets of objects with associated structural relations. We are interested in finding useful predictive and/or descriptive patterns. In this paper, the term probabilistic is used to refer to representations and forms of reasoning based on the probability calculus. The term statistical is used to refer to empirical estimation techniques. The term logic is used here to refer to representations and reasoning related to the predicate calculus such as those studied within the field of computational logic. The term relational is used for modeling data based on predicate logic and set theory as done in relational database. The primary advantage of using such representations is that it allows one to elegantly represent complex situations involving a variety of objects as well as relations among the objects. The term learning in the context refers to deriving the different aspects of a model in a probabilistic logic on the basis of data. Typically, one distinguishes various learning algorithms on the basis of the given data (fully or partially observable) or on the aspect being learned (parameter estimation or logical structure learning). The motivation for learning is that it is often easier to obtain data for a given application domain and learn the model than to build the model using traditional knowledge engineering techniques.
One approach to this problem is Probabilistic Inductive Logic Programming (PILP) (De Raedt and Kersting, 2004; De Raedt et al., 2008), which naturally extends traditional Inductive Logic Programming (ILP) (Muggleton and De Raedt, 1994) by introducing probabilities that can explicitly deal with uncertainty such as missing and noisy information. There have been some promising PILP frameworks and systems developed so far to help people build probabilistic logic models, such as Bayesian Logic Programs (BLPs) (Kersting and De Raedt, 2000), Stochastic Logic Programs (SLPs) (Muggleton, 1996), Independent Choice Logic (ICL) (Poole, 1997) and PRogramming In Statistical modeling (PRISM) (Sato, 1995), Markov Logic Networks (MLNs) (Richardson and Domingos, 2006), etc (De Raedt et al., 2008).
Although an increasing number of systems and applications have been published, there are still many challenges in the PILP research. The question investigated in this paper is whether PILP should always be used to learn from non-probabilistic examples. This is motivated by the fact that the data sets used by most PILP systems and applications have non-probabilistic class values, like those used in ILP systems. On the one hand, there is information lost by learning using just non-probabilistic data compared with the raw (possibly continuous) data. In many cases, however, we could extract more information, such as empirical probability or validity, in addition to categorical values. Such information can be further used to support learning as well as model evaluation in PILP. On the other hand, the ability of handling such probabilistic examples should be one of the distinct positive features of PILP against ILP. The main reason for the problem is the lack of an obvious source of probabilistic class values and corresponding methods of exacting probabilistic examples from raw data. In this context, we investigate the use of Abductive Stochastic Logic Programs and the PRISM system (Sato, 1995; Sato et al., 2008) for metabolic network inhibition learning and demonstrate that PILP models with higher predictive accuracy can be learned from probabilistic examples than non-probabilistic examples.
This paper is organized as follows. Section 2 provides background relating to an introduction of probabilistic examples, PILP, SLPs, abduction, PRISM and the biological application area of metabolic network inhibition as well as the previous study of abductive ILP. This is followed by a description of the abductive approach to SLPs used in this paper as well as the allied system PRISM. A general approach is described in Section 4 for extracting empirical probability labels from scientific data. This approach is employed in the experiments of Section 5 which apply abductive SLP learning and the PRISM system to the metabolic network inhibition problem. We show that significant accuracy increases are achieved by learning the PILP models from probabilistic examples. Section 6 concludes with a comparison to some related approaches and a discussion of the future work.
2 Motivation and Background
2.1 Interpretation of Probability and Probabilistic Examples
For the purposes of understanding the learned knowledge, it is vital to identify the interpretation of probability employed within any PILP application. We are not going to review the existing arguments of interpretations of probability in philosophy and statistics (Alan, 2007). Instead, we distinguish two types of probabilistic knowledge following Halpern's categorisation on first-order logics of probability (Halpern, 1989).
In order to analyse the semantics of first-order logics of probability, two approaches are considered in (Halpern, 1989). A type 1 probability structure is defined on the domain and is appropriate for giving semantics to formulae involving statistical information; By contrast, a type 2 probability structure puts probabilities on possible worlds and is appropriate for giving semantics to formulae describing degrees of belief. According to the categorisation, type 1 probabilities capture statistical information about the world by performing experiments or trials, in which objective domain frequencies or empirical probability distribution are gathered over objects, instances or sample spaces; whereas type 2 probabilities implicitly assume the existence of a number of possibilities or possible worlds (in some of which formulae are true while in others are false), with some subjective probability distribution over these worlds. Thus, the key difference between the two probability structures is that probabilities are defined over the domain in type 1 structures, while probabilities are defined over the domain in type 2 structures. Although they are two fundamentally different types of probabilities, Halpern remarks there is a sense in which we can translate between the two types of probability structures and furthermore combine the two modes of probabilistic reasoning in some situations (Halpern, 1989).
In addition, it is common that the probability distributions are defined over possible worlds (first-order models) which give for each closed logical formula the probability that it is true, but it is not necessary to define a distribution over the truth values of formulae in type 1 probability logic (Cussens, 2001). While possible worlds semantics are widely used in Bayesian approaches (Pearl, 1988) and PILP (Sato, 1995; Poole, 1997; Kersting and De Raedt, 2000), type 1 semantics have been applied in Probably Approximately Correct (PAC) Learning (Haussler, 1990) and Stochastic Logic Programs (SLPs) (Muggleton, 1996; Cussens, 2001).
Based on the above categorisation of probabilities, we define a probabilistic example in our study to be an example together with either a type 1 empirical frequency (probability defined on the domain) or a type 2 empirical probability (probability defined on the possible worlds). In mathematics, empirical probability of an event is the fraction of times we expect it to occur in an experiment (Stefan and Steven, 2004). In a general case, the empirical probability (of a sample) estimates the theoretical probability (of a population) by the law of large numbers: as the number of trials of an experiment increases, the empirical probability approaches the theoretical probability. It is fair to say that the normal empirical probabilities accord with type 1 probability semantics as they are domain-based. We introduce type 2 empirical probability in order to deal with the cases when we are unable to count empirical frequencies, but we could instead estimate the degree of belief or validity that some event happened in terms of possible worlds semantics. We demonstrate a method in the paper that can extract type 2 empirical probabilities from a small data set containing control and treated cases where the empirical frequencies are not countable due to the size of sample space.
Definition 1 (Probabilistic Example)
A probabilistic example is a tuple (e, Pe(e)), in which e is a ground logic atom and Pe(e)1 is either a (type 1) empirical frequency that is defined by counting the frequency e occurred in a sample space, or a (type 2) empirical probability which is defined by capturing the degree we believe e is true in some possible worlds.
Probabilistic examples are more accurate representation of what we know about the world than non-probabilistic examples. For example, a probabilistic example (concentration(citrate, down), 0.80) in metabolic network research means that either we believe the statement ‘the concentration level of metabolite citrate goes down’ is true with 80% validity or in terms of degree of belief (empirical probability), or we have observed ‘the concentration level of citrate has a down regulation’ in an experiment with a frequency of 80% (empirical frequency). This contrasts with a (positive) binary example concentration(citrate, down) used in ILP. In addition, another advantage of providing probabilistic examples results in implicit introduction of the corresponding complements for binary examples. For example, in the above example, a complementary probabilistic example (concentration(citrate, up), 0.20) can be assumed accordingly. Thus probabilistic examples enrich the observations we could extract from the raw data and provide extra support for learning. It is necessary to clarify that empirical probability is not prior probability, which is always used in Bayesian learning (Friedman, 1998) and is often the purely subjective assessment made by domain experts. Empirical probability could be thought as posterior probability conditional on the experimental data.
Although the empirical frequency and the empirical probability in the definition have different semantics, we fuse them in the context of abductive SLPs, that is, empirical probabilities extracted from a sample space are converted into empirical frequencies for abductive SLP learning. The motivation for this is, as detailed in section 3, that domain-based probability distribution and possible-world-based probability distribution are treated identically in terms of SLD-derivation in abductive SLPs.
2.2 Probabilistic ILP with Probabilistic Examples
Probabilistic ILP aims to provide a formal learning framework for probabilistic logic learning. It extends ILP to deal with uncertainty. To address our motivation, we use the following learning setting of PILP with probabilistic examples.
Definition 2 (Probabilistic ILP with Probabilistic Examples)
Given a probabilistic logic programming representation language ℒ, a set E = {(ei, Pe(ei))} of probabilistic examples over ℒ, and a background theory B, PILP finds a set of probabilistic hypotheses {(LP, λ)} over ℒ by applying some scoring function score(E, LP, λ, B) such that (LP, λ) stands for a logic program LP annotated with probabilistic parameters λ. The scoring function is some objective score that returns a posterior distribution over the models {(LP, λ)} and consists of the likelihood Pr(E|LP, λ, B) and/or a function that penalises the complexity of LP.
Following the traditional Bayesian learning (Friedman, 1998) paradigm, the scoring function defined above could be as simple as the likelihood, Pr(E|LP, λ, B), (e.g. for maximum likelihood estimation); or the posterior probability of the model, Pr(LP, λ|E, B), (e.g. for maximum a posteriori); or the scores that take into account prior probabilities and penalised functions, such as minimum description length (MDL) score (Rissanen, 1982) and Bayesian Information Criterion (BIC) score (Friedman, 1998). If we suppose that the examples are independent and identically distributed (i.i.d.)2, then where m is the cardinality of E.
This formulation is more general than the one described in (De Raedt and Kersting, 2004) which is to find a single best hypothesis. Firstly, we include probabilistic examples and define the hypothesis scoring function to have not only the examples {ei} but also their associated empirical probability values {Pe(ei)} as the arguments. Secondly, our goal is to select a set of candidate hypotheses using the Bayesian approach that finds a posterior distribution over hypotheses. Thirdly, the penalised part in the scoring function plays an important role in the learning as it overcomes the overfitting (to data) problem caused by only employing likelihood.
2.3 SLPs and Failure-Adjusted maximisation Algorithm
Stochastic logic programs (SLPs) (Muggleton, 1996) are one of the developed PILP frameworks that provide a natural way of associating probabilities with logical rules. SLPs were introduced originally as a way of lifting stochastic grammars to the level of first-order logic programs. SLPs were considered to be a generalisation of hidden Markov models and stochastic context-free grammars. SLPs have later been used to define distributions for sampling within ILP.
Definition 3 (Stochastic Logic Programs)
An SLP S is a definite logic program, where each clause C is a first-order range-restricted definite clause3 and some of the definite clauses are labelled/parameterised with non-negative numbers, l : C. S is said to be a pure SLP if all clauses have parameters, as opposed to an impure SLP if not all clauses have labels. The subset Sq of clauses in S whose head share the same predicate symbol q is called the definition of q. For each definition Sq, we use πq to denote the sum of the labels of the clauses in Sq. S is normalised if πq = 1 for each q and unnormalised otherwise.
For our purposes, SLPs are restricted to define probability distributions over definite clauses, where each l is set to be a number in the interval [0,1]. In a pure normalised SLP, each choice for a clause C has a parameter attached and the parameters sum to one, so they can therefore be interpreted as probabilities. Pure normalised SLPs are defined such that each parameter l denotes the probability that C is the next clause used in a derivation given that its head C+ has the correct predicate symbol. Impure SLPs are useful to define logic programs containing both probabilistic (or parameterised) and deterministic (or non-parameterised) rules4. Unnormalised SLPs can conveniently be used to represent other existing probabilistic models, such as Bayesian nets (Cussens, 2001).
Generally speaking, an SLP S has a distribution semantics (Muggleton, 2000), that is one which assigns a probability distribution to the atoms of each predicate in the Herbrand base of the clauses in S. Let n(S) denote the logic program formed by dropping all the probability labels from S. A stochastic SLD-resolution procedure will be used to define a probability distribution over the Herbrand base of n(S). The stochastic SLD-derivation of an atom a is as follows: suppose ← g is a unit goal with the same predicate symbol as a and without other function symbols and distinct variables; next suppose there exists a ground substitution θ such that gθ = a (since the clauses at n(S) are range restricted, θ is necessarily ground); now suppose the first atom in ← g can unify with the heads of m > 0 stochastic clauses {l1 : C1,…, lm : Cm} in which the clause li : Ci is chosen (by some selection function), then the probability of the choice is 5; and the probability of a derivation of is a the product of the probabilities of all the choices made in the derivation; moreover, the probability of the atom a is the sum of the probabilities of all the derivations of a. Such stochastic SLD-derivation of a goal is always represented as a stochastic SLD-tree. It is clear that a Markov chain, whose states are goals, is defined by a pure normalised SLP and an initial goal through a stochastic SLD-resolution. The clause parameters thus define transition probabilities between goals in the Markov chain.
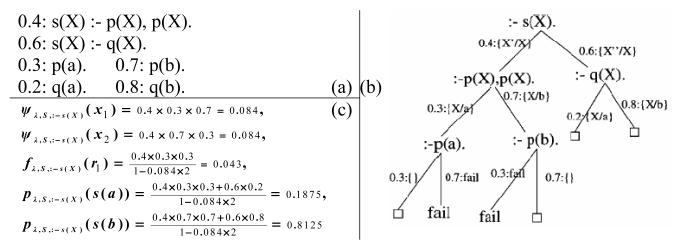
Furthermore, some quantitative results are shown in (Cussens, 2001), in which an SLP S with parameter λ = log l together with a goal g defines up to three related distributions in the stochastic SLD-tree of g: ψλ,S,g (x), fλ,S,g(r) and pλ,S,g(y), defined over derivations {x}, refutations {r} and atoms {y}, respectively. An example is illustrated in Fig. 1, in which the example SLP S defines a distribution, for a goal : –s(X), {0.1875, 0.8125} over the sample space {s(a), s(b)}. As stated in (Cussens, 2001), SLPs do not define distributions over possible worlds, i.e., pλ,S,G(y) defines a distribution over atoms, not over the truth values of atoms. Thus, we could claim that the distribution semantics of SLPs is in accordance with type 1 or domain frequency probabilistic logic (Halpern, 1989) and SLPs have not previously been provided with a possible worlds semantics.
Fig. 1.
(a)an example of SLP S (adapted from Cussens (2001)); (b)a stochastic SLD-tree for S with goal :-s(X), including 6 derivations in which 4 are refutations (end with □) and 2 are fail derivations (end with ‘fail’); (c)probabilities computed in S for the two fail derivations x1 and x2, for the leftmost refutation r1, and for the two atoms s(a) and s(b), respectively (Cussens, 2001).
Learning SLPs has been studied in (Cussens, 2001), which solves the parameter estimation problem by developing failure-adjusted maximisation (FAM) algorithm, and in (Muggleton, 2000, 2002a), which presents a preliminary approach to structure learning. The problem of SLP structure selection is still an open hard problem in the area that requires one to solve almost all the existing difficulties in ILP learning (De Raedt and Kersting, 2003).
FAM is designed to deal with SLP parameter learning from incomplete or ambiguous data in which the atoms in the data have more than one refutation that can yield them. It is an adjustment to the standard EM algorithm where the adjustment is explicitly expressed in terms of failure derivation. The algorithm maximises, at iteration h, the likelihood of parameters λh given the observed data y with empirical frequencies, i.e. Pe(y|λh), the probability of y given the current parameters. Since an SLP's parameters are its clausal probabilities, FAM works on the expected contribution a particular clause has in stochastic SLD-derivations with respect to the data at hand. This is ψλh[νi|y], the expected frequency for clause Ci given the observed data y and the hth iteration parameter estimation λh
where νi counts times Ci appeared in some derivation, Nk is the number of times datum yk occurred in the observed data, N = ΣkNk is the number of observed data, ψλh[νi|yk] is the expected number of times Ci was used in refutations yielding yk, ψλh[νi|fail] denotes the expected contribution of Ci to failed derivations, and Zλh is the probability of all the refutations (Cussens, 2001). Therefore, the first part corresponds to refutations while the second term to failed derivations. Broadly speaking, the equation gathers together the contributions of a particular clause Ci to derivations against the program, the current parameters and the data. The counts are used to estimate the probabilities for the parameterised clauses in each FAM iteration.
2.4 Abductive Logic Programming
Considering a logical approach to the problem of incremental development of scientific models, scientists have distinguished three forms of reasoning: deduction, abduction and induction. Several studies have been conducted on the comparison and integration of abduction and induction from the perspective of Artificial Intelligence (Kakas et al., 1992; Flach and Kakas, 2000). A basic assumption in the study of abduction is that a logical theory or model T can be separated into two disjoint sets of predicates: the observable predicates describe the empirical observations of the domain and the abducible predicates describe underlying relations in the model that are not observable directly, but can bring about observable information through T. In practice, observations are typically represented by ground atomic facts on the observable predicates, and abducibles are the ground atoms generated during reasoning on the abducible predicates that could complement the current theory T. These two types of predicates form the basis of abductive explanation for understanding the observations. In general, abduction generates, in the explanations, extensional knowledge that refers only to the abducible predicates and that is specific to some particular state of world; whereas induction generates intensional knowledge in the form of new general rules that can provide new links between predicates. The combination of abduction and induction has been deployed within ILP, e.g. the framework of theory completion and its implementation Progol 5.0 (Muggleton and Bryant, 2000; Muggleton, 2002b), application of abductive ILP to learning metabolic network inhibition (Tamaddoni-Nezhad et al., 2006).
A general approach of integrating abduction with induction is developed in (Flach and Kakas, 2000). Abduction is first used to transform the observations to an extensional hypothesis on the abducibles. The induction takes this as input and tries to generalize the extensional information to general rules for the abducible predicates now treating them as observables for its own purposes. The cycle can then be repeated by adding the learned information on the abducibles back into the model as new partial information on the incomplete abducible predicates. This will affect the abductive explanations of new observations to be used again in a subsequent phase of induction. Hence through the integration, the abductive explanations of the observations are added to the theory in a generalized form given by a process of induction on them. Adding an explanation to the model allows us to predict further observable information but the predictive power of abduction is restricted to come from the already known rules in the model.
A framework that supports abduction in logic programming is that of abductive logic programming (ALP) (Kakas et al., 1992; Kakas and Denecker, 2002).
Definition 4 (Abductive Logic Programming)
(I) An ALP theory or model T is a triple (LP, A, IC), in which a logic program LP contains definitional knowledge about the domain through a set of observable predicates and background predicates, a set of abducible predicates A appear only in the condition parts of the program rules with no definition in LP, and a set of integrity constraints formulae IC represent assertional knowledge about the domain, augmenting the model in LP but without defining any predicates. (II) Given an ALP theory, an abductive explanation for an observation O is a set Δ of ground abducible atoms on the predicates A such that LP∪Δ |= O and LP∪ |Δ |= IC.
An ALP system thus returns an abductive explanation Δ which represents a hypothesis that together with the model T explains how an observation O could hold. An abductive explanation partially completes the current model T by providing new knowledge (abducibles). This framework provides the background for the studies of abductive ILP in (Tamaddoni-Nezhad et al., 2006) and abductive SLPs in this paper.
2.5 PRISM and ICL
PRogramming In Statistical modeling (PRISM) (Sato, 1995) and Independent Choice Logic (ICL) (Poole, 1997) are two existing PILP formalisms supporting abduction. The common feature of these frameworks is that a purely probabilistic component (probabilistic facts or alternatives) and a purely logical component (logical rules) are connected to produce a hybrid model. Both of them, as well as SLPs, fall into the category of directed approaches where there is a nonempty set of formulae all of whose probabilities are explicitly stated (Cussens, 2007).
There are two disjoint sets of ground atomic formula in the languages: probabilistic facts in PRISM, similarly to the alternatives in ICL, that define a base distribution; and those come from using a set of logical rules that extend the base distribution to an induced distribution over the set of least models. The PRISM system represents probabilistic facts in the form of multiary random switches (msw). In ICL, an atomic choice specifies the truth value of an alternative and a total choice specifies atomic choices for all alternatives. The base distributions are defined over a set of mutually independent msw facts or atomic choices with the closed-world assumption (CWA). From a statistical point of view, both probabilistic facts and alternatives can be treated as random variables that have truth values and probabilities. Thus possible worlds semantics are explicitly invoked in the two formalisms, where a possible world is determined by a total choice in ICL and a conjunction of msw facts in PRISM.
In both frameworks, there is a strict separation between probabilistic facts, whose probabilities are explicitly given, and formulae whose probabilities have to be inferred from the probabilistic facts, the logical rules and the CWA (Cussens, 2007). The logical rules are non-probabilistic and are used to deterministically map the base distribution defined over facts to other atomic formulae. To compute the probability of a formula F which is not a probabilistic fact, it suffices to find the possible conjunctions of facts that entail F, each of which is a product of base probabilities, and then compute the sum of the probabilities of the conjunctions with the help of CWA. This is based on the distribution semantics defined in PRISM. Moreover, abduction is a key operation in finding the required conjunctions. The importance of abduction is reflected in the name probabilistic Horn abduction (PHA)(Poole, 1993), the original version of ICL. In PRISM, abduction is achieved by one of the two underlying probabilistic inferences: explanation search. An explanation for a probabilistic goal G is a conjunction E of the ground switch instances that occurs in a derivation path of G. Explanation search works as an underlying subroutine for probability calculation and parameter learning. In particular, the parameter estimation in the PRISM is exactly a process of abduction, where the base probabilities of a set of msw facts (abducibles defined by multi-valued switch declarations) are estimated from a set of ground atomic formulae (observables defined by target declarations). From a point of view of prediction, the learning could be done with a subset of examples (train data) and we could further calculate the probabilities for another subset of examples (test data) with the learned models using explanation search so as to evaluate the performance of modeling. An example of applying PRISM for such abductive learning and prediction can be found in the next section.
An explicit difference can be found between SLPs and PRISM/ICL, i.e. pure SLPs attach probabilities to first-order clauses/rules as well as facts, but the logical rules are restricted to be deterministic in PRISM/ICL. This further extends to the difference between their probabilistic semantics, i.e. PRISM and ICL have possible worlds semantics but SLPs define probabilities for proofs without much concern about the probabilities with which the atomic formulae are true. However, as shown later, both PRISM and abductive SLPs can achieve the same goal of abductive learning. To understand the representation problem better let us consider the case of learning metabolic network inhibition.
2.6 Learning Metabolic Network Inhibition
Metabolism provides a source of energy for cells and degrades toxic compounds in preparation for excretion. The graph of these interlinked chemical reactions is known as the metabolic network (Alm and Arkin, 2003). The reactions that take place in the network are catalysed by highly specialised proteins known as enzymes. One of the less understood phenomena in the metabolic network is inhibition. Some chemical compounds, known as inhibitors, can affect enzymes, impeding their function. This in turn affects the normal flux in the metabolic network, the result of which is reflected in the accumulation or depletion of certain metabolites. Inhibition is important because many substances designed to be used as drugs can have an inhibitory effect on other enzymes. Any system able to predict such inhibitory effect on the metabolic network would be useful in assessing the potential side-effects of drugs.
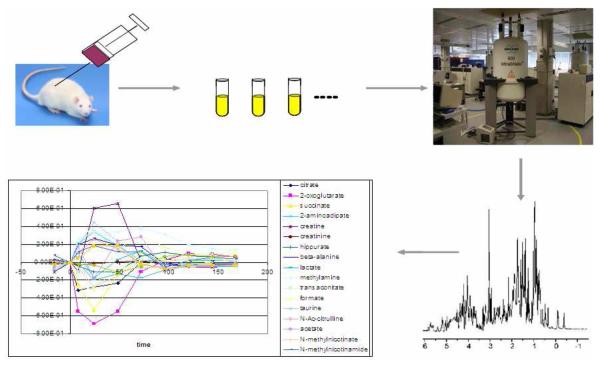
In the Systems Biology project (MetaLog Project, 2006), several machine learning techniques have been conducted to use experimental data on the accumulation and depletion of metabolites to model the inhibitory effect of various toxins, such as hydrazine, in the metabolic network of rats (Fig. 2). In order to measure the actions of toxin, a group of rats were injected with hydrazine and the changes on the concentrations of a number of chemical compounds are monitored during a period of time. Relative concentrations of chemical compounds are extracted from Nuclear Magnetic Resonance (NMR) spectra of urine which provide information concerning the flux of metabolite concentrations before, during and after administration of a toxin.
Fig. 2.
Description of the scientific experiments for machine learning metabolic network inhibition. The example data was derived from studies of the effects of toxins on rats using NMR time-trace analysis of their biofluids.
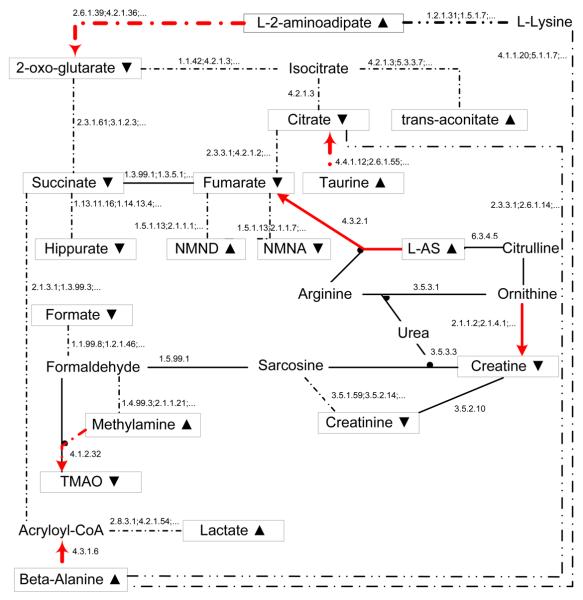
One of the applied machine learning approaches is abductive ILP (Tamaddoni-Nezhad et al., 2006), a variant of ILP supporting both abductive and inductive logic programming. In that work, the binary information on up/down regulations of metabolite concentrations following toxin treatment is combined with background knowledge representing a subset of the Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic diagrams. Based on the underlying ALP paradigm, an abductive ILP program is used to suggest the inhibitory effects occurring in the network, which includes a set of different types of predicates (Table 1), a set of general rules under which the effect of the toxin can increase or reduce the concentration of the metabolites, and some integrity constraints involving self-consistency requirements of the model. In general, we can have a set of metabolites on each side of the reaction and a set of different enzymes that can catalyze the reaction. An example of metabolic network and the learned inhibition are demonstrated in Fig. 3.
Table 1.
The predicates defined in the abduction ILP for learning metabolic network inhibition (Tamaddoni-Nezhad et al., 2006).
| Predicate | Type | Description |
|---|---|---|
|
concentration(Metabolite, Level, Time) |
observable | at some Time a Metabolite has a certain Level of concentra- tion (up or down) |
|
reactionnode(Metabolites1, Enzymes, Metabolites2) |
background | a metabolic pathway between Metabolites1 and Metabo- lites2 catalyzed by Enzymes |
| enzyme(Enzyme) | background | a (sequence of) Enzyme(s) |
| enzyme(Metabolite) | background | a (set of) Metabolite(s) |
|
inhibited(Enzyme, true, Metabolites1, Metabolites2, Time) |
abducible | at Time the reaction from Metabolites1 to Metabolites2 is inhibited by the toxin through an adverse effect on Enzyme that catalyzes the re- action |
|
inhibited(Enzyme, false, Metabolites1, Metabolites2, Time) |
abducible | at Time the reaction from Metabolites1 to Metabolites2 is not inhibited by the toxin through an adverse effect on Enzyme that catalyzes the re- action |
Fig. 3.
An example of rat metabolic network and the corresponding inhibition of hydrazine (at hour 8) learned by abductive ILP. Information on up/down changes in metabolite concentrations (boxed nodes) from NMR spectra is combined with KEGG metabolic diagrams. The nodes without boxes are the metabolites whose concentrations are not observed/observable. The enzymes associated with a single reaction (solid line) or a linear pathway (dotted line) are shown as a single enzyme or a sequence of enzymes. Colored arrows show the found inhibition with directions.
The key point in the abductive ILP study is that, as introduced in section 2.4, it supports the integration of abduction and induction and provides abductive explanations for observations in an ILP setting. Abduction is a procedure of finding explanations for observations and induction is a procedure of formulating general rules for the abducible predicates. There are three main reasons for us to revisit this application work of abductive ILP by using a probabilistic ILP approach. Firstly, we believe the data set provides us a test bed for developing a method that can extract probabilistic examples instead of non-probabilistic examples, as it contains raw data for control and treatment cases. Secondly, we attempt to reject a null hypothesis “learning probabilistic logic models from probabilistic examples does not provide better prediction than learning probabilistic logic models from non-probabilistic examples” with the empirical probabilities as the base line. Finally, we want to demonstrate that probabilistic logic models provide richer interpretability than pure logic models from the application point of view. Therefore, by adapting the pure logic program to an SLP program and extracting probabilistic examples from the same data, our study aims at learning probabilistic logic models of metabolic network inhibition from probabilistic examples.
3 Abductive SLPs
3.1 Abduction with SLPs based on a Possible Worlds Semantics
Despite their use in stochastic contexts, SLPs have not previously been provided with a (type 2) possible worlds semantics and their interpretation has generally been allied in the literature (Puech and Muggleton, 2003) to Halpern's (type 1) domain frequency based probabilistic models (Halpern, 1989). Abductive SLPs provide a setting to SLPs which supports abductive modeling and learning with SLPs through defining a probability distribution over the abductive hypotheses based on a possible worlds semantics (Arvanitis et al., 2006).
As stated in section 2.3, SLPs are originally set to have a distribution semantics (Sato, 1995), that assigns a probability distribution to the atoms in the Herbrand base of the clauses in an SLP program according to a stochastic SLD resolution strategy (Muggleton, 2002a). The stochastic SLD-derivation procedure gives SLPs a domain-based semantics which define probability distributions over ground atoms through building stochastic SLD-trees. However, being motivated by Markov models, we now provide a new setting which interprets the probabilities assigned to the clauses as conditional probabilities between possible worlds. When introducing abduction into SLPs (Arvanitis et al., 2006), given a clause p : H ← B being applied in a stochastic SLD-derivation, the probability p is interpreted as Pr(B | H), i.e. the conditional probability of the ground body B being true (in some possible worlds) given that the ground head H is true (in the same possible worlds). This setting corresponds to an explanatory semantics of conditional probability that explain the possible causes for a given result, in contrast with the normal causal semantics (Pr(H | B)) that infer the result given the causes, like the semantics defined in Bayesian Networks (Pearl, 1988) and BLPs (Kersting and De Raedt, 2000). Under the explanatory semantics, the possible explanations that are computed for an atom are based on possible worlds, i.e. each possible world (explanation) corresponds to a stochastic SLD-refutation (a branch in a proof tree) and the probability of the atom is the sum of the probabilities over all the possible worlds (explanations or proofs). In fact, all the stochastic SLD-refutations of an atom compose a subset of possible worlds (in which the atom is true), each of which is associated with a non-zero probability, while all the other possible worlds are set to have zero probabilities under a closed world assumption (CWA). When addressing this in logical reasoning and learning, the new setting also suggests the possibility of introducing abduction into SLPs which can find abductive explanations for observations. Therefore, SLPs with abduction setting are called abductive SLPs which provide SLPs with a possible worlds semantics.
3.2 A Worked Example
We now explain the idea with an example. Suppose we are given the following SLP and the domain is set to be {a, b}
We are now asked to provide explanations of the observation s(a)6. We can view the labels in the SLP above as providing probabilities associated with various abductive explanations of s(a) and s(b). Thus 0.6 is the probability associated with the explanation from the first clause above. If we abduce p(a) from s(a) then a CWA leads us to conclude that ¬q(a) holds in the world description in which this hypothesis is true7. Thus we have the following conditional probabilities
By the laws of conditional probability
Suppose we know a prior distribution over the observations8, i.e. Pr({s(a), s(b)}) = {0.8, 0.2}. Thus we get the following possible worlds along with their probabilities
This leads to an assignment in which all other possible worlds have probability 0. In this context, there are two possible explanations for s(a): p(a) with probability 0.48 and q(a) with probability 0.32. Suppose we now extend the SLP above with the following.
By applying the normal Markov chain assumption in SLPs, and employing the same arguments as above we get the following possible worlds with probabilities
Furthermore, the marginal probabilities of all the abducibles of the two observations are
Thus, we could conclude that the abducible u(a) is the abduced explanation for the observation s(a) with the highest probability 0.464 in the example. It is also worth noting that there is no overlapping between the possible worlds that abduce u(a) (or u(b)), as the worlds derived from the rule 0.5 : p(X) ← u(X) and the rule 0.7 : q(X) ← v(X), v(X) are two sets of different worlds that have no connections between each other.
3.3 Framework of Abductive SLPs
We now define the framework of abductive SLPs by three parts–abductive SLPs, stochastic abduction and learning setting of abductive SLPs.
Definition 5 (Abductive SLPs)
An abuductive SLP SA is a first-order SLP with abductive logic programming setting based on a possible worlds semantics. Let n(SA) denote the logic program formed by dropping all the probability labels from SA, then n(SA) is an ALP theory (as defined in Definition 4). Given an abductive SLP SA, a stochastic abductive explanation for an observation can be derived by applying the following defined stochastic abduction procedure.
Definition 6 (Stochastic Abduction with Abductive SLPs)
Suppose that SA is an abductive SLP, e is a first order ground atom (defined by some observable predicate) with a given prior probability Pr(e), δ(e, SA) is a ground stochastic SLD-derivation of e derived from SA involving a set of ground abducibles Ae (defined by some abducible predicates). We say that a model Me is a least Herbrand model of (SA, e, Ae) if it contains all and only the ground facts in δ and we have
where C is a (grounded) stochastic clause with probability Pr(C) in δ. From this, we have the probability of the possible world (e, Me)
Now suppose an arbitrary abducible a  Ae, then the (marginal) probability of a can be defined to be the sum of the probabilities of all the least models that have a in their abduced facts
Ae, then the (marginal) probability of a can be defined to be the sum of the probabilities of all the least models that have a in their abduced facts
In the definition, a stochastic SLD-derivation δ is a least Herbrand model of the observation e if and only if δ is a stochastic SLD-refutation of e that proves e to be true.
Based on the underlying process of stochastic abduction, abductive SLPs further provide a learning mechanism to learn a set of abducibles. Ideally, abductive SLPs should support structure selection that combines induction and abduction in the learning. However, as SLP structure learning is still a challenging problem in the area, we only consider SLP parameter estimation in our study which learns probabilities for a given set of abducibles. This can be done by applying some SLP parameter estimation algorithms, such as FAM (Cussens, 2001).
Definition 7 (Parameter Estimation Setting of Abductive SLPs)
Suppose B is a background knowledge theory in the form of logic program, E = {(e, Pe(e))} is a set of independently observed ground probabilistic examples, and A is a set of mutually independent abducibles (ground facts) based on some abducible predicates, abductive SLPs aim to learn a set of parameters λ for (A, B) such that (A, B, λ) composes an abductive SLP SA, A Λ B |= E and λ is chosen to maximise the likelihood of SA,
where γ(e, SA) represents the set of stochastic SLD-derivations of e from SA.
The above parameter learning setting is a special case of Definition 2, where the scoring function is set to be the maximum likelihood. Because FAM is not developed for the abduction purpose, i.e. it does not explicitly compute probabilities for abducibles, we have to treat abducibles as ground clauses and learn their probabilities using FAM by maximising the likelihood Pr(E|SA). In fact, FAM estimates a clausal probability for a clause using the same computation process as the stochastic abduction for an abducible through stochastic SLD-resolution and it supports learning from empirical probabilities/frequencies.
3.4 Possible Worlds Semantics vs. Distribution Semantics
Distribution semantics are originally introduced in Sato (1995) as a basic attitude towards the use of probability in logic or logic programming. The distribution approach defines a specific probability distribution which gives the probability that each logical formula is true. It is common that the probability distribution in question is defined over possible worlds (first-order models) which (by marginalization) give for each closed logical formula the probability that it is true (Cussens, 2001). Thus, distribution semantics are originally designed for representing Halpern's type 2 possible worlds probability logic.
SLPs were given a distribution semantics over Herbrand models or a proof-theoretic interpretation to the probability labels attached with stochastic clauses: whenever an SLD-resolution procedure has to choose between clauses, the choice is made according to probability labels. On the other hand, SLPs represent uncertain knowledge as procedural descriptions of sampling distributions, e.g. those defined in stochastic grammar and hidden Markov models (Muggleton, 2000). A pure SLP thus defines a distribution over instantiations of any top-level goal, which is a sample space of ground atoms, but not over the truth values of atoms. Although more complex SLPs can be used to encode other probabilistic models, such as Bayesian net and Markov nets (Cussens, 2001), the distributions encoded are over hypothesis spaces of logic program rather than possible worlds. In fact, for a given goal, an SLP defines an empirical distribution over a stochastic SLD-tree which is determined by an empirical distribution over a set of observations or atoms. Thus, the distribution semantics used in SLPs are similar to Halpern's type 1 domain frequency probability logic (Puech and Muggleton, 2003) and SLPs have not previously been provided with a possible worlds semantics.
Possible worlds semantics provide model-theoretic interpretation to the probabilities: some models or atoms or formulae are said to be true only in some possible worlds or states, which are determined by multiple (exclusive) joint instantiations of some facts. For example, in Poole (1997), “Possible worlds are built by choosing propositions from sets of independent choice alternatives”. It is common that the probability distributions in possible worlds semantics are often defined over the truth values of atoms or variables. One of the advantages of possible worlds semantics lies in the easy interpretation and understanding of probabilities. On the other hand, from the logic programming perspective, there is a need in SLPs to discuss probability distributions over the truth values of atoms and clauses.
Abductive SLPs are a framework that provides possible worlds semantics for SLPs with the help of abduction. On the one hand, abductive SLPs provide a new setting to SLPs: by introducing abduction and abductive explanation, the probability label of a clause can be interpreted by a conditional probability of its body given its head; and under possible worlds semantics, we could define and discuss probability distributions over the truth values of atoms. We have already shown in the previous sections how stochastic abduction works and how the distributions are computed over possible worlds in the underlying SLD-resolution proof procedures. Another advantage of the possible worlds semantics lies in that there is implicitly a closed world assumption set in the stochastic abduction procedure in which the atoms that are not in the derivations are considered false in the world of the derivations.
On the other hand, abductive SLPs do not define any new probability distributions, i.e. for a given goal or atom, the distribution defined under possible worlds semantics over a set of possible worlds is equivalent to that defined under distribution semantics over a set of stochastic SLD-derivations. This is based on a fact that a stochastic SLD-refutation in the traditional SLPs corresponds to a possible world or an abductive explanation in abductive SLPs, while some possible worlds are assumed to have zero probabilities under the CWA. In addition, the computation of the probability of an abducible (is true) is equivalent to the computation of the probability of an atom (without truth value) in SLPs, i.e. fλ,S,G(r). For example, in Fig. 1, a distribution is defined over a set of four refutations (r1, r2, r3 and r4 from left to right in Fig. 1(b)) in the SLP S:
When treating S as an abductive SLP, an equivalent distribution can be computed over a set of 16 possible worlds (for proving s(a) and s(b) to be true):
in which four possible worlds have probabilities and all the others are set zero probabilities under the CWA.
This is the reason why we could transform type 2 empirical validities into type 1 empirical frequencies in our study, as the same distribution is built over two equivalent spaces (possible worlds vs. derivations) that have different semantics (type 2 vs. type 1).
3.5 Abductive SLPs and PRISM
As both abductive SLPs and PRISM support abductive learning, we now show with an artificial example how they work respectively. We suppose a simple metabolic network in the example which contains three metabolites (a, b and c) and two pathways (one is between a and b through enzyme e1 and the other is between a and c through enzyme e2). We use the same background knowledge as that in the later experiment, which models the toxic inhibition (inhibited/49) in the network caused by the concentration up/down regulation changes (concentration/2). We assume two probabilistic examples (concentration levels of b and c) have been observed as training data. The inputs for both approaches are the background knowledge, the abducibles and the observed probabilistic examples. And the outputs will be the probability distributions learned for the abducibles by abductive learning and the probability predicted for the concentration level of a (test datum) by the probability predictions . In addition, abductive SLPs will also estimate the probabilities for the probabilistic clauses (concentration/2), which has to be treated as pure logical (non-probabilistic) rules in the modeling part of PRISM program.
The following SLP program shows the learning and prediction result using abductive SLPs by running FAM software Pe-pl 0.12 (Angelopoulos and Cussens, 2006).
%% abducibles, inhibition, with learned probabilities
0.3497 : inhibited(e1,a,b,t). 0.0999 : inhibited(e2,a,c,t).
0.1499 : inhibited(e1,b,a,t). 0.3997 : inhibited(e2,c,a,t).
0.0002 : inhibited(e1,a,b,f). 0.0002 : inhibited(e2,a,c,f).
0.0002 : inhibited(e1,b,a,f). 0.0002 : inhibited(e2,c,a,f).
%% probabilistic background knowledge with learned probabilities
0.4496 : concentration(X,down) :-
reactionnode(X,Enz,Y),inhibited(Enz,Y,X,t).
0.0004 : concentration(X,down) :-
reactionnode(X,Enz,Y),inhibited(Enz,Y,X,f),observed(Y,down).
0.5496 : concentration(X,up) :-
reactionnode(X,Enz,Y),inhibited(Enz,X,Y,t).
0.0004 : concentration(X,up) :-
reactionnode(X,Enz,Y),inhibited(Enz,Y,X,f),observed(Y,up).
%% deterministic (non-probabilistic) background knowledge
reactionnode(a,e1,b). reactionnode(b,e1,a).
reactionnode(a,e2,c). reactionnode(c,e2,a).
observed(b,down). observed(c,up).
%% observables, probabilistic examples (with empirical probabilities),
%% the train data
%(concentration(b,down),0.70), (concentration(b,up),0.30)
%(concentration(c,down),0.20), (concentration(c,up),0.80)
%% probabilities predicted for the test data
%Pr(concentration(a,down))=0.483, Pr(concentration(a,up))=0.517
In the above abductive SLP, the possible world semantics apply to not only the abductive learning but also the probability predictions, e.g. the probability Pr(concentration(a, down)) is computed by searching from sets of possible worlds (refutations) as done in abduction. The following PRISM program shows the learning and prediction results using PRISM 1.11.2 (Sato et al., 2008).
%% Declaration of targets and msws
target(concentration,2).% Observable predicate
target(failure,0).% Handling failures
data(user). % Data
values(inhibited,[[e1,a,b,t],[e2,a,c,t],[e1,b,a,t],[e2,c,a,t],
[e1,a,b,f],[e2,a,c,f],[e1,b,a,f],[e2,c,a,f]]). % Abducibles,msw values
%% Modeling part, logical background knowledge rules
failure :- not(success).
success :- concentration(_,_).
concentration(X,down) :-
reactionnode(X,Enz,Y),msw(inhibited,[Enz,Y,X,t]).
concentration(X,down) :-
reactionnode(X,Enz,Y),msw(inhibited,[Enz,Y,X,f]),observed(Y,down).
concentration(X,up) :-
reactionnode(X,Enz,Y),msw(inhibited,[Enz,X,Y,t]).
concentration(X,up) :-
reactionnode(X,Enz,Y),msw(inhibited,[Enz,Y,X,f]),observed(Y,up).
%% Utility part, other background knowledge
reactionnode(a,e1,b). reactionnode(b,e1,a).
reactionnode(a,e2,c). reactionnode(c,e2,a).
observed(b,down). observed(c,up).
%% Observations, probabilistic examples, train data
%learn([count(failure,1),count(concentration(b,down),70),
%count(concentration(b,up),30),count(concentration(c,down),20),
%count(concentration(c,up),80)])
%% Probabilities learned for abducibles by calling show_sw
%Switch inhibited: unfixed_p: [e1,a,b,t] (p: 0.3460) [e2,a,c,t] (p: 0.1075)
%[e1,b,a,t] (p: 0.1589) [e2,c,a,t] (p: 0.3876) [e1,a,b,f] (p: 0.0000005)
%[e2,a,c,f] (p: 0.000001) [e1,b,a,f] (p: 0.0000) [e2,c,a,f] (p: 0.0000)
%% Probabilities predicted for the test data by calling prob()
% prob(concentration(a,down))=0.5465,prob(concentration(a,up))=0.4535
From the learning results, we conclude that similar inhibition have been found by the two frameworks in terms of the probabilities learned for the abducibles. However, different predictions have been made by them for the test datum. The reason lies in the difference in the representations, learning algorithms and implementations. A distinct difference in the above example is that it is necessary to represent and learn the background knowledge clauses (concentration/2) as probabilistic rules in the abductive SLPs10. By contrast, the background knowledge clauses have to be modelled as purely logical rules in the PRISM code. Although deterministic clauses seem more natural in some cases, they have to be treated as parameterised probabilistic rules in the SLPs when multiple refutations are needed, otherwise the probabilities could not be correctly calculated. In such cases, if we consider the SLD-derivations as Markov chains, then the difference between the SLPs and the PRISM becomes whether the transitions in the Markov chains are attached with probabilities or not. The probability calculations of the states in the Markov chains are consequently different. In addition, abducibles are denoted by random switches in the PRISM, but by the ground atomic formulae in the SLPs, and probabilities are also estimated for the probabilistic rules. Please note that failures are handled by applying FAM algorithm in the recent versions of PRISM in order to relax its strict uniqueness condition – that exactly one atomic formula representing observed data is derivable from any instantiation of the base distribution.
4 Extracting Probabilistic Examples from Scientific Data
In this section we outline a method to extract probabilistic examples from scientific data divided into control and treated cases11 and exemplify its application to our rat metabolic network data set. Table 2 presents a pseudo code for the following explained algorithm applied to our rat metabolic network inhibition data set.
Table 2.
Algorithm of estimating empirical probabilities from control/treatment data of metabolic network inhibition.
| 1. | Initialize a matrix MR with column=2 and row=number of metabolites; % MR[α, 1] stores the state value of the concentration of α (up or down) and MR[α, 2] stores the extracted probability, Pe(concentration(α, MR[α, 1])) |
| 2. | for each metabolite α do
|
| 3. | Apply matrix MR in the abductive SLP learning. |
We have a scientific data set involving a set of data values collected from some control cases as well as a set of data points from some treated cases. All the data are mutually independent. In the ILP study, a positive example extracted from such data set is a ground atom stating that some attribute takes some non-probabilistic value by comparing the difference of the average of the values observed in the control cases and treated cases respectively. In our study, we attempt to extract, in addition to a non-probabilistic example, an empirical probability for the example which shows the degree we believe it holds certain value.
The method consists in constructing, for each metabolite α in the control case (step 2.1), a normal distribution Nα with parameters μ and σ calculated from a set of concentration values of α, Cα, in all the control cases (step 2.2 and 2.5). Then, for each concentration value of α, τα, that is observed in the treated cases (step 2.3), the integral from −∞ to τα is calculated in Nα (e.g. using the function pnorm(x, m, sd)12, step 2.5). Meanwhile, a binary state value (up or down) is set for α by comparing the difference between Mean(Cα) and Mean({τα}) (step 2.4). Finally, the average of the integrals (each in [0, 1]), ρα, is taken to be the extracted probability (step 2.5).
Next, we claim that ρα indicates to what extent the set of τα in the treated cases differ from the concentration values of α in the control cases. It follows that a value of ρα < 0.5 specifies α is less expressed in the treated cases compared to that in the control cases in terms of the concentration levels, ρα > 0.5 indicates α is more expressed, and ρα = 0.5 shows that the concentration of α observed in the treated cases has no difference from that in the control cases. Furthermore, we could say that Cα = ρα if ρα > 0.5 or Cα = 1 − ρα otherwise13, where Cα represents the confidence (or degree of belief) of the assertion ‘α is more or less expressed in the treated cases relative to the control cases’. From our point of view, Cα is the estimated type 2 empirical probability (or validity) of the concentration of α happened in the treated cases against the control cases, i.e. we believe the statement ‘the concentration level of metabolite α takes some value’ is true with some probability Cα. For example, a tuple (concentration(citrate, down), 0.9843) derived from the method corresponds to a probabilistic example that means ‘the concentration of metabolite citrate is observed in the given data set to have a down regulation with empirical probability 0.9843’.
In our sample data file, after some pre-processing, we had the raw data values of 20 rows (one per rat) and 20 columns (one per metabolite). The first 10 rows represent control rats (injected with a placebo) and the latter 10 represent treated rats which were injected with 30mg dose of hydrazine. Each column has information on the concentration of a given metabolite at the 8th hour after the injection14. The above method has been applied to the raw data set by developing a small R script. We are aware that using only 10 data points to build a normal distribution for control case is not ideal but have to treat it as an appropriate approximation with the data at hand15. The result matrix with the estimated concentration level and empirical probabilities for hydrazine are presented in column 2 and 3 of Table 4.
Table 4.
Experiment results of learning rat metabolic network inhibition (hydrazine hour 8) from probabilistic examples.
| Probabilistic Examples |
Predictions |
|||||
|---|---|---|---|---|---|---|
| Metabolite | Concen- tration |
Empirical Probs. |
SLPN | SLPP | PSMN | PSMP |
| citrate | down | 0.9843 | 0.6900 | 0.6860 | 1.0000 | 0.9999 |
| 2-og | down | 1.0000 | 0.5680 | 0.6900 | 0.9999 | 1.0000 |
| succinate | down | 0.9368 | 0.2590 | 0.2970 | 0.0726 | 0.9989 |
| l-2-aa | up | 0.9962 | 0.6580 | 0.8280 | 1.0000 | 1.0000 |
| creatine | down | 0.5052 | 0.3070 | 0.4430 | 0.0000 | 0.9985 |
| creatinine | down | 0.5798 | 0.3220 | 0.4930 | 0.0000 | 0.9998 |
| hippurate | down | 0.7136 | 0.3030 | 0.1660 | 0.0000 | 0.0000 |
| beta-alanine | up | 0.9659 | 0.5670 | 0.6860 | 0.9998 | 1.0000 |
| lactate | up | 0.9503 | 0.5400 | 0.5160 | 0.5227 | 0.4646 |
| methylamine | up | 1.0000 | 0.3010 | 0.5250 | 0.0000 | 0.9996 |
| trans-ac | down | 0.6488 | 0.3920 | 0.4410 | 0.3710 | 0.7741 |
| formate | down | 0.9368 | 0.3920 | 0.4230 | 0.7414 | 0.7297 |
| taurine | up | 0.7362 | 0.6500 | 0.8100 | 0.9987 | 0.9780 |
| acetate | up | 0.6727 | 0.5560 | 0.5390 | 0.4594 | 0.1869 |
| nmna | up | 0.5239 | 0.4890 | 0.4920 | 0.0006 | 0.0000 |
| nmnd | up | 0.6414 | 0.4890 | 0.4990 | 0.0032 | 0.0000 |
| tmao | up | 0.5166 | 0.3100 | 0.1120 | 0.0000 | 0.0000 |
| fumarate | up | 0.6970 | 0.2970 | 0.5020 | 0.1188 | 0.9998 |
| l-as | up | 0.6748 | 0.5040 | 0.5070 | 0.0002 | 0.0000 |
| glucose | up | 0.8096 | 0.5570 | 0.5310 | 0.0876 | 0.7617 |
| Predictive Accuracy | 68.31% | 72.74% | 56.27% | 70.02% | ||
| Root Mean Square Error | 36.34% | 32.29% | 52.54% | 38.89% | ||
| Significance of Difference (p-value) | 0.041 | 0.034 | ||||
5 Experiments - Learning Metabolic Network Inhibition
The experiments16 include two learning tasks – learning abductive SLPN and PRISM model PSMN from non-probabilistic examples, and learning abductive SLPP and PRISM model PSMP from probabilistic examples. Our learning algorithm is shown in Table 3.
Table 3.
Learning algorithm used in the study.
| 1. | Extract probabilistic examples E with type 2 empirical probabilities from metabolic network inhibition data. |
| 2. | Transform type 2 empirical probabilities into type 1 empirical frequencies in E. |
| 3. | Derive a background theory B from the abductive ILP study (Tamaddoni-Nezhad et al., 2006) and manually choose a set of abducibles, A. |
| 4. | Apply leave-one-out approach to learn 20 SLPP models and 20 SLPN models, each of which estimates probabilities for (A, B) from E using FAM implementation Pe-pl (Angelopoulos and Cussens, 2006). |
| 5. | Apply leave-one-out approach to learn 20 PSMP models and 20 PSMN models, each of which estimates probabilities for A from E using the PRISM system (Sato et al., 2008). |
| 6. | Evaluate the leave-one-out predictions made by abductive SLP models and PRISM models against probabilistic examples with empirical probabilities. |
| 7. | Interpret the significance of the abducibles based on their learned proba- bilities, e.g. abducibles are said to be significant if their probabilities are greater than a threshold. |
5.1 Hypotheses to Be Tested
The null hypotheses to be empirically investigated in the study are
– The predictive accuracy of an SLPP model does not outperform an SLPN model for predicting the concentration levels of metabolites in a given rat metabolic network inhibition (caused by a given toxin, such as hydrazine) experiment.
– The predictive accuracy of an PSMP model does not outperform an PSMN model for predicting the concentration levels of metabolites in a given rat metabolic network inhibition (caused by a given toxin, such as hydrazine) experiment.
Based on the above null hypotheses and our interests of study in this paper, the following restrictions and assumptions should be followed in the experiment – 1) only PILP models, i.e. the SLPP, SLPN, PSMP and PSMN models, are learned and evaluated; 2) empirical probabilities are used as metric to evaluate the predictive performance of the PILP models, as we believe they provide more accurate information than the non-probabilistic values; 3) we do not compare PILP models with the ILP models, as they use different evaluation metrics; 4) we only compare the predictive performance between SLPP and SLPN; 5) we only compare the predictive performance between PSMP and PSMN; 6) we do not compare the predictive performance between the abductive SLP models and the PRISM models as it is beyond the research purpose of this paper.
5.2 Materials and Inputs
The (estimated) type 2 empirical probabilities are extracted from the raw data consisting of the concentration level of 20 metabolites on 20 rats (10 control cases and 10 treated cases) after 8 hours of the injection of hydrazine. In particular, each observation inputted into SLPP is associated with a type 2 empirical probability ρ we have obtained in last section. In addition, our learning framework also allow us to provide the complementary observations with probability (1 − ρ) (like the negative examples in ILP). Both the FAM implementation Pe-pl (Angelopoulos and Cussens, 2006) and the PRISM system indirectly support the introduction of probabilities in the observation list by allowing the same observation to duplicate an arbitrary (integer) number of times (or frequencies). This makes us possible to transform the type 2 empirical probabilities extracted from the data set into the type 1 empirical frequencies. For instance, a (positive) non-probabilistic example would be simply inputted in SLPN as follows,
concentration(citrate, down)-1.
In addition, a corresponding probabilistic example could be inputted to SLPP in the following form,
concentration(citrate, down)-98.
concentration(citrate, up)-2.
This could be done by using predicate count/2 in the PRISM system (as shown in section 3.5). We use the numbers 98 and 2 to stand for the relative frequencies of the observation, which implicitly corresponds to ‘the concentration of metabolite citrate is down with an empirical frequency 98% and is up with frequency 2%’17. So, probabilistic examples are applied in the abductive SLP and the PRISM frameworks rather than positive and negative non-probabilistic examples in the standard ILP learning18.
A background theory B has been derived and adapted from the existing ILP model (see section 2.6). A set of abducibles A is manually chosen based on the abducible predicates (inhibited/4) and we are interested in finding the potential inhibitions (denoted by inhibited(enzyme, metabolite1, metabolite2, t)) in a given metabolic network involving the pathways between metabolites catalyzed by enzymes. Thus, A and B together with the initial parameters (in an uniform or random distribution) compose of the initial SLP that could be inputted for learning abductive SLPs, while A and B are needed to build up a PRISM program for learning the PRISM models.
5.3 Methods
We apply leave-one-out cross validation technique to do the prediction and evaluation, in which 20 SLPN models, 20 SLPP models, 20 PSMN models and 20 PSMP models are built respectively. Each model is trained by 19 metabolites and tested by the left out one. We perform the SLP learning by playing FAM using Pe-pl 0.12 (Angelopoulos and Cussens, 2006) with both non-probabilistic examples (SLPN) and probabilistic examples (SLPP) under Yap 5.1.1 (Costa et al., 2006). The corresponding PRISM models PSMN and PSMP are learned by PRISM 1.11.2 (Sato et al., 2008).
5.4 Results
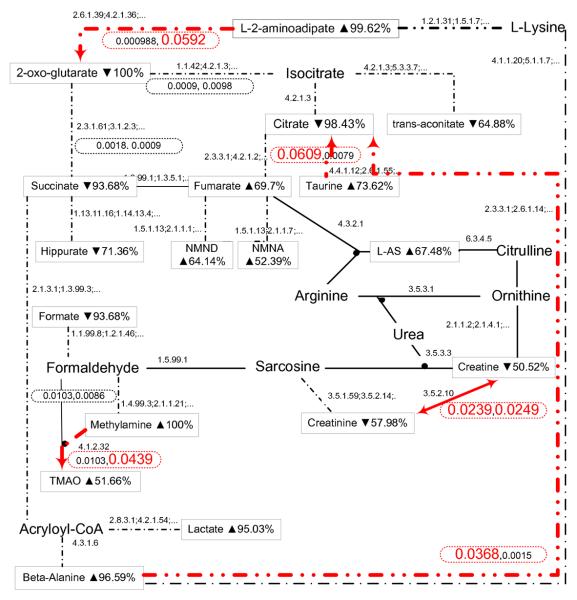
The following SLP shows part of the learned SLPP model and Fig. 4 illustrates a complete model built from all the probabilistic examples, in which we set a threshold (0.02) to decide which abducibles are significant (e.g. the significant inhibitions we have found) based on their learned probabilities.
%% abducibles
0.0592 : inhibited(2.6.1.39,l-2-aminoadipate,2-oxo-glutarate,t).
0.0010 : inhibited(2.6.1.39,2-oxo-glutarate,l-2-aminoadipate,t).
……
0.0358 : inhibited(2.3.3.1,beta-alanine,citrate,t).
0.0015 : inhibited(2.3.3.1,citrate,beta-alanine,t).
……
0.0239 : inhibited(3.5.2.10,creatinine,creatine,t).
0.0249 : inhibited(3.5.2.10,creatine,creatinine,t).
……
%% probabilistic background knowledge
0.3762 : concentration(X,down) :- reactionnode(X,Enz,Y), inhibited(Enz,Y,X,t).
0.0856 : concentration(X,down) :- reactionnode(X,Enz,Y), inhibited(Enz,Y,X,f), observed(Y,down).
0.4535 : concentration(X,up) :- reactionnode(X,Enz,Y), inhibited(Enz,X,Y,t).
0.0846 : concentration(X,up) :- reactionnode(X,Enz,Y), inhibited(Enz,Y,X,f), observed(Y,up).
%% non-probabilistic background knowledge
reactionnode(l-2-aminoadipate,2.6.1.39,2-oxo-glutarate).
reactionnode(2-oxo-glutarate,2.6.1.39,l-2-aminoadipate).
……
enzyme(2.6.1.39).
……
metabolite(l-2-aminoadipate). metabolite(2-oxo-glutarate).
……
observed(citrate,down). observed(2-oxo-glutarate,down).
……
The program for learning PRISM models has a similar form as the one shown in section 3.5. The abducibles with significant probabilities learned by the program are listed as follows,
[e2_6_1_39,l-2-aminoadipate,2-oxo-glutarate,t] (p: 0.096670606)
[e1_13_11_16,hippurate,succinate,t] (p: 0.044315196)
[e2_6_1,taurine,citrate,t] (p: 0.057226669)
[e3_5_2_10,creatinine,creatine,t] (p: 0.045813141)
[e3_5_2_10,creatine,creatinine,t] (p: 0.052581862)
[e4_1_2_32,methylamine,tmao,t] (p: 0.072195070)
[e2_6_1_14,beta-alanine,citrate,t] (p: 0.074273705)
in which the second pattern is not found by the SLPP model.
Fig. 4.
Metabolic network inhibition of hydrazine learned by abductive SLPs from probabilistic examples (SLPP). Each observed metabolite is associated with its concentration and the estimated empirical probability. The learned posterior probabilities for each inhibition (in two directions) are shown in the associated ellipse. For example, the left corner ellipse specifies a learned inhibition in the form of two SLP clauses: ‘0.000988:inhibited(2.6.1.39,2-og,l-2-aa,t).’ and ‘0.0592:inhibited(2.6.1.39,l-2-aa,2-og,t).’, which mean there is a significant inhibition from metabolite l-2-aa to metabolite 2-og with a probability 0.0592.
The probabilistic background knowledge in the program are interpreted as follows,
– the concentration level of metabolite X is down if, in the metabolic network, there is a reaction edge between X and metabolite Y through an enzyme Enz that has been inhibited from Y to X.
– the concentration level of metabolite X is up if, in the metabolic network, there is a reaction edge between X and metabolite Y through an enzyme Enz that has been inhibited from X to Y.
– the concentration level of metabolite X is down/up if, in the metabolic network, there is a reaction edge between X and metabolite Y through an enzyme Enz that has not been inhibited from Y to X and the concentration level of Y has been observed to be down/up.
The program models both inhibited reactions (inhibited(_,_,_ ,t)) and not-inhibited reactions (inhibited(_,_,_,f)) occurred in the metabolic network as well as the changes of metabolite concentrations. In our experiments we adapted the recursive model used in (Tamaddoni-Nezhad et al., 2006) to a non-recursive one because both the SLP learning software Pe-pl and the PRISM system failed to converge using the recursive programs. The recursion in the program stands for not-inhibited reactions occurring between a chain of metabolites in the network. Using some means to control the depth of the recursion, such as Peano numbers, Pe-pl played well for the recursive models with depth 0 (i.e. non-recursive model), but could not provide stable outputs for the recursive models with depth 1 and even crashed for some recursive models with depth 2. Therefore, we assumed the metabolic network exhibits a locality property, i.e. the status of a metabolite is mostly affected by its nearest neighbours, which we believe is not very far from the truth19. From the perspective of PILP, the learning aims to induce and abduce the probabilities for a set of inhibited reactions (abducibles) from a set of observed metabolite concentration levels (probabilistic examples) given a set of background knowledge rules. The background knowledge are represented by probabilistic clauses in abductive SLPs and by unparameterised logical clauses in PRISM, respectively.
5.5 Model Evaluation
The evaluation of the prediction models is made by calculating the predictive accuracy of SLPN, SLPP, PSMN and PSMP against the probabilistic examples respectively. As shown in Table 4, the prediction of a metabolite is the predicted probability of its concentration level (down or up) when it is a test datum in the leave-one-out prediction; the predictive accuracy of a model is defined to be (1 - the average absolute error of predictions over all the metabolites against empirical probabilities); by convention, we calculate the root mean square errors (RMSE) for the predictions against empirical probabilities; and the significance of difference is made by a one-tailed t-test20 (on the deviations from the empirical probabilities) between SLPN (or PSMN) and SLPP (or PSMP) predictions, i.e. the p-value by which we test the null hypothesis.
In particular, when treating the empirical probabilities as the evaluation baseline, SLPP outperforms SLPN by 72.74% against 68.31% in predictive accuracy (1-absolute error) and by 32.29% against 36.34% in RMSR with a significance level of 0.041 (p-value); PSMP outperforms PSMN by 70.02% against 56.27% in predictive accuracy and by 38.99% against 52.54% in RMSR with a significance level of 0.034. It is worth noting that in Table 4 the abductive SLP models appear to outperform the PRISM models. An explanation of this outcome is beyond the scope of this paper, and is believed to be based on differences in the representations and associated learning algorithms.
Based on these results, the null hypotheses to be tested in the experiments could be rejected, i.e. both the abductive SLP models and the PRISM models that are learned from probabilistic examples outperform the corresponding models learned from non-probabilistic examples in terms of prediction in the metabolic network inhibition experiments.
5.6 Interpretability
By comparing the learned SLPP model (illustrated in Fig. 4) with the previous ILP model (illustrated in Fig. 3), apart from the inhibition patterns found in both models, at least two promising new findings have been discovered in the SLPP model21. The inhibition from ‘beta-alanine’ to ‘citrate’ that was not shown in the ILP model has been confirmed to be crucial by the experts. Moreover, the inhibition between ‘creatine’ and ‘creatinine’ showed a contradictory result,
0.0239 : inhibited(3.5.2.10,creatinine,creatine,t).
0.0249 : inhibited(3.5.2.10,creatine,creatinine,t).
in which the learned probabilities of the two inhibited reactions are very close, i.e. the inhibition could happen in both directions (as shown in a bilateral arrow in Fig. 4). This can be further explained by their empirical probabilities,
concentration(’creatine’,’down’)-51.
concentration(’creatine’,’up’)-49.
concentration(’creatinine’,’down’)-58.
concentration(’creatinine’,’up’)-42.
which suggest that their down/up regulations are less expressed to decide the possible inhibition between them.
These findings have also been found in the PSMP model, which also has discovered an extra pattern. In addition, the PILP models learned not only the patterns but also the probabilities (the degrees of belief) of the patterns which improve the interpretability from the learned models.
6 Discussion and Conclusions
6.1 Related Work
We now conclude the discussion of the relationship between abductive SLPs and PRISM/ICL. First of all, clauses or rules are treated as probabilistic (associated with probability labels) in SLPs but purely logical in both PRISM and ICL. Logical rules are used to deterministically map a base probability distribution to an induced distribution in PRISM and ICL, however, there is no mechanism of choosing between rules that have the same head. We believe that the ability of dealing with probabilistic clauses is one of the distinct features of SLPs based on the discussion in section 3.5. Cussens (2007) presents some methods of translating impure SLPs into PRISM programs.
From the point of view of semantics or the interpretation of probability, traditional SLPs have a distribution semantics and interpret probabilities as sampling distributions or domain frequencies over atoms and Herbrand base; PRISM is a distribution approach which defines probability distributions over the truth values of logical formulae in possible worlds; ICL explicitly defines possible worlds by choosing propositions from sets of independent choice alternatives; and the framework of abductive SLPs is designed to introduce possible worlds semantics to SLPs through abduction, where the possible worlds are determined by stochastic SLD-refutations. Abduction is always applied in the frameworks with possible worlds semantics.
In terms of applying abduction, abductive SLPs provide a way to directly learn the parameters for a set of abducibles, i.e. a distribution over a set of ground atomic formulae. The PRISM system provides the explanation search function for abductive learning and probability calculations; and ICL assumes all the atomic choices as abducibles to find consistent explanations that imply the observations.
Despite of the above differences and comparison, as we have shown in the previous sections, both abductive SLPs and PRISM can be used to do abductive learning (abduction) and probability predictions (probability calculations). The experiment results appear to show that the abductive SLPs to abduction are a step forward compared to the previous work in PRISM. However, a further discussion of the topic is beyond the research scope and purpose of this paper. The most significant feature of using PRISM is the efficiency achieved by dynamic programming and sophisticated logic programming “tabling” technology.
At last, as stated in (Cussens, 2007), the distinction between possible worlds approaches and domain frequency approaches is not so fundamental since any probability distribution can be viewed as one over some set of possible worlds. We develop the idea of abductive SLPs in the paper in order to impose a possible worlds semantics on the traditional SLP formalism. Abductive SLPs provide not only abduction but also possible worlds semantics that are easy to understand.
6.2 Conclusions and Future Work
We revisit an application developed originally using ILP by replacing the underlying logic program description with PILP (SLPs and PRISM). Instead of learning logic models from non-probabilistic examples as done in ILP, the PILP approach applied in this paper is based on a general technique for introducing probability labels within a standard scientific experimental setting involving control and treated data. The estimation of empirical probabilities could introduce errors compared with the unknown real distribution of control data due to the limited number of data points. However, our method shown here aims to save some probabilistic information that may have lost in non-probabilistic examples, so that PILP makes better predictions.
It is worth noting that the goal of learning probabilistic logic models from probabilistic examples is to predict accurate (posterior) probabilities rather than the class labels (which is the target of standard ILP). To achieve the goal, we use a regression method which makes estimations of probabilities through abduction from probabilistic examples and then tests if the predictions of the test examples fit the empirical probabilities and models well. During the process, we not only introduce abductive logic programming setting into SLPs that provides possible worlds semantics and abductive explanations for goals, but also transform type 2 empirical probabilities extracted from raw data into type 1 empirical frequencies that can be used in SLP and PRISM parameter learning.
The future work, in theory, include further research of the relationship between different probabilistic semantics: model-theoretic or possible worlds, proof-theoretic, domain frequency and distribution semantics. In practise, it is necessary to do some extra work to investigate why the recursive models are not well applicable in the current PILP modeling and how to achieve this goal. Another area that needs more consensus in the machine learning community, although not directly related with the purpose of this paper, is the proper way to do cross validation for data sets where the independent and identically distributed (i.i.d.) assumption does not hold (i.e. the data is somewhat clustered). In our problem we simply used leave-one-out because our data set was very small. However cross validating by doing leave-one-out can yield an over estimation of the real predictive probability if the left out observation is correlated with the training observations. Albeit there is theoretical work about this problem from the statistical community (Martensa and Dardenne, 1998) there is no standard way to minimise this problem employed by the machine learning community.
In conclusion, the null hypotheses we have set in the paper and experiments were rejected on the bases of the abductive SLP models and the PRISM models we are using and the experimental results. Our results demonstrate that the PILP approach, e.g. SLPs and PRISM, not only leads to a significant decrease in error accompanied by improved insight from the learned result but also provides a way of learning probabilistic logic models from probabilistic examples.
Acknowledgements
The authors would like to acknowledge support from the Royal Academy of Engineering/Microsoft Research Chair on ‘Automated Microfluidic Experimentation using Probabilistic Inductive Logic Programming’; the BBSRC grant supporting the Centre for Integrative Systems Biology at Imperial College (Ref. BB/C519670/1); ESPRIT IST project ‘Application of Probabilistic Inductive Logic Programming II’ (Ref. FP-508861); and the funding from Wellcome Trust for the third author's PhD program.
Footnotes
We distinguish the empirical probability/frequency (denoted by Pe) from the normal probabilities (denoted by Pr) in the context.
The assumption of i.i.d. does not hold in some application areas and data sets where there exist correlations between examples, e.g. the data set used in this paper. However, this is still an open question and has no standard way to minimize the problem in the machine learning community.
A definite logical clause C is range-restricted if every variable in C+, the head of C, is found in C−, the body of C.
The desired meaning for unparameterised clauses in the impure SLPs is to see them as non-probabilistic domain knowledge acting as constraints (Cussens, 2001). The ability of combining such deterministic background knowledge with those probabilistic (parameterised) clauses is one of the central features of SLPs. However, one should satisfy an equivalence relation constraint to apply impure SLPs so that only one single refutation (with probability 1) could be derived from possibly multiple non-probabilistic rules in the underlying SLD-derivations. More details are discussed in Cussens (2001).
It is li if S is normalised.
We apply skolemisation to deal with existential quantifiers appeared in abducible predicates. For example, assuming a clause s(X) ← p(X, Y) (in which Y is a existentially quantified variable) and an observation s(a), a ground fact p(a, $y) could be abduced from, where $y is a skolem constant of Y.
It means that in the worlds that p(a) is true the explanation of q(a) is false. This shows a mutual exclusion of the two explanations of s(a) and implies that 0 : s(X) ← p(X), q(X), which could not be derived under the distribution semantics. It is also worth noting that the CWA here has the same meaning with the exclusiveness condition set in the PRISM, which states that with any parameter settings, for any observable goal G, the explanations (and sub-explanations) for G (and subgoals of G) are probabilistically exclusive to each other.
Please be aware that this distribution is the subjective prior knowledge (de-noted by Pr), not the objective empirical distribution (denoted by Pe).
It denotes inhibited if the fourth argument is set to be t or not-inhibited if the fourth argument is f.
It is necessary because such probabilistic rules in the impure SLP forms can derive more than one refutations for an observation (e.g. concentration(citrate,down)) in the SLD-derivation. As a counterexample, we used unparameterised background knowledge to learn the abductive SLPs for the above example and got the predictions: Pr(concentration(a, down)) = Pr(concentration(a, up)) = 0.5, which means no predictions at all.
The control cases are a set of data gathered from rats without toxin and the treated cases are a set of data gathered from rats with toxin injection.
pnorm(x, m, sd) is a function in the R language, which calculates the area to the left of x in a normal distribution with mean m and standard deviation sd, i.e. the cumulative distribution of the normal distribution.
We do so under an assumption that α takes a binary state value, e.g. up or down, and a threshold of 0.5 is set. It could be extended to categorical cases by setting multiple thresholds.
The data of metabolite concentrations are gathered at some time points. For our research purpose, we are using the non-temporal data collected after 8 hours of toxin injection. The methods in the paper could also be applied to process the data at other time points. Temporal data have been dealt with in (Tamaddoni-Nezhad et al., 2006).
Please note that experiments in some scientific areas, such as metabolic network inhibition, are very expensive.
The probabilistic examples and programs used in the experiments can be found at http://www.doc.ic.ac.uk/~cjz/AbductiveSLPs.
We could choose any integers x and y that satisfy , but the sum (x + y) is required equivalent for all the metabolites.
It is worth noting that the method of duplicating examples to represent frequency information can also be employed in ILP systems such as Progol. However the resulting learned logic programs will predict new examples as either true or false, compared with the frequency assignment given by a learned SLP. On the other hand, the simultaneous appearance of both concentration(citrate,down) and concentration(citrate,up) in ILP systems will be treated as noise that could be avoided.
The non-recursive assumption restricts the predictive ability of the PILP models to some extent, which might be one of the reasons why the predictive errors are relatively high. However, the assumption does not affect the hypotheses we test in the study. It will be the future work to further investigate this problem.
One-tailed test is used because of the hypotheses to be tested, in which SLPP (or PSMP) either outperforms SLPN (or PSMN) or does not.
There are also three inhibitions found in the ILP model but not shown significant in the SLPP model. They might be included if we reduce the significance threshold for the SLPP model, however, it is our future work to investigate the cases with the help of domain experts.
References
- Alan H. Interpretations of probability. In: Zalta EN, editor. The Stanford Encyclopedia of Philosophy (Winter 2007 Edition) Stanford University; 2007. http://plato.stanford.edu/archives/win2007/entries/probability-interpret. [Google Scholar]
- Alm E, Arkin AP. Biological networks. Curr. Opin. Struct. Biol. 2003;13(2):193–202. doi: 10.1016/s0959-440x(03)00031-9. [DOI] [PubMed] [Google Scholar]
- Angelopoulos N, Cussens J. Parameter estimation software implementing the f(ailure) a(adjusted) m(aximasation) algorithm for slps. 2006. http://scibsfs.bch.ed.ac.uk/nicos/sware/slps/pe/
- Arvanitis A, Muggleton S, Chen J, Watanabe H. Abduction with stochastic logic programs based on a possible worlds semantics; Short Paper Proceedings of the 16th International Conference on Inductive Logic Programming; University of Corunna; 2006. [Google Scholar]
- Costa VS, Damas L, Reis R, Azevedo R. Yap prolog user's maual. 2006. http://www.ncc.up.pt/vsc/Yap/
- Cussens J. Parameter estimation in stochastic logic programs. Machine Learning. 2001;44(3):245–271. [Google Scholar]
- Cussens J. Logic-based formalisms for statistical relational learning. In: Getoor L, Taskar B, editors. Introduction to Statistical Relational Learning. The MIT Press; 2007. pp. 269–290. [Google Scholar]
- De Raedt L, Frasconi P, Kersting K, Muggleton S. Probabilistic Inductive Logic Programming - Theory and Applications. Vol. 4911. Springer; Berlin / Heidelberg: 2008. Lecture Notes in Computer Science. [Google Scholar]
- De Raedt L, Kersting K. Probabilistic Logic Learning. ACMSIGKDD Explorations: Special issue on Multi-Relational Data Mining. 2003;5(1):31–48. [Google Scholar]
- De Raedt L, Kersting K. Probabilistic inductive logic programming. In: Ben-David S, Case J, Maruoka A, editors. Proceedings of the 15th International Conference on Algorithmic Learning Theory, volume 3244 of Lecture Notes in Computer Science; Springer-Verlag; 2004. [Google Scholar]
- Flach P, Kakas A. Abductive and Inductive Reasoning. Kluwer; 2000. Pure and Applied Logic. [Google Scholar]
- Friedman N. The bayesian structural em algorithm. In: Cooper G, Moral S, editors. Proceedings of the Fourteenth Annual Conference on Uncertainty in Artificial Intelligence (UAI-98); Madison, Wisconsin, USA: Morgan Kaufmann; 1998. pp. 129–138. [Google Scholar]
- Getoor L, Taskar B. Introduction to Statistical Relational Learning. The MIT Press; Cambridge, Mass: 2007. Adaptive Computation and Machine Learning. [Google Scholar]
- Halpern JY. An analysis of first-order logics of probability. Artificial Intelligence. 1989;46:311–350. [Google Scholar]
- Haussler D. Probably approximately correct learning; National Conference on Artificial Intelligence; 1990. pp. 1101–1108. [Google Scholar]
- Kakas A, Denecker M. Abduction in logic programming. In: Kakas A, Sadri F, editors. Computational Logic: Logic Programming and Beyond. Part I, volume 2407 of Lecture Notes in Artificial Intelligence. Springer-Verlag; 2002. pp. 402–436. [Google Scholar]
- Kakas AC, Kowalski RA, Toni F. Abductive logic programming. Journal of Logic and Computation. 1992;2(6):719–770. [Google Scholar]
- Kersting K, De Raedt L. Bayesian logic programs; Proceedings of the Work-in-progress Track at the 10th International Conference on Inductive Logic Programming; 2000. pp. 138–155. [Google Scholar]
- Martensa HA, Dardenne P. Validation and verification of regression in small data sets. Chemometrics and Intelligent Laboratory Systems. 1998;44(1-2):99–121. [Google Scholar]
- MetaLog Project (2004-2006) http://www.doc.ic.ac.uk/bioinformatics/metalog.
- Muggleton S. Stochastic logic programs. In: De Raedt L, editor. Advances in Inductive Logic Programming. IOS Press; 1996. pp. 254–264. [Google Scholar]
- Muggleton S. Learning stochastic logic programs. In: Getoor L, Jensen D, editors. Proceedings of the AAAI2000 workshop on Learning Statistical Models from Relational Data. AAAI; 2000. [Google Scholar]
- Muggleton S. Learning structure and parameters of stochastic logic programs. Electronic Transactions in Artificial Intelligence. 2002a:6. [Google Scholar]
- Muggleton S. Progol version 5.0. 2002b. http://www.doc.ic.ac.uk/shm/Software/progol5.0/
- Muggleton S, Bryant C. Proc. of the 10th International Workshop on Inductive Logic Programming (ILP-00) Springer-Verlag; Berlin: 2000. Theory completion using inverse entailment; pp. 130–146. [Google Scholar]
- Muggleton S, De Raedt L. Inductive logic programming: Theory and methods. Journal of Logic Programming. 1994;19(20):629–679. [Google Scholar]
- Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. Morgan Kaufmann; Los Altos: 1988. [Google Scholar]
- Poole D. Probabilistic Horn abduction and Bayesian networks. Artificial Intelligence. 1993;64(1):81–129. [Google Scholar]
- Poole D. The independent choice logic for modelling multiple agents under uncertainty. Artificial Intelligence. 1997;94(1-2):5–56. [Google Scholar]
- Puech A, Muggleton S. IJCAI03 Workshop on Learning Statistical Models from Relational Data. IJCAI; 2003. A comparison of stochastic logic programs and Bayesian logic programs. [Google Scholar]
- Richardson M, Domingos P. Markov logic networks. Mach. Learn. 2006;62(1-2):107–136. [Google Scholar]
- Rissanen J. A universal prior for integers and estimation by Minimum Description Length. Annals of Statistics. 1982;11:416–431. [Google Scholar]
- Sato T. A Statistical Learning Method for Logic Programs with Distribution Semantics; Proceedings of the 12th International Conference on Logic Programming (ICLP-1995); 1995. pp. 715–729. [Google Scholar]
- Sato T, Zhou N-F, Kameya Y, Izumi Y. Prism user's manual (version 1.11.2) 2008. http://sato-www.cs.titech.ac.jp/prism/
- Stefan W, Steven C. Finite Mathematics and Applied Calculus. 3 edition Brooks/Cole Publishing Co.; 2004. [Google Scholar]
- Tamaddoni-Nezhad A, Chaleil R, Kakas A, Muggleton S. Application of abductive ILP to learning metabolic network inhibition from temporal data. Machine Learning. 2006;64:209–230. DOI: 10.1007/s10994-006-8988-x. [Google Scholar]